Abstract
In the postgenomic era, the comprehensive proteomic analysis of metabolic and signaling pathways is inevitably faced with the challenge of large-scale identification and characterization of polypeptides with a particular enzymatic activity. Previous work has shown that a wide variety of enzymatic activities of microbial, plant, and animal origin can be assigned to individual polypeptides using in-gel activity staining (zymography). However, a number of limitations, such as special substrate requirements, the lack of a standard procedure, and difficulties in distinguishing enzymes with overlapping activities have precluded the widespread use of zymography as a routine laboratory method. Here we demonstrate that, by employing small-defined substrates that are covalently attached to the gel matrix, we can largely overcome the aforementioned problems and assay readily a number of different classes of enzymatic activities within gels after standard SDS-polyacrylamide electrophoresis. Moreover, this development is compatible with the two-dimensional separation of proteins and thus has great potential in the high-throughput screening and characterization of complex biological and clinical samples.
Zymography (in-gel activity staining), a two-stage technique involving protein separation by electrophoresis followed by in situ (in-gel) assay of enzymatic activities, has proved to be extremely useful for the detection of a wide range of microbial, animal, and plant enzymes—including numerous nucleases and proteases (for reviews, see Gabriel and Gersten 1992; Gersten and Gabriel 1992; Lantz and Ciborowski 1994; Cazenave and Toulme 2001; Scadden and Naaby-Hansen 2001). Certain advantages of zymography over conventional assays, such as the ability to assess the repertoire of enzymes that have a particular activity in nonfractionated cell extracts and to estimate the molecular weight and isoelectric point of the corresponding polypeptides and their isoforms, can be invaluable in identifying and monitoring specific and nonspecific activities in complex biological and clinical samples and developing purification schemes. The most straightforward and widely used zymographic assays are based on the degradation or modification of high-molecular-weight substrates (e.g., gelatin, RNA transcripts) that are retained within the gel and remained unchangeable except within the immediate vicinity of the enzyme(s) being assayed (Gersten and Gabriel 1992; Lantz and Ciborowski 1994; Cazenave and Toulme 2001; Scadden and Naaby-Hansen 2001). Suitable substrates, however, are only available for a limited number of classes of enzymes. A significant obstacle in developing assays for specific enzymes using high-molecular-weight substrates can be the existence of multiple sites that are recognizable by enzymes with different specificities. In this regard, small defined substrates, which are currently being synthesized in great variety and used in enzymological studies, have obvious advantages over macromolecules, but cannot be used directly in zymography as they are not retained efficiently within gels due to their small size. We demonstrate here that this barrier can be overcome by the covalent linking of a small, defined substrate to the gel matrix prior to electrophoresis.
RESULTS
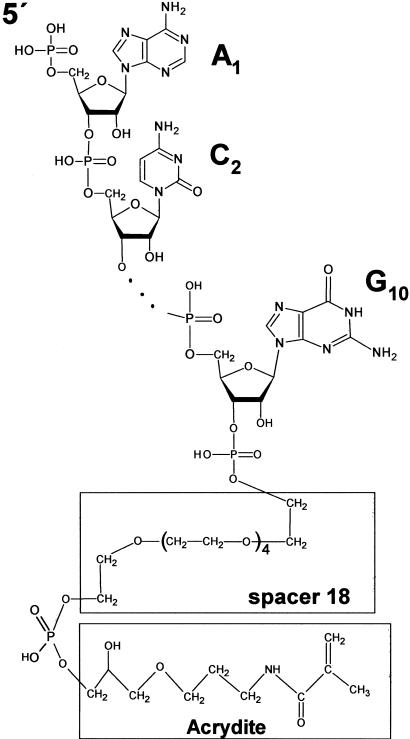
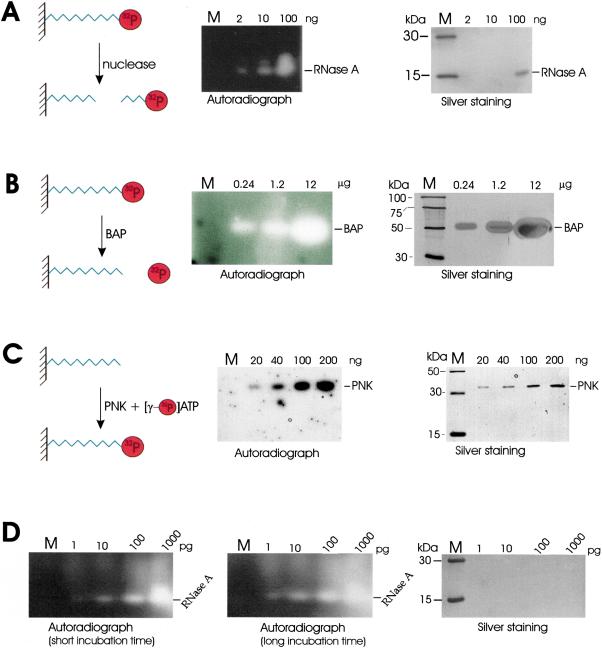
To illustrate the effectiveness of using synthetic molecules that are covalently attached to the gel matrix, we chose a short synthetic decaribonucleotide as a model substrate. The oligonucleotide was synthesised with the sequence ACAGUAUUUG linked at the 3′ end via an 18-link spacer to an acrylamide group (Fig. 1), which was incorporated using Acrydite phosphoramidite (Kenney et al. 1998). This acrylamide-oligonucleotide was radiolabeled at the 5′ end when required (see below), co-polymerized into standard SDS-polyacrylamide gels and used for the in-gel activity staining of several evolutionarily unrelated enzymes; mammalian RNase A, bacterial alkaline phosphatase, and T4 polynucleotide kinase. As shown in Figure 2 (panels A–C), we were able to detect the release of radiolabel from gel areas that corresponded to RNase A and alkaline phosphatase and the attachment of radiolabel to the 5′ end of hydroxylated substrate by polynucleotide kinase. The [γ-32P] ATP for the latter reaction was supplied in buffer after electrophoresis and the removal of SDS. Remarkably, these assays were at least as sensitive as silver staining without modification of standard gel electrophoresis and washing conditions (see Methods). Further increases in sensitivity can, however, be obtained by optimizing the conditions for each enzyme. We were able, for example, to increase the sensitivity of our RNase A assay by 1000-fold. This was achieved simply by omitting reducing reagent and raising the amount of immobilized substrate by fivefold (Fig. 2D). The detection of 1–10 pg of RNase A is comparable with the sensitivity of previously described zymograms that used poly(C) as substrate (Bravo et al. 1994).
Figure 1.
Chemical structure of acrydite-modified oligonucleotides Acr#1 (5′ ACAGUAUUUG 3′) and Acr#2 (5′ ACAGUGCCCG 3′). The acrydite and 18-link spacer are indicated by open square boxes. Three dots represent segments (underlined) of both oligonucleotides that are not shown.
Figure 2.
In-gel assay of ribonuclease A (A) alkaline phosphatase (B) and polynucleotide kinase (C) activities using an oligonucleotide substrate (5′ ACAGUAUUUG; Acr #1) chemically linked to the polyacrylamide matrix. The substrate used in A and B was radiolabeled at the 5′ end prior to incorporation into the gels, whereas in C the substrate was unlabeled. The diagrams that are part of each panel provide a schematic illustration of the corresponding reactions. BAP and PNK are abbreviations for bacterial alkaline phosphatase and polynucleotide kinase, respectively. The gels on the right of each panel have been silver stained and the sizes of marker bands (M) are indicated on the left of each gel. Autoradiographs on the left of each panel show the results of each in-gel assay. The amount of enzyme added to each lane is indicated at the top of the gels and autoradiographs. In D, the RNase A assay was repeated using fivefold more substrate in the absence of a reducing agent and followed the protocol of Bravo et al. (1994). The short and long incubation times were 30 and 90 min, respectively.
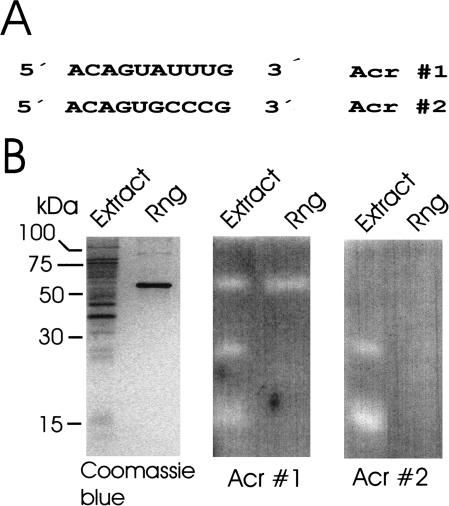
Synthetic oligoribonucleotides including the oligonucleotide 5′-ACAGUAUUUG are being used to probe the substrate specificity and enzymology of the RNase E/G family of ribonucleases (McDowall et al. 1995; Kaberdin et al. 2000), members of which have been shown to have central roles in RNA processing and decay (for review, see Coburn and Mackie 1999). We show (Fig. 3) that an oligonucleotide of the same sequence can be successfully used in an in-gel assay of RNase G activity: a zone of clearing was associated with the position of the 58-kD RNase G polypeptide present in a purified sample and an Escherichia coli cell lysate. Two additional zones of clearing were detected in the lysate lane at positions corresponding to polypeptides of around 27 and 15 kD, respectively. However, importantly, we were able to discriminate RNase G from these other activities using a second covalently-linked substrate 5′-ACAGUGCCCG 3′ that contained nucleotide changes (underlined) known from conventional assays to block cleavage by RNase G. Clearing remained associated with the polypeptides of 27 and 15 kD, but not with RNase G. The 27-kD polypeptide is likely to be the E. coli RNase I (Nicholson 1997), whereas the identity of the 15-kD polypeptide is the subject of further studies.
Figure 3.
In-gel discrimination of a sequence-specific nuclease activity using a pair of oligonucleotide substrates. The sequences of the two substrates (Acr#1 and Acr#2) that were covalently linked separately to gel matrix are shown in A. Aliquots of an extract of E. coli BL21(DE3) and purified recombinant RNase G (Rng) were run side by side in three SDS polyacrylamide gels shown in B. The gel on the left did not contain substrate and was stained with Coomassie blue to indicate the position of the 58-kD Rng polypeptide. The gels in the middle and on the right contained Acr#1 and Acr#2, respectively, and were activity stained as described for RNase A in Fig. 1.
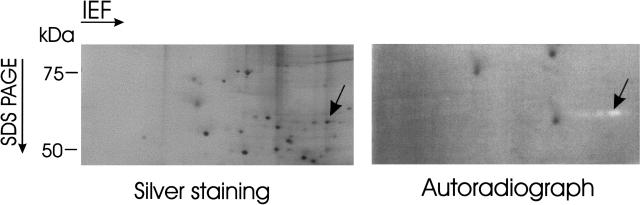
Given the increasingly important role of 2D gel techniques in the large-scale analysis of proteins, we next investigated the utility of covalently attached substrates in the assay of enzymatic activities following 2D gel electrophoresis using RNase G as an example. As shown in Figure 4, a number of spots that appear to correspond to different isoforms of RNase G (a polypeptide with molecular weight 58 kD and isoelectric point 5.6) were readily detected following 2D separation of E. coli cell extract and subsequent in-gel activity staining. Furthermore, the entire area of the 2D gel could be assayed as the distribution of covalently attached substrate remained uniform. In contrast, many noncovalently attached substrates move appreciably in an electric field resulting in an area at either the top or bottom of SDS/polyacrylamide gels where substrate is depleted. Without covalent attachment our radiolabeled oligonucleotide could not be detected within the gel following electrophoresis (data not shown).
Figure 4.
Silver and activity staining of E. coli proteins separated by 2D electrophoresis. The left half of this figure is a silver-stained gel that shows the position of polypeptides following 2D SDS PAGE. The right half is an autoradiography of the same gel that shows the results of an in-gel assay using as substrate the Acr#1 oligonucleotide radiolabeled at the 5′ end. An arrow in each half of the figure indicates the position of spots corresponding to nuclease digestion of the substrates.
DISCUSSION
We have shown here that small, defined oligonucleotides that have been covalently linked to the gel matrix are excellent substrates for zymography incorporating either standard SDS or 2D gel electrophoresis (Figs. 2–4). This approach can be adopted readily and expanded to assay a wide variety of activities that modify or cleave nucleic acids as Acrydite-containing oligonucleotides can be purchased (e.g., from Sigma-Genosys) or synthesized with relative ease by laboratories that have access to phosphoramidite chemistry. For example, enzymes such as helicases, polymerases, transcriptases, methylases, and certain endo-nucleases that have substrates with double-stranded segments can be assayed by hybridizing complementary segments to Acyridite-linked oligonucleotides prior to polymerization of the gel. Peptides can also be chemically synthesized, in most cases, by relatively straightforward approaches and procedures have been developed recently that allow peptides to be conjugated efficiently to mono- and oligonucleotides (Stetsenko and Gait 2000a; Stetsenko and Gait 2000b). Combined, these technologies provide a route for linking small, defined peptide substrates to the matrix of polyacrylamide gels thereby expanding our development of zymography to include protein-modifying enzymes such as proteases, phosphatases, and protein kinases. The latter should be particularly advantageous in the biochemical identification and analysis of activities that have important roles in apoptosis and signaling pathways. In principle, the chemical linkage of any type of small substrate (e.g., a carbohydrate) to the gel matrix could facilitate the identification of the corresponding enzyme activity by zymography.
Mass spectrometric techniques are being developed that are able to analyze femtomole to attomole levels of protein isolated from 2D gels (Bakhtiar and Nelson 2001). This level of sensitivity exceeds that of conventional techniques for staining proteins using Coomassie Blue, silver nitrate, zinc imidazole, or fluorescent dyes such as SYBRO Ruby. Zymography that utilizes synthetic substrates may be extremely sensitive without necessitating the use of radio-isotopes. Fluorescent markers (and quenchers) can be incorporated readily during the synthesis of, for example, oligonucleotide (Park et al. 2001) and peptide substrates (Chersi et al. 1997; Fulop et al. 2001) and used in conjunction with plate readers that quantitatively measure specific florescence signals within gels. In combination with the continuing development of mass spectrometric techniques, the above may lead to the identification and characterization of enzymes that are below the current limits of detection by nonenzymatic staining. Increased sensitivity will be particularly beneficial in the analysis of biological material that can only be obtained in limited amounts and is difficult to culture, for example, that from biopsies and swaps, and may lead not only to new research tools but also assays for monitoring disease and detecting infection and biological contamination. The use of polyacrylamide gels containing covalently linked oligonucleotides is compatible with the trypsinization of polypeptides within gel plugs and the subsequent analysis of the released products by mass spectrometry. Likewise, synthetic peptide substrates should not be problematic as any product release from them following trypsin digestion can be predicted and taken into account.
METHODS
Chemical Synthesis of Acrydite-Modified RNA Oligonucleotides
Oligoribonucleotides Acr #1(ACAGUAUUUG) and Acr #2 (ACAGUGCCCG) were synthesized using an ABI 391DNA synthesizer with a standard 1-μmole DNA assembly cycle and ABI reagents. To universal support (500 Å), Acrydite 2.0 phosphoramidite (Mosaic Technologies) was added, followed by a spacer phosphoramidite 18 {18-O-Dimethoxytritylhexaethyleneglycol,1-[(2-cyanoethyl)-(N,N-diisopropyl)] phosphoramidite}. All these reagents were purchased from Glen Research. The nucleotide portion was then added using 5′-O-dimethoxytrityl-2′-O-t-butyldimethylsilyl-ribonucleoside-3′-O-(2-cyanoethyl)-N,N-diisopropylphosphoramidites purchased from ChemGenes, Inc.
Oligonucleotides were deprotected with methanolic ammonia (30°C, 30 h) followed by triethylamine trihydrogen fluoride/DMSO treatment (1:1, 20°C, 16 h). Deprotected oligonucleotides were added to a DNAPac PA-100 (4 × 250) anion exchange column (Dionex) that had been equilibrated with 50 mM NaCl and were eluted with a gradient of 10% to 70% 1 M NaCl over 20 min. The peak fractions were further purified using a Jupiter C18 reverse phase (5 μm, 300 Å, 4 × 250) column (Phenomenex) equilibrated with 100 mM NH4OAc and eluted with a gradient of 0 to 35% 100 mM NH4OAc/acetonitrile (1:1) using a DX500 HPLC Chromatography System (Dionex). Following dialysis against de-ionized water, the masses of the purified oligonucleotides were confirmed using Q-Tof mass spectrometry in conjunction with maximum entropy analysis of the m/z spectrum.
Enzymes and Protein Preparations
The enzymes used in this study were bacterial alkaline phosphatase (MBI Fermentas), T4 polynucleotide kinase (MBI Fermentas) and ribonuclease A (Sigma-Aldrich). Aliquots of an extract of E. coli BL21(DE3) cells (Novagen) were prepared as described in Miczak et al. (1996) and recombinant RNase G was purified as described previously (Tock et al. 2000).
Activity Staining
SDS protein gels containing covalently attached oligonucleotides were prepared according to the standard protocol (Sambrook et al. 1989) except that the solution used to generate the resolving gel(s) was supplemented with acrylamide-modified oligonucleotide (∼0.3 pmole/mL) prior to polymerization. After electrophoresis, the gels were washed extensively with 25% (vol/vol) isopropanol to remove SDS (Blank et al. 1982) and then proteins were refolded using the procedure described by (Nilsson et al. 2002). The activity staining was performed at 30–37°C for 1 h, followed by 2-h incubation at ambient temperature. The buffers used for the assay of enzymatic activities were 20 mM Tris-HCl (pH 7.5) containing 10 mM MgCl2 for bacterial alkaline phosphatase, 20 mM Tris-HCl (pH 7.5) containing 5 mM magnesium acetate, 100 mM NH4Cl, 0.1 mM EDTA, 1 mM DTT for ribonucleases A and G, and 50 mM Tris-HCl (pH 7.6) containing 10 mM MgCl2, 5 mM DTT, 1 μCi/mL [γ-32P] ATP (Amersham Biosciences) for T4 polynucleotide kinase. To achieve higher sensitivity in the assay of RNase A (Fig. 2D), we increased the concentration of the immobilized substrate fivefold, excluded 2-mercaptoethanol and dithiothreitol from RNase A samples and performed the assay using the protocol of Bravo et al. (1994).
2D Electrophoresis of E. coli Proteins
E. coli cells BL21(DE3) were grown in M9 medium supplemented with glucose (Sambrook et al. 1989) to an optical density (600 nm) of ∼1.0, harvested by centrifugation (5000g, 10 min), resuspended in lysis buffer (Amersham Biosciences), incubated for 30 min at room temperature and centrifuged (37,000g, 5°C) for 1 h. The supernatant was precipitated using CHCl3/methanol (Wessel and Flugge 1984) and the protein pellet resuspended in 1× sample buffer (Amersham Biosciences). An aliquot containing 150 to 200 μg of protein was used for rehydration loading of the sample (∼12 h) onto an 18-cm-long Immobiline pH 4–7 DryStrip (Amersham Biosciences) followed by isoelectric focusing (IEF), which was performed according to the vendor's instructions using the IPGphor Isoelectric Focusing system (Amersham Biosciences). The polypeptides were separated in a SDS 12% (wt/vol) polyacrylamide gel containing 5′-radio-labeled Acr#1 linked to the gel matrix as described above.
Acknowledgments
This work was supported by grant no. F1707 from the Austrian Science Foundation to V.R.K. and by a Royal Society University Research Fellowship to K.J.M. We are extremely grateful to Dr. Chris Adams (Leeds) for synthesizing the oligonucleotides and Dr. Alison Ashcroft (Leeds) for mass spectrometry. We also acknowledge Ms. Lily Tong (Leeds) for confirming preliminary zymography results.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1277303.
References
- Bakhtiar, R. and Nelson, R.W. 2001. Mass spectrometry of the proteome. Mol. Pharmacol. 60: 405-415. [PubMed] [Google Scholar]
- Blank, A., Sugiyama, R.H., and Dekker, C.A. 1982. Activity staining of nucleolytic enzymes after sodium dodecyl sulfate-polyacrylamide gel-electrophoresis: Use of aqueous isopropanol to remove detergent from gels. Anal. Biochem. 120: 267-275. [DOI] [PubMed] [Google Scholar]
- Bravo, J., Fernandez, E., Ribo, M., de Llorens, R., and Cuchillo, C.M. 1994. A versatile negative-staining ribonuclease zymogram. Anal. Biochem. 219: 82-86. [DOI] [PubMed] [Google Scholar]
- Cazenave, C. and Toulme, J.-J. 2001. Gel renaturation assay for ribonucleases. In Methods of enzymology, vol. 341, pp. 126-141. Academic Press, New York. [DOI] [PubMed] [Google Scholar]
- Chersi, A., di Modugno, F., and Rosano, L. 1997. Selective “in synthesis” labelling of peptides by fluorochromes. Biochem. Biophys. Acta. 1336: 83-88. [DOI] [PubMed] [Google Scholar]
- Coburn, G.A. and Mackie, G.A. 1999. Degradation of mRNA in Escherichia coli: An old problem with some new twists. Prog. Nucl. Acid Res. Mol. Biol. 62: 55-108. [DOI] [PubMed] [Google Scholar]
- Fulop, L., Penke, B., and Zarandi, M. 2001. Synthesis and fluorescent labelling of β-amyloid peptides. J. Pept. Sci. 7: 397-401. [DOI] [PubMed] [Google Scholar]
- Gabriel, O. and Gersten, D.M. 1992. Staining for enzymatic activity after gel electrophoresis, I. Anal. Biochem. 203: 1-21. [DOI] [PubMed] [Google Scholar]
- Gersten, D.M. and Gabriel, O. 1992. Staining for enzymatic activity after gel electrophoresisII. Enzymes modifying nucleic acids. Anal. Biochem. 203: 181-186. [DOI] [PubMed] [Google Scholar]
- Kaberdin, V.R., Walsh, A.P., Jakobsen, T., McDowall, K.J., and von Gabain, A. 2000. Enhanced cleavage of RNA mediated by an interaction between substrates and the arginine-rich domain of E. coli ribonuclease E. J. Mol. Biol. 301: 257-264. [DOI] [PubMed] [Google Scholar]
- Kenney, M., Ray, S., and Boles, T.C. 1998. Mutation typing using electrophoresis and gel-immobilized Acrydite probes. Biotechniques 25: 516-521. [DOI] [PubMed] [Google Scholar]
- Lantz, M.S. and Ciborowski, P. 1994. Zymographic techniques for detection and characterization of microbial proteases. In Methods of enzymology, vol. 235, pp. 563-594. Academic Press, New York. [DOI] [PubMed] [Google Scholar]
- McDowall, K.J., Kaberdin, V.R., Wu, S.W., Cohen, S.N., and Lin-Chao, S. 1995. Site-specific RNase E cleavage of oligonucleotides and inhibition by stem-loops. Nature 374: 287-290. [DOI] [PubMed] [Google Scholar]
- Miczak, A., Kaberdin, V.R., Wei, C.L., and Lin-Chao, S. 1996. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc. Natl. Acad. Sci. 93: 3865-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson, A. 1997. Escherichia coli ribonucleases: Paradigms for understanding cellular RNA metabolism and regulation. In Ribonucleases:Structures and functions (eds. G. D'Alessio and J.F. Riordan), pp. 38-49. Academic Press, Inc., New York.
- Nilsson, R.-P., von Gabain, A., and Kaberdin, V.R. 2002. In-gel analysis of site-specific RNases. BioTechniques 33: 272-275. [DOI] [PubMed] [Google Scholar]
- Park, C., Kelemen, B.R., Klink, T.A., Sweeney, R.Y., Behlke, M.A., Eubanks, S.R., and Raines, R.T. 2001. Fast, facile, hypersensitive assays for ribonucleolytic activity. In Methods in enzymology, pp. 81-94. Academic Press, New York. [DOI] [PubMed]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning:A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Scadden, A.D.J. and Naaby-Hansen, S. 2001. Analysis of ribonucleases following gel electrophoresis. In Methods in enzymology (ed. A.W. Nicholson), vol.341, pp. 126-141. Academic Press, New York. [DOI] [PubMed] [Google Scholar]
- Stetsenko, D.A. and Gait, M.J. 2000a. Efficient conjugation of peptides to oligonucleotides by “native ligation.” J. Org. Chem. 65: 4900-4908. [DOI] [PubMed] [Google Scholar]
- Stetsenko, D.A. and Gait, M.J. 2000b. A new “native ligation” procedure for peptide-oligonucleotide conjugation. Nucl. Acids. 20: 801-804. [DOI] [PubMed] [Google Scholar]
- Tock, M.R., Walsh, A.P., Carroll, G., and McDowall, K.J. 2000. The CafA protein required for the 5′-maturation of 16 S rRNA is a 5′-end-dependent ribonuclease that has context-dependent broad sequence specificity. J. Biol. Chem. 275: 8726-8732. [DOI] [PubMed] [Google Scholar]
- Wessel, D. and Flugge, U.I. 1984. A method for the quantitative recovery of proteins in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138: 141-143. [DOI] [PubMed] [Google Scholar]