Abstract
Background
Public sharing of scientific data has assumed greater importance in the ‘omics’ era. Transparency is necessary for confirmation and validation, and multiple examiners aid in extracting maximal value from large datasets. Accordingly, database submission and provision of the Minimum Information About a Microarray Experiment (MIAME) are required by most journals as a prerequisite for review or acceptance.
Methods
In this study, the level of data submission and MIAME compliance was reviewed for 127 articles that included microarray-based microRNA profiling and that were published from July, 2011 through April, 2012 in the journals that published the largest number of such articles—PLOS ONE, the Journal of Biological Chemistry, Blood, and Oncogene—along with articles from nine other journals, including Clinical Chemistry, that published smaller numbers of array-based articles.
Results
Overall, data submission was reported at publication for less than 40% of all articles, and almost 75% of articles were MIAME-noncompliant. On average, articles that included full data submission scored significantly higher on a quality metric than articles with limited or no data submission, and studies with adequate description of methods disproportionately included larger numbers of experimental repeats. Finally, for several articles that were not MIAME-compliant, data re-analysis revealed less than complete support for the published conclusions, in one case leading to retraction.
Conclusions
These findings buttress the hypothesis that reluctance to share data is associated with low study quality and suggest that most miRNA array investigations are underpowered and/or potentially compromised by a lack of appropriate reporting and data submission.
Keywords: microRNA, biomarker, profiling, MIAME, database
Introduction
Irreproducibility of scientific findings is a cause for ongoing concern. The majority of apparently positive results are likely to be false positives (1). Confirmation of reported results was achieved for only six of 53 “landmark” preclinical oncology studies published in journals with impact factors of 5 or greater (2) and, separately, for ~20–25% of 67 published studies (3). With such dismal results for influential and presumably closely scrutinized studies, it is possible that opportunities for misinterpretation or misrepresentation are even greater when experimental design, data, or analyses are of low quality. The authors of a recent analysis (4) presented a parsimonious surrogate for poor quality: a “reluctance to share published research data,” presumably stemming from investigators’ “fear that reanalysis may expose errors in their work or may produce conclusions that contradict their own” (4).
For publication of research that involves array technology, mandatory full data deposition is already the norm, based on the Minimum Information About a Microarray Experiment (MIAME) standard (4). First published in 2001 and rapidly adopted by most journals (5), MIAME describes the minimum information that is needed to interpret “the results of the experiment unambiguously and potentially to reproduce the experiment” (4). This includes raw data, normalized data, sample annotation, experimental design, feature descriptors, and a detailed account of pre-processing and normalization. In truth, MIAME simply “stated the obvious” (5). Its guidelines should be common sense and, although designed for hybridization microarray results, the standard should also apply to quantitative PCR arrays and even next generation sequencing (NGS). In addition to interpretation and reproduction, MIAME compliance facilitates derivation of maximal scientific benefit from experiments that are often resource-intensive. For example, data may be used to answer questions not addressed by the original investigators, or to inform new study design. Databases such as the Gene Expression Omnibus (GEO) (6), CIBEX (7), and ArrayExpress (8) are curated, user-friendly, free-to-the-depositor options for MIAME-compliant data reporting. Of course, adherence to MIAME requires the efforts of individual editors, reviewers, and authors, and profiling studies are not uniformly MIAME compliant. Only ten of 18 Nature Genetics microarray studies that were examined in a 2009 study were found to link to MIAME-compliant data (9).
I noticed recently that many microRNA (miRNA) microarray profiling publications do not reference public data, suggesting that the miRNA field may be a special area of concern. Interest in microRNAs as regulators and biomarkers of clinical conditions (10,11) has led to a rapid increase of miRNA profiling studies and funding opportunities. The influx of new investigators has been facilitated by the relatively small number of canonical miRNAs in comparison with, for example, protein-coding transcripts, combined with the availability of off-the-shelf profiling systems and mail-in services. Even experience with analysis of large datasets may appear to be an unnecessary prerequisite to miRNA profiling, since data analysis is offered by services companies and vendors provide largely automated analysis workflows, obviating direct data manipulation by the investigator. While access to research options is positive, “black box” services and software also present potential pitfalls and may contribute to irreproducible results and confusion in the miRNA profiling field—as in any work that involves large datasets.
To assess the current level of MIAME compliance in miRNA array studies, I reviewed articles that reported array-based miRNA profiling in the four journals that published the largest number of such studies during a ten month period in 2011–2012. To provide a sampling of the wider literature as well, I examined all such articles published during a two-week period within these ten months (chosen because it included a publication in Clinical Chemistry).
Methods
Initial literature search
Candidate articles were first identified by a PubMed search, requiring:
a) Publication during the 10-month period from July 1, 2011 to April 30, 2012, inclusive, with publication date defined as the earliest listed date (usually e-publication date); b) Use in the title or abstract of at least one of the terms “miR”, “miRNA”, “microRNA”, “small RNA”, or “noncoding RNA,” and, in the article text, of at least one of the terms “array”, “microarray”, or “TLDA”; c) English language; and d) primary research publication type.
Candidate screening
PubMed and journal websites were used to examine articles in PLOS ONE, Blood, the Journal of Biological Chemistry (JBC), and Oncogene to remove false positives and identify true positives that may not have appeared in the original search. For example, articles that were removed included those that were: a) published outside the specified date range (for unknown reasons, a small number of extraneous results were returned); or b) false positives, containing keywords but not miRNA profiling (for example, articles that discussed miRNAs but reported transcriptome array results). All articles that were published during the two week period surrounding publication of a Clinical Chemistry miRNA profiling manuscript were also identified, and any articles duplicating those found above were discarded.
Validated article database
A database was created using Microsoft Excel. For each publication, the title, first author, publication date, academic editor (PLOS ONE only, and later removed as uninformative since most articles had a different editor) and URL were recorded, along with the following information:
Type of miRNA profiling platform: hybridization (hyb) or RT-qPCR array (qPCR).
Sample: tissue, cells, body fluid.
Validation of results using a separate method: yes or no.
Were the data deposited in a public database? If so, the accession code was recorded.
Did the authors specify the number of biological and technical array replicates?
Number (or range) of biological replicates per study condition.
Sufficient data processing description: e.g., threshold determination, signal cutoff or background determination, quality control?
Adequate data normalization description: controls, exact normalization methods. (For example, “We normalized the data to internal controls” would be insufficient unless the internal controls were specified, their values were reported, and the exact methods of control averaging and normalization were described.)
Sufficient description of statistical analyses to facilitate replication.
Specification of software programs and/or contracted service companies used to generate the data and analyses.
Use of a global normalization method.
Use of multiple comparison correction for significance testing (or other methods appropriate for large datasets).
Overall ruling on MIAME compliance (liberally interpreted as availability of raw and normalized data, description of technical and biological replicates, and some combination of information on data processing, normalization, and analysis): yes or no.
Notes.
Data submission and MIAME compliance were assessed for each article as it existed at the time of publication. Note that, for some articles, authors may have since deposited or provided links to data because of post-publication requests.
Assignment of quality score
An overall quality score was given to each study and comprised eight component scores. These scores were assigned based on study characteristics and factors important for independent replication of the results (Supplemental Table 1). Minimum and maximum possible overall scores were 0 and 19. A review of potential weaknesses of this scoring system is presented in the on-line Supplemental Text file that accompanies this article. The components of the scoring system were:
Component 1. Sample size (where ‘n’ was the smallest number of samples per experimental or control group; 0 to 5 points):
5: n based on a reported power calculation (no study received this score)
4: n=ten or more
3: n=three to nine (three is the minimum number for identification of outliers)
1: n=two replicates (minimal replication; does not allow identification of outliers)
0: n=one or not reported/no indication of replicates for all conditions
If different numbers of replicates were included for different conditions, the lower number was used to calculate the score. For example, two experimental samples compared with one control would receive a ‘0’, since no meaningful biological information can be derived from this comparison.
Components 2 (data processing), 3 (normalization), and 4 (statistical procedures), 0 to 3 points each
3: Procedures adequately described (e.g., background correction, thresholding, exclusion criteria for data processing)
1: Some procedures described, but incompletely or with insufficient detail to allow faithful replication
0: Procedures not described or inadequately described
Component 5 Software score (0 to 1)
1: all programs reported
0.5: some reported
0: not reported
Component 6. Global normalization score (0 or 1)
A point was awarded if a global normalization strategy was used. (Global normalization may be superior to normalization to just one or a small number of controls.)
Component 7. Multiple comparison correction score (0 or 2)
Two points were awarded if statistical procedures were chosen based on the presence of multiple comparisons. (It is questionable to consider an unadjusted p value of 0.05 “significant” when hundreds of comparisons are made.)
Component 8. Validation score (0 or 1)
One additional point was awarded if some form of result validation was provided: technical validation of array results using the same samples and a different technique or biological validation using different samples.
Statistics
Microsoft Excel, XLStat, and GraphPad Prism were used for statistical analysis. Comparison of multiple groups was done by one-way ANOVA with Tukey’s post-test for multiple comparisons.
Assessment of GEO datasets
Several GEO datasets, including but not limited to those of Allantaz, et al. (12), Chen, et al. (13), Mellios, et al. (14), and Bignami, et al. (15), were downloaded in full from the Gene Expression Omnibus and closely examined or re-analyzed. Additionally, post-publication requests for missing data were made for multiple articles, including but not limited to Huang, et al. (16), Gupta, et al. (17), Rahman, et al. (18), and Mohan, et al. (19). When data were made available, they were accessed and re-analyzed if possible, first following the authors’ described methods as faithfully as possible and then using different approaches, for example, as outlined in previous publications (20–23).
Results
Distribution of articles across journals
According to my PubMed-based literature search, more than 750 research articles that reported array-based miRNA profiling were first published during a 10-month period from July 1, 2011 through April 30, 2012. Articles that reported deep sequencing were omitted due to the relatively inchoate nature of reporting standards for these studies. The journals with the largest share of miRNA array articles were PLoS ONE (now officially titled and henceforth referred to as PLOS ONE), Blood, Oncogene, and the Journal of Biological Chemistry (Fig. 1A). As per the search results, 25 journals included five or more miRNA array articles. Many articles appeared in journals that published fewer than five such articles during the specified time period. This category included Clinical Chemistry, which published one miRNA microarray profiling study in the ten-month span.
Figure 1. Distribution of articles reporting miRNA microarray results and predominance of PLOS ONE in this subject area.
A, Share of miRNA array articles for journals that published five or more miRNA microarray articles from July, 2011 through April, 2012, along with Clinical Chemistry (far right column). B, Increasing percentage of all miRNA articles and share of miRNA array profiling studies published by PLOS ONE.
The most miRNA array profiling articles were published in PLOS ONE, with a share of over 14% of returned articles (Fig. 1A). PLOS ONE published more than six times as many miRNA array articles as the next most represented journal and approximately as many as the next 14 combined. Since the inception of PLOS ONE in late 2006, the journal has grown rapidly, and, in 2011, contributed over 2% of the total English-language, primary literature listings on PubMed (13,624 of 662,224). During the same time, the expansion of PLOS ONE miRNA articles has occurred even faster, reaching about 9% of all primary miRNA reports in 2011 (Fig. 1B). Among these reports, there has been a special concentration of array profiling publications (Fig. 1B), since PLOS ONE captured almost 13% of the share in 2011 and over 14% in the ten-month period examined here.
The inexact nature of literature searches and the different reporting habits of some journals mean that the original search likely returned some false positives and eliminated some true positives. Focusing on the top four journals, PubMed and journal websites were used to assemble and curate lists of articles that could be verified to report on array-based miRNA profiling. For three journals (PLOS ONE, Blood, and Oncogene), true positives corresponded to 60–76% of the original search results, while JBC was found to have published one article more than was indicated in the initial search. The ratios of articles in the first- and second-ranked journals, as well as in the top journal versus the next three combined, were unchanged.
Data submission and MIAME compliance policies
All four of the top-publishing journals have editorial policies that mandate or strongly encourage public data submission and/or MIAME compliance (Journal Policies section of the on-line Supplemental Text file that accompanies this article). Since these journals publish more miRNA profiling studies than most journals, the editorial staff and reviewers may be disproportionately practiced in handling such submissions. Each of these journals also has an impact factor higher than the mean for journals in the biological sciences, possibly indicating higher quality. These three factors might combine to skew the results of this study towards higher apparent MIAME compliance than actually exists in the wider literature, so I examined publications that appeared in journals other than the top four publishers during a two-week period in August/September, 2011. This period, subsequently referred to as “2 weeks”, was centered on the September 2 publication date of a Clinical Chemistry miRNA microarray profiling report (24).
Altogether, 127 verified miRNA array-reporting publications appeared in the top four journals or in the nine journals from the “2 weeks” period (Supplemental Table 2 and the references in the on-line Supplemental Text file that accompanies this article); each article was reviewed and categorized based upon indication of raw and/or normalized data deposition with a public database. Each article was also assigned a multi-component “quality score” as detailed in Methods, considering experimental design and assessment of components of the MIAME guidelines in addition to data submission. Where accession numbers were provided, the links were followed to examine the submission for completeness (all submissions were to either GEO or ArrayExpress). For a subset of submissions, raw and processed data were downloaded and spot-checked or re-analyzed. Finally, articles were judged to be MIAME-compliant or not.
Low levels of MIAME compliance and data submission across journals
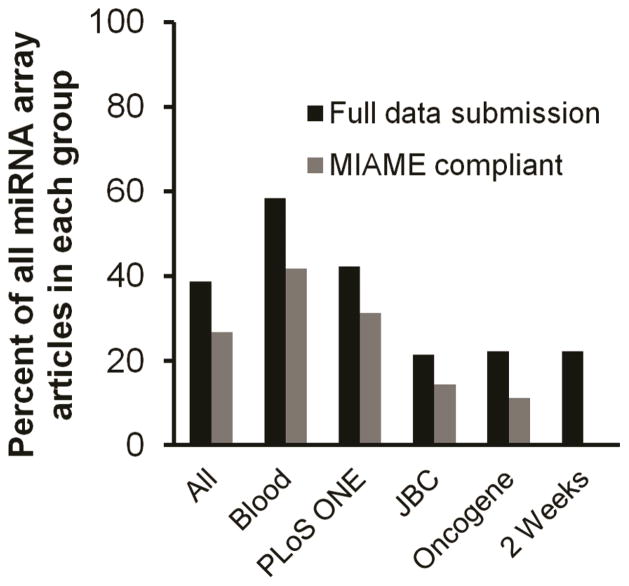
Of the 127 articles examined here, 93 (73%) were judged to be MIAME noncompliant. 15 articles (12%) included incomplete public database submissions (either raw or normalized data but not both), while 78 articles (61%) had no public database links. 34 of 127 articles (27%) were associated with full dataset submission and were deemed MIAME-compliant. Interestingly, the presence of an accession number or even a statement asserting that data were “fully MIAME compliant” (25) did not necessarily mean that data had been properly submitted or that the MIAME checklist had been followed. Accession numbers cited in some articles, even one stating that “(a)ll of the microarray data have been deposited in the Gene Expression Omnibus” (26) did not contain miRNA data [examples include but are not necessarily limited to (26–28)]. Among the four top-publishing journals, there were large differences in reporting and MIAME compliance (Fig. 2). Over 60% of Blood publications reported public database submissions, and over 40% were apparently MIAME compliant. More than 40% of articles in PLOS ONE reported data submission, and almost one third appeared to satisfy MIAME. JBC had the lowest level of public reporting, at just over 20%, while Oncogene had the lowest MIAME compliance, at around 11%. Among the “2 weeks” group, 20% reported data accession numbers and none adhered to MIAME (Fig. 2), supporting the hypothesis that journals with a record of publishing more articles that report array results also tend to publish articles of higher quality.
Figure 2. Rates of data submission and MIAME compliance.
Percentage of all articles examined here (“All”) or of articles in specific journals or the “2 Weeks” group, that included links to publicly available data (“Submission,” black) and that were deemed MIAME-compliant (gray).
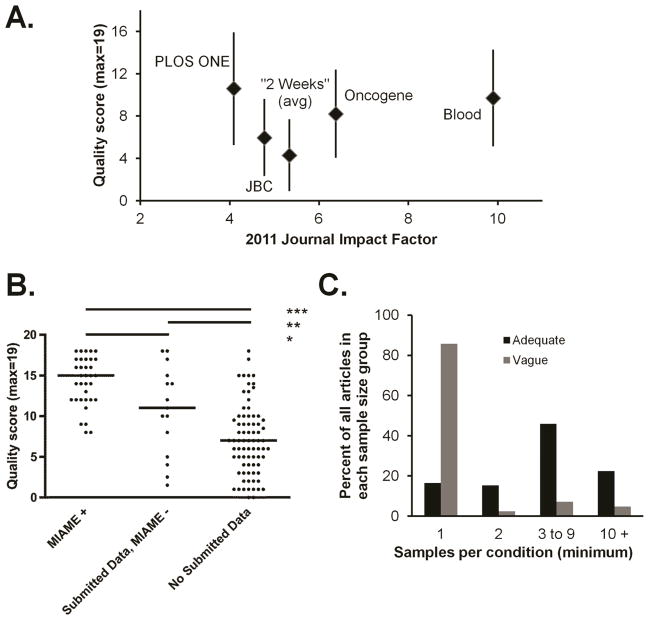
Association of quality score, impact factor, data submission, and MIAME compliance
For journals represented by more than one article, there was no clear association between mean quality score and impact factor (Fig. 3A). It is possible that such an association exists, but that my focus on a small number of journals precluded its identification. However, there was a clear association between quality score and data submission (Fig. 3B). Although several individual exceptions were found—some articles included full data submission but scored very low on the quality scale, while several apparently high-quality and meticulously reported studies did not submit data—the highest mean quality scores were associated with full data submission and MIAME compliance, followed by partial data submission and finally by no data submission. The mean scores were significantly different between categories (Fig. 3B, p<0.0001, ANOVA), and each pairwise category comparison was also significant (Fig. 3B, p<0.05, Tukey’s multiple comparison test).
Figure 3. Associations of impact factor, data submission, experimental design, and quality.
A, Mean quality scores (with standard deviation) for specific journals plotted against journal impact factor or, for the “2 weeks” group, against the group mean without the highest and lowest values. B, Quality score was plotted for articles with full, partial, or no data submission. P values (*<0.05, **<0.01, ***<0.001) are from one-way ANOVA with Tukey’s post-test for multiple comparisons. C, Studies that provided adequate descriptions of experimental design also tended to include larger numbers of experimental/biological repeats. Most studies with vague descriptions relied on microarray experiments with ‘n’ of one in at least one condition.
Experimental design reporting and sample size
Studies that involved larger sample groups also tended to include full descriptions of experimental design (Fig. 3C). The description of technical and biological replicates was recorded as adequate or vague for 85 and 42 studies, respectively; studies with no description of experimental design were also classified as “vague.” Furthermore, the number of biological replicates was recorded when sufficient information was available. For approximately one fifth of studies, it was not, and for most of these, it appeared that at least one examined condition had experimental n of 1. Most studies (36 of 42) with vague or no experimental design descriptions had only one experimental sample in at least one category (e.g., control or treated). In contrast, of the 85 articles found to provide an adequate description of technical and biological replicates, most (58) had n of 3 or more, while 14 had n of 1 and 13 included experimental duplicates.
Examples of high and low quality and apparently erroneous conclusions
When MIAME-compliant data were available, spot-checking of datasets generally revealed high quality and supported confidence in results. To provide one excellent example, I re-analyzed the datasets of Allantaz, et al. (12) and closely replicated the authors’ conclusions. In contrast, the true quality of MIAME-noncompliant articles may be difficult or impossible to judge when data are unavailable. However, in some cases, partial data were available for review or full datasets were uploaded post-publication, either at my request or at the behest of the journal. In the cases (15,17–19) I examined most closely, the original data were found to provide limited support for the conclusions of the articles (Supplemental Text file, Section III). One of these articles has since been retracted (17,29).
Discussion
The results of this investigation indicate that levels of data reporting and MIAME compliance in miRNA array articles are cause for concern despite journal policies that mandate data submission and/or MIAME compliance as a prerequisite for review or acceptance. It is not encouraging that Blood, the journal with the most stringently worded policies and the best marks in this study, had just 60% data submission and ~40% MIAME compliance. Reporting and quality issues were found for articles in journals with impact factors ranging from approximately one to 30, with no obvious association between impact factor and quality score, indicating the endemic nature of the problem. However, other associations were clear. MIAME noncompliant studies were twice as likely to arise from array experiments with n of one. Articles with vague descriptions of experimental design were disproportionately those with few experimental replicates. Studies with fully submitted data received significantly higher mean quality scores than articles with partial submitted data or no data deposition.
For at least several articles, the quality issues may have affected the conclusions. Among articles chosen for more in-depth follow-up, one has since been retracted and the conclusions of others appeared to have a tenuous connection with the data. An apparent retraction rate of one out of 83 articles without submitted data is much higher than the mean retraction rate for scientific publications; however, it is as impossible to make firm conclusions from this observation as it is to make biological conclusions from an underpowered array study. Additional monitoring is therefore needed.
This study has several weaknesses. I focused mainly on articles in the four journals that published the largest number of microarray-based miRNA profiles during the study period. This could skew results in favor of apparent article quality because of the higher-than-average impact factors of these journals and the relative familiarity of the journal staff with processing array submissions. These journals could also be “dumping grounds” for low-quality studies; however, no evidence to support this theory was uncovered here. By chance, the “two weeks” period fell at the end of the traditional summer vacation season, when relatively fewer publications appear. As a result, the small number of articles in this group may not have been representative of the wider literature. The study also examined only English-language articles, but PubMed-listed non-English articles would have represented fewer than 2% of miRNA microarray results.
My interpretation of the MIAME criteria was somewhat liberal in that I did not require complete fulfillment of every point to consider a study compliant. A more stringent interpretation would diminish even further the number of studies found to be MIAME compliant, already a minority. On the other hand, the MIAME criteria are not laws of physics, and some projects may have greater need of adherence than others. A study designed to indicate what is present in a given sample, rather than relative quantities between multiple samples, might not require strict normalization [e.g., (32)]. Nevertheless, to avoid another layer of subjective judgments, I applied the same criteria to all studies.
The “quality” metric I used here to assess thoroughness of reporting and appropriate sample size, processing, and analysis, may be imperfect and, in some respects, subjective. The choice of numerical values and weighting of criteria is debatable. For example, I awarded an extra point to all studies that presented some form of independent validation, but this was done whether two miRNAs or 40 were measured, whether in the same samples analyzed by array or in other samples. It could also be argued that the focus on array quality was unwarranted for “small n” array studies that included non-array validation. However, because few statistically meaningful conclusions may be drawn from array studies with n of one, performing validation studies based on such results is unlikely to be more efficient than selecting follow-up candidates randomly. Additional important factors (e.g., for two-color hybridizations, performance of dye-swap experiments) were not considered. Finally, since I performed this study alone and over several sessions, it is possible that my application of criteria and my assignment of scores were imperfectly uniform.
Despite these weaknesses, I believe that the study is reliable and that the overall quality of miRNA microarray articles may be overestimated because of my almost exclusive focus on the most prolific journals and my fairly liberal assessment of MIAME compliance.
In the interest of maximizing the utility of miRNA biomarker studies and the efficiency of the scientific review process, I make the following recommendations that, if implemented, might help to ensure needed improvements in the quality of miRNA microarray-based studies.
On the part of journals and reviewers, renewed adherence to existing data submission policies or implementation of mandatory submission policies where they do not exist. Specific endorsement of MIAME is encouraged if not already included in journal policies. Although this recommendation applies to all journals, the publishers specifically reviewed in this report—and especially PLOS ONE— could make the greatest contributions because of the large numbers of publications for which they are responsible.
At least one scientist with experience with large dataset analysis should be involved in the review process for manuscripts reporting miRNA (and other) profiling results. This individual should verify the raw and normalized data or, ideally, perform a rapid analysis check. A review should not be considered complete until this is done.
On the part of researchers, acceptance of the need for public submission of data and encouragement of maximal use of public data. This is particularly important in academic science. Unless I have personally and fully funded my lab and research out-of-pocket, my data do not belong to me. They belong to my institution and to the taxpayer, and I have no right to withhold them to prevent another lab from analyzing my data in a way I did not consider. Indeed, “integrators” of existing data—informaticists who can provide insight into what appears to be an expensively expansive morass of under- or low-powered studies—should be encouraged and fostered, as eloquently stated in a recent plaidoyer by Andrew Moore of BioEssays (30).
Availability or introduction of a letter-to-the-editor publication category for journals that do not already offer this publication type (e.g., PLOS ONE) to facilitate open, public communication about missing data and methodological information. Although online comments may be helpful in some cases, and are certainly less of an editorial burden, they carry correspondingly little weight and are not available to the reader in the same way as a letter or a formal correction.
Emphasis on statistically meaningful experiments. Scientists who lack experience with large dataset generation and processing must recognize the need to collaborate with biostatisticians on experimental design and analysis, even when apparently attractive profiling services and vendor-supplied start-to-finish analysis software programs are available. Performing and publishing a study with ‘n’ of 1, or a study in which data are improperly processed, normalized, and analyzed, is scientifically uninformative and a waste of valuable resources, especially when precious patient samples are involved and in an era in which important public health concerns are juxtaposed with talk of funding sequestration.
Researchers should remain closely involved in all stages of their projects. In many cases of low quality and inadequate reporting, array-based profiling and data analysis were performed by a remote company [e.g., (19,31–37)]. This arrangement, in which the (usually academic) researcher is not involved with data generation or analysis, and may not even have full access to the raw data, may be necessary in some cases. However, it also seems to create high risk for misunderstandings and errors. In addition to the communication disconnect, the goals and motivations of academic researcher and company are simply not aligned in the same way as those of the researcher and, say, an academic collaborator or core facility.
Caveat lector. With under 40% data submission, just over 25% MIAME compliance, widespread data normalization issues, lack of multiple comparison corrections, and fully half of all experiments conducted with ‘n’ of 1 or 2, most published claims about miRNA profiles are probably erroneous and, I would predict, will not be independently verified. Exceptions might include large, well-designed cohort studies such as those reviewed by Nair, et al. (38). It may be wise not to draw conclusions from published miRNA profiling unless the results are independently experimentally verified or at least derived from a high-quality, publicly available dataset.
Supplementary Material
Abbreviations
- MIAME
Minimum Information About a Microarray Experiment
Footnotes
This is an un-copyedited authored manuscript copyrighted by the American Association for Clinical Chemistry (AACC). This may not be duplicated or reproduced, other than for personal use or within the rule of ‘Fair Use of Copyrighted Materials’ (section 107, Title 17, U.S. Code) without permission of the copyright owner, AACC. The AACC disclaims any responsibility or liability for errors or omissions in this version of the manuscript or in any version derived from it by the National Institutes of Health or other parties. The final publisher-authenticated version of the article will be made available at http://www.clinchem.org 12 months after its publication in Clinical Chemistry.
References
- 1.Ioannidis JP. Why most published research findings are false. PLoS Med. 2005;2:e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Begley CG, Ellis LM. Drug development: Raise standards for preclinical cancer research. Nature. 2012;483:531–3. doi: 10.1038/483531a. [DOI] [PubMed] [Google Scholar]
- 3.Prinz F, Schlange T, Asadullah K. Believe it or not: how much can we rely on published data on potential drug targets? Nat Rev Drug Discov. 2011;10:712. doi: 10.1038/nrd3439-c1. [DOI] [PubMed] [Google Scholar]
- 4.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–71. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 5.Brazma A. Minimum Information About a Microarray Experiment (MIAME)--successes, failures, challenges. ScientificWorldJournal. 2009;9:420–3. doi: 10.1100/tsw.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ikeo K, Ishi-i J, Tamura T, Gojobori T, Tateno Y. CIBEX: center for information biology gene expression database. C R Biol. 2003;326:1079–82. doi: 10.1016/j.crvi.2003.09.034. [DOI] [PubMed] [Google Scholar]
- 8.Brazma A, Parkinson H, Sarkans U, Shojatalab M, Vilo J, Abeygunawardena N, et al. ArrayExpress--a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2003;31:68–71. doi: 10.1093/nar/gkg091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ioannidis JP, Allison DB, Ball CA, Coulibaly I, Cui X, Culhane AC, et al. Repeatability of published microarray gene expression analyses. Nat Genet. 2009;41:149–55. doi: 10.1038/ng.295. [DOI] [PubMed] [Google Scholar]
- 10.Fabbri M. miRNAs as molecular biomarkers of cancer. Expert Rev Mol Diagn. 2010;10:435–44. doi: 10.1586/erm.10.27. [DOI] [PubMed] [Google Scholar]
- 11.Krutovskikh VA, Herceg Z. Oncogenic microRNAs (OncomiRs) as a new class of cancer biomarkers. Bioessays. 2010;32:894–904. doi: 10.1002/bies.201000040. [DOI] [PubMed] [Google Scholar]
- 12.Allantaz F, Cheng DT, Bergauer T, Ravindran P, Rossier MF, Ebeling M, et al. Expression Profiling of Human Immune Cell Subsets Identifies miRNA-mRNA Regulatory Relationships Correlated with Cell Type Specific Expression. PLoS One. 2012;7:e29979. doi: 10.1371/journal.pone.0029979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, Zhang X, Lentz C, Abi-Daoud M, Pare GC, Yang X, et al. miR-193b Regulates Mcl-1 in Melanoma. Am J Pathol. 2011;179:2162–8. doi: 10.1016/j.ajpath.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mellios N, Sugihara H, Castro J, Banerjee A, Le C, Kumar A, et al. miR-132, an experience-dependent microRNA, is essential for visual cortex plasticity. Nat Neurosci. 2011;14:1240–2. doi: 10.1038/nn.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bignami F, Pilotti E, Bertoncelli L, Ronzi P, Gulli M, Marmiroli N, et al. Stable changes in CD4+ T-lymphocyte microRNA expression following exposure to HIV-1. Blood. 2012 doi: 10.1182/blood-2011-09-379503. [DOI] [PubMed] [Google Scholar]
- 16.Huang L, Lin JX, Yu YH, Zhang MY, Wang HY, Zheng M. Downregulation of six microRNAs is associated with advanced stage, lymph node metastasis and poor prognosis in small cell carcinoma of the cervix. PLoS One. 2012;7:e33762. doi: 10.1371/journal.pone.0033762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta A, Nagilla P, Le HS, Bunney C, Zych C, Thalamuthu A, et al. Comparative expression profile of miRNA and mRNA in primary peripheral blood mononuclear cells infected with human immunodeficiency virus (HIV-1) PLoS One. 2011;6:e22730. doi: 10.1371/journal.pone.0022730. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Rahman S, Quann K, Pandya D, Singh S, Khan ZK, Jain P. HTLV-1 Tax mediated downregulation of miRNAs associated with chromatin remodeling factors in T cells with stably integrated viral promoter. PLoS One. 2012;7:e34490. doi: 10.1371/journal.pone.0034490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohan KV, Devadas K, Sainath Rao S, Hewlett I, Atreya C. Identification of XMRV infection-associated microRNAs in four cell types in culture. PLoS One. 2012;7:e32853. doi: 10.1371/journal.pone.0032853. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Sisk JM, Clements JE, Witwer KW. miRNA Profiles of Monocyte-Lineage Cells Are Consistent with Complicated Roles in HIV-1 Restriction. Viruses. 2012;4:1844–64. doi: 10.3390/v4101844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Witwer KW, Clements JE. Evidence for miRNA expression differences of HIV-1-positive, treatment-naive patients and elite suppressors: a re-analysis. Blood. 2012;119:6395–6. doi: 10.1182/blood-2012-02-412742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Witwer KW, Sarbanes SL, Liu J, Clements JE. A plasma microRNA signature of acute lentiviral infection: biomarkers of CNS disease. AIDS. 2011;204:1104–14. doi: 10.1097/QAD.0b013e32834b95bf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Witwer KW, Watson AK, Blankson JN, Clements JE. Relationships of PBMC microRNA expression, plasma viral load, and CD4+ T-cell count in HIV-1-infected elite suppressors and viremic patients. Retrovirology. 2012;9:5. doi: 10.1186/1742-4690-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palacin M, Reguero JR, Martin M, Diaz Molina B, Moris C, Alvarez V, Coto E. Profile of microRNAs differentially produced in hearts from patients with hypertrophic cardiomyopathy and sarcomeric mutations. Clin Chem. 2011;57:1614–6. doi: 10.1373/clinchem.2011.168005. [DOI] [PubMed] [Google Scholar]
- 25.Lang MF, Yang S, Zhao C, Sun G, Murai K, Wu X, et al. Genome-wide profiling identified a set of miRNAs that are differentially expressed in glioblastoma stem cells and normal neural stem cells. PLoS One. 2012;7:e36248. doi: 10.1371/journal.pone.0036248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z, Huang H, Li Y, Jiang X, Chen P, Arnovitz S, et al. Up-regulation of a HOXA-PBX3 homeobox-gene signature following down-regulation of miR-181 is associated with adverse prognosis in patients with cytogenetically abnormal AML. Blood. 2012;119:2314–24. doi: 10.1182/blood-2011-10-386235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saba R, Gushue S, Huzarewich RL, Manguiat K, Medina S, Robertson C, Booth SA. MicroRNA 146a (miR-146a) is over-expressed during prion disease and modulates the innate immune response and the microglial activation state. PLoS One. 2012;7:e30832. doi: 10.1371/journal.pone.0030832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang MH, Lin BR, Chang CH, Chen ST, Lin SK, Kuo MY, et al. Connective tissue growth factor modulates oral squamous cell carcinoma invasion by activating a miR-504/FOXP1 signalling. Oncogene. 2012;31:2401–11. doi: 10.1038/onc.2011.423. [DOI] [PubMed] [Google Scholar]
- 29.Retraction: Comparative expression profile of miRNA and mRNA in primary peripheral blood mononuclear cells infected with human immunodeficiency virus (HIV-1) PLoS One. 2012;7:8. doi: 10.1371/annotation/d28d38b2-41a3-42a6-b421-68f9460a676d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore A. Have we produced enough results yet, Sir? BioEssays. 2012;34:163. [Google Scholar]
- 31.Liu S, Guo W, Shi J, Li N, Yu X, Xue J, et al. MicroRNA-135a contributes to the development of portal vein tumor thrombus by promoting metastasis in hepatocellular carcinoma. J Hepatol. 2012;56:389–96. doi: 10.1016/j.jhep.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 32.Tilghman SL, Bratton MR, Segar HC, Martin EC, Rhodes LV, Li M, et al. Endocrine disruptor regulation of microRNA expression in breast carcinoma cells. PLoS One. 2012;7:e32754. doi: 10.1371/journal.pone.0032754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pasha Z, Haider H, Ashraf M. Efficient non-viral reprogramming of myoblasts to stemness with a single small molecule to generate cardiac progenitor cells. PLoS One. 2011;6:e23667. doi: 10.1371/journal.pone.0023667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tano N, Kim HW, Ashraf M. microRNA-150 Regulates Mobilization and Migration of Bone Marrow-Derived Mononuclear Cells by Targeting Cxcr4. PLoS One. 2011;6:e23114. doi: 10.1371/journal.pone.0023114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu J, Kang Y, Liao WM, Yu L. MiR-194 regulates chondrogenic differentiation of human adipose-derived stem cells by targeting Sox5. PLoS One. 2012;7:e31861. doi: 10.1371/journal.pone.0031861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raychaudhuri S. MicroRNAs overexpressed in growth-restricted rat skeletal muscles regulate the glucose transport in cell culture targeting central TGF-beta factor SMAD4. PLoS One. 2012;7:e34596. doi: 10.1371/journal.pone.0034596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biyashev D, Veliceasa D, Topczewski J, Topczewska JM, Mizgirev I, Vinokour E, et al. miR-27b controls venous specification and tip cell fate. Blood. 2012;119:2679–87. doi: 10.1182/blood-2011-07-370635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nair VS, Maeda LS, Ioannidis JP. Clinical outcome prediction by microRNAs in human cancer: a systematic review. J Natl Cancer Inst. 2012;104:528–40. doi: 10.1093/jnci/djs027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.