Abstract
We screened all intergenic regions in the human genome to identify pseudogenes with a combination of homology searches and a functionality test using the ratio of silent to replacement nucleotide substitutions (KA/KS). We identified 19,724 regions of which 95% ± 3% are estimated to evolve neutrally and thus are likely to encode pseudogenes. Half of these have no detectable truncation in their pseudocoding regions and therefore are not identifiable by methods that require the presence of truncations to prove nonfunctionality. A comparative analysis with the mouse genome showed that 70% of these pseudogenes have a retrotranspositional origin (processed), and the rest arose by segmental duplication (nonprocessed). Although the spread of both types of pseudogenes correlates with chromosome size, nonprocessed pseudogenes appear to be enriched in regions with high gene density. It is likely that the human pseudogenes identified here represent only a small fraction of the total, which probably exceeds the number of genes.
Pseudogenes are complete or partial copies of genes unable to code for functional polypeptides (for review, see Vanin 1985; Mighell et al. 2000). According to the theory of neutral evolution (Kimura 1968), pseudogenes are unconstrained by selection. Therefore, over time they randomly accumulate mutations (insertions, deletions, and substitutions) that often cause disruptions of the original reading frame. Two types of pseudogenes are generally formed by independent mechanisms that are believed to have different implications in gene evolution. “Nonprocessed” pseudogenes arise usually after partial or complete segmental duplication of genes and subsequent loss of function by mutations. Only a tiny fraction of the duplicated genes will remain functional, yet they are believed to be the major source for the formation of new gene functions or expression profiles (Prince and Pickett 2002). A small fraction of nonprocessed pseudogenes can also be due to niche losses or can correspond to null allelic variants (Menashe et al. 2003). “Processed” pseudogenes are formed through retrotransposition of mature RNAs. They integrate randomly into the genome and therefore lack upstream promoters. Because of early termination of the reverse transcription, many of the processed pseudogenes contain either no or only partial coding regions (Pavlicek et al. 2002). Nevertheless, a few cases of expressed intronless genes with a likely retrotranspositional origin (retrogenes) have been described in several organisms (Brosius 1999). The identification of both types of pseudogenes is of great importance as it provides the opportunity to determine the rate and age of gene duplication events. Furthermore, the neutral character of all pseudogenic regions makes them suitable to determine different forms and rates of neutral sequence evolution among different regions in the genome and even among different organisms. The identification of pseudogenes has also become a necessary component of the primary genome annotation in Metazoans, mainly because of their significant (up to 20%) misincorporation into gene collections (International Human Genome Sequencing Consortium 2001; Mounsey et al. 2002; Mouse Genome Sequencing Consortium 2002). Although pseudogene analysis was included in some animal sequencing projects (e.g., mouse and mosquito), the total number of human pseudogenes and the genomic location for most of them are still uncertain. Several groups have proposed estimates of the pseudogene content in the human genome from the extrapolation of analyses of restricted parts of the genome (Goncalves et al. 2000; Harrison et al. 2002). Although the present annotation of human Chromosomes 21 and 22 indicates the presence of 59 (Hattori et al. 2000) and 234 (Collins et al. 2003) pseudogenes, respectively, a detailed analysis of these chromosomes showed the presence of at least 149 (52% processed) and 244 (46% processed) pseudogenes using stop codons and frameshifts as indicators of nonfunctionality (Harrison et al. 2002). This was extrapolated to a predicted total of ∼20,000 human pseudogenes. In a different approach, the number of processed pseudogenes identified in a limited number of human genomic fragments under the same “presence of truncation” criteria led to an estimate of 23,000–33,000 processed pseudogenes in humans (Goncalves et al. 2000). Because these numbers were based on the assumption of 75,000–100,000 genes in humans, a lower estimate of 9000–11,000 total processed pseudogenes would result when considering only 30,000–35,000 human genes (International Human Genome Sequencing Consortium 2001). Both approaches have confined the identification of pseudogenes to intergenic regions that contain either stop codons or frameshifts in their potential coding regions, predicted by homology to known protein sequences. Because these truncations are likely to appear as a result of the random degeneration of the sequence, we believe that this criterion is only applicable to a fraction of all detectable pseudogenes and cannot evaluate those with apparently intact coding regions, for example, with replacements of functionally essential amino acids, or disrupted or missing promoters. Note that even the detection of truncations in this type of analysis does not always imply the absence of function, as these can be artificially created as a result of misplacing intron–exon boundaries, or they are naturally spliced out from the mature RNA.
To be independent of the presence of stop codons or frame-shifts, we developed a methodology for pseudogene detection that is based on their neutral rate of evolution. We applied parts of this methodology to the Drosophila, Anopheles (Zdobnov et al. 2002), and mouse (Mouse Genome Sequencing Consortium 2002) genomes and to human Chromosome 7 (Hillier et al. 2003) and found a significant fraction of pseudogenes (30%–40%) with no truncations in their coding regions. Our approach takes into account the ratio of silent (synonymous, KS) to amino acid replacement (nonsynonymous, KA) substitutions, which indicates selective constraints on candidate regions (Li et al. 1981). KA/KS ratios of pseudogenes and those of the vast majority of genes are generally different, as mutations in genes causing amino acid replacements with functional consequences are selected against, in contrast to mutations occurring in pseudogenes.
Here we present the details and recent implementations of our methodology, and the results of its application to the human genome. This survey also includes a new and reliable protocol for the distinction between processed and nonprocessed pseudogenes that does not depend on the accuracy of the predicted coding region (e.g., detection of introns).
RESULTS
Homology Searches
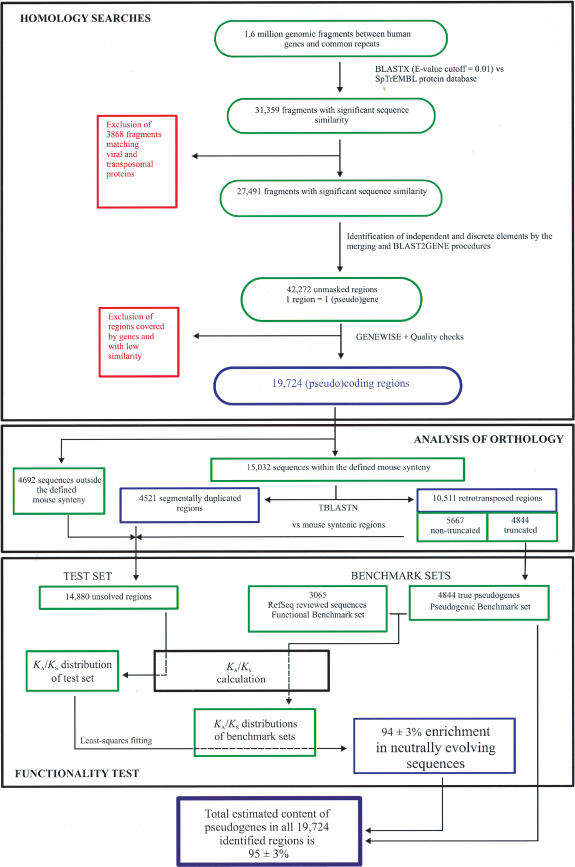
We identified candidate pseudogenic regions using a combination of homology searches and filters to increase the signal and to minimize artifacts (Fig. 1). From 1.6 million genomic regions between predicted genes or common human repeats, we detected 31,359 candidate regions with significant sequence similarity to known proteins (SpTrEMBL database) using BLASTX (E-value cutoff = 0.01; Altschul et al. 1997). Of these, we excluded 3868 regions that are likely to have emerged from viral or transposon-related proteins. The two following steps aimed at the prediction and discrimination of overlooked genes and pseudogenes that can be assembled from the local regions with similarity to known proteins. For this purpose, we identified the boundaries for all detectable candidate elements by a two-step procedure: First, we merged neighboring regions that match the same or a similar reference protein, and then we evaluated the frequency and the protein regions that can be aligned to each DNA region to delimit independent (pseudo)genes. This procedure not only reduces overpredictions due to possible fragmentation of (pseudo)genes, but also dramatically improves the identification of tandem gene duplications, even in gene-rich regions. These two steps generated 42,272 independent genomic regions that mainly contain pseudogenes and overlooked genes, but also known or predicted genes (10,344). The latter fraction was inevitably retrieved in parallel with tandemly duplicated pseudogenes or fused to overlooked exons detected in the first BLASTX round. After removing the regions overlapping with ENSEMBL genes, we extracted the (pseudo)coding sequence of the remaining regions by comparing them with its most similar protein sequence using GENEWISE (Birney and Durbin 1997). Additional quality checks were based on the GENEWISE scores (those lower than 35 were rejected) and the required consistency of the resulting global alignment with local BLAST similarity (E-value cutoff = 0.001). The result was a total set of 19,724 intergenic (pseudo)genes distributed on all chromosomes.
Figure 1.
General overview of the strategy for pseudogene search and evaluation. Our analysis can be divided into three different parts: homology search, analysis of orthology for the selection of KA/KS benchmark sets, and the functionality test based on KA/KS. Green, red, and blue boxes denote the intermediate steps, the excluded sequences, and the final results for each of the sections, respectively. See text for details.
As all predicted and known genes were excluded from our analysis, one would expect that most of the regions that we detected are (parts of) pseudogenes. Nevertheless, at this stage we cannot discount the possibility that a considerable fraction of overlooked (parts of) genes was also included, because as many as 9927 (50%) of our predictions contain no truncation compared with their best-matching protein. To test the functionality of all predicted (pseudo)genes, we analyzed the associated level of selective constraints indicated by their KA/KS ratio.
Obtaining KA/KS Benchmark Collections
There are different factors that can compromise the accuracy of the KA/KS calculation on any (but particularly a large scale) sequence analysis: (1) the number and the selection of the reference sequences; (2) the quality of their alignment with the test sequence; (3) the genomic context of the sequence of interest (Bustamante et al. 2002); (4) the actual protocol for the KA/KS calculation; and (5) the arguable but inevitable assumption that synonymous substitutions are always neutral, whereas nonsynonymous substitutions are always deleterious. Consequently one can expect a certain degree of deviation of the KA/KS ratios calculated on real data from the theoretically expected values (1 for pseudogenes and ≪1 for most genes). For this reason, we constructed benchmark sets of true genes and pseudogenes to obtain their representative KA/KS values.
For the benchmark set of true functional genes, we took the 3065 nonredundant (<50% amino acid identity) human cDNAs from the reviewed section of the RefSeq database (Pruitt and Maglott 2001). Because there is no available human pseudogene collection with comparable size and annotation accuracy, we extracted from our identified candidate regions a subfraction with clear signs of nonfunctionality, that is, retrotransposed (processed) pseudogenes with obvious truncations. The selection of these sequences is not straightforward, as some of our identified regions might correspond to single exons.
In previous analyses, processed pseudogenes have been identified because of the absence of introns, the presence of downstream poly(A) tracks or flanking inverted repeats (Goncalves et al. 2000; Harrison et al. 2001), by measuring their uninterrupted coverage of the known matching protein (normally >70%; Venter et al. 2001; Mouse Genome Sequencing Consortium 2002; Zdobnov et al. 2002; Zhang et al. 2002), or by the presence in the same genome of an intron-containing region with significant sequence similarity (Goncalves et al. 2000; Mouse Genome Sequencing Consortium 2002; Zdobnov et al. 2002). Although these criteria can provide reasonable approximations, they are often inapplicable or inconclusive because of the respective requirement of (1) a reliable prediction of the (pseudo)coding sequence; (2) the conservation of the poly(A) track; (3) the identification of the intron-containing region in the living paralog; and (4) the annotation of the complete matching protein.
In the context of the annotation of human Chromosome 7 (Hillier et al. 2003), we distinguished retrotransposed from segmentally duplicated regions using an approach that is not constrained by the points described above. Here we develop this approach further. The identification of retrotransposed regions is based on the comparative analysis with the corresponding orthologous region in the mouse genome. The vast majority of the pseudogenes identified here qualify for this orthology test, as they were formed after the human–mouse split, that is, they align better to a different region in human than anywhere in the mouse genome. The orthology criterion relies on the fact that retrotransposed mRNAs are expected to integrate randomly into the genome and are unlikely to be located next to the living paralogous gene. This is in contrast to the regions with segmentally duplicated genes that are likely either to remain in the proximity of the paralogous gene (tandem duplications), or are not represented in the orthology maps (interspersed duplications). By comparing the translations of 15,032 of our candidates located within regions with defined mouse orthology with TBLASTN (E-value cutoff = 10-8), we found no significant sequence similarity for 10,511 (70%) of these regions and therefore considered them as processed. In agreement with these results, 9316 (89%) of these processed regions appear as intronless according to the predictions provided by GENEWISE. We then manually examined the remaining fraction (11%). The vast majority of these pseudogenes only contain a single putative intron that cannot be found in the respective parental genes. The respective intron often correlates with transposons or other DNA that apparently inserted after retrotransposition and GENEWISE annotates as intron to maximize the similarity to the reference sequence. Further studies will be needed to try to quantify the insertion of DNA in time.
As there should be no biases in the regions without defined mouse orthology and as we, indeed, found no significant differences as to the length, the number of truncations, and the presence and number of introns between our candidate pseudogenes in or out of orthologous blocks, we assume that the same fraction of retrotransposition events accounts for all 19,724 regions.
From all the candidates predicted to represent processed pseudogenes, we further selected a subgroup with at least one truncation in their coding region to define the “pseudogene benchmark set” comprising 4844 true pseudogenes that will be used to calibrate the KA/KS values associated with neutrally evolving regions. Although we cannot exclude the possibility that a small fraction of nonprocessed pseudogenes might present exceptionally low KA/KS values, the vast majority of the pseudogenes should be represented adequately by this benchmark set.
Detecting Nonfunctionality From KA/KS Distributions
After identifying these pseudogenes, the functionality of the remaining 14,880 regions (the “final test set”) remained to be evaluated. We therefore applied a procedure similar to the one we used for the identification of pseudogenes in the mouse (Mouse Genome Sequencing Consortium 2002), Anopheles, and Drosophila (Zdobnov et al. 2002) genomes, as well as in human Chromosome 7 (Hillier et al. 2003). It consists of evaluating the level of neutral evolution associated with a particular sequence set by finding the best fitting of its KA/KS distribution to the corresponding functional and pseudogenic benchmark KA/KS distributions. The calculation of the KA/KS ratio for a particular sequence always requires its comparison to a homologous reference sequence. Thus, we compared each of our candidate regions with their ancestral sequence, which was previously inferred using two additional homologous functional sequences (see Methods for details). It is important to note that like other approximations that also permit comparisons of more than two sequences (e.g., codeml; Yang 1997) and in contrast to direct pairwise comparison approaches, our procedure allows the calculation of lineage-specific KA/KS ratios. We believe this is essential particularly when identifying and assigning nucleotide substitutions from the comparison of regions with different patterns of evolution (e.g., pseudogenes and functional genes).
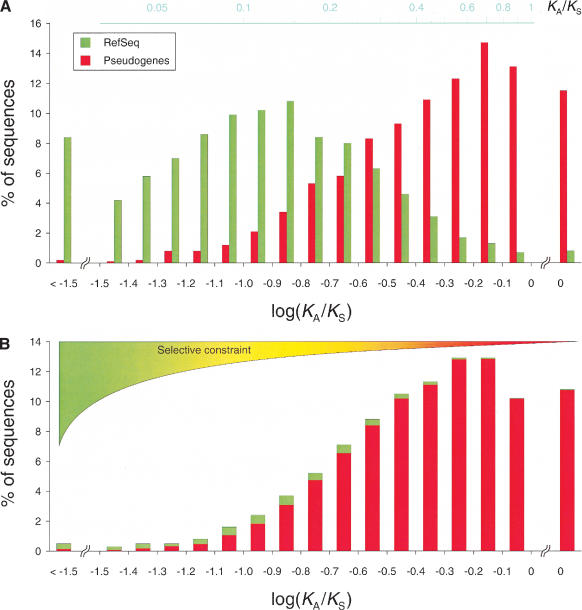
Initially we calculated the KA/KS ratios associated with the functional and pseudogenic benchmark sets. As expected, the distributions obtained are distinct and characteristic because most of the KA/KS values associated with the genes and pseudogenes are indicative of higher and lower (absent for the majority) selective constraints, respectively (Fig. 2A). We further demonstrate by cross-validation that these two distributions permit a reliable protocol to estimate the fraction of pseudogenes included in any given set of sequences with an associated error no larger than 3% (see Methods). Thus, after evaluating KA/KS values for our test set, the comparison of the resulting distribution (Fig. 2B) with both benchmark distributions using the leastsquares fitting procedure showed an enrichment of 94% ± 3% in neutrally evolving sequences, for example, pseudogenes. By adding the pseudogenes initially defined as the pseudogene benchmark set, we estimate that virtually all (95% ± 3%) of the intergenic regions identified in this study are indeed pseudogenes.
Figure 2.
KA/KS distributions of benchmark and candidate sets. The KA/KS distributions (as log KA/KS) associated with the functional (green) and pseudogenic (red) benchmark sets (A) as well as the test sequence set (B) are shown. An average of 40% of the sequences analyzed in this study satisfied our requirements for the KA/KS calculation. The subsets of sequences with KA/KS values (1659 for the functional, 1703 for the pseudogenic benchmark sets, and 3291 for the test set) are expected to be representative for each of the corresponding complete sets, as what determines whether a KA/KS value can be calculated for a sequence (availability of homologous sequences and restrictions on the KA/KS calculation; see Methods) is likely to equally affect genes and pseudogenes. By using the least-squares fitting against the benchmark distributions, we evaluated the fraction of pseudogenic (red) and functional (green) sequences for each of the bins of the test distribution and combined them to determine that up to 95% of the sequences analyzed correspond to pseudogenes.
A classification of the pseudogenes using InterPro domains (Mulder et al. 2003) did not reveal any particular overrepresented functional class (see Table 1 for the top 10 classes), but the most frequent pseudogenes come from multigene families with large copy numbers. Exceptions, such as GAPDH in rodents, where we identified ∼400 copies (Mouse Genome Sequencing Consortium 2002), are not observed in humans. The most frequent group is formed of pseudogenes derived from ribosomal protein genes, for which we see 1300 copies. This is less than the 2000 pseudogenes reported in a recent study (Zhang et al. 2002), but as part of a wider analysis we detected a few more than 600 ribosomal pseudogenes included in the ENSEMBL gene collection (data not shown).
Table 1.
Domain-Based InterPro Analysis of Human Pseudogenes
| InterPro | Name | Occurrences |
|---|---|---|
| IPR000276 | Rhodopsin-like GPCR superfamily | 434 |
| IPR001428 | UTPases | 388 |
| IPR003006 | Immunoglobulin/major histocompatibility complex | 252 |
| IPR000504 | RNA-binding region RNP-1 | 171 |
| IPR001909 | KRAB box | 153 |
| IPR003593 | ATPase | 104 |
| IPR000719 | Eukaryotic protein kinase | 104 |
| IPR001664 | Intermediate filament protein | 101 |
| IPR001147 | Ribosomal protein L21e | 96 |
| IPR000910 | HMG1/2 (high mobility group) box | 78 |
Distribution of Pseudogenes in the Human Genome
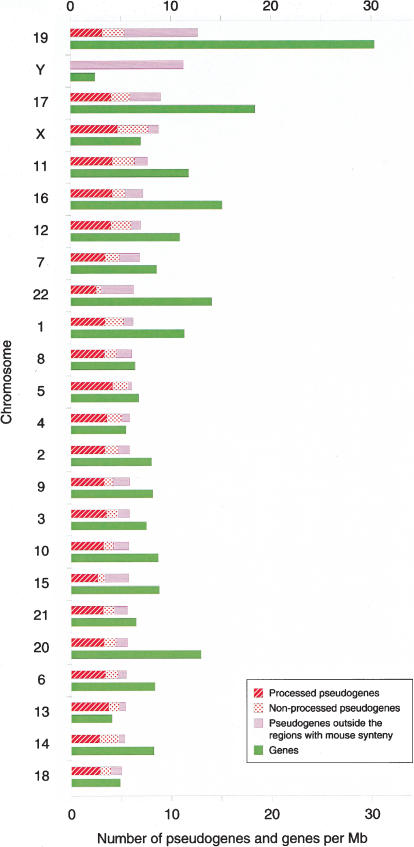
The estimated high specificity of the method allows their further genome-wide analysis. We also assume that the discrimination between processed and nonprocessed pseudogenes is not affected, as it not only relies on KA/KS analysis, but also on the orthology context. Thus, we are able to study the distributions of both pseudogene types within and across chromosomes (Fig. 3). The average density of pseudogenes detected is 6.5 per megabase for the whole genome, which is comparable with the density of genes (9/Mb). There is little deviation among chromosomes, except for Chromosome 19 and the Y-chromosome, where we detected nearly twice as many pseudogenes per megabase: 12 and 11, respectively. Note that whereas Chromosome 19 has the highest gene-to-pseudogene ratio, the Y-chromosome presents the lowest.
Figure 3.
Distribution of genes and the different types of pseudogenes for each of the human chromosomes. We have displayed for each human chromosome the number of pseudogenes (separated in different types; see chart legend for details) and genes per megabase. Chromosomes have been ordered according to the density of pseudogenes (highest on top).
Processed pseudogenes have a similar distribution pattern among chromosomes as has been previously described for a subset derived from ribosomal genes (Zhang et al. 2002). We detected a strong correlation between the number of processed pseudogenes and the size of the chromosomes (r = 0.97), with an overall density of 4/Mb. Although processed pseudogenes cover the entire euchromatin, there are regions where they accumulate above the average density (see supplemental material available online at http://www.bork.embl-heidelberg.de/Docu/Human_Pseudogenes/). Some of these regions are often located close to the telomeres. As expected, the position of processed pseudogenes does not correlate with gene-rich regions (r = 0.36).
The number of nonprocessed pseudogenes also correlates with the size of the chromosomes (r = 0.78) at a similar level as genes (annotated in ENSEMBL) do (r = 0.67). Despite an unexpected weak overall correlation between these pseudogenes and genes (r = 0.34 in a 2-Mb window), they often cluster in gene-rich regions. This can be illustrated by the annotated T-cell receptor (TCR) β locus on Chromosome 7. In the 0.7-Mb TCR β region, we detected 11 of the 19 annotated nonprocessed pseudogenes that are located in between the 74 genes (accession no. NG_001333). Compared with the average density of 1.7 nonprocessed pseudogenes per megabase, this is a 10-fold enrichment.
In agreement with the distribution of pseudogenes derived from ribosomal protein genes (Zhang et al. 2002), and in contrast to functional genes (International Human Genome Sequencing Consortium 2001), we found no significant correlation of the CG content (measured using 100- and 2000-kb windows) and the number of processed or nonprocessed pseudogenes (data not shown).
DISCUSSION
We identified ∼20,000 pseudogenes in the human genome. The strategy used in this study ensures that each pseudogenic region represents a single event of gene or exon duplication and that regions matching to the same protein are fused. Therefore, the number of regions should correspond to the number of (pseudo)-genes in between annotated genes. Nearly all of the regions appear as neutrally evolving and therefore nonfunctional according to their associated KA/KS values. Only ∼1000 regions are estimated to evolve under selective pressure and might correspond to (parts of) functional genes missed in the first-pass genome annotation, or even to nonprocessed pseudogenes that were subjected to prolonged purifying selection before their pseudogenization. Yet we cannot exclude that a few cases of overlooked genes that are fast evolving are classified here as neutrally evolving pseudogenes.
Although nearly all human pseudogenes identified here match a mouse genomic region, the vast majority of them (92%) align better to another human gene (likely the functional paralog), indicating that they formed after the split of human and mouse. The remaining 8% could be due to pseudogenes that arose before the human–mouse split, overlooked genes, null alleles (Menashe et al. 2003), or to niche losses. This observation implies that the genome orthology between mouse and human can be used to obtain the first reliable genome-wide discrimination between detectable processed and nonprocessed pseudogenes. This, in turn, is an essential prerequisite to evaluate the impact of pseudogenes on gene and protein evolution. Of the pseudogenes identified here, 72% arose through retrotransposition, whereas 28% were formed by segmental duplication. This last fraction appears slightly higher than estimated in a previous analysis of human Chromosome 7 (18% nonprocessed pseudogenes; Hillier et al. 2003). This difference is probably due to the higher sensitivity of our study in regions with clustered genes and pseudogenes, as we implemented a special procedure for the identification of duplicated regions (see Methods). Although the number of both types of pseudogenes correlates with the size of the chromosomes, their intrachromosomal distribution differs. Processed pseudogenes are more abundant in regions adjacent to telomeres, and their distribution does not correlate with the distribution of genes within chromosomes. This is in disagreement with the idea that regions with relaxed chromatin, that is, with higher transcriptional activity, are more exposed to the integration of retrotransposed elements (Cost and Boeke 1998). Although there are regions with a higher density of processed pseudogenes, this cannot be necessarily coupled to the presence of genes. The latter effect is much more apparent for nonprocessed pseudogenes that are enriched in many (but not all) genedense regions. The distinction between the two types of pseudogenes enables both the evaluation of the rate and age of gene duplication events, and the study of the processes leading to the formation of new gene functions or expression profiles.
Although the discovery of almost 20,000 human pseudogenes represents the largest collection so far in any genome, the real number of human pseudogenes must be considerably higher. Our approach identifies neither pseudogenes derived from the duplication of non-protein-coding genes nor copies of partially retrotransposed mRNAs that include only the 3′-UTR and no (or little) coding region. The latter seem abundant in the genome (Pavlicek et al. 2002), and their detection is essential for a complete study of the retrotransposition of mRNAs. Because we initially masked all known and predicted genes provided by the ENSEMBL annotation pipeline, we excluded pseudogenes misannotated as genes from our analysis. The current annotation of genomes can include up to 20% processed and nonprocessed pseudogenes in their gene collections (International Human Genome Sequencing Consortium 2001; Mounsey et al. 2002; Mouse Genome Sequencing Consortium 2002; D. Torrents, M. Suyama, and P. Bork, unpubl.), which implies that we could have missed up to 5000 of these regions just by this effect. Masking genes also excludes pseudogenes located within introns. As ∼10% of the processed pseudogenes identified in human Chromosome 7 are located within introns of unrelated genes (Hillier et al. 2003), we might have missed as many as 2000 intronic pseudogenes. A small fraction of pseudogenes will be missed by masking identified human repeats (3% of the annotated pseudogenes in human Chromosome 22 are located in repetitive regions). Last but not least, homology detection has its limits, and we don't see or include (see filtering in Results and Methods) pseudogenes that diverged beyond the limits of reliable alignment and statistical significance for sequence similarity. Obviously, we detect only the tip of the iceberg with our present methods, and it is reasonable to speculate that a considerable fraction of the human genome has evolved from pseudogenes.
Taken together, it is very likely that the number of detectable human pseudogenes will exceed 30,000 and thus most likely also the number of genes in the human genome.
METHODS
Homology Searches
We obtained the human genome DNA sequence (build30) from NCBI (ftp://ftp.ncbi.nih.gov/genomes/) already masked for common human repeats and excluded all regions covered by the known and predicted genes found in the ENSEMBL database (Hubbard et al. 2002). The initial homology searches comprised the comparison of all nonmasked fragments (>100 nt) with a nonredundant protein database comprising EMBL CDS translations + PDB + SWISS-PROT + PIR annotations (NRDB) using BLASTX (Altschul et al. 1997) with an initial E-value cutoff of 0.01. We excluded all regions similar to transposon or viral proteins by analyzing the description lines for all the hits provided by the database and discarding those with manually selected characteristic words such as “gag,” “pol,” reverse transcriptase, transposase, viral protein, and others; as well as regions with InterPro domains (Mulder et al. 2003) distinctive of transposon or retroviral proteins (e.g., IPR000477 from reverse transcriptases). The remaining fragments were further processed with the BLAST2GENE protocol (M. Suyama, D. Torrents, and P. Bork, in prep.), which includes different steps to identify all similar (pseudo)genes located in neighboring regions (i.e., in clusters): (1) Within each chromosome, all DNA fragments that share common protein hits were identified; (2) unmasked DNA region located between the first (upstream) and the last (downstream) of these fragments was extracted; (3) each of these large regions was compared with the common protein using BLASTX; (4) starting from the 5′-end of the DNA and from the N terminus of the protein, the peptide regions that are consecutively aligned to the DNA were evaluated to finally delimit subregions of DNA that can be completely or partially aligned to the protein only once. We next extracted the corresponding (pseudo)coding sequences included in these regions together with the position of detectable frameshifts and stop codons by comparing each of the divided regions again with the same protein using GENEWISE (accepting scores >35). At this stage we re-evaluated and discarded a possible remaining overlap with ENSEMBL predicted genes and finally compared each of our predicted coding regions to the NRDB database with BLASTX, now accepting E-values < 0.001.
Distinction Between Processed and Nonprocessed Regions
To differentiate between retrotransposed and segmentally duplicated pseudogenes, we took 217 conserved blocks as the orthologous correspondence between the mouse and human genomes (Mouse Genome Sequencing Consortium 2002). These orthologous blocks have an N50 of 23.2 Mb and cover ∼90% of each genome. Based on their location, we next assigned the translation of each of our pseudogenes to the corresponding mouse DNA block and compared them with TBLASTN (Altschul et al. 1997). We considered it a positive match whenever the associated E-value was <10-8 for any of the matching subregions.
KA/KS Calculations
Each target sequence (sequence A) was first compared with a nonredundant protein database (EMBL CDS translations + PDB + SWISS-PROT + PIR) using BLAST. Peptide and DNA sequences were automatically collected for each BLAST hit using the Sequence Retrieval System 5.0 (Etzold et al. 1996). To ensure a minimum of divergence among sequences, we compared them all pairwise and excluded all redundancy above a 95% amino acid identity. Protein matches identical or with <50% identical residues to our target sequence were also excluded. We then selected the remaining first and second sequences (sequences B and C) from the BLAST homologous list (sorted by their E-values) and performed a BLAST and GENEWISE comparison between each of them and the target DNA sequence. We chose those regions of A, B, and C capable of being aligned by BLAST and GENEWISE to construct a protein multiple alignment (CLUSTAL W; Thompson et al. 1994) and removed those regions presenting gaps in any of the aligned sequences. On the basis of such protein alignment, we obtained the corresponding codon alignment. We further excluded all the alignments shorter than 100 nt as uninformative. Next we reconstructed the hypothetical ancestral nucleotide sequence of A, B, and C using PAMP (PAML package; Yang 1997), which was then used as a reference to infer the synonymous and nonsynonymous substitutions specific for A using YN00 (PAML package). Although it is not required for our calculation, in most of the cases, sequences A and B are closer than A, or B to C. In parallel we obtained KA/KS values from the protein-based codon alignment described above using the maximum-likelihood-based CODEML program (model = 1; PAML package). Generally, the application of similar protocols in evolutionary studies implies the comparison of the whole coding region of the problem sequence with close paralogs and orthologs (Ohta and Ina 1995). Because this condition would dramatically limit the number of sequences in our set suitable for analysis, we used more distant homologs whenever closest sequences were not available. To partially compensate for a possible loss of sensitivity caused by the larger evolutionary distance and to avoid uninformative high KS values, we restricted this analysis to the most conserved regions among all three sequences. These regions are expected to be under a higher selective pressure in functional genes and hence to evolve much more slowly. In addition to the filters already included in each of the programs we used, we did not accept KA/KS calculations based on either a high or low number of substitutions (KS > 1 and KS < 0.1), which was expected to equally affect low (of genes) and high (of pseudogenes) KA/KS values. Less than 10% of the sequences analyzed in this work yielded a KS > 1. Positive selection, theoretically observed by KA/KS < 1, is suspected in very few cases (Endo et al. 1996) and is therefore not considered here.
Cross-Validation and Analysis of KA/KS Distributions
We evaluated the fraction of neutrally evolving regions included in a given set of sequences by comparing the distribution of its associated KA/KS values with those of the functional and pseudogenic benchmark sets. This estimation was applied in the cross-validation study, as well as in the 14,880 candidate regions as follows: For a given test distribution, we sequentially set different combinations of pseudogene (P) and gene (G) fractions (increasing/decreasing by 1). Each P/G distribution was then drawn using corresponding pseudogenic and functional KA/KS distributions as a pattern. From all possible P/G combinations, we adopted as the best estimation the one that best fitted the observed KA/KS distribution, for example, minimizing the sum of error squares.
This procedure, as well as the discrimination power of both benchmark distributions, was assessed by cross-validation as follows: From each benchmark set, we randomly extracted subsets (1000 sequences in total) of functional and pseudogenic elements in different and known proportions (iterated 100 times for each combination) and used them as known test sets. The pseudogene content for each of these test combinations was then estimated by least-squares fitting using all remaining sequences as the training set. All estimated and known pseudogene contents were then compared by extracting the associated expected error, which was <3% for all tested combinations (Table 2).
Table 2.
Cross-Validation for the Discrimination Between Genes and Pseudogenes from KA/KS Benchmark Distributions
|
Known test fractions
|
Training set
|
Estimated fractions of pseudogenes
|
|||
|---|---|---|---|---|---|
| Functional | Pseudogene | Functional | Pseudogene | Averagea | SDb |
| 1000 | 0 | 659 | 1703 | 16.1 | 15 |
| 900 | 100 | 759 | 1603 | 109.6 | 24.8 |
| 800 | 200 | 859 | 1503 | 205.4 | 20.5 |
| 700 | 300 | 959 | 1403 | 310.2 | 23.7 |
| 600 | 400 | 1059 | 1303 | 401.9 | 19.7 |
| 500 | 500 | 1159 | 1203 | 500.8 | 21.3 |
| 400 | 600 | 1259 | 1103 | 600.5 | 24.2 |
| 300 | 700 | 1359 | 1003 | 694.7 | 24.1 |
| 200 | 800 | 1459 | 903 | 793.2 | 26.1 |
| 100 | 900 | 1559 | 803 | 893.2 | 26.4 |
| 0 | 1000 | 1659 | 703 | 983.9 | 20 |
Average estimation of the 100 iterations
Standard Deviation from the complete test set
In addition to our ancestral-based calculation, we also evaluated the distributions of the KA/KS values associated with the pseudogenic and functional benchmark sets under an approach based on maximum likelihood (CODEML included in PAML package) with either three sequences (model = 1) or in pairwise (model = -2). The distributions derived from the three-sequence alignment calculation were almost identical to those obtained with our method (only the associated error provided by the cross-validation was slightly higher: 4.2%). However, as expected, the distribution associated with the pseudogenic benchmark set when the values were obtained from pairwise comparison is shifted toward lower values (KA/KS median = 0.17, in contrast to KA/KS median = 0.4 obtained with our method) and overlapped considerably with the KA/KS distribution of genes.
Acknowledgments
We thank Caroline Hadley and Patrick Aloy for manuscript revision.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1455503.
Footnotes
[Supplemental information as well as the sequences identified in this work can be found at http://www.bork.embl-heidelberg.de/Docu/Human_Pseudogenes/.]
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney, E. and Durbin, R. 1997. Dynamite: A flexible code generating language for dynamic programming methods used in sequence comparison. Proc. Int. Conf. Intell. Syst. Mol. Biol. 5: 56-64. [PubMed] [Google Scholar]
- Brosius, J. 1999. RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene 238: 115-134. [DOI] [PubMed] [Google Scholar]
- Bustamante, C.D., Nielsen, R., and Hartl, D.L. 2002. A maximum likelihood method for analyzing pseudogene evolution: Implications for silent site evolution in humans and rodents. Mol. Biol. Evol. 19: 110-117. [DOI] [PubMed] [Google Scholar]
- Collins, J.E., Goward, M.E., Cole, C.G., Smink, L.J., Huckle, E.J., Knowles, S., Bye, J.M., Beare, D.M., and Dunham, I. 2003. Reevaluating human gene annotation: A second-generation analysis of chromosome 22. Genome Res. 13: 27-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cost, G.J. and Boeke, J.D. 1998. Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry 37: 18081-18093. [DOI] [PubMed] [Google Scholar]
- Endo, T., Ikeo, K., and Gojobori, T. 1996. Large-scale search for genes on which positive selection may operate. Mol. Biol. Evol. 13: 685-690. [DOI] [PubMed] [Google Scholar]
- Etzold, T., Ulyanov, A., and Argos, P. 1996. SRS: Information retrieval system for molecular biology data banks. Methods Enzymol. 266: 114-128. [DOI] [PubMed] [Google Scholar]
- Goncalves, I., Duret, L., and Mouchiroud, D. 2000. Nature and structure of human genes that generate retropseudogenes. Genome Res. 10: 672-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, P.M., Echols, N., and Gerstein, M.B. 2001. Digging for dead genes: An analysis of the characteristics of the pseudogene population in the Caenorhabditis elegans genome. Nucleic Acids Res. 29: 818-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, P.M., Hegyi, H., Balasubramanian, S., Luscombe, N.M., Bertone, P., Echols, N., Johnson, T., and Gerstein, M. 2002. Molecular fossils in the human genome: Identification and analysis of the pseudogenes in chromosomes 21 and 22. Genome Res. 12: 272-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori, M., Fujiyama, A., Taylor, T.D., Watanabe, H., Yada, T., Park, H.S., Toyoda, A., Ishii, K., Totoki, Y., Choi, D.K., et al. 2000. The DNA sequence of human chromosome 21. Nature 405: 311-319. [DOI] [PubMed] [Google Scholar]
- Hillier, L.W., Fulton, R.S., Fulton, L.A., Graves, T.A., Pepin, K.H., Wagner-McPherson, C., Layman, D., Maas, J., Jaeger, S., Walker, R., et al. 2003. The DNA sequence of human chromosome 7. Nature 424: 157-164. [DOI] [PubMed] [Google Scholar]
- Hubbard, T., Barker, D., Birney, E., Cameron, G., Chen, Y., Clark, L., Cox, T., Cuff, J., Curwen, V., Down, T., et al. 2002. The Ensembl genome database project. Nucleic Acids Res. 30: 38-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Kimura, M. 1968. Evolutionary rate at the molecular level. Nature 217: 624-626. [DOI] [PubMed] [Google Scholar]
- Li, W.H., Gojobori, T., and Nei, M. 1981. Pseudogenes as a paradigm of neutral evolution. Nature 292: 237-239. [DOI] [PubMed] [Google Scholar]
- Menashe, I., Man, O., Lancet, D., and Gilad, Y. 2003. Different noses for different people. Nat. Genet. 34: 143-144. [DOI] [PubMed] [Google Scholar]
- Mighell, A.J., Smith, N.R., Robinson, P.A., and Markham, A.F. 2000. Vertebrate pseudogenes. FEBS Lett. 468: 109-114. [DOI] [PubMed] [Google Scholar]
- Mounsey, A., Bauer, P., and Hope, I.A. 2002. Evidence suggesting that a fifth of annotated Caenorhabditis elegans genes may be pseudogenes. Genome Res. 12: 770-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Mulder, N.J., Apweiler, R., Attwood, T.K., Bairoch, A., Barrell, D., Bateman, A., Binns, D., Biswas, M., Bradley, P., Bork, P., et al. 2003. The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res. 31: 315-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, T. and Ina, Y. 1995. Variation in synonymous substitution rates among mammalian genes and the correlation between synonymous and nonsynonymous divergences. J. Mol. Evol. 41: 717-720. [DOI] [PubMed] [Google Scholar]
- Pavlicek, A., Paces, J., Zika, R., and Hejnar, J. 2002. Length distribution of long interspersed nucleotide elements (LINEs) and processed pseudogenes of human endogenous retroviruses: Implications for retrotransposition and pseudogene detection. Gene 300: 189-194. [DOI] [PubMed] [Google Scholar]
- Prince, V.E.and Pickett, F.B. 2002. Splitting pairs: The diverging fates of duplicated genes. Nat. Rev. Genet. 3: 827-837. [DOI] [PubMed] [Google Scholar]
- Pruitt, K.D.and Maglott, D.R. 2001. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 29: 137-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanin, E.F.1985. Processed pseudogenes: Characteristics and evolution. Annu. Rev. Genet. 19: 253-272. [DOI] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- Yang, Z. 1997. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555-556. [DOI] [PubMed] [Google Scholar]
- Zdobnov, E.M., von Mering, C., Letunic, I., Torrents, D., Suyama, M., Copley, R.R., Christophides, G.K., Thomasova, D., Holt, R.A., Subramanian, G.M., et al. 2002. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science 298: 149-159. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., Harrison, P., and Gerstein, M. 2002. Identification and analysis of over 2000 ribosomal protein pseudogenes in the human genome. Genome Res. 12: 1466-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- ftp://ftp.ncbi.nih.gov/genomes/; NCBI.
- http://www.bork.embl-heidelberg.de/Docu/Human_Pseudogenes/; authors' Web site.