Abstract
We propose a novel method to identify functionally related genes based on comparisons of neighborhoods in gene networks. This method does not rely on gene sequence or protein structure homologies, and it can be applied to any organism and a wide variety of experimental data sets. The character of the predicted gene relationships depends on the underlying networks;they concern biological processes rather than the molecular function. We used the method to analyze gene networks derived from genome-wide chromatin immunoprecipitation experiments, a large-scale gene deletion study, and from the genomic positions of consensus binding sites for transcription factors of the yeast Saccharomyces cerevisiae. We identified 816 functional relationships between 159 genes and show that these relationships correspond to protein–protein interactions, co-occurrence in the same protein complexes, and/or co-occurrence in abstracts of scientific articles. Our results suggest functions for seven previously uncharacterized yeast genes: KIN3 and YMR269W may be involved in biological processes related to cell growth and/or maintenance, whereas IES6, YEL008W, YEL033W, YHL029C, YMR010W, and YMR031W-A are likely to have metabolic functions.
The function of many genes is still unknown; even for the well studied yeast Saccharomyces cerevisiae, about one-third of all genes are still uncharacterized (Ball et al. 2001). Functions of uncharacterized proteins are usually inferred computationally on the basis of sequence similarities, common structural motifs, gene order, gene fusion events, or similarities in gene expression (Bork and Koonin 1998; Enright et al. 1999; Marcotte et al. 1999; Ge et al. 2001; Ponting 2001; Kemmeren et al. 2002; Valencia and Pazos 2002; Wu et al. 2002; Huynen et al. 2003). Here we introduce a simple and general statistical method for functional predictions based on scoring the similarity of gene neighborhoods in various gene networks. It allows us to utilize recently published biological data from high-throughput technologies. This method allows us to perform functional predictions for proteins independent of homologies in protein structure or sequence and provides a way to characterize proteins that have not been studied previously.
Many biological data sets can be represented as gene networks, where nodes represent genes or proteins, and the connections between the nodes represent relationships between these entities. Directed relationships such as “protein A activates gene B” are represented by arcs (A B), whereas symmetric relationships such as “protein A and protein B bind to each other” are represented by edges (A—B; Schwikowski et al. 2000; Walhout and Vidal 2001; Gerstein et al. 2002; Schlitt and Brazma 2002; von Mering et al. 2002).
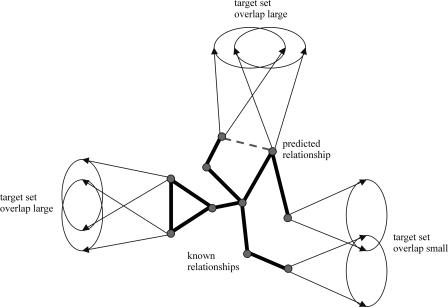
We compared the neighborhoods of genes in networks derived from microarray experiments on gene deletion mutants (Hughes et al. 2000), the localization of transcription factor binding sites (Pilpel et al. 2001), and chromatin immunoprecipitation (ChIP) experiments for the yeast Saccharomyces cerevisiae (Ren et al. 2000; Iyer et al. 2001; Simon et al. 2001; Lee et al. 2002). By neighborhood of a gene A we mean the set of genes that are directly connected to gene A in the network. If two genes share many neighbors in a network, it suggests that these genes might be functionally related (Fig. 1).
Figure 1.
Illustration of the correspondence between functionally related genes and similarity of the target sets. Pairs of functionally unrelated genes have smaller target-set overlaps. Large overlaps can be used to predict a functional relationship between the respective genes (top).
Validation of functional relationships is problematic, because various aspects and meanings are subsumed under the term “function” of a gene or protein. This is mainly due to different experimental approaches that focus either on the effects of mutations or on biochemical activities (Ashburner et al. 2000). Unlike in protein structure prediction, there are no established standards for the evaluation of functional predictions (Blaschke et al. 2002).
We use three approaches to validate the predicted functional relationships: We compare the gene pairs that are predicted to be related (1) with protein–protein interaction data, (2) with protein complexes, and (3) with a literature network. Many biological functions involve protein–protein interactions, and several large protein–protein interaction data sets are available (Uetz et al. 2000; Ito et al. 2001; Gavin et al. 2002; Ho et al. 2002). These data sets are a valuable resource, although they may contain a large number of false positives and are far from being complete (Bader and Hogue 2002; Edwards et al. 2002; von Mering et al. 2002). For protein complexes, a manually annotated data set of high quality is available from the Munich Information Centre on Protein Sequences (MIPS; http://mips.gsf.de/; Mewes et al. 2002). Protein function is not restricted to protein–protein interactions, and for that reason we included knowledge from published scientific articles in our verification procedure. We analyzed the frequency of co-occurrences of gene names in abstracts of scientific articles on S. cerevisiae to construct a literature network. Similar approaches have been used before under the assumption that functionally related genes occur more often in the same abstract than unrelated genes do (Blaschke et al. 1999; Jenssen et al. 2001).
Here we describe how the comparison of gene neighborhoods from different gene networks can be used to identify functionally related genes. We provide evidence that gene pairs with similar network neighborhoods occur more frequently together in article abstracts and more frequently encode proteins that interact physically than do genes with dissimilar neighborhoods. Our method allowed us to identify 816 functional relationships between 159 genes and to assign biological process annotation to seven previously uncharacterized genes. We examine some of the predictions in detail, and show that for the networks studied here the predicted functions concern biological processes rather than biochemical activities.
RESULTS
Our aim was to study the similarity of genes or proteins by assessing the similarity of their neighborhoods in gene networks (Fig. 2). Here we studied relationships between genes/proteins in six different networks of three different types for the yeast Saccharomyces cerevisiae (Table 1):
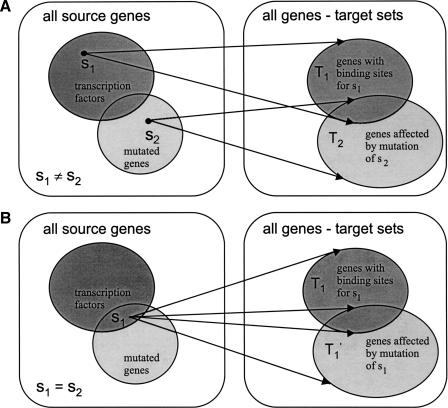
Figure 2.
Transcription factors with known binding sites and mutated genes form two sets of source genes (lefthand side). (A) The set T1 on the right represents all the genes in the genome that have binding sites for selected transcription factor s1 in their putative promoter regions (i.e., the target set of s1). The set T2 represents all the genes whose expression levels are changed in the deletion mutant of gene s2 (i.e., the target set of s1). If the target sets T1 and T2 overlap more than expected by chance, we can hypothesize that the two genes s1 and s2 are related. (B) The case when we compare two target sets from different networks, but for one gene s1.
Table 1.
Number of Source Genes, Total Number of Genes, Number of Connections, and the Ratio of Connections per Source Gene in Each Comprehensive Target Network
|
ChIP network
|
||||||
|---|---|---|---|---|---|---|
| Network | in silico network | mutant network | ren | simon | iyer | lee |
| Source genes | 38 | 187 | 2 | 9 | 3 | 83 |
| Genes | 5583 | 5555 | 130 | 567 | 207 | 2351 |
| Connections | 23446 | 27252 | 131 | 1208 | 453 | 4235 |
| Connections per source gene | 617.0 | 145.7 | 65.5 | 134.2 | 151.0 | 51.0 |
The maximal possible number of target genes in each network is the complete gene set of the yeast Saccharomyces cerevisiae (∼6200 genes).
Mutant network: An arc from a gene A to gene B means that in a mutant where A is deleted, the expression level of B is significantly changed (Rung et al. 2002). The network is derived from microarray studies of yeast mutants by Hughes et al. (2000).
In silico network: An arc from gene A to B means that A is a transcription factor, and its binding site is predicted in the putative promoter of B (Palin et al. 2002). The network is derived from the data of Pilpel et al. (2001), who matched binding sites against all upstream sequences in the entire yeast genome computationally. We included only the empirically known binding sites.
Four different ChIP networks: These were constructed from genome-wide transcription factor localization data based on ChIP experiments (Ren et al. 2000; Iyer et al. 2001; Simon et al. 2001; Lee et al. 2002). In ChIP networks, an arc from gene A to gene B means that transcription factor A was empirically found to bind to the putative promoter region of B.
All networks listed above are represented as directed graphs. In a directed graph, a node can have incoming and outgoing arcs, and thus we can divide the neighborhood of a node depending on the orientation of the arcs. We call the genes with outgoing arcs source genes, and for every source gene s1 we define the target set T1 as the set of genes which have incoming arcs from s1 (see Figs. 1, 2). All of the networks described above are asymmetric: Although source genes are an a priori selected subset of the genome (particular for each network), the whole genome is tested for targets. We call such networks comprehensive target networks.
For every pair of source genes s1 and s2, we test whether their target sets T1 and T2 intersect more than expected by chance, using the hypergeometric distribution (Sokal and Rohlf 1995) and Holm's correction (Holm 1979) for multiple testing (which leads to some P-values being greater than 1).
We performed 23,758 target-set comparisons for 15,061 source gene pairs within and between the networks (Table 2). For 816 (5.4%) source gene pairs, we found a strong target-set similarity (P ≤ 0.01). We provide the results of our target-set comparisons for all source gene pairs within our Supplemental data (full-table-long.txt), available at www.genome.org.
Table 2.
Number of Target Set Comparisons Which Have Been Performed (Total) and the Number and Proportion of Highly Similar Target Sets (P ≤ 0.01)
| Source gene pairs | Total | P ≤ 0.01 | % |
|---|---|---|---|
| s1 = s2, from different networks | 46 | 17 | 36.9 |
| s1 ≠ s2, from the same network | 7838 | 741 | 9.5 |
| s1 ≠ s2, from different networks | 10405 | 143 | 1.3 |
| All pairs s1, s2 | 15061 | 816 | 5.4 |
The source genes s1 and s2 are chosen from the same or from different networks.
When we compared target sets for the same source gene from different networks, we found that 34 out of 80 target-set pairs are highly similar. The similarities occur more frequently between the ChIP networks and between the in silico network and the ChIP networks. According to this comparison, the ChIP networks are similar to each other, and to the in silico network, whereas the mutant network is most different from the others. This is consistent with the small intersection of the mutant network and the ChIP networks: They share 16 source genes, but only 78 connections, although there are on average between 51 and 145 connections per source gene in both networks (Table 1).
To test whether the target-set similarity can be used to identify functionally related genes, we used three additional networks as reference networks:
Protein–protein interaction networks: Two proteins are connected by an edge if they physically interact. We integrated protein–protein interaction data from several large-scale experiments, including yeast two-hybrid screens (Uetz et al. 2000; Ito et al. 2001) and complex purifications with subsequent protein identification by mass spectrometry (TAP; Gavin et al. 2002; HMS-PCI; Ho et al. 2002). We used two different networks, ppi1 and ppi2. In ppi1, each identified interaction is represented by an edge, whereas ppi2 contains an edge only if two or more experiments identified the same interaction. Thus, edges in network ppi2 are more reliable, but consequently ppi2 is much sparser than ppi1.
mips network: This was derived from manually annotated complexes at MIPS (Mewes et al. 2002); two proteins are connected by an edge if they are components of the same complex. A similar network was used by von Mering et al. (2002) to assess the quality of recent large-scale experiments on protein–protein interaction in yeast; unfortunately the full reference network which was used is no longer publicly available.
Cocitation networks: Here, two genes are connected by an edge if they have been cocited in at least a specified number of abstracts (n). Again we constructed two different networks, mi2 and mi3. For mi2, we required cocitations in at least two abstracts (n=2), and for the more stringent but sparser network mi3, we required cocitations in at least three abstracts (n=3).
Functionally related genes are connected in these reference networks, and therefore we validated the results of the target-set comparison by comparing them with the connectivity in the reference networks. The proportion of genes with similar target sets increases four- to eightfold if we consider only gene pairs present in the reference networks, instead of all possible source gene pairs (Table 3). This indicates that functionally related genes, that is, genes connected in the reference networks, have similar target sets. In order to test our hypothesis, we ranked all source gene pairs according to their best target-set similarity, that is, from high similarity (low P-values) to low similarity (high P-values). All source gene pairs with a reported interaction were counted as true positive (tp) if their corrected P-value was smaller than a chosen threshold or as false negative (fn) if their corrected P-value was greater than this threshold. Pairs lacking an interaction were counted as false-positive (fp) if their corrected P-value was smaller than the chosen threshold or as true negative (tn) if their corrected P-value was greater than the threshold. We calculated the true-positive rate (sensitivity) as tp/(tp+fn) and the false-positive rate (1 - specificity) as fp/(fp+tn) at each row of the ranking, using the P-value of the respective row as a threshold. An ROC curve displays the true-positive rate versus the false-positive rate in Figure 3. Ideal prediction methods have a high true-positive rate and a low false-positive rate, with ROC curves getting close to the upper left corner of the plot, whereas randomized predictions would produce ROC curves close to the diagonal from the lower left corner to the upper right corner (Witten and Eibe 1999).
Table 3.
Source Gene Pairs With High Target Set Similarity Correspond More Frequently to Edges in Reference Networks (ppi1, ppi2, mips, mi2, mi3)
|
All pairs
|
Confirmed pairs
|
||||||
|---|---|---|---|---|---|---|---|
| Reference network | Total-all | P ≤ 0.01 | % all | Total-conf | P ≤ 0.01 | % conf | Increase % conf/% al |
| A | B | C | D | E | F | G | |
| ppi1 | 10762 | 631 | 5.86 | 151 | 37 | 24.50 | 4.18 |
| ppi2 | 5804 | 341 | 5.88 | 38 | 10 | 26.32 | 4.48 |
| mips | 1283 | 75 | 5.85 | 31 | 15 | 48.39 | 8.28 |
| mi2 | 8546 | 474 | 5.55 | 267 | 63 | 23.60 | 4.25 |
| mi3 | 6701 | 400 | 5.97 | 172 | 50 | 29.07 | 4.87 |
Columns A–C refer to all source gene pairs, where both genes are present in the particular reference network; columns D–F refer only to source gene pairs which are connected in the particular reference network (= confirmed source gene pairs). The proportion of pairs with highly similar target sets is increased between four- and eight-fold for the confirmed source gene pairs.
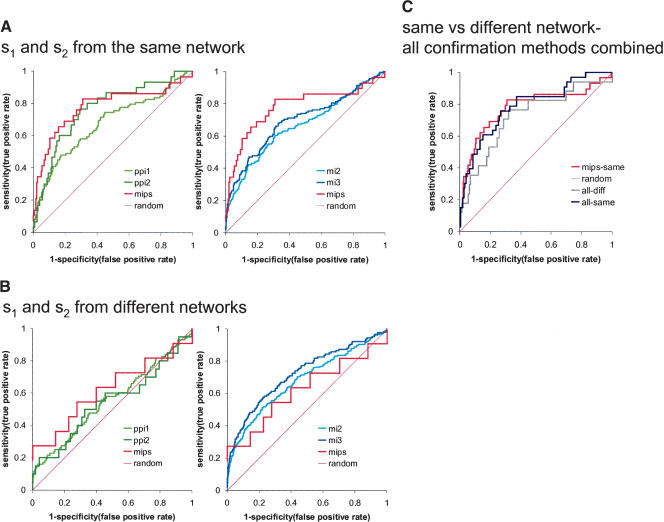
Figure 3.
ROC plots of true-positive rate (sensitivity) vs. false-positive rate (1 - specificity) for the prediction of protein–protein interaction (ppi1, ppi2), protein complexes (mips), and“co-citation” (mi2, mi3). The source genes s1, s2 are chosen from same (A) or different networks (B). (C) An ROC plot using the union of ppi2, mips, and mi3 as verification network, with source genes s1, s2 chosen from the same network (all-same) or different networks (all-diff).
The ROC curves in Figure 3 show the false-positive rate and the true-positive rate for our prediction method with respect to the different reference networks. A true-positive rate of 82% with a corresponding false-positive rate of 32% is found when using a verification network that is a union of ppi2, mips, and mi3 (Fig. 3C). If we use the more stringent reference sets ppi2 or mi3, the quality of our predictions is better (i.e., the ROC curve is further away from the diagonal). This effect may be due to high error rates in the reference sets; the accuracy of the protein–protein interaction network increases if several methods report the same interactions (Edwards et al. 2002; von Mering et al. 2002). We conclude that target-set similarity can be used as an indication of protein–protein interactions (pp1, ppi2, mips) and of cocitations (Fig. 3). This means that we can predict protein–protein interaction or a functional relationship based on target-set similarity.
If we base the predictions on target-set comparisons between different networks, we greatly expand the number of source gene pairs for which we perform target-set comparisons, but the false-positive rate also increases (Fig. 3B). This increase in false positives is higher for protein–protein interactions than for cocitations.
The data indicate that for the identification of protein–protein interactions, a comparison of source genes within the ChIP networks yield the best results. However, comparisons of target sets in the mutant network perform best for the identification of interactions in MIPS complexes and literature data. Generally, comparisons between different networks perform worse than comparisons within the same network (see netComparison.pdf in our Supplemental data). It should be noted that there is not enough data available for a reliable analysis of which network combinations yield the best predictions.
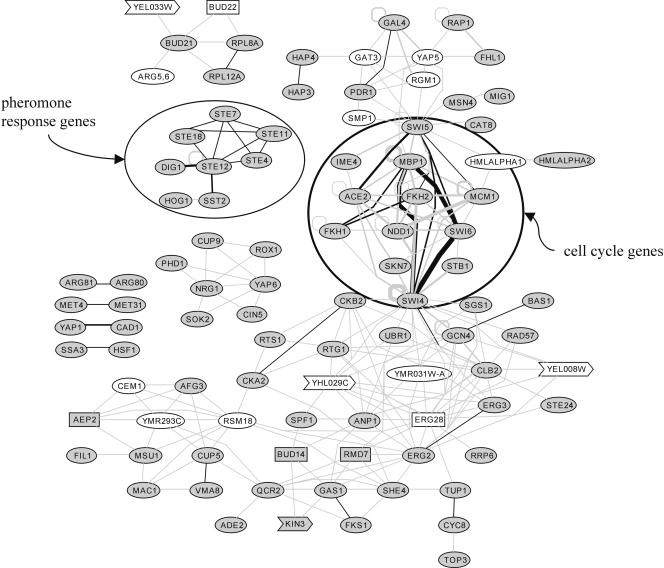
The correlation between target-set similarity and functional similarity is evident in the graph representation of the predictions (Fig. 4, fig4.txt in Supplemental data). Genes involved in the same biological processes such as pheromone response or cell-cycle control are linked by several target-set similarities, and are therefore close to each other in the graph. Applying a guilt-by-association approach, we used proximity in the graph to infer gene function (Oliver 2000): We predict function (using a P-value threshold of 10-12) for four genes (KIN3, YEL008W, YEL033W, and YHL029C) which are currently not assigned to a biological process in SGD.4 YEL033W is connected to only one other gene, BUD21, which is involved in rRNA processing; KIN3 shows strong target-set similarity to GAS1, a 1,3-β-glucanosyltransferase involved in cell wall organization and biogenesis, and to BUD14, which is involved in bud site selection according to SGD (http://www.yeastgenome.org/). This would imply that Kin3p may be involved in cell growth, budding, or related processes.
Figure 4.
Visualization of the source gene pairs with highly similar target sets as a graph. Genes involved in the same biological processes are often connected and are thus close in the resulting graph; e.g., pheromone response genes or cell-cycle genes (encircled). Genes sharing highly similar target sets (P ≤ 10-12) are connected by gray edges or, if there is also a corresponding edge in one of the reference networks ppi2, mi3, and mips, by black edges. The thicker edges indicate that the respective source gene pair had significantly similar target sets in several network comparisons. White nodes: genes which are not present in any of the reference networks; these genes therefore are not adjacent to any confirmed edges. Gray nodes: genes present in at least one of the reference networks. Rectangular nodes: genes with unknown molecular function (BUD14, ERG28, RMD7, BUD22, and AEP2). Arrow-shaped nodes: genes of unknown biological process (KIN3, YEL008W, YEL033W, YHL029C).
It is difficult to find terms describing a set of genes appropriately and objectively; therefore we use the “SGD Gene Ontology Term Mapper” (http://db.yeastgenome.org/cgi-bin/SGD/GO/goTermMapper). SGD uses the Gene Ontology (GO) terms from the Gene Ontology Consortium to annotate yeast genes (Dwight et al. 2002; Blake and Harris 2003). GO terms are organized hierarchically, which allows an investigator to find higher-level terms starting from a more specific GO term. GO Term Mapper does exactly this; starting from the GO annotation for each gene in the group, it identifies GO terms which are shared by the whole group (or at least the majority of the genes). When we query GO Term Mapper for the genes that have similar target sets as KIN3, it returns “cell growth and/or maintenance” as a common annotation, which is consistent with the conclusion we reached above. The same approach applied to the target sets of YEL008W and YHL029C suggests that these genes are involved in metabolism (Table 4). Four additional uncharacterized genes show high target-set similarities (at a P-value threshold of 0.01) to several other source genes. Using GO Term Mapper as described above, IES6, YMR010W, and YMR031W-A can be mapped to “metabolism,” whereas YMR269W can be mapped to “cell growth and/or maintenance” (see predictions.pdf in Supplemental data).
Table 4.
Part of the Data Shown in Figure 4
| Source gene 1 | Network 1 | Source gene 2 | Network 2 | P value | ppi | mi | mips | goTermMapper | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genes for which the biological process is unknown (SGD) | ||||||||||||||||
| YEL033W | mutant | BUD21 | mutant | 2.18E-14 | ||||||||||||
| KIN3 | mutant | GAS1 | mutant | 9.16E-26 | cell growth/maintenance | |||||||||||
| KIN3 | mutant | BUD14 | mutant | 1.12E-20 | ||||||||||||
| YEL0008W | mutant | CLB2 | mutant | 1.41E-20 | metabolism | |||||||||||
| YEL008W | mutant | ERG2 | mutant | 2.08E-18 | ||||||||||||
| YEL008W | mutant | ERG28 | mutant | 3.64E-25 | ||||||||||||
| YEL008W | mutant | SWI4 | mutant | 1.12E-21 | ||||||||||||
| YEL008W | mutant | GCN4 | mutant | 1.61E-15 | ||||||||||||
| YHL029C | mutant | ANP1 | mutant | 1.28E-26 | metabolism | |||||||||||
| YHL029C | mutant | CKB2 | mutant | 9.49E-18 | ||||||||||||
| YHL029C | mutant | CLB2 | mutant | 7.38E-43 | ||||||||||||
| YHL029C | mutant | ERG2 | mutant | 1.57E-34 | ||||||||||||
| YHL029C | mutant | ERG28 | mutant | 1.18E-50 | ||||||||||||
| YHL029C | mutant | ERG3 | mutant | 1.12E-20 | ||||||||||||
| YHL029C | mutant | GCN4 | mutant | 3.44E-30 | ||||||||||||
| YHL029C | mutant | RTG1 | mutant | 3.44E-32 | ||||||||||||
| YHL029C | mutant | SPF1 | mutant | 1.52E-13 | ||||||||||||
| YHL029C | mutant | SWI4 | mutant | 2.56E-64 | ||||||||||||
| YHL029C | mutant | UBR1 | mutant | 8.29E-25 | ||||||||||||
| Genes with similar target sets as SWI6 | ||||||||||||||||
| SWI6 | ChIP (Simon) | FKH2 | ChIP (Simon) | 5.02E-76 | no | no | cell cycle | |||||||||
| SWI6 | ChIP (Simon) | MBP1 | ChIP (Lee) | 1.14E-65 | yes | yes | yes | cel growth/maintenance | ||||||||
| SWI6 | ChIP (Lee) | MBP1 | ChIP (Lee) | 8.60E-54 | yes | yes | yes | metabolism | ||||||||
| SWI6 | ChIP (Simon) | SWI4 | ChIP (Iyer) | 3.24E-51 | no | yes | ||||||||||
| SWI6 | ChIP (Lee) | SWI4 | ChIP (Lee) | 4.82E-49 | no | yes | ||||||||||
| SWI6 | ChIP (Simon) | ACE2 | ChIP (Simon) | 3.28E-48 | no | yes | ||||||||||
| SWI6 | ChIP (Simon) | SWI6 | ChIP (Lee) | 6.01E-46 | no | no | no | |||||||||
| SWI6 | ChIP (Lee) | SWI6 | ChIP (Simon) | 6.01E-46 | no | no | no | |||||||||
| SWI6 | ChIP (Simon) | NDD1 | ChIP (Simon) | 4.12E-45 | no | |||||||||||
| SWI6 | ChIP (Simon) | NDD1 | ChIP (Lee) | 1.54E-28 | no | |||||||||||
| SWI6 | ChIP (Simon) | SWI5 | ChIP (Simon) | 7.51E-28 | no | yes | ||||||||||
| SWI6 | ChIP (Simon) | ACE2 | ChIP (Lee) | 1.71E-20 | no | yes | ||||||||||
| SWI6 | ChIP (Simon) | FKH2 | ChIP (Lee) | 4.99E-20 | no | no | ||||||||||
| SWI6 | ChIP (Simon) | SKN7 | ChIP (Lee) | 3.48E-19 | no | |||||||||||
| SWI6 | ChIP (Lee) | SWI4 | ChIP (Iyer) | 5.79E-18 | no | yes | ||||||||||
| SWI6 | ChIP (Simon) | MCM1 | ChIP (Simon) | 1.94E-17 | no | no | no | |||||||||
| SWI6 | ChIP (Lee) | MBP1 | in-silico | 1.03E-16 | yes | yes | yes | |||||||||
| SWI6 | ChIP (Simon) | STB1 | ChIP (Lee) | 1.82E-16 | no | no | ||||||||||
| SWI6 | ChIP (Simon) | SWI5 | ChIP (Lee) | 2.52E-15 | no | yes | ||||||||||
The target set similarity for the particular source gene/network combination is indicated by the P value. yes; the particular interaction is confirmed by a reference network, no: both genes are present in the respective reference network, but no interaction is reported; ppi2, protein-protein interaction network, mi3, co-citation network, mips, mips network; the last column contains the GO term describing the set of genes according to GO Term Mapper http://genome-www4.stanford.edu/cgi-bin/SGD/GO/goTermMapper).
Lastly, we examined some source gene pairs with high target-set similarity in detail to illustrate the nature of our predictions: There are 14 source gene pairs for which both genes are present in the protein interaction network ppi2, but no interaction between them is reported in this network, although they are connected in cocitation network mi3. Of these 14 pairs, six have highly similar target sets (P ≤ 0.01). The pair with the lowest P-value (linked by 11 abstracts in the cocitation network) is MBP1-SWI4. Both genes encode related transcription factors, and each of them can form a complex with Swi6p: MBF (Swi6p-Mbp1p) and SBF (Swi6p-Swi4p; Koch et al. 1993). The second pair consists of the homologous transcription factors Ace2p and Swi5p (linked by three articles in cocitation network mi3; Stillman et al. 1994; Measday et al. 2000). The next two pairs are between genes involved in pheromone signaling: Ste12p-Ste4p (four abstracts) and Ste4p-Ste7p (four abstracts). The pheromone signal in yeast is transmitted from the receptor via a G-protein-complex (Ste4p and others) and a MAP kinase cascade (Ste7p and others) to the transcription factor Ste12p (Sprague and Thorner 1992). The two proteins of the fifth pair—repressor Sum1p and activator Ndt80p (three abstracts)—compete for the transcriptional control of genes containing a middle sporulation element (MSE) in their promoters (Xie et al. 1999; Lindgren et al. 2000). Mig1p (three abstracts) was shown to repress the expression of the SUC2 gene synergistically with the Ssn6p-Tup1p repressor complex (Alepuz et al. 1997). Thus, all six source gene pairs with high target-set similarity are in fact functionally related, but do not show protein–protein interactions.
DISCUSSION
We conclude that the comparison of target sets in gene networks can be used to find functionally related proteins: We predict 816 relations for 159 genes (P ≤ 0.01). The nature of the predicted functional relationships is dependent on the nature of the comprehensive target networks. The Gene Ontology consortium differentiates between three major subcategories “cellular localization,” “biological process,” and “molecular function” (Ashburner et al. 2000). For the networks studied here, the predicted functions concern biological processes rather than molecular functions. In this respect our method is similar to other nonhomology methods (Marcotte 2000). We demonstrated that the products of genes with similar network neighborhoods often interact physically, are likely to be part of the same protein complex, and/or are often reported together in the literature. These results are in agreement with the recent finding that protein–protein interactions correlate with protein–DNA interactions (Manke et al. 2003). The proposed method can be used to identify functionally related candidate genes using a guilt-by-association approach.
Our method can be used for the comparison of data from a variety of methods. Large-scale experiments can vary extensively in terms of data quality, as has been described by several groups (Edwards et al. 2002; von Mering et al. 2002). We employ a well established statistical method to cope with the high error rates in the underlying data. We do not believe that simple intersections or unions of networks constructed from large-scale high-throughput approaches are a reliable way to integrate these data of variable quality. Our method allows one to score interactions by comparing them to previously observed data obtained by the same or different experimental techniques.
With the proposed method we did not identify all functional relationships reported in the reference networks. It therefore remains an open question as to how many of the errors are due to limitations of the available data or due to the method. There are several reasons why not all of the target-set pairs derived from the same source gene, or from two genes having a known functional relationship, were highly similar. One reason is that we combined experimental data from different types of experiments, and certain interactions are only observable under very specific conditions not necessarily attained in a given experiment. For example, some transcription factors may bind DNA only if they are phosphorylated.
One advantage of this method is that we can use and integrate a wide variety of different experimental data sets, as long as they can be represented as comprehensive target networks. Even small data sets can be successfully included; unlike clustering of microarray data, there is no need for extensive experiments consisting of tens of microarray hybridizations to provide biologically meaningful results. Our method is versatile; in the present study, for instance, we were able to explore which transcription factor deletions lead to predicted effects on the basis of the localization of its binding sites. We can also look for transcription factors which act in combination with other factors and elucidate possible upstream regulatory mechanisms.
Although sequence information may be important for the design of the experiments which underlie the comprehensive target networks, this is not a prerequisite for our method, which is completely independent of sequence or structural homology. A limitation of this method is that the data sets used for our predictions must be represented as comprehensive target sets. This means that, for example, large-scale protein-interaction networks cannot be used, because of the way these experiments are performed. Only positive interactions are reported, and we do not know which protein interactions do not occur. In contrast, the data sets we included for the predictions always report a signal for all genes in the genome. Therefore, within the limitations of the experimental methods, we always have information regarding the individual behavior of all genes.
The possibility of integrating data derived from different experimental methods and conditions allows the exploration of the complexity of cellular regulatory mechanisms. It is feasible to perform repeated analysis of data from different experimental conditions and then use the variations in conditions to explain the changes in interactions predicted. This would lead to a dynamic rather than a static view of protein function.
METHODS
Construction of the Networks
The mutant network was constructed with data from Hughes et al. (2000). Target sets T contain genes whose expression level changed significantly; that is, the ratio of gene expression in the mutant divided by the background standard deviation in the wild-type strain has an absolute value larger than 2.5 (Rung et al. 2002).
The in silico network was compiled from data reported by Pilpel et al. (2001) on the occurrence of known binding sites in putative promoter regions of yeast genes.
The four ChIP networks were constructed from data published by Ren et al. (2000), Simon et al. (2001), Iyer et al. (2001), and Lee et al. (2002) derived for two, nine, three, and 106 transcription factors, respectively.
Experimental data on yeast protein–protein interactions was retrieved from the following databases and publicly available data sets: DIP (Xenarios et al. 2001), MINT (Zanzoni et al. 2002), MDS (Ho et al. 2002), and cellzome (Gavin et al. 2002). Although DIP and MINT contain binary interactions, the data from the Gavin and Ho studies contain sets of proteins from a number of immunoprecipitations. These were broken down into a complete set of binary interactions.
The MIPS network was derived from manually annotated complexes at MIPS (Mewes et al. 2002) and provided to us by Christian von Mering (von Mering et al. 2002).
The cocitation network: Using a synonym dictionary for gene/protein names in yeast, we scanned over 70,000 journal abstracts from Medline for co-occurrences of genes/proteins, using the SRS server (http://srs.ebi.ac.uk). We compiled a synonym dictionary containing the complete set of unique yeast ORF identifiers, the corresponding gene names, and their synonyms from publicly available information in the following databases: SGD (http://www.yeastgenome.org/), MIPS (http://mips.gsf.de/proj/yeast/), and EBI Proteome Analysis Database (http://www.ebi.ac.uk/proteome/). Each Medline entry was required to contain at least one ORF/gene name or one of its associated synonyms in the text body of the abstract or in the title; in addition, we required the MESH term `Saccharomyces cerevisiae' to limit the search to our chosen model organism. A co-occurrence between two different gene/protein identifiers was counted if they or any of their respective synonyms were found in the same abstract. This resulted in 41,129 associations, among which about 10285 pairs were co-occurring at least twice for 3616 genes. All networks are available from our Web supplement.
Network Comparison
Assessing the similarity between target sets using the hypergeometric distribution: The null hypothesis for testing the similarity of target sets T1 and T2 is that the genes in the sets are picked from the genome independently, randomly with equal probabilities. Under this null hypothesis, the number of genes in the intersection of T1 and T2 is distributed according to the hypergeometric distribution with the size of the genome, the size of T1, and the size of T2 as parameters (Palin et al. 2002). With this distribution we can compute the probabilities of observing an intersection at least this large, given that the null hypothesis is true. The pairwise P-values need to be corrected, because we evaluate multiple hypothesis tests. For the adjustment of the P-values, we used the sequential Holm's correction (Holm 1979). In order to be more stringent we only compared set target sets with 10 or more genes.
Acknowledgments
We thank Michael Ashburner, Aria Baniahmad, Cath Brooksbank, Frank Holstege, Patrick Kemmeren, Helen Parkinson, and Steve Russell for helpful discussion of the manuscript and Christian von Mering for the MIPS reference set. The project is funded by the European Commission as the TEMBLOR, contract no. QLRI-CT-2001-00015 under the RTD programme “Quality of Life and Management of Living Resources.” K.P. and E.U. are supported by a grant from the Academy of Finland.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1111403.
Footnotes
[Supplemental material is available online at www.genome.org.]
The SGD database has been recently updated and the KIN3 gene is now assigned to the biological process “chromosome segregation” based on an experimental analysis performed by Chen et al. 2002. This annotation is corresponding well with our functional prediction. “Chromosome segregation” is a child process of “DNA replication andchromosome cycle,” which itself is a child process of “cell cycle” according to SGD and GO.
References
- Alepuz, P.M., Cunningham, K.W., and Estruch, F. 1997. Glucose repression affects ion homeostasis in yeast through the regulation of the stress-activated ENA1 gene. Mol. Microbiol. 26: 91-98. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., Ball, C.A., Blake, J.A., Botstein, D., Butler, H., Cherry, J.M., Davis, A.P., Dolinski, K., Dwight, S.S., Eppig, J.T., et al. 2000. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25: 25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader, G.D. and Hogue, C.W. 2002. Analyzing yeast protein–protein interaction data obtained from different sources. Nat. Biotechnol. 20: 991-997. [DOI] [PubMed] [Google Scholar]
- Ball, C.A., Jin, H., Sherlock, G., Weng, S., Matese, J.C., Andrada, R., Binkley, G., Dolinski, K., Dwight, S.S., Harris, M.A., et al. 2001. Saccharomyces Genome Database provides tools to survey gene expression and functional analysis data. Nucleic Acids Res. 29: 80-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake, J. and Harris, M. 2003. The Gene Ontology (GO) Project: Structured vocabularies for molecular biology and their application to genome and expression analysis. In Current protocols in bioinformatics.(eds. A. Baxevanis, et al.), J. Wiley, New York. [DOI] [PubMed]
- Blaschke, C., Andrade, M.A., Ouzounis, C., and Valencia, A. 1999. Automatic extraction of biological information from scientific text: Protein–protein interactions. Proc. Int. Conf. Intell. Syst. Mol. Biol. 7: 60-67. [PubMed] [Google Scholar]
- Blaschke, C., Hirschman, L., and Valencia, A. 2002. Information extraction in molecular biology. Brief Bioinform. 3: 154-165. [DOI] [PubMed] [Google Scholar]
- Bork, P. and Koonin, E.V. 1998. Predicting functions from protein sequences—Where are the bottlenecks? Nat. Genet. 18: 313-318. [DOI] [PubMed] [Google Scholar]
- Chen, Y., Riley, D.J., Zheng, L., Chen, P.L., and Lee, W.H. 2002. Phosphorylation of the mitotic regulator protein Hecl by Nek2 kinase is essential for faithful chromosome segregation. J. Biol. Chem. 277: 494088-49416. [DOI] [PubMed] [Google Scholar]
- Dwight, S.S., Harris, M.A., Dolinski, K., Ball, C.A., Binkley, G., Christie, K.R., Fisk, D.G., Issel-Tarver, L., Schroeder, M., Sherlock, G., et al. 2002. Saccharomyces Genome Database (SGD) provides secondary gene annotation using the Gene Ontology (GO). Nucleic Acids Res. 30: 69-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, A.M., Kus, B., Jansen, R., Greenbaum, D., Greenblatt, J., and Gerstein, M. 2002. Bridging structural biology and genomics: Assessing protein interaction data with known complexes. Trends Genet. 18: 529-536. [DOI] [PubMed] [Google Scholar]
- Enright, A.J., Iliopoulos, I., Kyrpides, N.C., and Ouzounis, C.A. 1999. Protein interaction maps for complete genomes based on gene fusion events. Nature 402: 86-90. [DOI] [PubMed] [Google Scholar]
- Gavin, A.C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J.M., Michon, A.M., Cruciat, C.M., et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141-147. [DOI] [PubMed] [Google Scholar]
- Ge, H., Liu, Z., Church, G.M., and Vidal, M. 2001. Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae. Nat. Genet. 29: 482-486. [DOI] [PubMed] [Google Scholar]
- Gerstein, M., Lan, N., and Jansen, R. 2002. Proteomics. Integrating interactomes. Science 295: 284-287. [DOI] [PubMed] [Google Scholar]
- Ho, Y., Gruhler, A., Heilbut, A., Bader, G.D., Moore, L., Adams, S.L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., et al. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180-183. [DOI] [PubMed] [Google Scholar]
- Holm, S. 1979. A simple sequentially rejective multiple test procedure. Scandinavian Journal of Statistics 6: 65-70. [Google Scholar]
- Hughes, T.R., Marton, M.J., Jones, A.R., Roberts, C.J., Stoughton, R., Armour, C.D., Bennett, H.A., Coffey, E., Dai, H., He, Y.D., et al. 2000. Functional discovery via a compendium of expression profiles. Cell 102: 109-126. [DOI] [PubMed] [Google Scholar]
- Huynen, M.A., Snel, B., Mering, C., and Bork, P. 2003. Function prediction and protein networks. Curr. Opin. Cell Biol. 15: 191-198. [DOI] [PubMed] [Google Scholar]
- Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M., and Sakaki, Y. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. 98: 4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer, V.R., Horak, C.E., Scafe, C.S., Botstein, D., Snyder, M., and Brown, P.O. 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409: 533-538. [DOI] [PubMed] [Google Scholar]
- Jenssen, T.K., Laegreid, A., Komorowski, J., and Hovig, E. 2001. A literature network of human genes for high-throughput analysis of gene expression. Nat. Genet. 28: 21-28. [DOI] [PubMed] [Google Scholar]
- Kemmeren, P., van Berkum, N.L., Vilo, J., Bijma, T., Donders, R., Brazma, A., and Holstege, F.C. 2002. Protein interaction verification and functional annotation by integrated analysis of genome-scale data. Mol. Cell 9: 1133-1143. [DOI] [PubMed] [Google Scholar]
- Koch, C., Moll, T., Neuberg, M., Ahorn, H., and Nasmyth, K. 1993. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science 261: 1551-1557. [DOI] [PubMed] [Google Scholar]
- Lee, T.I., Rinaldi, N.J., Robert, F., Odom, D.T., Bar-Joseph, Z., Gerber, G.K., Hannett, N.M., Harbison, C.T., Thompson, C.M., Simon, I., et al. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298: 799-804. [DOI] [PubMed] [Google Scholar]
- Lindgren, A., Bungard, D., Pierce, M., Xie, J., Vershon, A., and Winter, E. 2000. The pachytene checkpoint in Saccharomyces cerevisiae requires the Sum1 transcriptional repressor. EMBO J. 19: 6489-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manke, T., Bringas, R., and Vingron, M. 2003. Correlating protein–DNA and protein–protein interaction networks. J. Mol. Biol. 333: 75-85. [DOI] [PubMed] [Google Scholar]
- Marcotte, E.M. 2000. Computational genetics: Finding protein function by nonhomology methods. Curr. Opin. Struct. Biol. 10: 359-365. [DOI] [PubMed] [Google Scholar]
- Marcotte, E.M., Pellegrini, M., Ng, H.L., Rice, D.W., Yeates, T.O., and Eisenberg, D. 1999. Detecting protein function and protein–protein interactions from genome sequences. Science 285: 751-753. [DOI] [PubMed] [Google Scholar]
- Measday, V., McBride, H., Moffat, J., Stillman, D., and Andrews, B. 2000. Interactions between Pho85 cyclin-dependent kinase complexes and the Swi5 transcription factor in budding yeast. Mol. Microbiol. 35: 825-834. [DOI] [PubMed] [Google Scholar]
- Mewes, H.W., Frishman, D., Guldener, U., Mannhaupt, G., Mayer, K., Mokrejs, M., Morgenstern, B., Munsterkotter, M., Rudd, S., and Weil, B. 2002. MIPS: A database for genomes and protein sequences. Nucleic Acids Res. 30: 31-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, S. 2000. Guilt-by-association goes global. Nature 403: 601-603. [DOI] [PubMed] [Google Scholar]
- Palin, K., Ukkonen, E., Brazma, A., and Vilo, J. 2002. Correlating gene promoters and expression in gene disruption experiments. Bioinformatics 18: 172-180. [DOI] [PubMed] [Google Scholar]
- Pilpel, Y., Sudarsanam, P., and Church, G.M. 2001. Identifying regulatory networks by combinatorial analysis of promoter elements. Nat. Genet. 29: 153-159. [DOI] [PubMed] [Google Scholar]
- Ponting, C.P. 2001. Issues in predicting protein function from sequence. Brief Bioinform. 2: 19-29. [DOI] [PubMed] [Google Scholar]
- Ren, B., Robert, F., Wyrick, J.J., Aparicio, O., Jennings, E.G., Simon, I., Zeitlinger, J., Schreiber, J., Hannett, N., Kanin, E., et al. 2000. Genome-wide location and function of DNA binding proteins. Science 290: 2306-2309. [DOI] [PubMed] [Google Scholar]
- Rung, J., Schlitt, T., Brazma, A., Freivalds, K., and Vilo, J. 2002. Building and analysing genome-wide gene disruption networks. Bioinformatics 18: 202-210. [DOI] [PubMed] [Google Scholar]
- Schlitt, T. and Brazma, A. 2002. Learning about gene regulatory networks from gene deletion experiments. Comp. Funct. Genom. 3: 499-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwikowski, B., Uetz, P., and Fields, S. 2000. A network of protein–protein interactions in yeast. Nat. Biotechnol. 18: 1257-1261. [DOI] [PubMed] [Google Scholar]
- Simon, I., Barnett, J., Hannett, N., Harbison, C.T., Rinaldi, N.J., Volkert, T.L., Wyrick, J.J., Zeitlinger, J., Gifford, D.K., Jaakkola, T.S., et al. 2001. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell 106: 697-708. [DOI] [PubMed] [Google Scholar]
- Sokal, R.R. and Rohlf, F.J. 1995. Biometry—The principles and practice of statistics in biological research, 3rd ed. W.H. Freeman and Company, New York.
- Sprague, G.F.J. and Thorner, J.W. 1992. Pheromone response and signal transduction during the mating process of Saccharomyces cerevisiae. In The molecular and cellular biology of the yeast Saccharomyces: Gene expression (eds. E.W. Jones, J.R. Pringle and J.R. Broach), pp. 657-744. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- Stillman, D.J., Dorland, S., and Yu, Y. 1994. Epistasis analysis of suppressor mutations that allow HO expression in the absence of the yeast SWI5 transcriptional activator. Genetics 136: 781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz, P., Giot, L., Cagney, G., Mansfield, T.A., Judson, R.S., Knight, J.R., Lockshon, D., Narayan, V., Srinivasan, M., Pochart, P., et al. 2000. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403: 623-627. [DOI] [PubMed] [Google Scholar]
- Valencia, A. and Pazos, F. 2002. Computational methods for the prediction of protein interactions. Curr. Opin. Struct. Biol. 12: 368-373. [DOI] [PubMed] [Google Scholar]
- von Mering, C., Krause, R., Snel, B., Cornell, M., Oliver, S.G., Fields, S., and Bork, P. 2002. Comparative assessment of large-scale data sets of protein–protein interactions. Nature 417: 399-403. [DOI] [PubMed] [Google Scholar]
- Walhout, A.J. and Vidal, M. 2001. Protein interaction maps for model organisms. Nat. Rev. Mol. Cell Biol. 2: 55-62. [DOI] [PubMed] [Google Scholar]
- Witten, I.H. and Eibe, F. 1999. Data mining: Practical machine learning tools and techniques with JAVA implementations. Morgan Kaufman, London.
- Wu, L.F., Hughes, T.R., Davierwala, A.P., Robinson, M.D., Stoughton, R., and Altschuler, S.J. 2002. Large-scale prediction of Saccharomyces cerevisiae gene function using overlapping transcriptional clusters. Nat. Genet. 31: 255-265. [DOI] [PubMed] [Google Scholar]
- Xenarios, I., Fernandez, E., Salwinski, L., Duan, X.J., Thompson, M.J., Marcotte, E.M., and Eisenberg, D. 2001. DIP: The Database of Interacting Proteins: 2001 update. Nucleic Acids Res. 29: 239-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, J., Pierce, M., Gailus-Durner, V., Wagner, M., Winter, E., and Vershon, A.K. 1999. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 18: 6448-6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanzoni, A., Montecchi-Palazzi, L., Quondam, M., Ausiello, G., Helmer-Citterich, M., and Cesareni, G. 2002. MINT: A Molecular INTeraction database. FEBS Lett. 513: 135-140. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://db.yeastgenome.org/cgi-bin/SGD/GO/goTermMapper; goTermMapper from SGD.
- http://mips.gsf.de/ and http://mips.gsf.de/proj/yeast/; MIPS.
- http://www.yeastgenome.org/; SGD.
- http://srs.ebi.ac.uk; SRS server.
- http://www.ebi.ac.uk/proteome/; EBI Proteome Analysis Database.