Abstract
Cortisol is synthesized by 11β-hydroxysteroid dehydrogenase type 1, inhibitors of which may treat disease associated with excessive cortisol levels. The crystal structures of 11β-hydroxysteroid dehydrogenase type 1 that have been released may aid drug discovery. The crystal structures have been analyzed in terms of the interactions between the protein and the ligands. Despite a variety of structurally different inhibitors the crystal structures of the proteins are quite similar. However, the differences are significant for drug discovery. The crystal structures can be of use in drug discovery, but care needs to be taken when selecting structures for use in virtual screening and ligand docking.
A significant problem facing society is the increase in obesity. Often observed in obese people is metabolic syndrome, a disease characterized by a variety of symptoms including congestive heart failure, hypertension, atherogenic lipidemia, glucose intolerance, insulin resistance and Type II diabetes [1]. These symptoms, widely recognized as being risk factors for cardio vascular disease [2], are also associated with high levels of the glucocorticoid hormone cortisol [3,4]. Glucocorticoid hormones play essential roles in a range of physiological processes including the regulation of carbohydrate, lipid and bone metabolism, maturation and differentiation of cells, and modulation of inflammatory responses and stress [5-7]. They exert their effect primarily through binding to glucocorticoid receptors, leading to altered target gene transcription. Symptoms similar to those of metabolic syndrome are observed in patients suffering from Cushing’s syndrome, which is marked by increased glucocorticoid levels [8]. The similarity of these symptoms suggests that the suppression of glucocorticoid activity may be a treatment for the individual indications of metabolic syndrome [9] despite the fact that in metabolic syndrome circulating glucocorticoid levels are not usually elevated [10]. Therefore, it is speculated that intercellular, but particularly intracellular local levels of glucocorticoid regulated by prereceptor metabolism are responsible for metabolic abnormalities. The increasing prevalence of metabolic syndrome has highlighted the need for novel treatments. There is now growing evidence that the oxidoreductase enzyme 11β-hydroxysteroid dehydrogenase type 1 (11β-HSD1) provides a novel and attractive target for manipulation of glucocorticoid action.
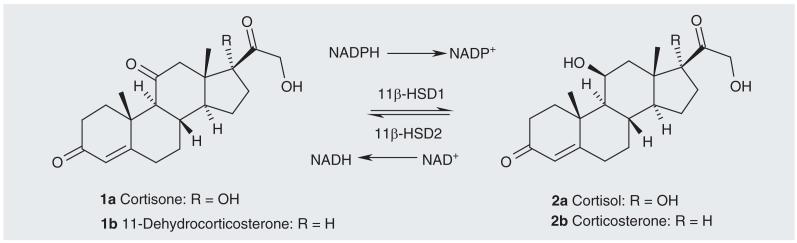
The physiological in vivo role of 11β-HSD1 is that of a reductase, although in vitro it can also function as a dehydrogenase. In the liver and fat tissue of humans, in a reaction catalyzed by 11β-HSD1, the active glucocorticoid cortisol (2a) is produced by the reduction of inactive cortisone (1a) with the concomitant conversion of NADPH to NADP+ (Figure 1). The reduction is favored over the oxidation because of the high NADPH concentration in the liver and fat tissue. The reverse (oxidation) reaction is catalyzed in vivo by 11β-hydroxysteroid dehydrogenase type 2 (11β-HSD2), which uses NAD+ as the cofactor. Both these enzymes are from the short-chain dehydrogenase/reductase super family [11] and are found located in microsomes. The 11β-HSD1 isoform is highly expressed in liver and adipose tissue, resulting in high concentrations of the active compound in these tissues [12,13], whereas the inactivating 11β-HSD2 isoform is found mainly in mineralocorticoid target tissues, such as the kidney and colon where it prevents occupation of the mineralocorticoid receptor, which may lead to hypernatremia, hypokalemia and hypertension [14,15]. Mice overexpressing 11β-HSD2 in fat tissue (resulting in a greater rate of cortisol oxidation and, therefore, low cortisol levels) are more insulin sensitive, glucose tolerant and resistant to weight gain than normal mice [16]. In agreement with this, it has been demonstrated that 11β-HSD1 knockout mice (which, therefore, also have low cortisol levels) are resistant to metabolic syndrome, resist stress-induced hyperglycemia, and have decreased cholesterol and triglyceride levels [17,18]. Conversely, overexpression of 11β-HSD1 in mouse liver and adipose tissue (leading to high cortisol levels) leads to a metabolic syndrome-like phenotype with insulin-resistant diabetes, hyperlipidemia and visceral obesity being observed [19,20]. Inhibition of 11β-HSD1 without inhibiting 11β-HSD2 should lower cortisol levels and reduce the symptoms of metabolic syndrome. The biological, physiological and pathophysiological roles of 11β-HSD1 have been reviewed [3], as has the targeting of the prereceptor metabolism of cortisol as a therapy in obesity and diabetes [21]. Most data on the consequences of selective 11β-HSD1 inhibition are available from studies in rodents, but the field has recently benefited from early studies in humans.
Figure 1. The reactions catalyzed by 11β-hydroxysteroid dehydrogenase types 1 and 2.
In the reaction catalyzed by the human enzymes R = OH (1a, 2a). In the reaction catalyzed by the rodent enzymes R = H (1b, 2b).
Clinical data originally suggested that inhibition of 11β-HSD1 with the nonselective inhibitor carbenoxolone, which also inhibits 11β-HSD2, increases hepatic insulin sensitivity and decreases glucose production [22]. However, while this had some worth as a proof-of-concept in humans, inhibition of 11β-HSD2 has several clinical disadvantages, making the design of selective enzyme inhibitors a necessity. Most recently, Incyte has reported results for Phase II clinical trials of their compound INCB13739 (the structure of which has yet to be released). In patients with Type II diabetes where metformin monotherapy was failing to provide adequate glycemic control, INCB13739 reduced fasting plasma glucose levels and, in hyperlipidemic patients, total cholesterol, low-density lipoprotein cholesterol and triglycerides were all significantly reduced [23]. Relative to placebo, body weight decreased after treatment with INCB13739.
The surge of research and clinical interest in this area has resulted in many companies and academic groups developing selective 11β-HSD1 inhibitors from a variety of structural classes [24-27], six of which are briefly described.
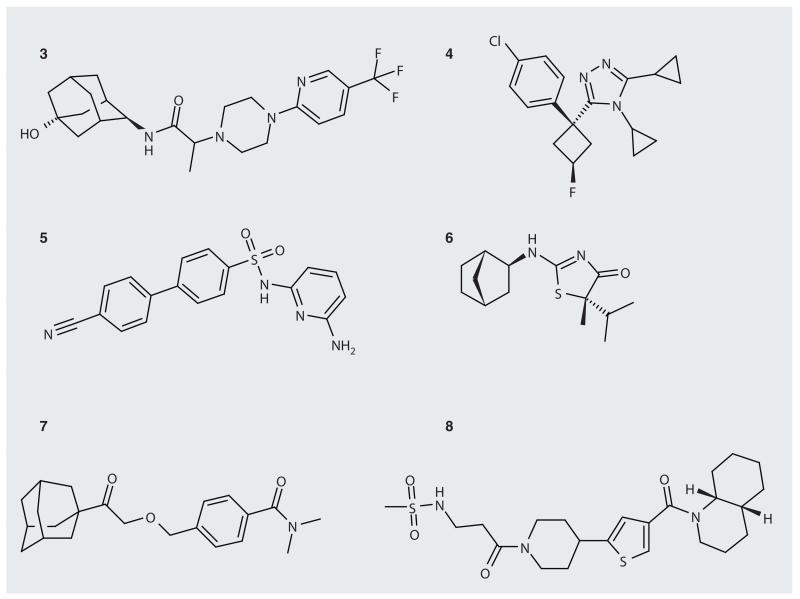
Compound 3 (Abbott; Figure 2) inhibits human 11β-HSD1 with Ki = 5 nM and the mouse and rat enzymes with Ki = 15 nM and 4 nM, respectively [28]. Inhibition of 11β-HSD2 from all three species is poor (Ki >100 μM). Inhibition of 11β-HSD1 in HEK cells occurs with IC50 = 29 nM. The metabolic stability of the compound was tested and Clint was found to be 3 l/h.kg, 7 l/h.kg and 5 l/h.kg in human, mouse and rat liver microsomes, respectively. In vivo pharmacokinetic profiles in mice and rats have been reported. Suitably high plasma concentrations were obtained in mice, but a high clearance rate led to a half-life of less than 1 h. The volume of distribution predicted good intracellular tissue penetration. Similar behavior was observed in rats, but the clearance rate was lower and the half-life was longer.
Figure 2. Some 11β-hydroxysteroid dehydrogenase type 1 inhibitors.
Compound 4 (Merck) inhibits human 11β-HSD1 with IC50 = 5 nM and the mouse enzyme with IC50 = 16 nM [29]. In mice it is approximately 100% bioavailable and, in a mouse pharmacodynamic assay, was found to inhibit the conversion of cortisone to cortisol (after oral dosing at 10 mg/kg) by over 90% for at least 4 h.
Compound 5 (Pfizer) is selective for human 11β-HSD1 with Ki <1 nM as opposed to Ki = 750 nM for the mouse enzyme [30]. This difference between human and rodent enzymes is reflected in cellular assays where the EC50 for HEK293 cells is 5 nM but for rat hepatoma cells is 14,500 nM. In HEK293 cells the compound poorly inhibits 11β-HSD2, causing only 1.5% inhibition at 10 μM. In a rat, this compound had an excellent pharmacokinetic profile with low clearance, long half-life and good oral bioavailability.
Compound 6 (Amgen) inhibits human 11β-HSD1 with Ki = 12.8 nM, is selective over 11β-HSD2 (IC50 >10 μM) and is active in cells with IC50 = 10.1 nM [31]. In an ex vivo experiment following oral gavage the compound reduced 11β-HSD1 activity by between 33 and 55% in inguinal fat, depending on the dose. In obese mice the compound was found to reduce blood glucose and plasma insulin levels, and reduce the weight of the mice. Pharmacokinetic profiles suggested there is good bioavailability in mice, rats and dogs, but in cynomolgous monkeys bioavailability is low, possibly attributable to first-pass hepatic metabolism consistent with low microsomal stability.
Compound 7 (Sterix) inhibits human 11β-HSD1 with IC50 = 56 nM [32]. In metabolism studies with human liver microsomes it was found to be stable (87% remaining after 30 min) with a half-life of 59 min, a clearance rate of 11 μl/min/mg and no detectable metabolites. The inhibition of cytochromes 1A2, 2C9 and 2D6 was at a low level with IC50 >100 μM and the inhibition of three others, 2C19, 3A4-BFC and 3A4-BQ, at IC50 = 20, 22 and 86 μM, respectively.
Recent work has shown that, as well as having a role in metabolic syndrome, glucocorticoids play a role in cognitive function. Hippocampal expression of 11β-HSD1 increases with aging in mice and correlates with spatial memory defects [33]. Mice deficient in 11β-HSD1 are protected from age-related spatial memory impairments. Treatment of aged normal mice with a selective 11β-HSD1 inhibitor (UE1961, 8) resulted in improved spatial memory performance [34].
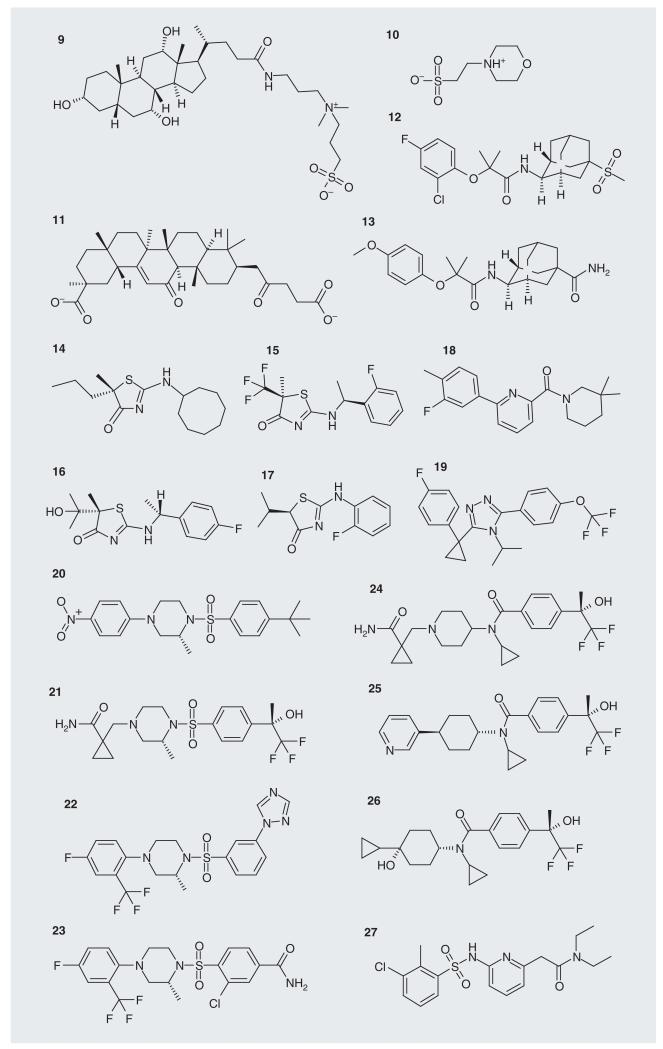
The modern approach to inhibitor development normally requires crystal structures of the target enzyme for use in structure-based design work. Details of eighteen crystal structures of human 11β-HSD1 (Table 1) have been released [35-48]; these have a variety of inhibitors bound (9-26; Figure 3). In rodents, 11β-HSD1 catalyzes the reduction of 11-dehydrocorticosterone (1b) to corticosterone (2b; Figure 1). There are two structures of the mouse protein available (Table 1) [49] and three structures of the guinea pig protein (Table 1) [30,50,51]. One of the mouse structures has a ligand in the substrate binding site (2b), as does one of the guinea pig structures (27).
Table 1. Protein Data Bank codes, ligands and sources of crystal H structures of 11β-HSD1.
| PDB code | Ligand | Source | Ref. |
|---|---|---|---|
| Human 11β-HSD1 | |||
| 1XU7 | 9 | Syrrx | [35] |
| 1XU9 | 9, 10 | Syrrx | [35] |
| 2BEL | 11 | Biovitrum | _† |
| 2ILT | 12 | Abbott | [36] |
| 2IRW | 13 | Abbott | [37] |
| 2RBE | 17 | Amgen & Biovitrum | [38] |
| 3BYZ | 14 | Amgen & Biovitrum | [39] |
| 3BZU | 15 | Amgen | [40] |
| 3CH6 | 18 | Bristol-Myers Squibb | [41] |
| 3CZR | 9, 20 | Amgen | [42] |
| 3D3E | 25 | Amgen | [43] |
| 3D4N | 21 | Amgen | [43,44] |
| 3D5Q | 19 | Amgen | [44] |
| 3EY4 | 16 | Amgen | [45] |
| 3FCO | 26 | Amgen | [46] |
| 3FRJ | 24 | Amgen | [47] |
| 3H6K | 23 | Wyeth | [48] |
| 3HFG | 22 | Wyeth | [48] |
| Mouse 11β-HSD1 | |||
| 1Y5M | - | Amgen | [49] |
| 1Y5R | 2b | Amgen | [49] |
| Guinea pig 11β-HSD1 | |||
| 1XSE | - | Biovitrum | [50] |
| 3DWF | - | University of Birmingham (UK) | [51] |
| 3G49 | 27 | Pfizer | [30] |
No paper has been published describing the 2BEL structure.
11β-HSD1: 11β-hydroxysteroid dehydrogenase type 1; PDB: Protein Data Bank.
Figure 3. Ligands in the crystal structures of 11β-hydroxysteroid dehydrogenase type 1.
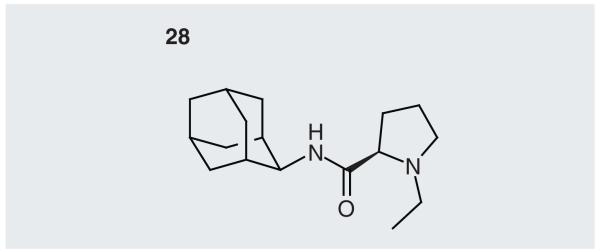
A study from Pfizer has described a fourth structure of the guinea pig protein, Protein Data Bank (PDB) code 3LZ6, for which, at the time of writing, the coordinates have not yet been released [52]. This structure has in it an inhibitor, 28 (Figure 4), an adamantyl amide derivative with Ki = 1.4 nM against the human enzyme and Ki = 0.63 nM against the mouse enzyme. Another structure of 11β-HSD1 has been deposited in the PDB under the code 3GMD, but the coordinates have not yet been released. The article describing the structure has not yet been published, and, in what little information that is available (from the PDB), the species from which the enzyme comes is not mentioned, although, from the title of the structure deposited in the PDB, the structure may contain a 7-azaindole-pyrrolidine inhibitor.
Figure 4. Ligand in the 3LZ6 crystal structure.
This article reviews the publicly available crystal structures of 11β-HSD1, starting with a brief overview of the whole structure before detailing the differences between the substrate binding sites. A concluding section discusses the use of these structures in virtual screening and computational drug discovery and design. The Supplementary Material gives a detailed break-down of the sequence and structure of each of the PDB entries, and details the interactions each protein molecule makes with any ligands in the cofactor and/or substrate binding sites. A recent review has focused on the same topic, but from a different angle and may be useful in conjunction with this review [53].
Sequence & structure of human 11β-HSD1
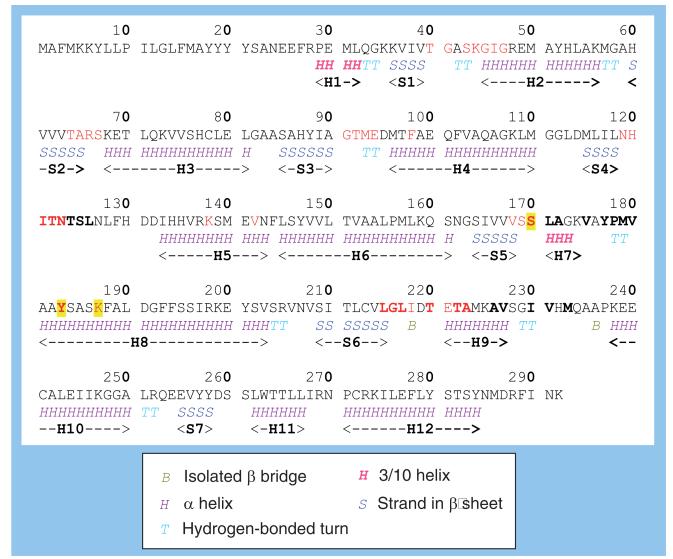
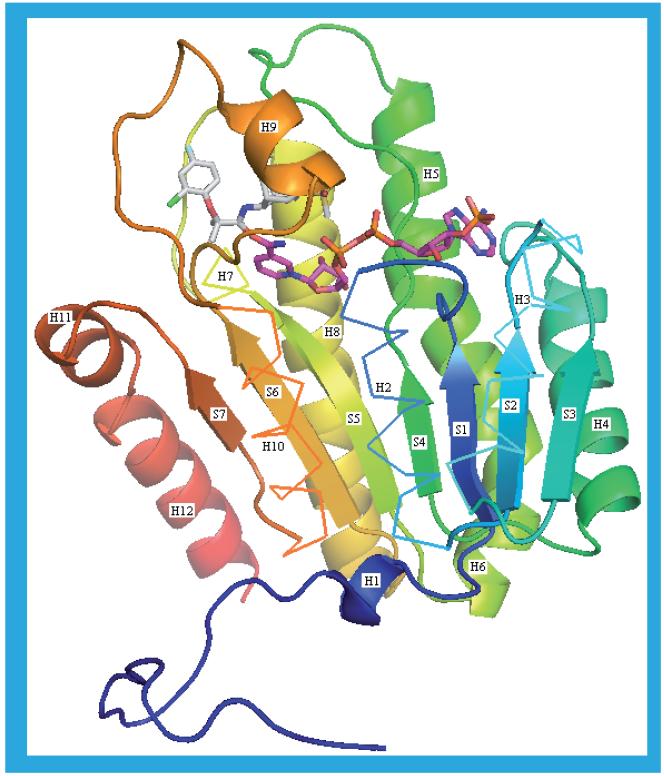
Figures 5 & 6 show the sequence, secondary-structure assignment and overall fold of human 11β-HSD1. This enzyme is a member of the short-chain dehydrogenase/reductase family that contains a structurally conserved nucleotide-binding Rossmann fold [54]. 11β-HSD1 has a seven-stranded parallel β sheet (S1-7) and 12 helices (H1-12). Only the 2BEL structure has the wild-type sequence shown in Figure 5; the 3CH6 structure is of the L262R/F278E double mutant and all the other structures have the C272S mutation. These mutations are remote from the substrate and cofactor binding sites and any impact on ligand binding is likely to be minimal. The residues involved in catalysis are S170, Y183 and K187. The positively charged side chain of K187, in close proximity to the Y183 hydroxyl oxygen, may facilitate hydride transfer. S170 may participate in catalysis either by a direct interaction with Y183 or as part of a hydride relay network. This promotes electrophilic attack on the substrate carbonyl oxygen, thus enabling the carbon atom to accept a hydride from the NADPH.
Figure 5. The amino acid sequence and secondary structure of human 11β-hydroxysteroid dehydrogenase type 1.
SwissProt ID code: P28845. Those residues that form the NADP(H) binding site are shown in red and those bordering the substrate binding site are shown in bold. The three residues involved in catalysis are highlighted in yellow. The secondary structure assignment is by DSSP [58] and is for the 2ILT structure. The helices (H1–12) and sheets (S1–7) are identified by the same numbers used in Figure 6.
Figure 6. The secondary and tertiary structure of human 11β-hydroxysteroid dehydrogenase type 1.
The 2ILT structure is shown. The helices (H1–12) and sheets (S1–7) are identified by the same numbers used in Figure 5. For clarity, helices H2, −3, −7 and −10 are shown as ribbons rather than cartoons. The protein is colored blue at the N-terminus through to red at the C-terminus. The NADP(H) is shown with purple carbons and the ligand, 12, in the substrate binding site with white carbons. This figure, in common with all the other molecular figures in this paper, was created in PyMOL [102].
The 18 crystal structures of human 11β-HSD1 have, between them, 67 11β-HSD1 molecules. Only the 2ILT structure has one protein molecule in the crystallographic asymmetric unit: the 3CZR, 3FCO and 3FRJ structures are dimers, the 2IRW structure is an octamer and the remaining 13 structures are tetramers. There is some difference in the 3D structure of these mole cules: when the protein Cα atoms are overlaid on the 2ILT structure there is an average root mean square deviation (RMSD) of 1.526 Å and a spread of 0.341–4.390 Å (Table 2). However, the substrate and cofactor binding sites are much more similar with an average Cα RMSD of 0.468 Å and a spread of 0.186–1.624 Å (Table 2). In the remainder of this article the various protein molecules are identified in the following manner: molecule ‘A’ of the 1XU7 structure is signified by 1XU7_a and molecule ‘B’ of the same structure by 1XU7_b, and so on.
Table 2. Root mean square deviation values for Ca overlays.
| Structure | RMSD (Å) | Structure | RMSD (Å) | Structure | RMSD (Å) |
|---|---|---|---|---|---|
| 1XU7_a | 2.139 | 2RBE_c | 0.341 | 3D4N_c | 2.463 |
| 1.624 | 0.355 | 0.404 | |||
|
| |||||
| 1XU7_b | 1.925 | 2RBE_d | 0.915 | 3D4N_d | 2.452 |
| 0.364 | 0.505 | 0.416 | |||
|
| |||||
| 1XU7_c | 1.944 | 3BYZ_a | 0.543 | 3D5Q_a | 2.051 |
| 0.345 | 0.452 | 0.329 | |||
|
| |||||
| 1XU7_d | 1.928 | 3BYZ_b | 0.528 | 3D5Q_b | 1.906 |
| 0.304 | 0.443 | 0.376 | |||
|
| |||||
| 1XU9_a | 2.227 | 3BYZ_c | 0.366 | 3D5Q_c | 0.974 |
| 1.621 | 0.328 | 0.302 | |||
|
| |||||
| 1XU9_b | 2.519 | 3BYZ_d | 0.877 | 3D5Q_d | 1.940 |
| 0.414 | 0.440 | 0.426 | |||
|
| |||||
| 1XU9_c | 2.524 | 3BZU_a | 0.865 | 3EY4_a | 0.654 |
| 0.439 | 0.456 | 0.553 | |||
|
| |||||
| 1XU9_d | 2.487 | 3BZU_b | 1.436 | 3EY4_b | 0.483 |
| 0.332 | 0.343 | 0.484 | |||
|
| |||||
| 2BEL_a | 1.176 | 3BZU_c | 0.663 | 3EY4_c | 0.383 |
| 0.797 | 0.269 | 0.458 | |||
|
| |||||
| 2BEL_b | 3.045 | 3BZU_d | 0.620 | 3EY4_d | 0.909 |
| 0.815/0.816 | 0.387 | 0.509 | |||
|
| |||||
| 2BEL_c | 4.390 | 3CH6_a | 0.837 | 3FCO_a | 1.646 |
| 0.444 | 0.223 | 0.585 | |||
|
| |||||
| 2BEL_d | 2.997 | 3CH6_b | 3.906 | 3FCO_b | 1.609/1.608 |
| 0.633 | 0.218 | 0.462/0.461 | |||
|
| |||||
| 2IRW_a | 0.820 | 3CH6_d | 3.873 | 3FRJ_a | 1.969 |
| 0.241 | 0.186 | 0.566 | |||
|
| |||||
| 2IRW_b | 0.822 | 3CH6_e | 0.857 | 3FRJ_b | 2.480 |
| 0.237 | 0.231 | 0.714 | |||
|
| |||||
| 2IRW_c | 0.404 | 3CZR_a | 2.490 | 3H6K_a | 0.817 |
| 0.259 | 0.612 | 0.441 | |||
|
| |||||
| 2IRW_d | 0.791 | 3CZR_b | 2.005 | 3H6K_b | 0.740 |
| 0.262 | 1.003 | 0.256 | |||
|
| |||||
| 2IRW_e | 0.387 | 3D3E_a | 1.975 | 3H6K_c | 0.729 |
| 0.279 | 0.891 | 0.451 | |||
|
| |||||
| 2IRW_f | 0.820 | 3D3E_b | 1.975 | 3H6K_d | 0.561 |
| 0.309 | 0.457 | 0.360 | |||
|
| |||||
| 2IRW_g | 0.837 | 3D3E_c | 2.021 | 3HFG_a | 0.673 |
| 0.257 | 0.743 | 0.538 | |||
|
| |||||
| 2IRW_h | 0.831 | 3D3E_d | 2.014 | 3HFG_b | 1.465 |
| 0.250 | 0.748 | 0.333 | |||
|
| |||||
| 2RBE_a | 0.669 | 3D4N_a | 1.936 | 3HFG_c | 0.594 |
| 0.587 | 0.408 | 0.414 | |||
|
| |||||
| 2RBE_b | 0.380 | 3D4N_b | 2.472 | 3HFG_d | 0.561 |
| 0.353 | 0.304 | 0.373 | |||
For each protein molecule (indicated by the lowercase letter) in each crystal structure of human 11β-HSD1, the Cα atoms were superimposed on those of 2ILT_a and the RMSD was calculated using the Sybyl software [101]. 2ILT_a was used as the reference structure simply because it is the only structure of 11β-HSD1 with only one protein chain in the structure. The numbers in normal type are for the whole protein and the numbers in bold are for the residues around the substrate and cofactor binding sites (T40-G47, T64-S67, G91-F98, N119-L126, K138-V142, V168-K187 and L215-M233). Note that for the whole protein 3FCO_b has three Cα atoms with alternate conformations, giving rise to different values for the RMSD. The 2BEL_b, 3CZR_a and 3CZR_b structures have, respectively, two, three and one Cα atoms with alternate conformations, but in these structures the differences, at least to four significant figures, make no difference to the RMSD when superimposed on 2ILT_a. For the binding-site residues, the 2BEL_b structure has different RMSD values for the two conformations. RMSD: Root mean square deviation.
Cofactor binding
All the crystal structures of human 11β-HSD1 have NADP(H) in the cofactor binding site. This site is in the form of a cleft at the C-terminal end of strands 1, 2, 4 and 5 of the β sheet with the adenine ring at the end of S2 and the nicotinamide ring at the end of S5 (Figure 6). The residues that form the cleft are T40, G41, S43, K44, G45, I46, G47, T64, A65, R66, S67, G91, T92, M93, E94, F98, N119, H120, I121, T122, N123, K138, V142, V168, S169, S170, Y183, K187, L215, G216, L217, I218, T220, E221, T222 and A223. The cleft is approximately 17 Å long and at the nicotinamide end overlaps the substrate binding site.
In the article describing the 1XU7 and 1XU9 structures the cofactor is identified as having geometry consistent with being in the ionized form (NADP+) [35], and the authors of the paper describing the 3BZU structure state that the structure resolution was insufficient to determine the ionization state [40]. The papers describing the remaining structures neither discuss the cofactor geometry nor experimentally identify the ionization state: the cofactor in these papers is variously named as NADP and NADP+. In all the structures the adenine and nicotinamide rings are almost perpendicular to the plane of the ribose rings with the adenine adopting the anti configuration and the nicotinamide being in the syn configuration.
When the Cα atoms of all 67 protein complexes are overlaid the NADP(H) molecules overlay very well (Figure 7). Given the similarity of the structures it is likely that the cofactors in all the structures exist in the ionized, NADP+ form. The analysis, below, of the interactions between the protein and the NADP(H), is for the 2ILT structure but, as detailed in the Supplementary Material, the NADP(H) in all the structures can potentially form the same hydrogen bonds to the protein. In addition, several structures, have water-mediated hydrogen bonds. As well as the hydrogen bonds to the protein, several of the ligands in the substrate binding site form hydrogen bonds to the NADP(H); these are discussed later.
Figure 7. Overlay of NADP(H) molecules.
The Cα atoms of all 67 protein chains in the 18 structures of human 11β-hydroxysteroid dehydrogenase type 1 were overlaid using Sybyl [101] and the NADP(H) molecules extracted. The NADP(H) from the 2ILT structure is shown as semitransparent green sticks with the other structures shown as lines colored according to atom type with the carbons in purple.
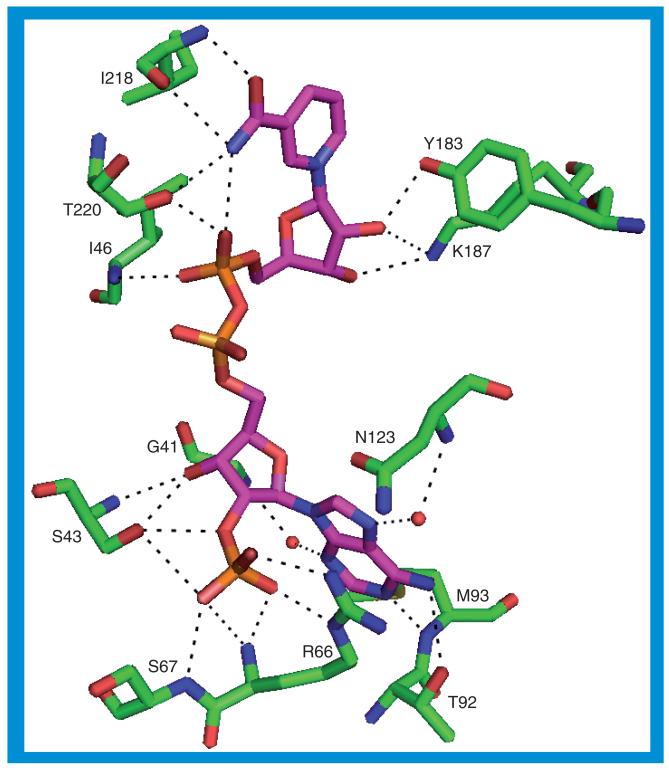
There are numerous stacking and packing interactions between the protein and the NADP(H), and between the various protein structures more than 20 possible hydrogen bonds can be identified. Some of these hydrogen bonds are mutually exclusive and others are not present in every structure because the orientation of amino acid side chains or NADP(H) phosphate oxygens is incompatible with hydrogen-bond formation. The possible hydrogen bonds in the 2ILT structure are illustrated in Figure 8 [36]. In the hydrogen bonds listed in the following paragraph, the NADP(H) atoms involved in those bonds are identified by their naming in the 2ILT structure; the names of the NADP(H) oxygen atoms involved in specific bonds can change between the different structures, for example, atom O1X in one structure may be named O2X in another structure despite their being in the same or similar position.
Figure 8. The hydrogen bonds between 11β-hydroxysteroid dehydrogenase type 1 and NADP(H) for the 2ILT structure.
The other 11β-hydroxysteroid dehydrogenase type 1 structures have a similar range of interactions between the protein and the NADP(H).
The hydrogen bonds between the protein and the adenine moiety are M93 N–N1A and T92 OG1–N6A. The adenine moiety stacks with the side chain of R66 and packs against the side chain of H120. Possible hydrogen bonds between the protein and the adenine phosphorylribose include R66 NH1–O1X, S43 OG–O2X, R66 N–O2X, S67 N–O2X, S67 OG–O2X, R66 N–O3X, R66 NE–O3X, S43 OG–O2B, S43 N–O3B and S43 OG–O3B. The adenine phosphorylribose is sandwiched between the side chains of K44 and H120. The nicotinamide ring is not involved in any hydrogen bonds but the carboxamide moiety may form three hydrogen bonds to the protein: I218 O–N7N, T220 OG1–N7N and I218 N–O7N. Possible hydrogen bonds between the protein and the nicotinamide ribose include Y183 OH–O2D, K187 NZ–O2D and K187 NZ–O3D. The nicotin amide ribose packs against the side chains of I46 and V168. There are two possible hydrogen bonds between the protein and the phosphates linking the ribose rings: T220 OG1–O1N and I46 N–O2N. The NADP(H) also has an internal hydrogen bond from the nicotinamide N7N to the phosphate O1N.
Ligand binding
The substrate binding site is a predominantly hydrophobic pocket situated at the C-terminal end of S5 and S6, and sandwiched between H8 and H9 (Figure 6) with a flexible loop comprising residues V227-Q234 capping the pocket. It is approximately 12 Å long, although approximately half that length overlaps the NADP(H) nicotinamide and ribose rings. The crystal structure ligands found in the substrate binding site make contact with residues I121, T122, N123, T124, S125, L126, S170, L171, A172, V175, Y177, P178, M179, V180, Y183, G216, L217, T220, T222, A223, A226, V227, V231 and M233. Both ends of the pocket are open to solvent, so ligands that are too long to fit into the substrate binding site can extend out of it, and, from the crystal structures, those that do, make contact with surface residues Q234, S260, S261, T264 and Y280. There is a ligand in the substrate binding site of all the protein structures except for 3FRJ_a [47]. In the following discussion the interactions of the ligand with the protein in the A molecule are analyzed (except the 3FRJ structure where the B molecule is analyzed) and mention of the interactions in the other molecules is made only if the interactions are significantly different. Ligand atoms are identified by their names in the PDB files. In all cases, details of the hydrogen-bond interactions in every structure are tabulated in the Supplementary Material.
1XU7
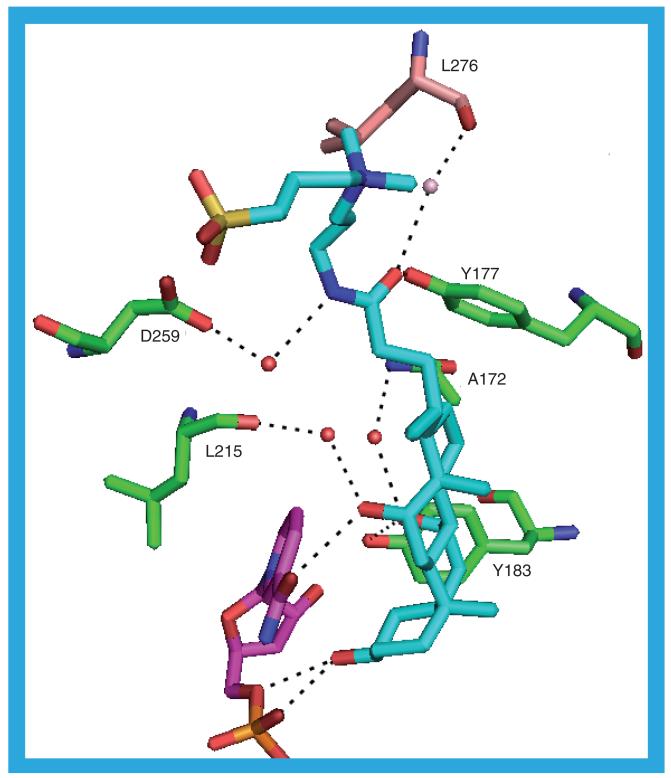
There are four 11β-HSD1 molecules in this structure all with the same ligand, 3-([3-chol-amidopropyl]dimethylammonio)-1-propane-sulfonate (CHAPS; 9), in the substrate binding site. When the four protein molecules are super-imposed (average Cα RMSD = 0.866 Å) the steroid cores of the ligands also super imposed very well. There is a little variability in the position of the tail, which runs along the surface of the protein making contacts with L171, Y177, V231, Q234, S260, S261 and T264. The hydrogen bonds involving the ligand in the 1XU7_a structure are shown in Figure 9 [35]. There are two hydrogen bonds directly from the protein to the ligand: Y177 OH–O1 and Y183 OH–O3. The ligand O1 atom is also involved in a water-mediated hydrogen bond involving a 1XU7_b-associated water molecule to L276 in 1XU7_b: L276 O–HOH–O1. There are a further three possible water-mediated hydrogen bonds: A172 N–HOH–O3, D259 OD2–HOH–N1 and L215 O–HOH–O4. Between the NADP+ and the ligand there are three possible hydrogen bonds: O7N–O4, O5D–O2 and O2N–O2. The steroid component of the ligand has surface contact with I121, T124, S125, L126, L171, A172, Y177, V180, Y183, G216, L217, T222, A223, A226 and V227.
Figure 9. The possible hydrogen bonds between the protein, ligand (9) and cofactor in the 1XU7_a structure.
Note that L276 and the water to which it is hydrogen bonded are in the 1XU7_b structure. 1XU7_a protein: Green carbons; 1XU7_b protein: Buff carbons; CHAPS: Cyan carbons; NADP+: Purple carbons; 1XU7_a water molecules: Red spheres; 1XU7_b water molecules: Pink spheres.
At the interface between the protein molecules there are two areas of electron density that have each been filled with the steroid core, but not the tail, of a CHAPS molecule, one associated with 1XU7_a and the other with 1XU7_c. These are remote from, and unlikely to have any influence on the structure of the substrate binding site and will not be discussed further here.
1XU9
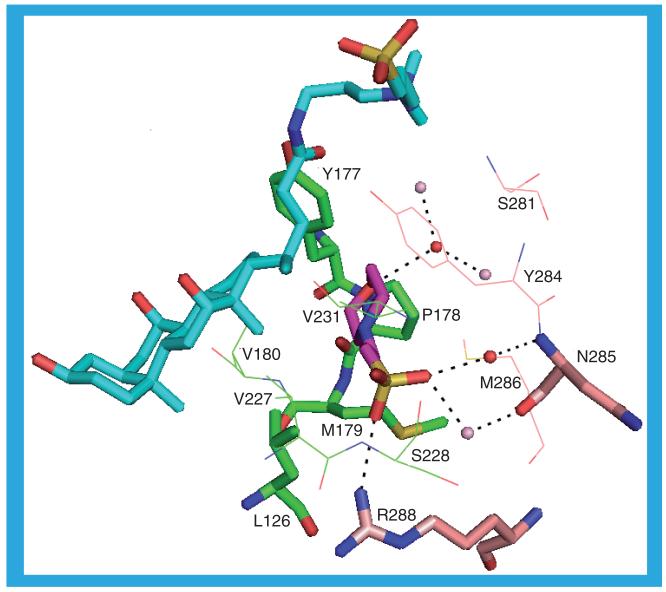
Grown under identical conditions to 1XU7, 1XU9 crystals exist in a different form. The protein structure, however, is identical to that of 1XU7. The same ligand, CHAPS, is in the substrate binding site and it makes the same hydrogen bonds as the ligands in 1XU7, except that there is no water-mediated hydrogen bond to L276 in another molecule. There is no second CHAPS molecule associated with any of the protein molecules but there is a molecule of 2-(N-morpholino)-ethanesulfonic acid (MES; 10) in 1XU9_a (but not the other 1XU9 molecules) [35]. This lies along the interface between 1XU9_a and 1XU9_b (Figure 10). It contacts, in 1XU9_a, the ligand in the substrate binding site and residues L126, Y177, P178, M179, V180, V227, S228 and V231, and in 1XU9_b it contacts S281, Y284, N285, M286 and R288. The morpholine oxygen forms a hydrogen bond to a water molecule from 1XU9_a that is involved in a hydrogen-bonding network with two water molecules from 1XU9_b. One of the sulfate oxygens forms a hydrogen bond to NH1 of R288 in 1XU9_b and a second sulfate oxygen may be involved in two water-mediated hydrogen bonds to N285 in 1XU9_b, one via a 1XU9_a-associated water molecule to the backbone nitrogen, and the other via a 1XU9_b associated water molecule to the backbone carbonyl oxygen.
Figure 10. The interactions between 2-(N-morpholino)-ethanesulfonic acid (10) and 1XU9_a and 1XU9_b.
2-(N-morpholino)-ethanesulfonic acid: Purple carbons; 1XU9_a protein: Green carbons; 1XU9_b protein: Buff carbons; 1XU9_a substrate binding site ligand (CHAPS): Cyan; 1XU9_a water molecules: Red spheres; 1XU9_b water molecules: Pink spheres. For clarity, some residues are shown only as lines rather than sticks.
The 3CZR_b structure also has CHAPS in the substrate binding site [42]. The interactions of the ligand with the protein are similar to those seen in the 1XU7 and 1XU9 structures. However, residues V227-Q234, which form a loop over the binding site are in a slightly different conformation.
2BEL
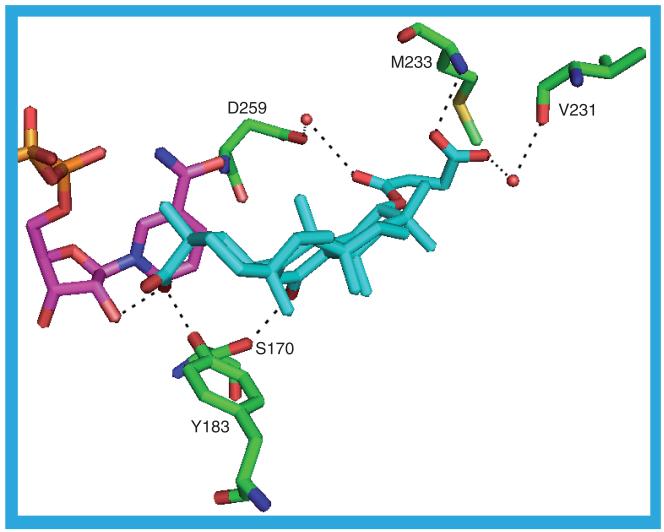
No paper describing the 2BEL structure has been published. The ligand in this structure, carbenoxolone (11; Ki = 11 nM [55]), has the same pose in all four protein chains. In 2BEL_a there are three possible hydrogen bonds between the ligand and the protein: M233 N–O33, S170 OG–O11 and Y183 OH–O34 (Figure 11). The NADP(H) is hydrogen bonded to the ligand: O2D–O34. There are two water-mediated hydrogen bonds between the protein and the ligand: V231 O–HOH–O32 and D259 OD1–HOH–O29. The ligand packs against the NADP(H) nicotinamide and ribose rings and, on the same face, contacts residues L217, T222, A223, A226, V227 and I230. The opposite face of the ligand contacts I121, T124, L126, S170, L171, A172, Y177, V180, Y183 and L215. The carboxyl group at the end of the ligand tail pushes up against M233. In 2BEL_b and 2BEL_c the four terminal atoms in the ligand tail are missing, resulting in different hydrogen bonds possibly forming in this region. A water molecule forms a hydrogen bond to the ligand O29. This water is possibly involved in one, or both, of the two hydrogen bonds to the protein: D259 N–HOH and D259 OD2–HOH. The hydrogen bonds to V231 and M233 are absent. The same four atoms that are missing from the ligand in 2BEL_b and 2BEL_c are also missing from 2BEL_d, as is the water molecule that forms the hydrogen bond to O29.
Figure 11. The possible hydrogen bonds between the protein, ligand (11) and cofactor in the 2BEL_a structure.
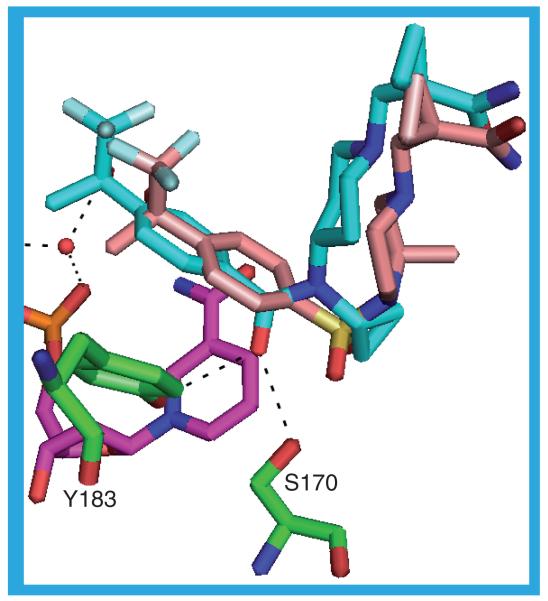
2ILT & 2IRW
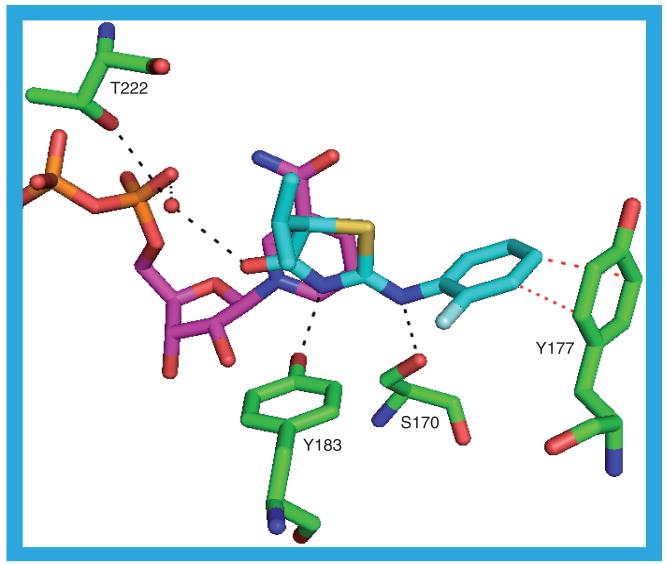
The inhibitors in the 2ILT and 2IRW crystal structures share a common core structure: a substituted phenyl ring connected to a substituted adamantyl moiety through a 2-methoxy-N,2-dimethylpropanamide linker. The inhibitors adopt the same pose in the substrate binding site, which itself is almost identical between the two structures. The adamantyl moiety packs against the NADP(H) nicotinamide ring and contacts T124, S125, L126, V180, Y183, L217, A223, A226 and V227. The phenoxy ring lies in a hydrophobic pocket formed by L126, Y177, M179, V180, L217, V231 and M233. The distance between the phenoxy moiety and the Y177 side chain is a little too great, and the angle not quite right, for there to be a π-stacking interaction. In the linker the amide carbonyl may be involved in two hydrogen bonds to the protein (S170 OG–O1 and Y183 OH–O1; Figure 12) and the gem dimethyl moiety contacts L171, A172, Y177, G216 and L217. The inhibitor in the 2ILT structure, 12 (Ki = 7 nM), has a sulfone group attached to the adamantane. One of the sulfone oxygens may be involved in a rather long hydrogen bond to the protein: T124 N–O2. Both sulfone oxygens could be hydrogen bonded to a water molecule, which itself may be involved in forming a rather long hydrogen bond to one of N123 OD1 or to one of the NADP(H) phosphate oxygens, O1A. The inhibitor in the 2IRW structure, 13 (IC50 = 6 nM), replaces the sulfone with a carboxamide, the NH of which might form a hydrogen bond to one of the NADP(H) phosphate oxygens, O1N [36,37].
Figure 12. The possible hydrogen bonds between the protein, ligand (12) and cofactor in the 2ILT structure.
2RBE, 3BYZ, 3BZU & 3EY4
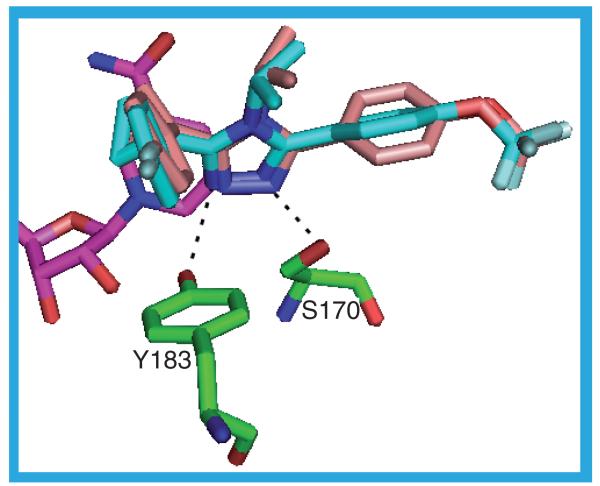
The ligands in these four structures all fall into the same structural class, being 2,5-substituted derivatives of thiazol-4-one. Despite this similarity there are two distinct binding poses. In the 3BYZ, 14 (Ki = 28 nM), 3BZU, 15 (IC50 = 14 nM) and 3EY4, 16 (Ki = 65 nM) structures the ligands bind with the thiazol-4-one ring adjacent to the catalytic tyrosine, Y183, and the 2-substituent lying over the NADP(H) nicotinamide ring. The thiazol-4-one ring in all three structures is superimposable and contacts the protein through residues L126, V180, Y183 and V227. The same two hydrogen bonds between the protein and the ligand are formed in all three structures: Y183 OH–N17 and A172 N–O (Figure 13). The double bond in the thiazol-4-one ring is endocyclic with the NH at the 2-position acting as the donor to the Y183 side chain oxygen. None of these structures has any water-mediated hydrogen bonds: there are no waters in the 3BYZ and 3EY4 structures and the waters in the 3BZU structure are not positioned appropriately to engage in hydrogen bonds to the ligand. In the 3BYZ structure the methyl and propyl groups at the thiazolone 5-position are positioned in a hydrophobic pocket formed by L126, A172, L171, Y177, M179, V180, L217, I230 and V231. The cyclooctyl moiety linked through a nitrogen to the thiazolone 2-position is enclosed by residues I121, T122, N123, T124, S125, L126, V180, Y183, T222, A223, A226 and V227. The NH and one of the CH2 groups of the cyclooctyl ring contact the NADP(H) nicotinamide ring. The NH and methyl group linking the two rings in the 3BZU and 3EY4 structures mimic this interaction by packing against the NADP(H) nicotinamide ring. The fluorophenyl ring in both 3BZU and 3EY4 makes contacts with T124, S125, L126, V180, A223, A226 and V227 and has an edge on interaction with the ring of Y183 at a distance of approximately 4 Å. The thiazol-4-one 5-position substituents in 3BZU contact L171, Y177, G216, L217 and M233 and the equivalent contacts in the 3EY4 structure are to the same residues plus A172, V180 and V231 [38-40,45].
Figure 13. The possible hydrogen bonds between the protein, ligand (14) and cofactor in the 3BYZ_a structure.
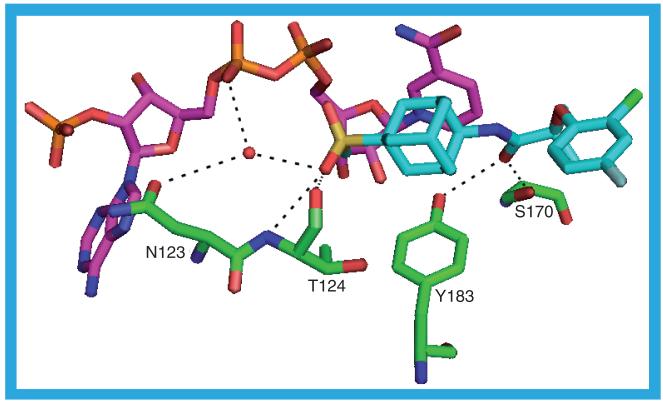
In the 2RBE structure the ligand, 17 (Ki = 17 nM), binds the opposite way round with the thiazolone ring packing against the NADP(H) nicotinamide ring at a distance of approximately 4 Å and making contacts to the protein through residues I121, Y183 and L217. The isopropyl group at the thiazol-4-one 5-position makes contacts with T124, S125, L126, A223, A226 and V227. It is the lack of a second substituent at the 5-position that enables this ligand to adopt this pose in the binding site: a second substituent would clash with the nicotinamide ring, hence the alternative pose adopted by the other three ligands with the thiazol-4-one core structure. The fluorophenyl ring has an edge-on interaction with the ring of Y177 at a distance of approximately 4 Å (Figure 14). It also has contacts with L171, A172, V180, G216, L217, V231 and M233. There is a water-mediated hydrogen bond from the ligand to one, or both, of the proteins and one of the NADP(H) phosphate oxygens: T222 OG1–HOH–O14 and NADP(H) O1N–HOH–O14. Hydrogen-bond lengths in the 2RBE protein chains are shown in the Supplementary Material, although in all those other than 2RBE_a those involving T222 are rather long.
Figure 14. The possible hydrogen bonds between the protein, ligand (17) and cofactor in the 2RBE_a structure.
The interaction of the ligand with Y177 is also shown (red lines).
There are two hydrogen bonds between the ligand and the protein: S170 OG–N7 and Y183 OH–N12 but it is not entirely clear what is acting as donor or acceptor. On the grounds that the side chain of S170 is oriented syn to residues L171 and A172 and that, therefore, the S170 side chain oxygen lone pairs can interact with the backbone NH moieties of L171 and A172, the authors of the paper describing this structure claim that the serine is the donor to N7 [38]. They go on to argue that N7 can act as an acceptor because the double bond involving the thiazol-4-one C8 atom (at the 2-position in the thiazol-4-one ring) rather than being the more normal endocyclic bond to N12 (at the 3-position in the thiazol-4-one ring) is exocyclic to N7, the nitrogen external to the thiazol-4-one ring to which it is attached at the 2-position. While this is possible there are three arguments against it. First, the postulated hydrogen bond involving the L171 NH would be very strained: assuming a normal planar peptide bond, and there is no reason to think it would not be planar, the NH is nowhere near pointing towards either the S170 oxygen or the probable position of the lone pair. Second, for the S170 OG–N7 hydrogen bond to form with the serine acting as the donor, the C–O–H angle would be 124.61°, which is a larger deviation from ideal than the C–O–H angle of 101.85° observed if Y183 were to be the donor in the Y183 OH–N12 hydrogen bond, which would leave N7 as a donor. Third, calculations predict that the compound with an endocyclic double bond is more stable by approximately 2 kcal/mol than the compound with an exocyclic double bond. (The ligand was extracted from the 2RBE_a structure, given the appropriate double bonds and protonation states, and the energy calculated using Sybyl [101].) None of the arguments either way are conclusive. In the other 11β-HSD1 structures the serine can act as a donor or acceptor depending on the nature of the ligand. Consequently, in this structure it is unclear whether the serine is acting as a donor or acceptor, or whether the double bond in the thiazol-4-one ring is endocyclic or exocyclic.
3CH6
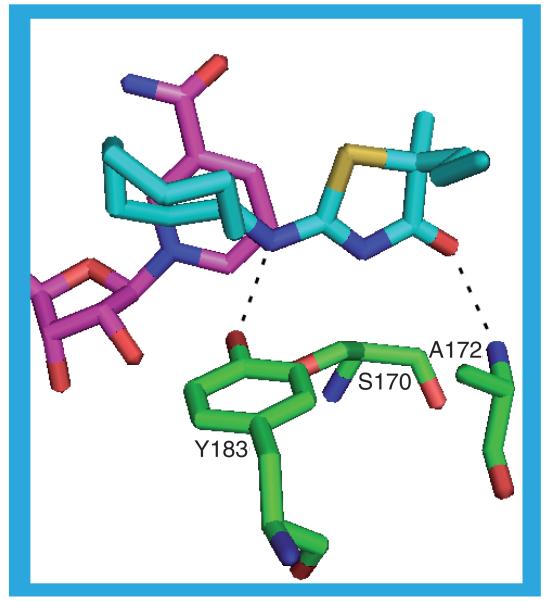
The ligand in the 3CH6 structure, 18 (IC50 = 0.1 nM), may form two hydrogen bonds to the protein, both through the carbonyl oxygen: S170 OG–O23 and Y183 OH–O23. The piperidine ring and the carbonyl pack against the nicotinamide ring. The gem-dimethyl group on the piperidine ring packs against I121, T124, S125, V180 and Y183 with the piperidine ring contacting L126, L217, A223, A226 and V227. The pyridine ring contacts A172, Y177, G216, L217 and M233. The phenyl ring is sandwiched between Y177, P178, M179 and V180 on one side and L126, I230, V231 and M233 on the other. In 3CH6_a and 3CH6_e the fluorine attached to the phenyl ring points back over the pyridine ring in the approximate direction of the piperidine ring but in 3CH6_b and 3CH6_d the phenyl ring is rotated by 180° so that the fluorine points away from the piperidine. This, however, has no effect on the interactions the ligand makes with the protein [41].
3D5Q
The ligand in the 3D5Q structure, 19 (IC50 = 1.6 nM), adopts an ‘L’-shaped pose with two of the triazole nitrogens forming hydrogen bonds to S170 OG and Y183 OH (Figure 15). The angle for the hydrogen bond involving S170 is a little strained in 3D5Q_a and in the other protein molecules it is even more so due to the orientation of the S170 side chain. The triazole and cyclopropyl rings pack against the nicotinamide ring and make contact with the protein through residues S170, Y183 and A223. The isopropyl group attached to the triazole ring is surrounded by the hydrophobic residues L126, V180 and V227. The fluorophenyl ring is flanked on one side by V180 and Y183 and on the other by T124, S125, L126 and A226. Only one face of the trifluoromethoxyphenyl ring is in contact with the protein, mainly through A172 but also L171 and Y177. The orientation of this ring with respect to the triazole ring varies across the four 3D5Q molecules – the plane of the ring in 3D5Q_c is twisted approximately 43° relative to the plane of the ring in 3D5Q_a – but the same three residues are in contact with the ring and the different orientations have little effect on the positions of the ring substituents. The trifluoromethoxy group makes contacts with L171, A172, V175 and Y177. The loop over the substrate binding site, A226–A235, has several residues missing in 3D5Q_a, 3D5Q_c and 3D5Q_d [44].
Figure 15. The possible hydrogen bonds between the protein, ligand (19) and cofactor in the 3D5Q_a structure.
The ligand in the 3D5Q_a structure is in cyan. The buff ligand is that from the 3D5Q_c structure and shows the rotation of the phenyl ring and the minimal impact of this rotation on the position of the ring substituents.
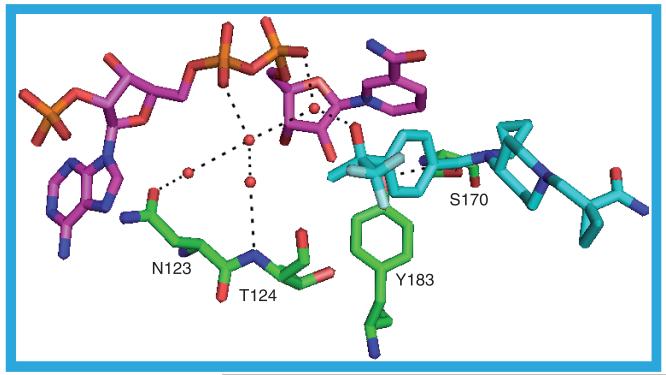
3CZR
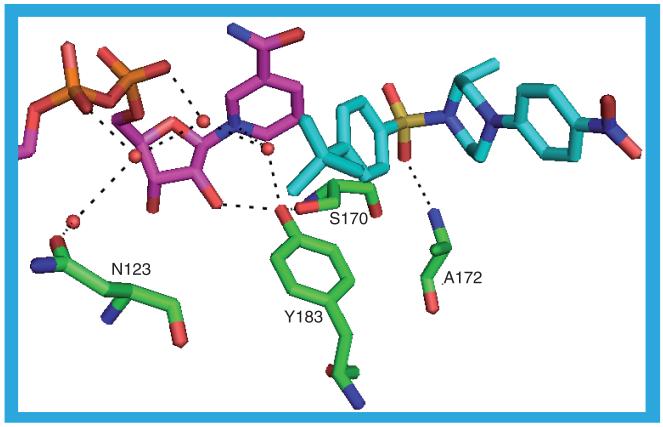
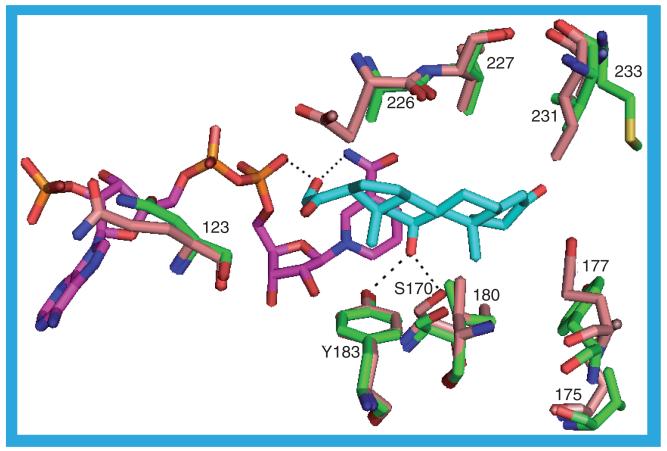
As mentioned previously 3CZR_b has CHAPS (9) in the substrate binding site. However, in the binding site of 3CZR_a is a molecule of 20 (IC50 = 3 nM). Despite the different ligands the structures of the two protein chains are very similar with the greatest difference lying in residues S228-A235, which form a loop over the substrate binding site: in 3CZR_a this is sufficiently disordered so that there are no coordinates for residue I230. The ligand in 3CZR_a adopts a ‘V’ shape in the binding site with the sulfonyl group forming the base of the V. There is only one hydrogen bond between the protein and the ligand, from a backbone nitrogen to a sulfonyl oxygen: A172 N–O1. With most ligands, at least one of S170 OG and Y183 OH hydrogen bond to the ligand, but in 3CZR_a S170 OG hydrogen bonds to Y183 OH, which itself is hydrogen bonded to a water molecule sandwiched between the NADP(H) nicotinamide ring and the ligand butylphenyl moiety. This water molecule could be hydrogen bonded to the nicotinamide ring nitrogen and is one of four water molecules that form a chain along the NADP(H) binding site, making hydrogen bonds to each other, the NADP(H) and the protein (Figure 16). The butylphenyl moiety contacts T124, V180, Y183, L217, T222, A223 and A226. One of the sulfonyl oxygens pushes up against the hydride-donating carbon of the nicotinamide ring. The sulfonyl moiety is surrounded by residues S170, L171, A172, L215, G216 and L217. The methyl-piperazine group lies between M233 on one side and L171, Y177 and V180 on the other. The nitrophenyl group projects to the surface of the protein through a gap between residues L126, P178, M179, V227 and M233. The 3CZR structure is one of four to contain a ligand with a core of (R)-2-methyl-1-(phenylsulfonyl)piperazine, the others being 3D4N, 3HFG and 3H6K [42].
Figure 16. The possible hydrogen bonds between the protein, ligand (20) and cofactor in the 3CZR_a structure.
The network of hydrogen-bonded water molecules stretching along the NADP(H) binding site is shown.
3D4N
The core of the ligand in 3D4N, 21 (IC50 = 1.2 nM), overlays that in 3CZR_a and makes the same interactions. The butyl phenyl moiety in the 3CZR inhibitor is replaced with an (S)-1,1,1-trifluoro-2-phenylpropan-2-ol group that occupies approximately the same volume and makes the same contacts as the butylphenyl group. However, it is capable of forming a hydrogen bond from the hydroxyl group to a water molecule that can in turn form hydrogen bonds to one of the NADP(H) phosphate oxygens and to the water sandwiched between the NADP(H) nicotinamide ring and the ligand phenyl ring. In 3D4N_a residue T124 has alternate side chain conformations due to rotation about the Cα–Cβ bond but this has little effect on the contacts made with the ligand. The nitrophenol group is replaced with a 2-cyclopropyl-propanecarboxamide group that extends to the surface of the protein between residues Y177, P178 and M179 on one side and L126, I230, V231 and M233 on the other. M179 in 3D4N_b and 3D4N_d has alternate side chain conformations but both make contact with the cyclopropyl group. The water-mediated hydrogen bond to the NADP(H) does not exist in the 3D4N_d structure as the water is missing [43,44].
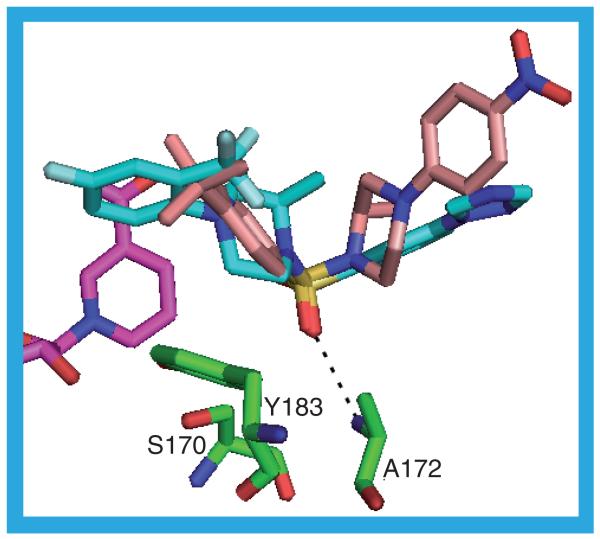
3HFG & 3H6K
3HFG and 3H6K are the third and fourth structures containing a ligand with the (R)-2-methyl-1-(phenylsulfonyl)piperazine core. In both of these structures the ligand again adopts a V-shaped pose but it is bound the opposite way round to the ligands in 3CZR and 3D4N (Figure 17). The sulfonyl group is in the same position as it is in the 3CZR and 3D4N structures and the hydrogen bond from A172 to a sulfonyl oxygen is the only one between the protein and the ligand. The 4-fluoro-2-(trifluoromethyl)phenyl moiety lies edge on to the NADP(H) nicotinamide ring at a distance of approximately 4 Å with the trifluoromethyl group pointing away from the nicotinamide ring. The trifluoromethyl group contacts T124, S125, L126, V180, A226 and V227, with the fluorophenyl ring to which it is attached, contacting I121, T124, Y183, T222 and A223. The methylpiperazine is surrounded by A172, V180 and Y183 on one side and L126, L217 and V227 on the other. In 3HFG, 22 (IC50 = 26 nM [56]), the phenyl-3-triazole moiety projects towards the protein surface, is planar and makes contacts with the protein on only one face of the plane. The phenyl ring is in contact with L171, Y177 and G216 and the triazole ring contacts Y177, P178 and V180. The surface loop over the substrate binding site is flexible to the point that only in 3HFG_a is the amino acid sequence visible: in the other three HFG-protein chains this loop has residues missing. In 3H6K, 23 (IC50 = 10 nM), the phenyl-3-triazole moiety is replaced with a 2-chloro-4-carboxamido-phenyl group. This protrudes to the surface of the protein between residues L171, A172, Y177 and V180 on one side and L217 on the other [48].
Figure 17. The possible hydrogen bonds between the protein, ligand (22) and cofactor in the 3HFG_a structure.
The 3HFG ligand is shown in cyan. The 3CZR ligand (20) is shown in buff.
3FRJ
The inhibitor in the 3FRJ_b substrate binding site is the same as that in the 3D4N substrate binding site, except that the (R)-2-methyl-1-(phenylsulfonyl)piperazine core is replaced by an N-cyclopropyl-N-(piperidin-4-yl)benzamide core. The ligand, 24 (IC50 = 14 nM), adopts a V shape with the amide forming the base of the V and making contacts with S170, L171, A172, Y177, Y183, L215, G216 and L217. The amide oxygen can form a hydrogen bond to S170 OG or possibly Y183 OH. The cyclopropyl is situated where the sulfonyl group in the 3D4N structure is found. This pushes the piperidine and benzyl rings out of the binding site relative to the positions of the piperazine and phenyl rings in 3D4N (Figure 18). This has knock-on effects on the positions of the (S)-1,1,1-trifluoro-2-phenylpropan-2-ol and cyclopropyl-propanecarboxamide groups. The benzamide ring is further away from the nicotinamide ring and there is no water sandwiched between the two rings. It contacts V180, Y183, L217 and A223. The (S)-1,1,1-trifluoro-2-phenylpropan-2-ol makes contacts with T124, S125 and L126 on one side and with T222, A223 and A226 on the other. The hydroxyl group can form a hydrogen bond to a water molecule that is hydrogen bonded to an NADP(H) phosphate oxygen and that is part of a network of four water molecules that stretch along the NADP(H) binding site making hydrogen bonds to each other, the NADP(H) and the protein (Figure 19). The piperidine ring makes contacts with L126, Y177, V180, L217, V227 and M233. The cyclopropyl-propanecarboxamide moiety protrudes to the surface of the protein through a gap between L126, Y177, P178 and M179 on one side and V231 and M233 on the other. There may be a long water-mediated hydrogen bond from the carboxamide oxygen to the backbone carbonyl of Y280 in 3FRJ_a. There is no inhibitor bound to the 3FRJ_a structure. Residues I230 and V231 are missing from the loop over the substrate binding site, and the conformations of the adjacent residues differ from those in 3FRJ_b, as do the conformations of residues N123–L126 [47].
Figure 18. The possible hydrogen bonds between the protein, ligand (24) and cofactor in the 3FRJ_b structure.
The 3FRJ ligand is shown in cyan. The 3D4N ligand (21) is shown in buff.
Figure 19. The water-mediated hydrogen bond network along the NADP(H) binding site in the 3FRJ_b structure.
3D3E
The ligand in the 3D3E structure, 25 (IC50 = 1.4 nM), is similar to that in 3FRJ_b; the differences are that the piperidine ring in 3FRJ is replaced with a cyclohexyl ring and the cyclopropyl-propanecarboxamide moiety is replaced with a 3-pyridyl ring. The substituted benzamide ring makes the same interactions as does the equivalent part of the inhibitor in 3FRJ_b, except that there is no water-mediated hydrogen bond from the hydroxyl group to the NADP(H) phosphate oxygen (due to there being only a few water molecules in this structure). The core is unable to make a hydrogen bond to S170 because the orientation of the side chain is unfavorable for such an interaction but the hydrogen bond to Y183 could form. The cyclohexyl ring is positioned slightly differently to the piperidine ring but makes the same interactions. The 3-pyridyl ring makes substantial contacts with P178 and M179, and lesser contacts with L126, Y177 and V227 [43].
3FCO
The 3FCO ligand, 26 (IC50 = 14 nM), has the same core structure as that in 3D3E: N-cyclohexyl-N-cyclopropylbenzamide. The same substituent is attached to the phenyl ring: the core and phenyl ring substituents make the same interactions as those in 3D3E. The cyclohexyl substituent, a 3-pyridyl ring in the 4-position in 3D3E is replaced with a hydroxyl group and a cyclopropyl at the 4-position in 3FCO. In 3FCO_b the hydroxyl can form a hydrogen bond to a water molecule but this water is not present in 3FCO_a. The cyclohexyl and substituents make contacts with L126, Y177, P178, M179, V180, L217, V227 and M233. The carbonyl oxygen is within hydrogen bonding distance of S170 and Y183. The residues in this structure with alternate conformations are not involved in ligand binding [46].
Sequences & structures of guinea pig & mouse 11β-HSD1
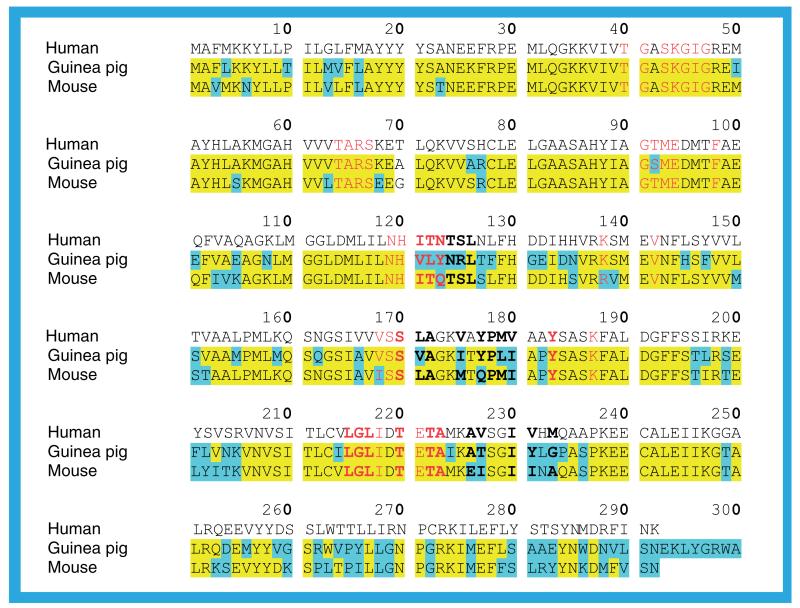
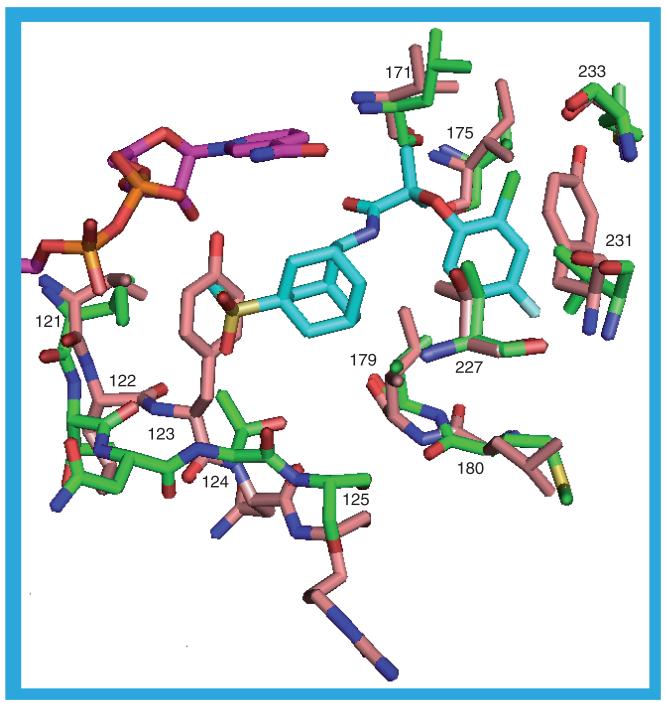
There are three crystal structures of the guinea pig 11β-HSD1 and two of the mouse protein. An alignment of the sequences with that of the human protein (Figure 20) shows that for the 292 superimposable residues common to all three sequences the guinea pig protein has 75% sequence identity to both the human and the mouse protein and the mouse protein has 79.1% sequence identity to the human protein. There are no gaps in, or insertions into, any of the sequences, only an extra eight residues at the C-terminus of the guinea pig protein. Given the high sequence identity and lack of gaps and insertions it is not surprising that the rodent structures superimpose on the human structure with low RMSDs for the Cα atoms (Table 3). For the 36 residues that border the NADP(H) binding site in the crystal structures of the human protein the sequence identity to the human protein is higher than for the whole protein, but for the 26 that border the substrate binding site the sequence identity is lower. For the guinea pig protein the NADP(H) binding site has 88.9% identity (four residues different) and the substrate binding site has 53.8% identity (12 residues different): the equivalent figures for the mouse protein are 91.7% (three residues different) and 69.2% (eight residues different) for the NADP(H) and substrate binding sites, respectively.
Figure 20. A sequence alignment of the guinea pig and mouse proteins with human 11β-hydroxysteroid dehydrogenase type 1.
Residues in the rodent sequences identical to the human sequence are highlighted in yellow; residues that are different are highlighted in cyan. Those residues that, in the human protein, form the NADP(H) binding site are shown in red and those bordering the substrate binding site are shown in bold. Human sequence (Homo sapiens) SwissProt P28845. Guinea pig sequence (Cavia porcellus) SwissProt Q6QLL4. Mouse sequence (Mus musculus) SwissProt P50172. The alignment was performed by Multalin [103].
Table 3. Root mean square deviations I for Ca overlays.
| Structure | RMSD (Å) |
|---|---|
| 1XSE_a | 1.146 |
| 1XSE_b | 1.147 |
| 3DWF_a | 0.982 |
| 3DWF_b | 0.957 |
| 3DWF_c | 1.032 |
| 3DWF_d | 0.962 |
| 3G49_a | 1.003 |
| 3G49_b | 1.018 |
| 3G49_c | 0.983 |
| 3G49_d | 1.027 |
| 1Y5M_a | 0.989 |
| 1Y5M_b | 1.002 |
| 1Y5R_a | 1.001 |
| 1Y5R_b | 1.035 |
For each protein chain (indicated by the lowercase letter) in each crystal structure, the Cα atoms were superimposed onto those of the human 2ILT_a structure and the root mean square deviation was calculated using the Sybyl software [101]. 2ILT_a was used as the reference structure simply because it is the only structure of 11β-HSD1 with only one protein molecule in the crystal structure. The 1XSE, 3DWF and 3G49 structures are of the guinea pig protein and the 1Y5M and 1Y5R structures are of the mouse protein.
Cofactor binding
When the Cα atoms of the guinea pig and mouse proteins are superimposed on those of the human protein, the NADP(H) molecules overlay very well. In the 3DWF (guinea pig) structure [51] the cofactor is identified as being in the ionized state (NADP+) but in the other rodent structures the ionization state is not explicitly identified. The hydrogen bonds between the rodent proteins and the NADP(H) are listed in the Supplementary Material. There are only three differences between the human and mouse proteins among the residues that form the NADP(H) binding site. N123, K138 and V168 in the human protein become Q123, R138 and I168 in the mouse protein. None of these changes is likely to have a major impact on NADP(H) binding as the side chains are either oriented away from the NADP(H) or are conformationally flexible and have only slight contact with the NADP(H).
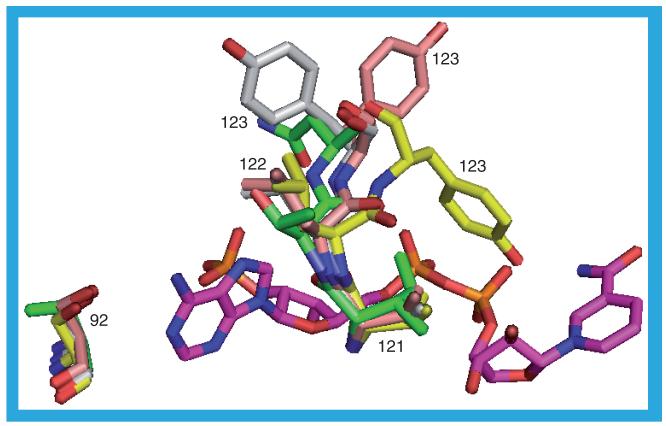
Of the residues bordering the NADP(H), four differ between the human and guinea pig proteins. The mutation of T92 (adjacent to the adenine ring) in the human protein to S92 should have minimal impact on cofactor binding; the hydroxyl group of both residues is oriented in the same direction towards the adenine with which they make a surface contact. The other three differences are residues 121–123, which in the human structures, also contact the ligand in the substrate binding site. The mutation of I121 in the human protein to the smaller L121 in the guinea pig protein results in the guinea pig protein having slightly less contact with the NADP(H). The side chains of T122 (human) and L122 (guinea pig) are oriented away from the NADP(H) and the backbone atoms are in sufficiently similar positions that any difference in their interaction with the NADP(H) is minimal. The big difference lies in the interactions of residue 123 with the NADP(H) (Figure 21). In the human protein this is an asparagine, the amide group of which is in contact with the adenine ring. In the guinea pig protein this residue is a tyrosine, but in the different protein structures it has three different orientations. The conformation adopted by Y123 in 3DWF_b and 3DWF_d is similar to that of the asparagine in the human protein in that it interacts with the adenine ring. In the 1XSE_a, 1XSE_b, 3DWF_a, 3DWF_c and 3G49_b structures the side chain points towards the nicotinamide ring (with which it makes contact) and makes contact, also, with the ribose and phosphate groups. The hydroxyl group forms a hydrogen bond to the nicotinamide N7N atom. Tyrosine 123 in the 3G49_a, 3G49_c and 3G49_d structures is oriented away from the NADP(H) having just slight contact with one of the phosphate groups.
Figure 21. Differences between the human and guinea pig interactions with NADP(H).
The NADP(H), shown in purple, is that in the human 2ILT structure. The human protein residues are shown in green: T92, I121, T122 and N123. The equivalent residues from the guinea pig structures are in yellow (1XSE_a), white (3DWF_b) and buff (3G49_a): S92, V121, L122 and Y123.
Ligand binding
1XSE
The 1XSE_a and 1XSE_b guinea pig structures are very similar, with a Cα RMSD value of 0.103 Å; the orientations of the side chains are also very similar. There is no ligand in the substrate binding site. Along one side of the substrate binding site there is a sequence of five residues, 121–125, which differs from the sequence of the human protein: ITNTS in the human protein becomes VLYNR in the guinea pig protein. A comparison of the human (2ILT) and guinea pig structures shows that in this region the backbone has a different structure. In the 1XSE structure residues 121–123 are closer to the NADP(H) than are the equivalent residues in the human structure, but residues 124–125 are further away. However, the major difference lies in the position of the side chain of residue 123. N123 in the human structure points towards the adenine ribose phosphate, but in the guinea pig structure Y123 points towards the nicotinamide ring. In this orientation it clashes with the ligand in several of the human structures (Figure 22). At the opposite end of the substrate binding site, V231 in the human protein becomes Y231 in the guinea pig protein. This greatly reduces the size of the opening at the end of the pocket so ligands are less likely to be able to reach the surface of the protein [50].
Figure 22. Differences in the substrate binding sites of 1XSE_a (guinea pig) and 2ILT (human).
The NADP(H) (purple carbons), ligand (cyan carbons) and protein (green carbons) are from the human 2ILT structure: I121, T122, N123, T124, S125, L171, V175, M179, V180, V227, V231 and M233. The guinea pig 1XSE_a structure is in buff: V121, L122, Y123, N124, R125, V171, I175, L179, I180, T227, Y231 and G233.
3DWF
The 3DWF structure of the guinea pig protein has four 11β-HSD1 molecules in the asymmetric unit. The substrate binding site in 3DWF_c and 3DWF_d is empty, but in both 3DWF_a and 3DWF_b there is a glycerol molecule. The glycerol in the two proteins has a different conformation, but in neither structure is there more than minimal contact with the residues forming the substrate binding site; the only hydrogen bond is in 3DWF_b to S170 OG. The F278E mutation in this protein is more than 25 Å from the substrate binding site so should have little influence on ligand binding [51].
3G49
The 3G49 structure of the guinea pig protein also has four molecules in the asymmetric unit. Neither 3G49_b nor 3G49_c have a ligand in the substrate binding site but there is an inhibitor, 27 (Ki = 287 nM), in both 3G49_a and 3G49_d. There are two hydrogen bonds between the protein and the ligand: A172 N–O12 and Y183 OH–N14. The pyridine ring stacks against the nicotinamide ring and makes contact with the protein through residues L126, I180, Y183, L217, A223 and T227. The diethylacetamide moiety pushes up against the NADP(H) ribose and phosphate groups and makes contact with V121, Y123, N124, Y183, T222 and A226. The sulfonamide moiety contacts S170, V171, A172, Y177, G216 and L217. The chlorine of the chloromethylphenyl group pushes up against Y231 with the ring stacking with Y177 and making contact with I182 and L217 [30].
1Y5M
The two protein molecules in the 1Y5M mouse protein structure each have a molecule of octane half in the substrate binding site and half protruding to the surface of the protein. It makes contact with residues L126, L171, Q177, I180, L217, I227, I231 and A233 [49].
1Y5R
In both protein molecules in the 1Y5R crystal structure of the mouse protein there is a molecule of corticosterone (2b), the product of the reaction catalyzed by murine 11β-HSD1. There are two possible hydrogen bonds between the corticosterone and the protein: S170 OG–O2 and Y183 OH–O2 (Figure 23). There are also two possible hydrogen bonds between the NADP(H) and the ligand: NADP(H) N7N–O4 and NADP(H) O1N–O4. The corticosterone is in contact with residues I121, T124, S125, L126, S170, L171, A172, Q177, I180, Y183, G216, L217, T222, A223, E226, I227 and I231, as well as with the NADP(H). Among the 26 residues that border the substrate binding site there are eight that differ between the human and mouse proteins (Figure 23). From the crystal structures of the human and mouse proteins the change to residue 123 should have little impact on ligand binding. However, the same residue in the guinea pig protein can impact on ligand binding. At the opposite end of the substrate binding site the changes to residues 175 and 233 might be significant for those ligands that are long enough to get to the surface of the protein. The changes to residues 180, 227 and 231 are ones of size: they change the size and shape of the binding site. Residues 177 and 226 in the human protein are tyrosine and alanine, respectively, which become glutamine and glutamic acid, respectively, in the mouse protein. These changes could impact on ligand binding due to the differences in both size and charge/electrostatics [49].
Figure 23. Differences in the substrate binding sites of the human and mouse enzymes.
The human 2ILT structure is shown as the green protein. The mouse 1Y5R_a structure is shown as the buff protein, purple NADP(H) and cyan ligand (corticosterone, 2b). For both structures S170 and Y183 are shown. All the other residues shown differ between the structures. Human: N123, V175, Y177, V180, A226, V227, V231 and M233. Mouse: Q123, M175, Q177, I180, E226, I227, I231 and A233.
Structures for use in ligand docking & virtual screening
The main reason for crystallizing 11β-HSD1 has been to aid the development of inhibitors for therapeutic use. As illustrated previously, the crystal structures of rodent 11β-HSD1 are sufficiently different from those of the human enzyme that designing a compound to inhibit the rodent enzyme will probably not produce an optimal inhibitor of the human enzyme. This is, perhaps, best shown by the comparative inhibition data (for some of the compounds previously discussed) shown in Table 4. However, because many in vivo pharmacokinetic and pharmacodynamic studies are carried out in rodents, it is useful for inhibitors of human 11β-HSD1 to also inhibit the rodent enzyme.
Table 4. Comparative inhibition data for human and murine 1ip-hydroxysteroid dehydrogenase types 1 and 2.
| Inhibitor | hHSD1 | hHSD2 | mHSD1 | mHSD2 | Ref. |
|---|---|---|---|---|---|
| 12 | Ki = 7 nM | 26,000 | Ki = 4 nM | >100,000 | [36] |
| 13 | 6 | 18,000 | 4 | 55,000 | [37] |
| 20 | 3 | – | 53 | – | [42] |
| 22 | 26 | – | 10 and 42† | – | [56] |
| 23 | 10 | – | 10 | – | [48] |
| 28 | Ki = 1.4 nM | – | Ki = 0.63 nM | – | [52] |
Unless otherwise stated the Inhibition data are IC50 values expressed as nM.
Two IC50 values are given for 22 with mHSDI.
h: Human; HSD: Hydroxysteroid dehydrogenase; m: Murine.
The crystal structures of the human protein vary in quality. The ranges of values for some of the crystallographic structure quality indices are shown in Table 5 and the details for each of the structures are shown in the Supplementary Material. These data, along with the B-factors and occupancy data (particularly for those atoms around the substrate binding site), may influence the choice of structure for use in ligand docking and virtual screening.
Table 5. The ranges of values for some crystallographic structure quality indices.
| Resolution | Completeness | RCryst | RFree | Bond-length deviation from ideal | Bond-angle deviation from ideal |
|---|---|---|---|---|---|
| Highest | Highest | Lowest | Lowest | Smallest | Smallest |
| 1.55 Å | 100.0% | 15.682% | 18.085% | 0.005 Å | 1.088° |
| 1XU9 | 3FRJ | 1XU9 | 1XU9 | 3D4N | 3H6K |
|
| |||||
| Lowest | Lowest | Highest | Highest | Largest | Largest |
| 3.1 Å | 66.2% | 23.7% | 31.9% | 0.024 Å | 2.31° |
| 2IRW | 3H6K | 2IRW | 3EY4 | 3FRJ | 3BYZ |
The structure associated with each of the values is shown.
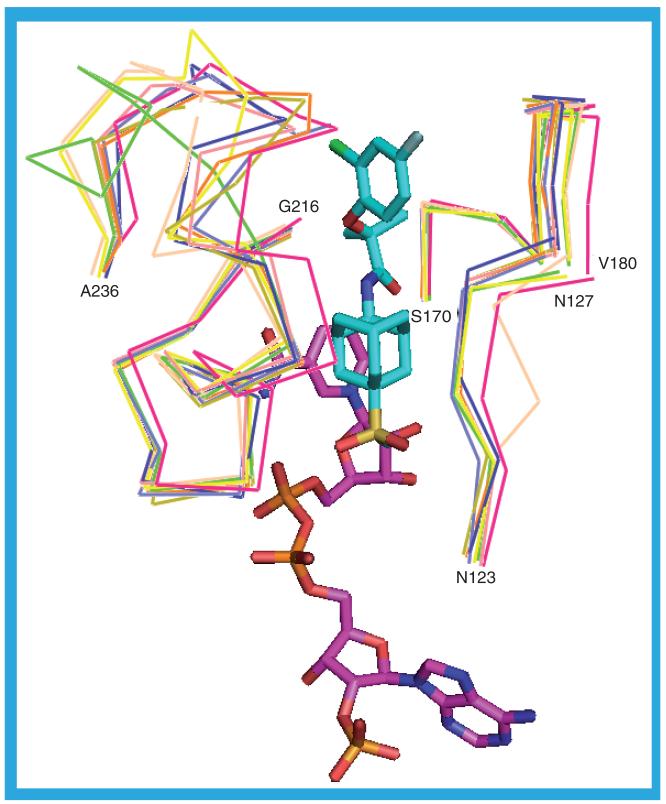
Any of the structures of human 11β-HSD1 could be used for ligand docking and virtual screening although it may be worth careful consideration before using any of those structures with residues missing from the flexible loop over the substrate binding site (residues V227–Q234) or with atoms missing from those residues. Those structures that merit this careful consideration are 1XU9_c, 1XU9_d, 2BEL_b, 2BEL_c, 2BEL_d, 3BZU_c, 3CZR_a, 3D3E_a, 3D3E_b, 3D3E_c, 3D3E_d, 3D4N_a, 3D4N_c, 3D4N_d, 3D5Q_a, 3D5Q_c, 3D5Q_d, 3FCO_a, 3FCO_b, 3FRJ_a, 3HFG_a, 3HFG_b, 3HFG_c and 3HFG_d. Figure 24 illustrates the range of movement in the flexible loop by showing Cα traces of those residues that form the opposing walls of the substrate binding site. There is a little variability in the position of the Cα atoms of those residues that form the right hand wall (N123–N127 and S170–V180) but much greater variability in the position of the residues that form the left hand wall (G216–A236) particularly those in the flexible loop. This variability is likely to be due, at least in part, to the different ligands in the various structures and is likely to influence the pose adopted by any docked ligand.
Figure 24. Overlay of the Cα trace of those residues that form the opposing walls of the substrate binding site showing the variability in the flexible loop.
Nine structures are shown: 1XU7_a, 2BEL_a, 2IRW_a, 2RBE_a, 3BYZ_d, 3CH6_b, 3CZR_b, 3EY4_c and 3EY4_d. The ligand (cyan carbons) and NADP(H) (purple carbons) from the 2ILT_a structure are shown.
The conformations of the side chains bordering the substrate binding site vary between the structures, which is likely to lead to the different structures having different docked-ligand poses and/or docking scores. Given the role it plays in forming hydrogen bonds to many of the inhibitors the orientation of the S170 side chain is significant: in some structures it is oriented along the substrate binding site with the hydroxyl pointing towards Y183 and the NADP(H) (e.g., Figure 16), but in others it is pointing the opposite way along the pocket (e.g., Figure 14). Particular care should be taken with those structures where residues with alternate conformations border the substrate binding site; one conformation should be chosen. (How do the docking programs handle an input protein structure with two [alternate] conformations? Do they recognize that two conformations are present and flag up an error? Do they use the first conformation in the input file, or the conformation with the greater occupancy, or do they assume both conformations are present and exclude the entire volume occupied by both conformations from the accessible docking space? Do the alternate conformations of charged residues more remote from the docking site have different electrostatic influences on the binding site resulting in different docked poses and/or docking scores?) Details of those structures and residues with alternate conformations are listed in the Supplementary Material but those residues with alternate conformations in, or adjacent to the substrate binding site, which may, therefore, influence the conformation of any docked ligand are T124 in 1XU9_a and 3D4N_a, M179 in 1XU9_b, 1XU9_d, 3D4N_b and 3D4N_d, Y177 in 3BZU_a and 3BZU_c, and, possibly, N127 in 3D5Q_c.
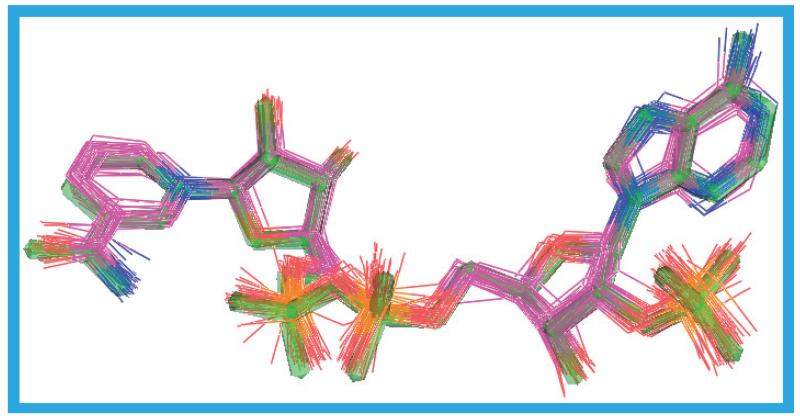
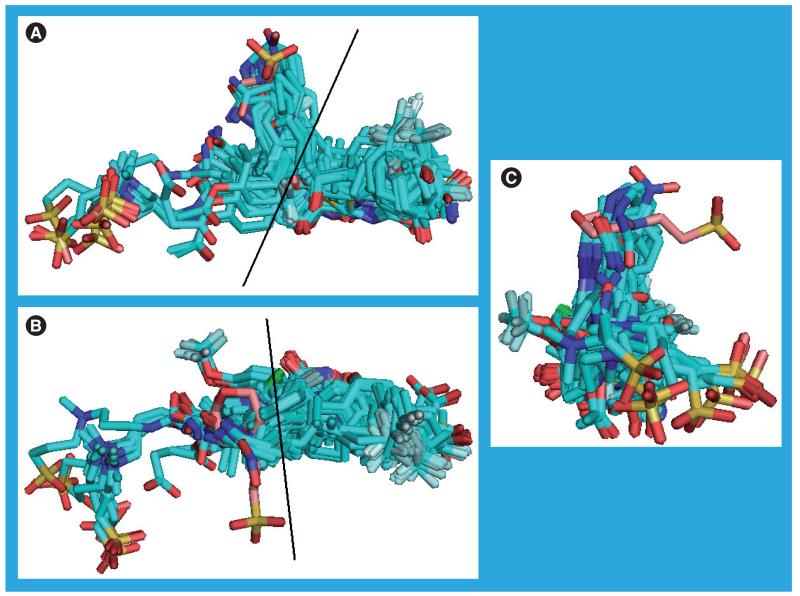
The various inhibitors in the crystal structures of the human protein vary in size and potency. Excluding CHAPS (9) and MES (10) the molecular weights range from 252 to 570 Da, the surface areas from 453 to 791 Å2, the polar surface areas from 31 to 202 Å2 and the volumes from 733 to 1483 Å3 (all calculated from the crystal structure conformations using the Sybyl software [101]). The potencies of twelve of the compounds are quoted in terms of IC50 and range from 0.1 to 63 nM, and the potencies of four others are quoted in terms of Ki and range from 7 to 210 nM. The ligands in all 67 crystal structure molecules are shown superimposed in Figure 25. Except for the smallest inhibitors, all the substituted thiazol-4-ones (14–17), extend to the surface of the protein where 12, 13, 18 and 20–26 occupy part of the space taken up by the MES morpholine ring in 1XU9_a. None of these ligands take up the whole width of the pocket occupied by MES, suggesting that novel ligands may be able to exploit this space to improve potency, selectivity and/or ADMET properties. Carbenoxolone (11) follows the path of CHAPS out of the pocket but 19, the ligand in the 3D5Q structure, induces a unique structure in the protein. In 3D5Q Y177 occupies the space taken up by the MES morpholine ring in 1XU9_a and the trifluoromethoxy group in 19 occupies the space taken by Y177 in all the other structures. Ligands 12, 13, 18 and 20–26 occupy broadly similar parts of the substrate binding site. Consequently, any compound being designed to mimic the inter actions of these compounds may dock poorly into the 3D5Q structure because of a clash with Y177.
Figure 25. The substrate binding site ligands.
Those ligands in, or adjacent to the substrate binding sites of all 67 human crystal structure protein chains are shown in three orientations. These figures demonstrate that the accessible width and depth of the substrate binding site is greater than that accessed by most of the ligands. The buff-colored molecule is the 2-(N-morpholino)-ethanesulfonic acid from structure 1XU9_a and is entirely outside the substrate binding site. In (A) and (B) the portion of the molecules to the right of the black line lies in the substrate binding site, with the remainder on the protein surface. In (C) the portion of the molecule on the surface of the protein is in the foreground. To create this image all the Cα atoms in the protein chains were superimposed and only the ligands displayed.
Future perspective
Crystal structures of drug targets have aided the discovery of many inhibitors. The availability of crystal structures of 11β-HSD1 with inhibitors bound will aid the discovery and design of more potent and selective inhibitors. Additional structures will help clarify those aspects of the protein and ligand structures that are important for potency. However, a potent inhibitor is not necessarily a drug. This requires a consideration of ADMET properties that may necessitate modification of the inhibitor. Structures of the ligand in the protein may help guide these modifications so that they have minimal impact on potency and specificity.
Recent development
Since the submission of this article the co ordinates of the 3GMD structure mentioned in the main text have been released. The structure, which is of the mouse protein, contains the inhibitor described. Also, two further structures have been deposited in the PDB. The 3PDJ structure is of the human protein in complex with a 4,4-disubstituted cyclohexylbenzamide inhibitor [57]. The 3OQ1 and 3QQP structures are in complex with a diarylsulfone and urea inhibitor, respectively, but for neither structure is the species origin of the protein specified.
Supplementary Material
To view the dynamic 3D structures of Figures 6-19 & 21-25 please visit the journal website at: www.FuTureScience.com/Doi/Suppl/10.4155/Fmc.10.282. 3D structures supported by Jmol viewer.
Executive summary.
-
■
Excessive cortisol production is associated with metabolic syndrome. Cortisol is produced by 11β-HSD1, the inhibition of which may alleviate the symptoms of metabolic syndrome.
-
■
There are several crystal structures of 11β-HSD1 from three species available. Given the sequence differences, the value of using the structures of the mouse and guinea pig proteins in structure-based inhibitor discovery and design is questionable, although the inhibition of these enzymes may be helpful in rodent pharmacokinetic and pharmacodynamic studies.
-
■
The crystal structures of the human protein vary in quality but are broadly similar. The differences are sufficient that care needs to be taken in selecting the structure(s) to be used in drug-discovery work.
Glossary
- 11β-hydroxysteroid dehydrogenase type 1
The enzyme that catalyzes the reduction of cortisone to cortisol.
- Virtual screening
The large-scale docking of many ligands into the active site of an enzyme in order to identify possible inhibitors
- Ligand docking
The computational docking of ligands into the active site of an enzyme in order to analyze possible binding poses
Footnotes
Financial & competing interests disclosure
Barry VL Potter has been funded previously by Sterix Ltd, a member of the Ipsen Group, to conduct work on the develop ment of inhibitors of 11β-HSD1. The authors thank the Wellcome Trust for a VIP award that substantially facilitated the compilation of this work and for Programme Grant support (082837). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
■ of interest
- 1.Morton NM. Obesity and corticosteroids: 11β-hydroxysteroid type 1 as a cause and therapeutic target in metabolic disease. Mol. Cell. Endocrinol. 2010;316:154–164. doi: 10.1016/j.mce.2009.09.024. [■ Reviews the evidence that 11β-hydroxysteroid dehydrogenase type 1 (11β-HSD1) is a good target for metabolic disease intervention.] [DOI] [PubMed] [Google Scholar]
- 2.Grundy SM. Obesity, metabolic syndrome, and cardiovascular disease. J. Clin. Endocrinol. Metab. 2004;89:2595–2600. doi: 10.1210/jc.2004-0372. [DOI] [PubMed] [Google Scholar]
- 3.Tomlinson JW, Walker EA, Bujalska IJ, et al. 11beta-hydroxysteroid dehydrogenase type 1: a tissue-specific regulator of glucocorticoid response. Endocrine Rev. 2004;25:831–866. doi: 10.1210/er.2003-0031. [DOI] [PubMed] [Google Scholar]
- 4.Walker BR. Cortisol – cause and cure for metabolic syndrome? Diabet. Med. 2006;23:1281–1288. doi: 10.1111/j.1464-5491.2006.01998.x. [DOI] [PubMed] [Google Scholar]
- 5.Friedman JE, Yun JS, Patel YM, McGrane MM, Hanson RW. Glucocorticoids regulate the induction of phosphoenolpyruvate carboxykinase (GTP) gene transcription during diabetes. J. Biol. Chem. 1993;268:12952–12957. [PubMed] [Google Scholar]
- 6.Wang M. The role of glucocorticoid action in the pathophysiology of the metabolic syndrome. Nutr. Metab. 2005;2:3. doi: 10.1186/1743-7075-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hasselgren P-O. Glucocorticoids and muscle catabolism. Curr. Opin. Clin. Nutr. Metab. Care. 1999;2:201–205. doi: 10.1097/00075197-199905000-00002. [DOI] [PubMed] [Google Scholar]
- 8.Arnaldi G, Angeli A, Atkinson AB, et al. Diagnosis and complications of Cushing’s syndrome: a consensus statement. J. Clin. Endocrinol. Metab. 2003;88:5593–5602. doi: 10.1210/jc.2003-030871. [DOI] [PubMed] [Google Scholar]
- 9.Wamil M, Seckl JR. Inhibition of 11β-hydroxysteroid dehydrogenase type 1 as a promising therapeutic target. Drug Discov. Today. 2007;12:504–520. doi: 10.1016/j.drudis.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Fraser R, Ingram MC, Anderson NH, Morrison C, Davies E, Connell JMC. Cortisol effects on body mass, blood pressure, and cholesterol in the general population. Hypertension. 1999;33:1364–1368. doi: 10.1161/01.hyp.33.6.1364. [DOI] [PubMed] [Google Scholar]
- 11.Stewart PM, Whorwood CB. 11β-hydroxysteroid dehydrogenase activity and corticosteroid hormone action. Steroids. 1994;59:90–95. doi: 10.1016/0039-128x(94)90082-5. [DOI] [PubMed] [Google Scholar]
- 12.Oppermann UCT, Persson B, Jörnvall H. The 11β-hydroxysteroid dehydrogenase system, a determinant of glucocorticoid and mineralocorticoid action: function, gene organization and protein structures of 11β-hydroxysteroid dehydrogenase isoforms. Eur. J. Biochem. 1997;249:355–360. doi: 10.1111/j.1432-1033.1997.t01-1-00355.x. [DOI] [PubMed] [Google Scholar]
- 13.Seckl JR, Walker BR. 11β-hydroxysteroid dehydrogenase type 1 – a tissue-specific amplifier of glucocorticoid action. Endocrinology. 2001;142:1371–1376. doi: 10.1210/endo.142.4.8114. [DOI] [PubMed] [Google Scholar]
- 14.Stewart PM, Valentino R, Wallace AM, Burt D, Shackleton CHL, Edwards CRW. Mineralocorticoid activity of liquorice: 11β-hydroxysteroid dehydrogenase deficiency comes of age. Lancet. 1987;330:821–824. doi: 10.1016/s0140-6736(87)91014-2. [DOI] [PubMed] [Google Scholar]
- 15.Agarwal AK, Mune T, Monder C, White PC. NAD+-dependent isoform of 11β-hydroxysteroid dehydrogenase. J. Biol. Chem. 1994;269:25959–25962. [PubMed] [Google Scholar]
- 16.Kershaw EE, Morton NM, Dhillon H, Ramage L, Seckl JR, Flier JS. Adipocyte-specific glucocorticoid inactivation protects against diet-induced obesity. Diabetes. 2005;54:1023–1031. doi: 10.2337/diabetes.54.4.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morton NM, Paterson JM, Masuzaki H, et al. Novel adipose tissue-mediated resistance to diet-induced visceral obesity in 11β-hydroxysteroid dehydrogenase type 1-deficient mice. Diabetes. 2004;53:931–938. doi: 10.2337/diabetes.53.4.931. [DOI] [PubMed] [Google Scholar]
- 18.Kotelevtsev Y, Holmes MC, Burchell A, et al. 11β-Hydroxysteroid dehydrogenase type 1 knockout mice show attenuated glucocorticoid-inducible responses and resist hyperglycemia on obesity or stress. Proc. Natl. Acad. Sci. USA. 1997;94:14924–14929. doi: 10.1073/pnas.94.26.14924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paterson JM, Morton NM, Fievet C, et al. Metabolic syndrome without obesity: hepatic overexpression of 11β-hydroxysteroid dehydrogenase type 1 in transgenic mice. Proc. Natl. Acad. Sci. USA. 2004;101:7088–7093. doi: 10.1073/pnas.0305524101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masuzaki H, Paterson J, Shinyama H, et al. A transgenic model of visceral obesity and the metabolic syndrome. Science. 2001;294:2166–2170. doi: 10.1126/science.1066285. [DOI] [PubMed] [Google Scholar]
- 21.Gathercole LL, Stewart PM. Targeting the prereceptor metabolism of cortisol as a novel therapy in obesity and diabetes. J. Steroid Biochem. Molec. Biol. 2010;122:21–27. doi: 10.1016/j.jsbmb.2010.03.060. [■ Reviews the role of cortisol in disease as well as the reports of in vivo pharmacological inhibition of 11β-HSD1.] [DOI] [PubMed] [Google Scholar]
- 22.Walker BR, Connacher AA, Lindsay RM, Webb DJ, Edwards CR. Carbenoxolone increases hepatic insulin sensitivity in man: a novel role for 11-oxosteroid reductase in enhancing glucocorticoid receptor activation. J. Clin. Endocrinol. Metab. 1995;80:3155–3159. doi: 10.1210/jcem.80.11.7593419. [DOI] [PubMed] [Google Scholar]
- 23.Rosenstock J, Banarer S, Fonseca VA, et al. The 11β-hydroxysteroid dehydrogenase type 1 inhibitor INCB13739 improves hyperglycemia in patients with Type 2 diabetes inadequately controlled by metformin monotherapy. Diabetes Care. 2010;33:1516–1522. doi: 10.2337/dc09-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Webster SP, Pallin TD. 11β-hydroxysteroid dehydrogenase type 1 inhibitors as therapeutic agents. Expert Opin. Ther. Patents. 2007;17:1407–1422. [■ Reviews the development of 11β-HSD1 inhibitors.] [Google Scholar]
- 25.Hughes KA, Webster SP, Walker BR. 11-β-hydroxysteroid dehydrogenase type 1 (11β-HSD1) inhibitors in Type 2 diabetes mellitus and obesity. Expert Opin. Investig. Drugs. 2008;17:481–496. doi: 10.1517/13543784.17.4.481. [■ Reviews the development of 11β-HSD1 inhibitors.] [DOI] [PubMed] [Google Scholar]
- 26.Su X, Vicker N, Potter BVL. Inhibitors of 11β-hydroxysteroid dehydrogenase type 1. Prog. Med. Chem. 2008;46:29–130. doi: 10.1016/S0079-6468(07)00002-1. [■ Reviews the development of 11β-HSD1 inhibitors.] [DOI] [PubMed] [Google Scholar]
- 27.Boyle CD, Kowalski TJ. 11β-hydroxysteroid dehydrogenase type 1 inhibitors: a review of recent patents. Expert Opin. Ther. Patents. 2009;19:801–825. doi: 10.1517/13543770902967658. [■ Reviews the development of 11β-HSD1 inhibitors.] [DOI] [PubMed] [Google Scholar]
- 28.Rohde JJ, Pliushchev MA, Sorensen BK, et al. Discovery and metabolic stabilization of potent and selective 2-amino-N-(adamant-2-yl) acetamide 11β-hydroxysteroid dehydrogenase type 1 inhibitors. J. Med. Chem. 2007;50:149–164. doi: 10.1021/jm0609364. [DOI] [PubMed] [Google Scholar]
- 29.Zhu Y, Olson SH, Graham D, et al. Phenylcyclobutyl triazoles as selective inhibitors of 11β-hydroxysteroid dehydrogenase type 1. Bioorg. Med. Chem. Lett. 2008;18:3412–3416. doi: 10.1016/j.bmcl.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 30.Siu M, Johnson TO, Wang Y, et al. N-(Pyridin-2-yl) arylsulfonamide inhibitors of 11β-hydroxysteroid dehydrogenase type 1: discovery of PF-915275. Bioorg. Med. Chem. Lett. 2009;19:3493–3497. doi: 10.1016/j.bmcl.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 31.Véniant MM, Hale C, Hungate RW, et al. Discovery of a potent, orally active 11β-hydroxysteroid dehydrogenase type 1 inhibitor for clinical study: identification of (S)-2-((1S,2S,4R)-bicyclo[2.2.1]heptan-2-ylamino)-5-isopropyl-5-methylthiazol-4(5H)-one (AMG 221) J. Med. Chem. 2010;53:4481–4487. doi: 10.1021/jm100242d. [DOI] [PubMed] [Google Scholar]
- 32.Su X, Pradaux-Caggiano F, Thomas MP, et al. Discovery of adamantyl ethanone derivatives as potent 11β-hydroxysteroid dehydrogenase type 1 (11β-HSD-1) inhibitors. ChemMedChem. 2010;5:1577–1593. doi: 10.1002/cmdc.201000081. [DOI] [PubMed] [Google Scholar]
- 33.Holmes MC, Carter RN, Noble J, et al. 11β-hydroxysteroid dehydrogenase type 1 expression is increased in the aged mouse hippocampus and parietal cortex and causes memory impairments. J. Neurosci. 2010;30:6916–6920. doi: 10.1523/JNEUROSCI.0731-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sooy K, Webster SP, Noble J, et al. Partial deficiency or short-term inhibition of 11β-hydroxysteroid dehydrogenase type 1 improves cognitive function in aging mice. J. Neurosci. 2010;30:13867–13872. doi: 10.1523/JNEUROSCI.2783-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hosfield DJ, Wu Y, Skene RJ, et al. Conformational flexibility in crystal structures of human 11β-hydroxysteroid dehydrogenase type 1 provide insights into glucocorticoid interconversion and enzyme regulation. J. Biol. Chem. 2005;280:4639–4648. doi: 10.1074/jbc.M411104200. [■ References [35-52] describe the crystal structures that are the subject of this review.] [DOI] [PubMed] [Google Scholar]
- 36.Sorensen B, Winn M, Rohde J, et al. Adamantane sulfone and sulphonamide 11-β-HSD1 inhibitors. Bioorg. Med. Chem. Lett. 2007;17:527–532. doi: 10.1016/j.bmcl.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 37.Patel JR, Shuai Q, Dinges J, et al. Discovery of adamantane ethers as inhibitors of 11β-HSD-1: synthesis and biological evaluation. Bioorg. Med. Chem. Lett. 2007;17:750–755. doi: 10.1016/j.bmcl.2006.10.074. [DOI] [PubMed] [Google Scholar]
- 38.Yuan C, St. Jean DJ, Jr, Liu Q, et al. The discovery of 2-anilinothiazolones as 11β-HSD1 inhibitors. Bioorg. Med. Chem. Lett. 2007;17:6056–6061. doi: 10.1016/j.bmcl.2007.09.070. [DOI] [PubMed] [Google Scholar]
- 39.Johansson L, Fotsch C, Bartberger MD, et al. 2-amino-1,3-thiazol-4(5H)-ones as potent and selective 11β-hydroxysteroid dehydrogenase type 1 inhibitors: enzyme-ligand co-crystal structure and demonstration of pharmacodynamic effects in C57BI/6 mice. J. Med. Chem. 2008;51:2933–2943. doi: 10.1021/jm701551j. [DOI] [PubMed] [Google Scholar]
- 40.Hale C, Véniant M, Wang Z, et al. Structural characterization and pharmacodynamic effects of an orally active 11β-hydroxysteroid dehydrogenase type 1 inhibitor. Chem. Biol. Drug Des. 2008;71:36–44. doi: 10.1111/j.1747-0285.2007.00603.x. [DOI] [PubMed] [Google Scholar]
- 41.Wang H, Ruan Z, Li JJ, et al. Pyridine amides as potent and selective inhibitors of 11β-hydroxysteroid dehydrogenase type 1. Bioorg. Med. Chem. Lett. 2008;18:3168–3172. doi: 10.1016/j.bmcl.2008.04.069. [DOI] [PubMed] [Google Scholar]
- 42.Sun D, Wang Z, Di Y, et al. Discovery and initial SAR of arylsulfonylpiperazine inhibitors of 11β-hydroxysteroid dehydrogenase type 1 (11β-HSD1) Bioorg. Med. Chem. Lett. 2008;18:3513–3516. doi: 10.1016/j.bmcl.2008.05.025. [DOI] [PubMed] [Google Scholar]
- 43.Julian LD, Wang Z, Bostick T, et al. Discovery of novel, potent benzamide inhibitors of 11β-hydroxysteroid dehydrogenase type 1 (11β-HSD1) exhibiting oral activity in an enzyme inhibition ex vivo model. J. Med. Chem. 2008;51:3953–3960. doi: 10.1021/jm800310g. [DOI] [PubMed] [Google Scholar]
- 44.Tu H, Powers JP, Liu J, et al. Distinctive molecular inhibition mechanisms for selective inhibitors of human 11β-hydroxysteroid dehydrogenase type 1. Bioorg. Med. Chem. 2008;16:8922–8931. doi: 10.1016/j.bmc.2008.08.065. [DOI] [PubMed] [Google Scholar]
- 45.Fotsch C, Bartberger MD, Bercot EA, et al. Further studies with the 2-amino-1,3-thiazol-4(5H)-one class of 11β-hydroxysteroid dehydrogenase type 1 inhibitors: reducing pregnane X receptor activity and exploring activity in a monkey pharmacodynamic model. J. Med. Chem. 2008;51:7953–7967. doi: 10.1021/jm801073z. [DOI] [PubMed] [Google Scholar]
- 46.McMinn DL, Rew Y, Sudom A, et al. Optimization of novel di-substituted cyclohexylbenzamide derivatives as potent 11β-HSD1 inhibitors. Bioorg. Med. Chem. Lett. 2009;19:1446–1450. doi: 10.1016/j.bmcl.2009.01.026. [DOI] [PubMed] [Google Scholar]
- 47.Rew Y, McMinn DL, Wang Z, et al. Discovery and optimization of piperidyl benzamide derivatives as a novel class of 11β-HSD1 inhibitors. Bioorg. Med. Chem. Lett. 2009;19:1797–1801. doi: 10.1016/j.bmcl.2009.01.058. [DOI] [PubMed] [Google Scholar]
- 48.Wan Z-K, Chenail E, Xiang J, et al. Efficacious 11β-hydroxysteroid dehydrogenase type 1 inhibitors in the diet-induced obesity mouse model. J. Med. Chem. 2009;52:5449–5461. doi: 10.1021/jm900639u. [DOI] [PubMed] [Google Scholar]
- 49.Zhang J, Osslund TD, Plant MH, et al. Crystal structure of murine 11β-hydroxysteroid dehydrogenase 1: an important therapeutic target for diabetes. Biochemistry. 2005;44:6948–6957. doi: 10.1021/bi047599q. [DOI] [PubMed] [Google Scholar]
- 50.Ogg D, Elleby B, Norström C, et al. The crystal structure of guinea pig 11β-hydroxysteroid dehydrogenase type 1 provides a model for enzyme-lipid bilayer interactions. J. Biol. Chem. 2005;280:3789–3794. doi: 10.1074/jbc.M412463200. [DOI] [PubMed] [Google Scholar]
- 51.Lawson AJ, Walker EA, White SA, Dafforn TR, Stewart PM, Ride JP. Mutations of key hydrophobic surface residues of 11β-hydroxysteroid dehydrogenase type 1 increase solubility and monodispersity in a bacterial expression system. Protein Sci. 2009;18:1552–1563. doi: 10.1002/pro.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng H, Hoffman J, Le P, et al. The development and SAR of pyrrolidine carboxamide 11β-HSD1 inhibitors. Bioorg. Med. Chem. Lett. 2010;20:2897–2902. doi: 10.1016/j.bmcl.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 53.Wang Z, Wang M. Structure and inhibitor binding mechanisms of 11β-hydroxysteroid dehydrogenase type 1. Curr. Chem. Biol. 2009;3:159–170. [■ Reviews the structures of 11β-HSD1 using a different approach from that adopted in this article, with which it is usefully read in conjunction.] [Google Scholar]
- 54.Rao ST, Rossmann MG. Comparison of super-secondary structures in proteins. J. Mol. Biol. 1973;76:241–256. doi: 10.1016/0022-2836(73)90388-4. [DOI] [PubMed] [Google Scholar]
- 55.Hult M, Shafqat N, Elleby B, et al. Active site variability of type 1 11β-hydroxysteroid dehydrogenase revealed by selective inhibitors and cross-species comparisons. Mol. Cell. Endocrinol. 2006;248:26–33. doi: 10.1016/j.mce.2005.11.043. [DOI] [PubMed] [Google Scholar]
- 56.Xiang J, Wan ZK, Li HQ, et al. Piperazine sulfonamides as potent, selective, and orally available 11β-hydroxysteroid dehydrogenase type 1 inhibitors with efficacy in the rat cortisone-induced hyper insulinemia model. J. Med. Chem. 2008;51:4068–4071. doi: 10.1021/jm8004948. [DOI] [PubMed] [Google Scholar]
- 57.Sun D, Wang Z, Caille S, et al. Synthesis and optimization of novel 4,4-disubstituted cyclohexylbenzamide derivatives as potent 11β-HSD1 inhibitors. Bioorg. Med. Chem. Lett. 2011;21:405–410. doi: 10.1016/j.bmcl.2010.10.129. [DOI] [PubMed] [Google Scholar]
- 58.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
Websites
- 101.Sybyl. Tripos www.tripos.com
- 102.PyMOL. DeLano Scientific LLC www.delanoscientific.com
- 103.Multalin http://bioinfo.genotoul.fr/multalin/multalin.html
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
To view the dynamic 3D structures of Figures 6-19 & 21-25 please visit the journal website at: www.FuTureScience.com/Doi/Suppl/10.4155/Fmc.10.282. 3D structures supported by Jmol viewer.