Abstract
We have sequenced an 81-kb genomic region from the honey bee, Apis mellifera, associated with a quantitative trait locus (QTL) sting-2 for aggressive behavior. This sequence represents the first extensive study of the honey-bee genome structure encompassing putative genes in a QTL for a behavioral trait. Expression of 13 putative genes, as well as two transcripts that were present in a honey-bee EST database, was confirmed through reverse transcription analysis of mRNA from the honey-bee head. Whereas most transcripts exhibited little or no variation between European and Africanized honey-bee alleles, one transcript demonstrated significant nonsynonymous substitutions, deletions, and insertions. All 13 putative genes lacked similarity to known invertebrate or vertebrate proteins or transcripts. This observation may be reflective of the processes that determine the genomic evolution of an insect with social behavior and/or haplo-diploidy and are an indication of the unique nature of the honey-bee genome. These results make this sequence an invaluable research tool for the ongoing honey-bee whole-genome sequencing effort.
The evolution of complex social behavior and its genetic basis has intrigued both behavioral ecologists as well as molecular geneticists. The identification of genes that regulate behavior will facilitate molecular genetic studies as well as the evolutionary and ecological analyses of social behavior (Page Jr. and Erber 2002;Robinson 2002). With its highly organized and complex society, the honey bee, Apis mellifera, serves as a good model organism for the study of complex behavioral traits.
Single genes, as well as quantitative trait loci (QTL), have been linked to various behaviors. In the honey bee, behavioral traits such as learning (Brandes 1991;Chandra et al. 2001) and hygiene (Rothenbuhler 1964;Lapidge et al. 2002) have been shown to have a genetic basis. For example, selective breeding and genetic linkage methodology was used to identify quantitative trait loci (QTLs) involved with foraging behavior in honey bees (Hunt et al. 1995;Page Jr. et al. 2000). Variation in these genomic regions affects the amount of pollen stored in honey-bee colonies and influences whether foragers will collect pollen or nectar. These pollen-hoarding QTLs influence response thresholds to sucrose of individual bees, confirming that allelic variation influences the behavior of individual bees in their society (Page Jr. et al. 2000).
Africanized honey bees are hybrids of African (A. mellifera scutellata) and European (A. mellifera ligustica) honey-bee subspecies. These hybrids originated in Brazil in 1956 (Kerr 1967) and spread into the United States in 1990 (Sugden and Williams 1991). Beside being able to out-compete European bees in warmer climates (Collins and Kubasek 1982), Africanized bees are highly defensive and are much more aggressive toward humans and animals (Collins and Rinderer 1991; Stort and Gonçalves 1991). The defense of a honey-bee colony is the consequence of both guarding and responding behaviors and is exhibited by nonreproductive female worker bees and not by male drones. Female guard bees, at the colony entrance, identify and remove bees that are foreign to the nest, whereas responder bees usually fly out and sting the target that serves as the defensive stimulus. Breed and Rogers (1991) demonstrated that individual differences in guarding behavior are at least partially under genetic control. Furthermore, an increase in the level of African parentage will increase colony defensive behavior (Guzman-Novoa and Page Jr. 1993, 1994). Such studies illustrate that this behavior is genetically dominant as a colony-level phenotype (Guzman-Novoa et al. 2002). Five putative QTL-influencing colony stinging responses have been mapped (Hunt et al. 1998). Although originally identified on the basis of colony-level response, three of these QTL, including sting-2, were shown to influence guarding behavior of individual workers (Guzman-Novoa et al. 2002; Arechavaleta-Velasco et al. 2003).
Sting-2 was mapped to the honey-bee linkage group III (Hunt and Page Jr. 1995) on the basis of a colony-level behavioral trait, namely, the number of stings per minute. The sts/a11-.31 marker is at the LOD-score peak corresponding to the most likely position of gene/s influencing this behavior and that of the linked markers tested in QTL confirmation studies. In reciprocal backcross families, alleles of sts/a11-.31 inherited from a defensive parent were found to associate with defensive guarding behavior (Arechavaleta-Velasco et al. 2003). Only this marker (along with the Z8 cosegregating marker) was significantly associated with the behavioral phenotype.
With the goal of characterizing a genomic segment of A. mellifera and identifying candidate sting-2 genes, we have sequenced the genomic region encompassing the sts/a11-.31 marker. The isolation of the sting-2 gene will have several important consequences, such as providing a means for following the African allele into wild and apiary populations and facilitating the breeding of less aggressive bees (Guzman-Novoa et al. 2002). This study facilitates the identification of this gene by providing both candidate transcripts for further analyses as well as sequence data that can be used in the selection of new markers for sting-2. Data and experiences from this study may have implications for the honey-bee whole-genome sequencing and annotation.
METHODS
Isolation and Sequencing of a BAC Clone Associated With the sting-2 Locus
PCR product from the sts/a11-.31 marker was used to probe a genomic bacterial artificial chromosome (BAC) library of the honey bee (Tomkins et al. 2002). Twelve positive clones were resolved by pulse-field gel electrophoresis on a BioRAD CHEF mapper, and the largest BAC clone (Am36L17) was chosen for subsequent sequencing and analysis. The strategy was used by Voss et al. (1995) to sequence Am36L17. Two random libraries were constructed by partially digesting the BAC clone with Sau3A1 or Tsp509 I, and cloning 2-5 kb fragments into pLitmus28i (NEB). Two 9-12-kb partial libraries were constructed similarly. A directional library, constructed by cloning all completely digested EcoR1 fragments, served as a scaffold to assemble the BAC sequence. Direct BAC sequencing was used to anchor the ends of the sequence. Plasmids cloned from all libraries were sequenced from both ends of the inserts with standard M13 forward and reverse primers using ABI Big Dye Terminator v2, and reactions were analyzed on the ABI Prism 3700 DNA Analyzer. Sequencing data was evaluated, trimmed, and assembled using SEQMAN II software package (DNASTAR Inc.;Swindell and Plasterer 1997). Gaps were filled by primer walking (see Supplemental data for all primer sequences).
Sequence Analysis
The assembled sequence was analyzed against Repbase Update, using Repeatmasker and CENSOR (Jurka et al. 1996) to search for repetitive elements and transposon sequences. The assembled sequence was annotated with both ab initio gene prediction and algorithms on the basis of sequence similarity. GENESCAN 1.0 (Burge and Karlin 1997), GENEID 1.1 (Parra et al. 2000), and FGENES 1.0 (Salamov and Solovyev 2000) were used with default parameters and the human training data set. To avoid overprediction, genes were only accepted if they were predicted by at least two algorithms, or if they were predicted by one algorithm and were also similar to known ESTs, cDNAs, or proteins. The similarity-based methods used were BLASTX, BLASTN, and BLASTP (Altschul et al. 1997) against the nr and EST databank (NCBI) and BLASTN against the Honey bee Brain EST Project (University of Illinois). EST searches were also run against Human, Anopheles gambiae and Drosophila melanogaster ESTs from the Gene2EST server and the Berkeley Drosophila Genome Project Server. Protein domain analysis was performed using SMART and INTERPRO. ARTEMIS (Release 4) (Rutherford et al. 2000) was used to view the analyses. Stringency parameters were similar to those used in Thomasova et al. (2002).
RT-PCR Expression Analysis
Adult European honey bees were obtained from Greg Hunt, Purdue University. Africanized honey bees (Africanized matriline/Africanized patriline) were obtained from an extremely aggressive colony (Mona Chambers, Carl Hayden Bee Research Centre, USDA-ARS). Total RNA was isolated from 30 European and Africanized worker and 30 European drone heads using TRIzol (GIBCO-BRL). Primer pairs were designed within an exon in each putative ORF. Control primers were designed to the Apis glutamate transporter (Am-EAAT) (Kucharski et al. 2000;AF144379). RNA was analyzed with RT-PCR using the SuperScript One-Step RT-PCR system with Platinum Taq using 35 cycles to confirm the presence of a gene product and 20 cycles for a preliminary expression level comparison. RT-PCR products were gel purified (QIAquick Gel Extraction Kit, QIAGEN) and sequenced using primers from the RT-PCR reactions. Sequences were analyzed as above. Africanized and European Honey bee sequences were compared using CLUSTALW (Jeanmougin et al. 1998) and viewed with the GeneDoc program (Nicholas et al. 1997).
RESULTS
Sequencing of the sting-2 Region
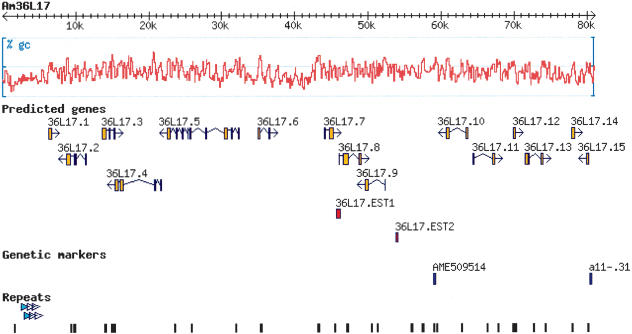
Sequences from libraries generated from the BAC clone Am36L17 were assembled into a single contig with a total length of 81,151 bp. About 90% of Am36L17 had over fivefold sequencing coverage. This genomic sequence had a G+C content of ∼40%. The assembled sequence had 30 simple repeats (di and tri and tetranucleotide;Table 1;Supplemental data). In addition to simple repeats, Am36L17 had a 371-bp sequence that repeated 5.4 times in tandem (Fig. 1). No transposon sequences were found in the assembled Am36L17 sequence. Similarity was found to an A. mellifera microsatellite marker AME509514 (accession AJ509514; Fig. 1).
Table 1.
Summary of Am36L17 Sequencing and Assembly
| Feature | Am36L17 |
|---|---|
| Length (bp) | 81,151 |
| Number trace sequences | 3,914 |
| Coverage above threshold (bp) (five equivalents) | 73,689 (90%) |
| Coverage below threshold (bp) | 4,959 (6%) |
| Average coverage | 26.83× |
| One strand sequenced (bp) | 2,462 (3%) |
| Sequenced once (bp) | 41 (0.05%) |
| GC content | 39.81% |
| Number of simple repeats of eight or more | 30× |
| Number of large repeats | 1 (371 bp × 5.4) |
Figure 1.
Annotation of Am36L17. The Am36L17 BAC clone was assembled into a single sequence with a GC content of ∼40%. All putative transcripts determined by gene-prediction methods (orange) are named numerically (36L17 1-15); 36L17.EST1 and 36L17.EST2 (red) were identified by their presence in the honey-bee brain EST database. The genetic marker linked to aggressive behavior in honey bees and used to isolate this BAC clone is a11-.31. The microsatellite sequence is AME509514. The position of simple repeats and the 371-bp repeat are identified (black bars and triangles, respectively).
Annotation Features
Fifteen putative protein coding regions (36L17.1 to 15) were predicted using gene-finding algorithms, whereas a similarity-based search of the Honey bee Brain Expressed Sequence Tag (EST) database identified 2 ESTs, 36L17.EST1 (Bee EST ID BB170031B10C10), and 36L17.EST2 (Bee EST ID BB160014B20E03). The 15 predicted transcripts in this region represent an approximate density of one gene prediction in 6 kb. Predicted transcripts were compared with nucleotide and protein databases for putative functional assignment. With the exception of the Apis microsatellite marker and the honey-bee brain EST sequences, BLASTX and BLASTP searches with predicted transcripts failed to generate hits with E values less than 0.002. There was no similarity between transcripts on a nucleotide or amino acid level. Protein domain analyses performed failed to demonstrate similarity to any known domains in these transcripts.
Reverse Transcription Analysis
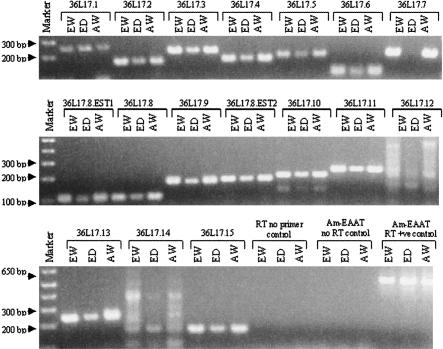
RT-PCR was utilized to confirm the accuracy of gene predictions and validate the expression of these transcripts in honey-bee heads. With the exception of 36L17.12 and 36L17.14, all predictions, including the segments corresponding to the EST hits, generated single reverse transcription amplification (RT) products of the expected sizes (Fig. 2). Transcripts 36L17.12 and 36L17.14 produced multiple RT-PCR products of varying sizes, indicating a failure of the RT-PCR reaction, nonspecific reaction products, or an absence of expression of these transcripts in honey-bee heads.
Figure 2.
Reverse transcription analysis of putative transcripts. RT-PCR was performed on RNA obtained from European worker (EW), European drone (ED), and Africanized worker (AW) heads. Primers were designed to an exon in each transcript. The putative transcripts from 36L17.1 to 36L17.15, 36L17.EST1, and 36L17.EST2 were visualized after 20 cycles. Positive control primers to the Apis glutamate transporter (Am-EAAT +ve control) produced the expected size fragment of 525 bp. The negative control using Taq Polymerase (Am-EAAT-No RT control) confirmed the lack of DNA contamination. With the exception of 36L17.12 and 36L1714, all predictions, including the segments corresponding to the EST hits, generated single RT products of the expected sizes in Africanized and European workers. 36L17.12 and 36L17.14 produced multiple RT-PCR products of varying sizes, indicating a possible failure of the RT-PCR reaction, nonspecific reaction products, or an absence of expression of that particular transcript in the bee head. With the exception of 36L17.7, all products seen in European workers were seen in European drones. RT-PCR products from most transcripts appeared to be present at a lower level in drones in comparison with workers in several cases. In contrast, 36L17.1 appeared to have a slightly elevated level of transcript in the drone sample in comparison with that of the workers. All transcripts seen in European worker heads appear to be expressed in their Africanized counterparts at similar levels.
RT-PCR was also performed on RNA from European drone heads to look for differences in expression between workers and drones. A total of 14 of the 15 transcripts that yielded an RT product in European honey-bee worker heads also generated products in the drone heads (Fig. 2). The 36L17.7 transcript was absent in European drones. The two putative transcripts that did not generate an expected product in worker bees (36L17.12 and 36L17.14) did not yield expected products in drones.
To facilitate a preliminary semiquantitative expression level analysis for each transcript, expression levels of transcripts were normalized to that of the control transcript after 20 RT-PCR cycles (Fig. 2). The control Apis glutamate transporter transcript—Am-EAAT (Kucharski et al. 2000), expressed in the honey-bee brain, is a highly conserved member of the EAAT carrier family. Consistent levels of Am-EAAT were seen in both European and African workers and European drones. RT-PCR products from most transcripts appeared to be present at a lower level in drones in comparison with workers. In contrast, 36L17.1 appeared to have a slightly elevated level of transcript in the drone sample in comparison with that of the workers. All 15 transcripts present in European workers appear to be expressed in their Africanized counterparts at similar levels.
RT-PCR products were purified and sequenced to facilitate nucleotide comparison of Africanized and European worker transcripts. In addition to products from 15 predicted genes, primers designed to 36L17.EST1 and 36L17.EST2 enabled the complete sequencing of these transcripts. On the basis of sequence similarity, all sequences analyzed were confirmed to be from the particular transcript in question. Products generated from 36L17.12 and 36L17.14 did not produce viable sequencing reactions. Products from the European honey-bee sequences were identical to sequences of the Am36L17 BAC clone.
Nucleotide differences were seen between European and Africanized sequences for four transcripts. Three of the transcripts that differed from their European honey-bee counterparts exhibited differences at only one nucleotide site (36L17.2, 36L17.8, and 36L17.11), whereas 36L17.EST1 exhibited a number of differences in the Africanized transcripts (Supplemental data). RT-PCR products from the control set of primers to Am-EAAT were also sequenced, and the nucleotide sequence was identical in both subspecies. These results were confirmed in three separate RT-PCR reactions using three different RNA preparations in each case.
DISCUSSION
We have sequenced and analyzed an 81-kb tract of A. mellifera genomic DNA, identified and preliminarily analyzed genes in the sting-2 locus for aggressive behavior, as well as made important observations that may affect the honey-bee whole-genome assembly.
Highly stringent parameters were optimized and used for this first sequence assembly effort in the honey bee. A low frequency of randomly dispersed sequence mate pairs with orientation or size inconsistencies (thought to be artifacts of the assembly or library generation procedure) were discarded, as their absence did not change the assembly. Nucleotide differences that differentiated the first and fifth repeat, as well as a restriction analysis of a plasmid clone spanning the 5.4× tandem repeat structure region validated the assembly algorithms computation of at least 5.4 repeats. The stringent measures taken ensure the accuracy of this assembly and support all ensuing predictions and conclusions.
A gene density of one gene in ∼6 kb demonstrates a possible higher number of genes in this region of the Hymenopteran genome than that of the Dipterans, An. gambiae (1/11 kb; F.H. Collins, pers. comm.) and D. melanogaster (1/13 kb;Adams et al. 2000) in general, when related to the genome sizes of 270 Mb, 268 Mb, and 180 Mb, respectively. Interestingly, none of the predicted transcripts had any similarity to known protein domains or genes in genomic databases. This unexpected result may be due to the fact that the Apis genome has greatly diverged from the sequenced insect genomes, namely D. melanogaster and An. gambiae (or vice versa). This is supported by studies of Apis mitochondrial DNA, in which mitochondrial genes are in the same relative positions as are their counterparts in Drosophila mitochondrial DNA, but have evolved at a significantly greater overall rate than have those of Drosophila in the 280 million years of evolutionary separation (Crozier and Crozier 1993;Crozier et al. 1989). Whitfield et al. (2002), in a study of bee-brain ESTs, conclude that about one-fourth of the genes expressed in the honey-bee brain may be highly diverged in primary structure. Kucharski and Maleszka (2002) not only identified a number of novel honey-bee genes, but also demonstrated that some honey-bee cDNAs had clear matches to proteins from the human genome, but had no orthologs in the Drosophila genome. This similarity seen between vertebrate and honey-bee sequences is reflected in the results of this study in which ab initio gene annotation using the human data set as a reference was found to be more successful than using the Drosophila data set. This finding has significant ramifications for the Apis whole-genome annotation. Differences that may be seen between the honey bee and the two Dipteran genomes may be reflective of fundamental processes associated with the evolution of traits unique to the honey bee, such as social behavior and haplo-diploidy. We conclude that the honey-bee transcripts identified here may not only demonstrate a high level of sequence variation from other organisms, but may also represent a genomic region or genes lost from Drosophila and Anopheles during evolution. Alternatively, they may represent novel honey-bee genes, or genes that have not been sequenced in other species. This finding has important implications for the annotation of the honey-bee genome, as the identification of genes on the basis of sequence similarity alone could result in a significant under-representation of the transcriptome. Comparisons with other genomic databases, such as those of vertebrates and other social organisms, may be necessary to uncover genes not identified in sequenced insect genomes.
In addition, both 36L17.EST1 and 36L17.EST2 were identified on the basis of their similarity to ESTs in the Bee Brain EST database, and their expression confirmed through RT-PCR. However, only part of 36L17.EST1 was recognized by the gene-finding algorithms, suggesting that novel transcripts in the Apis genome may be overlooked using standard training datasets. This validates the importance of EST projects (Arias and Sheppard 1996; Whitfield et al. 2002) as an annotation tool for whole-genome sequencing efforts.
Differing gene expression patterns, environment, and sex contribute to the development of bee castes and their behavior (Hepperle and Hartfelder 2001). Evans and Wheeler (2001) demonstrated that gene-expression patterns differed between queen larvae and worker larvae. The gene expression profile of an individual may therefore reflect its social caste. Although there may be many genes with sex-limited expression patterns in honey bees, an elevated expression of a worker-bee transcript, when compared with that in drones, may demonstrate a larger likelihood that the gene is involved with defensive behavior. In this preliminary expression profile study, most drone RT-PCR products appear to be present at lower levels in comparison with those of workers, supporting the above hypothesis. Additionally, expression levels were higher in 36L17.1 and absent in 36L17.7 drone RT-PCR profiles. Although these expression profiles are preliminary and need to be further examined with Real Time RT-PCR, these observations highlight the importance of characterizing gene-expression profiles between individuals as well as between caste members in the study of complex honey-bee social behaviors (Kucharski and Maleszka 2002). Hunt et al. (1999) suggest that the intense defensive response of Africanized bees may involve a lower threshold response to stimuli that elicit defensive behavior rather than a constitutively higher expression level of a particular transcript. The similar expression levels of most transcripts seen between Africanized and European worker bees seem to support this hypothesis.
The identification of genes that vary in structure and expression may lead to new ideas about the mechanisms that govern the expression of a particular social behavior (Robinson 2002). Allelic variation between Africanized and European bees may correlate with variation in social behaviors, including aggression. Most European transcripts identified here were identical to the sequence from the BAC clone, suggesting that there is little variation in the European honey-bee population used, or that these genes are under strong selective pressure. Sequence polymorphism is seen between Africanized and European transcripts in four sequences (Supplemental data). However, we feel that the sample size is not large enough to make any definitive predictions.
This genomic segment is well represented in the whole-genome shotgun (wgs) trace files, as well as trace files obtained by the Clone-Array Pooled Shotgun Sequencing (CAPPS;Cai et al. 2001) method. It can serve as a scaffold to extend sequences as well as to validate current assemblies. The RT-PCR data presented here validates the expression of novel honey-bee transcripts and can be compared with and utilized to incorporate other novel transcripts in the annotation of the honey-bee genome. This independently sequenced and analyzed sequence serves as an important genomic annotation and assembly tool.
The sequencing and analysis of this 81-kb genomic region from the honey bee, Apis mellifera, associated with a quantitative trait locus (QTL) sting-2 for aggressive behavior, has resulted in several important findings. This sequence represents the first extensive study of the honey-bee genome structure encompassing putative genes in a QTL for a behavioral trait. The analysis of 13 putative behavioral genes indicates the presence of unique Apis genes that may be specific to the evolution of social behavior or haplo-diploidy. Real Time RT-PCR will be utilized to further investigate expression-level differences seen between aggressive and nonaggressive honey-bee social casts and between the two subspecies. This genomic study provides sequences data, annotation, and preliminary expression analyses, serving as an invaluable research tool for behavioral studies as well as the honey-bee whole-genome sequencing effort.
Acknowledgments
We thank Mona Chambers (Carl Hayden Bee Research Center, USDA-ARS) for providing Africanized honey bees, and A. Dana, J. Hogan, A. Sarkar, and K. Merz for helpful advice. This work was supported by the Indiana 21st Century Research and Technology Fund Grant 042700-0207, and by grants from the Hayward Genetics Foundation and the National Science Foundation (0110842-IBN).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1634503.
Footnotes
[Supplemental material is available online at www.genome.org. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: G. Hunt and M. Chambers.]
References
- Adams, M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185-2195. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 17: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arechavaleta-Velasco, M.E., Hunt, G.J., and Emore, C. 2003. Quantitative trait loci that influence the expression of guarding and stinging behaviors of individual honey bees. Behav. Genet. 33: 357-364. [DOI] [PubMed] [Google Scholar]
- Arias, M.C. and Sheppard, W.S. 1996. Molecular phylogenetics of honey bee subspecies (Apis mellifera L.) inferred from mitochondrial DNA sequence. Mol. Phylogenet. Evol. 5: 557-566. [DOI] [PubMed] [Google Scholar]
- Brandes, C. 1991. Genetic differences in learning behavior in honey bees (Apis mellifera capensis). Behav. Genet. 21: 271-294. [DOI] [PubMed] [Google Scholar]
- Breed, M.D. and Rogers, K.B. 1991. The behavioral genetics of colony defense in honey bees: Genetic variability for guarding behavior. Behav. Genet. 21: 295-303. [DOI] [PubMed] [Google Scholar]
- Burge, C. and Karlin, S. 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78-94. [DOI] [PubMed] [Google Scholar]
- Cai, W.W., Chen, R., Gibbs, R.A., and Bradley, A. 2001. A clone-array pooled strategy for sequencing large genomes. Genome Res. 11: 1619-1623. [DOI] [PubMed] [Google Scholar]
- Chandra, S.B.C., Hunt, G.J., Cobey, S., and Smith, B.H. 2001. Quantitative Trait Loci associated with reversal learning and latent inhibition in honey bees (Apis mellifera). Behav. Genet. 31: 275-285. [DOI] [PubMed] [Google Scholar]
- Collins, A.M. and Kubasek, K.J. 1982. Field test of honey bee (Hymenoptera:Apidae) colony defensive behavior. Ann. Entomol. Soc. Am. 75: 383-397. [Google Scholar]
- Collins, A.M. and Rinderer, T.E. 1991. Genetics of defensive behavior I. In The “African” honey bee (eds. M. Spivak et al.), pp. 309-328. Westview Press, Boulder, CO.
- Crozier, R.H. and Crozier, Y.C. 1993. The mitochondrial genome of the honey bee Apis mellifera: Complete sequence and genome organization. Genetics 133: 97-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozier, R.H., Crozier, Y.C., and Mackinlay, A.G. 1989. The CO-I and CO-II region of honey bee mitochondrial DNA: Evidence for variation in insect mitochondrial evolutionary rates. Mol. Biol. Evol. 6: 399-411. [DOI] [PubMed] [Google Scholar]
- Evans, J.D. and Wheeler, D.E. 2001. Expression profiles during honey bee caste determination. Genome Biol. 2: Research 0001.1-0001.6. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman-Novoa, E. and Page Jr., R.E. 1993. Backcrossing Africanized honey bee queens to European drones reduces colony defensive behavior. Ann. Entomol. Soc. Am. 86: 352-355. [Google Scholar]
- Guzman-Novoa, E. and Page Jr., R.E. 1994. Genetic dominanace and worker interactions affect honey bee colony defense. Behav. Ecol. 5: 91-97. [Google Scholar]
- Guzman-Novoa, E., Hunt, G.J., Uribe, J.L., Smith, C., and Arechavaleta-Velasco, M.E. 2002. Confirmation of QTL effects and evidence of genetic dominance of honey bee defensive behavior: Results of colony and individual behavioral assays. Behav. Genet. 32: 95-102. [DOI] [PubMed] [Google Scholar]
- Hepperle, C. and Hartfelder, K. 2001. Differentially expressed regulatory genes in honey bee caste development. Naturwissenschaften 88: 113-116. [DOI] [PubMed] [Google Scholar]
- Hunt, G.J. and Page Jr., R.E. 1995. Linkage map of the honey bee, Apis mellifera, based on RAPD markers. Genetics 139: 1371-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, G.J., Page Jr., R.E., Fondrk, M.K., and Dullum, C.J. 1995. Major quantitative trait loci affecting honey bee foraging behavior. Genetics 141: 1537-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, G.J., Guzman-Novoa, E., Fondrk, M.K., and Page Jr., R.E. 1998. Quantitative trait loci for honey bee stinging behavior and body size. Genetics 148: 1203-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, G.J., Collins, A.M., Rivera, R., Page Jr., R.E., and Guzman-Novoa, E. 1999. Quantitative trait loci influencing honey bee alarm pheromone levels. J. Hered. 90: 585-589. [DOI] [PubMed] [Google Scholar]
- Jeanmougin, F., Thompson, J.D., Gouy, M., Higgins, D.G., and Gibson, T.J. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23: 403-405. [DOI] [PubMed] [Google Scholar]
- Jurka, J., Klonowski, P., Dagman, V., and Pelton, P. 1996. CENSOR—A program for identification and elimination of repetitive elements from DNA sequences. Computers and Chem. 20: 119-122. [DOI] [PubMed] [Google Scholar]
- Kerr, W.E. 1967. The history of the introduction of African bees to Brazil. S. Afr. Bee. J. 39: 3-5. [Google Scholar]
- Kucharski, R. and Maleszka, R. 2002. Evaluation of differential gene expression during behavioral development in the honey bee using microarrays and northern blots. Genome Biol. 3: Research0007.1-0007.9. Epub. [DOI] [PMC free article] [PubMed]
- Kucharski, R., Ball, E.E., Hayward, D.C, and Maleszka, R. 2000. Molecular cloning and expression analysis of a cDNA encoding a glutamate transporter in the honey bee brain. Gene 242: 399-405. [DOI] [PubMed] [Google Scholar]
- Lapidge, KL., Oldroyd, B.P., and Spivak, M. 2002. Seven suggestive quantitative trait loci influence hygienic behavior of honey bees. Naturwissenschaften 89: 565-568. [DOI] [PubMed] [Google Scholar]
- Nicholas, K.B., Nicholas Jr., HB., and Deerfield II., D.W. 1997. GeneDoc: Analysis and visualization of genetic variation, EMBNEW.NEWS 4: 14. [Google Scholar]
- Page Jr., R.E. and Erber, J. 2002. Levels of behavioral organization and the evolution of division of labor. Naturwissenschaften 89: 91-106. [DOI] [PubMed] [Google Scholar]
- Page Jr., R.E., Fondrk, M.K., Hunt, G.J., Guzman-Novoa, E., Humphries, M.A., Nguyen, K., and Greene, A.S. 2000. Genetic dissection of honey bee (Apis mellifera L.) foraging behavior. J. Hered. 91: 474-479. [DOI] [PubMed] [Google Scholar]
- Parra, G., Blanco, E., and Guigo, R. 2000. GeneID in Drosophila. Genome Res. 10: 511-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, G.E. 2002. Development. Sociogenomics takes flight. Science 297: 204-205. [DOI] [PubMed] [Google Scholar]
- Rothenbuhler, W.C. 1964. Behavior genetics of nest cleaning in honey bees. IV. Responses of F1 and backcross generations to disease-killed brood. Am. Zool. 12: 578-583. [DOI] [PubMed] [Google Scholar]
- Rutherford, K., Parkhill, J., Crook, J., Horsnell, T., Rice, P., Rajandream, M.A., and Barrell, B. 2000. Artemis: Sequence visualization and annotation. Bioinformatics 16: 944-945. [DOI] [PubMed] [Google Scholar]
- Salamov, A.A. and Solovyev, V.V. 2000. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 10: 516-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stort, A.C. and Gonçalves, L.S. 1991. Genetics of defensive behavior II. In The “African” honey bee (eds. M. Spivak et al.), pp. 329-356. Westview Press, Boulder, CO.
- Sugden, E.A. and Williams, K.R. 1991. October 15: The day the bee arrived. Gle. Bee Cult. 119: 18-21. [Google Scholar]
- Swindell, S.R. and Plasterer, T.N. 1997. SEQMAN. Contig assembly. Methods Mol. Biol. 70: 75-89. [PubMed] [Google Scholar]
- Thomasova, D., Ton, L.Q., Copley, R.R., Zdobnov, E.M., Wang, X., Hong, Y.S., Sim, C., Bork, P., Kafatos, F.C., and Collins, F.H. 2002. Comparative genomic analysis in the region of a major Plasmodium-refractoriness locus of Anopheles gambiae. Proc. Natl. Acad. Sci. 99: 8179-8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkins, J.P., Luo, M., Fang, G.C., Main, D., Goicoechea, J.L., Atkins, M., Frisch, D.A., Page Jr., R.E., Guzmán-Novoa, E., Yu, Y., et al. 2002. New genomic resources for the honey bee (Apis mellifera L.): Development of a deep-coverage BAC library and a preliminary STC database. Genet. Mol. Res. 1: 306-316. [PubMed] [Google Scholar]
- Voss, H., Schwager, C., Wiemann, S., Zimmermann, J., Stegemann, J., Erfle, H., Voie, AM., Drzonek, H., and Ansorge, W. 1995. Efficient low redundancy large-scale DNA sequencing at EMBL. J. Biotechnol. 41: 121-129. [DOI] [PubMed] [Google Scholar]
- Whitfield, C.W., Band, M.R., Bonaldo, M.F., Kumar, C.G., Liu, L., Pardinas, J.R., Robertson, H.M., Soares, M.B., and Robinson, G.E. 2002. Annotated expressed sequence tags and cDNA microarrays for studies of brain and behavior in the honey bee. Genome Res. 12: 555-566. [DOI] [PMC free article] [PubMed] [Google Scholar]