Abstract
Steady-state levels of mRNA in cells theoretically depend on the rate and efficiency of transcription and posttranscriptional processing, on mRNA stability, on transcriptional interference from other genes, and on poorly defined long-range chromatin effects.Although each of these cellular processes has been studied in detail for a few genes, it is not possible to predict expression levels by simply examining gene sequences.In this report, we have used a bioinformatics approach to identify critical factors that influence expression levels. To simplify the problem, we have limited our analysis to the collection of genes expressed in all tissues, because such genes provide a unique opportunity to distinguish the role of general genomic features that constrain gene expression from the effect of tissue-specific factors.Using correlation and regression techniques, we have investigated the dependence between expression level and morphological parameters (distance to neighbors, gene, mRNA or 3′-UTR length, number of exons, etc.) that can be directly related to transcription, posttranscriptional processing, mRNA stability, or transcriptional interference.We found that, on a genome-wide scale, highly expressed genes are significantly farther from their closest neighboring genes, are smaller, contain a moderate number of exons, and produce shorter mRNAs with shorter 3′-UTRs.This confirms that transcriptional and posttranscriptional processes are highly interrelated and implies that transcriptional interference plays a role in determining steady-state levels of mRNA in cells.
The steady-state levels of mRNA in cells depend on the rate of transcriptional initiation and elongation, on the efficiency of splicing and termination of transcription, on the rate of export to the cytoplasm, and on the stability of the mRNA in the cytoplasm. Transcriptional interference, broadly defined as the perturbation of one transcription unit by another, is also believed to affect expression levels. Several mechanisms including steric or topological constraints induced by transcription, competition for cis-acting sequence, production of antisense RNA, and epigenetic phenomena linked to DNA methylation or histone modifications have been implicated in transcriptional interference. In addition, some genes might require large regulatory sequences for their regulation, precluding the presence of other genes nearby.
Although some of these mechanisms and cellular processes have been studied in great detail for a few genes, it is generally not possible to predict expression levels by simply examining gene sequences. In this report, we have used a bioinformatics approach to try and identify critical factors that influence expression levels, using sequence and expression data available on the internet.
One major difficulty in relating sequence and expression information is the large number of tissue-specific effectors (e.g., transcription and splicing factors) that, through extremely complex pathways, determine the differential regulation of gene expression during development and differentiation. To simplify the problem, we have limited our study to the minimal human housekeeping transcriptome (MHKT), a collection of genes expressed in all tissues. The MHKT provides a unique opportunity to distinguish the role of general genomic features that constrain gene expression from the effect of tissue-specific factors, because the peculiarities of each individual tissue should average out.
Our approach uses correlation and regression techniques to investigate, on a genome-wide scale, the dependence between expression level and morphological parameters such as distance to closest neighboring gene, gene, mRNA and 3′-UTR length, and number of exons. We then attempt to relate these morphological parameters to one or more cellular processes using simple hypotheses such as: (1) If elongation rates limit expression levels, longer genes should be expressed at lower levels than shorter genes. (2) If splicing efficiency limits expression level, genes with few exons should be expressed at higher levels than genes with many exons. (3) If transcriptional interference limits expression levels, genes far from their neighbors should be expressed at higher levels than genes close to their neighbors.
The influences of the rate of transcriptional initiation and of mRNA stability on expression are harder to capture by measuring simple morphological parameters. Stability and turnover of mRNA is a complex, highly regulated process that occurs either by progressive degradation from the 3′-end or following endonuclease activities (Vreken et al. 1991; Mitchell and Tollervey 2000; Moore 2002). Both pathways often involve cis-acting elements located in the 3′-UTR (Pesole et al. 2001; Mignone et al. 2002). We therefore attempted to evaluate the influence of mRNA stability by investigating the relationship between expression levels and 3′-UTR length. Because long mRNA might provide more target sites for degradation, we also considered mRNA length. We did not attempt to assess the role of the rate of transcriptional initiation.
Our analyses revealed that, on a genome-wide scale, highly expressed genes are smaller and produce shorter mRNAs with shorter 3′-UTRs. Interestingly, we also found that highly expressed genes are significantly farther from their closest neighbors. These findings confirm the importance of transcriptional and posttranscriptional processing in determining expression levels and, importantly, provide evidence for a role of transcriptional interference in determining steady-state levels of mRNA in cells.
RESULTS
We obtained expression information from the database created by Velculescu et al. (1999). Using serial analysis of gene expression (SAGE), a method that provides absolute quantitation of steady-state mRNA levels per cell, these investigators analyzed 3.5 million transcripts representing 19 different tissues or cell lines, and reported that whereas >50% of all human genes were widely expressed, only ∼1000 genes were expressed in every tissue they examined. The latter group of genes, whose expression ranges from >1000 to <10 copies of mRNA per cell, can therefore be defined as the MHKT.
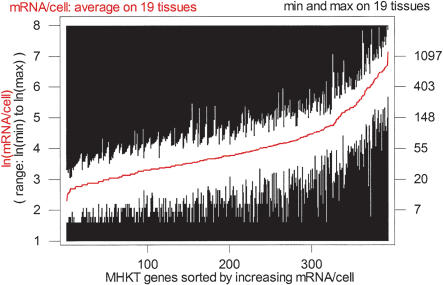
Through data preparation steps, we completely updated the list of genes in the MHKT using the most recent data from the human genome project, isolated a subset of 393 genes for which high-quality DNA sequence and RNA expression data were available, and extracted structural parameters such as gene length, number of exons, 3′-UTR length, and distance to the nearest neighbors on both sides. The complete list of genes used in this study is available online (Supplemental file 1, available at www.genome.org or at http://bio.cse.psu.edu/). The average and range of expression of these 393 genes in the 19 tissues is plotted in Figure 1.
Figure 1.
Range of expression of the genes of the MHKT. Average (red curve) and min-to-max range (white bars) of expression for the 393 genes of the MHKT in 19 tissues (from Velculescu et al. 1999).
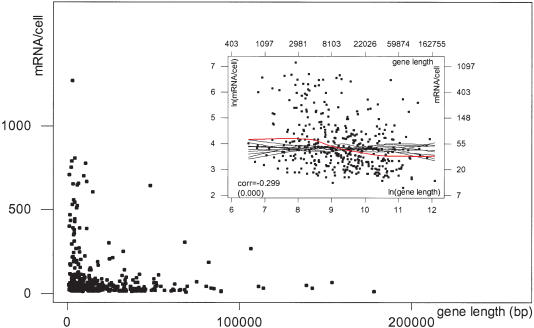
Inspection of the 393 “high-quality” MHKT genes revealed an association between gene length and level of mRNA expression: virtually all genes expressed at >200 copies of mRNA per cell are shorter than 10,000 bp, whereas most of the genes longer than 10,000 bp are expressed at low levels (Fig. 2). This strong influence of gene length on mRNA expression was verified and quantified on the whole data set by a detailed statistical analysis. On the natural log scale, we found a strong negative association between the number of copies of mRNA per cell and gene length. The correlation coefficient is -0.299, with a p-value equal to 0 to the third decimal approximation. Examination of the “lowess smooth,” a curve that captures the shape of the relationship between two variables (see Methods), revealed that the strongest effect of gene length occurs for genes between 4000 and 15,000 bp in length (red curve in Fig. 2, inset). To assess the significance of the observed pattern, lowess smooths were computed on random permutations of the data (black curves in Fig. 2, inset), showing that the observed dependence was unlikely to be caused by chance alone.
Figure 2.
Dependence of expression on gene length. Expression (mRNA/cell) of 393 genes in the MHKT plotted against gene length. (Inset) Natural log scatter plot of the same data. The logarithmic transformation regularizes the data and helps in visualizing dependence patterns. Axes on top and on the right provide readings before logarithmic transformation of the numbers. The red curve represents a lowess smooth of the data, and the black curves lowess smooths from random permutations. Both plots reveal a strong negative association between expression and gene length. (corr) Correlation coefficient. p-values are in parentheses.
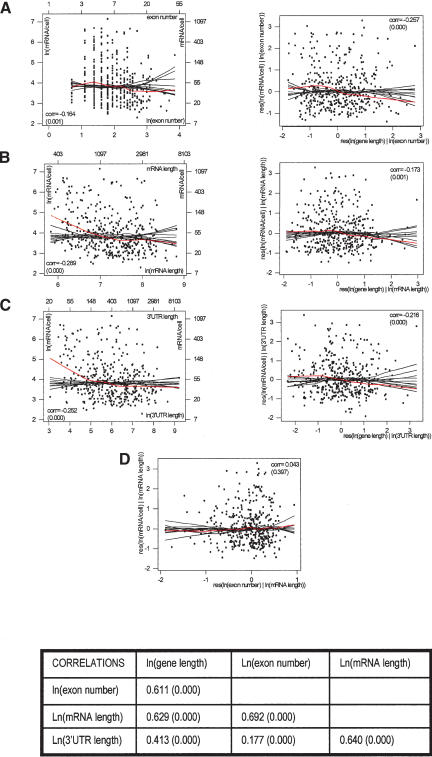
A simple explanation for the negative association between expression and gene length is that the rate of transcriptional elongation constitutes a limiting step. However, this negative association may also reflect other effects, such as those of splicing and mRNA stability. To investigate these effects, and whether or not they account for substantial portions of the effect of gene length, we considered the number of exons as a splicing-related parameter, and mRNA and 3′-UTR length as stability-related parameters (assuming that long mRNAs are more susceptible to degradation because they offer more target sites for nucleases, and that unstable mRNAs are characterized by long 3′-UTRs). All three of these parameters have strong positive correlations with gene length and with one another (table at the bottom of Fig. 3).
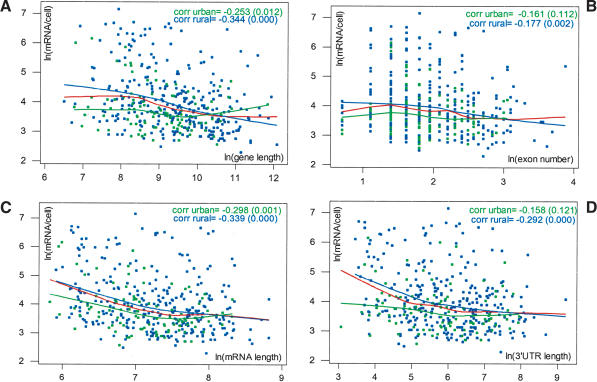
Figure 3.
Relationship between expression and morphological parameters. (A-C, left panels) Scatter plots of ln(mRNA/cell) versus ln(exon number), ln(mRNA length), and ln(3′-UTR length), respectively. The red curves represent lowess smooths, and the black curves lowess smooths from random permutations. mRNA expression has strong negative associations with all these parameters. (A-C, right panels) Added variable plots for ln(mRNA/cell) versus ln(gene length) after correcting for ln(exon number), ln(mRNA length), and ln(3′-UTR length), respectively. Red and black curves are again lowess smooths on original data and random permutations; (res) residual. All three of these parameters account for a substantial portion of the negative association between expression and gene length, but leave a substantial remainder effect, lending support to a role of transcriptional elongation. The table at the bottom of the figure contains correlations between parameters. Because of the very high positive correlation between exon number and mRNA length, the effects of splicing and mRNA stability and their contributions to the effect of gene length, although individually observable, are confounded. This is shown by panel D, which contains the added variable plots for ln(mRNA/cell) versus ln(exon number), after correcting for ln(mRNA length). (corr) Correlation coefficient. p-values are in parentheses.
We found a negative association between expression and number of exons. On the natural log scale, the correlation coefficient is -0.164 (p-value 0.001). However, the lowess smooth shows some curvature at low exon numbers (red curve in Fig. 3A, left panel), indicating a complex effect. For genes with less than four exons, the association is actually slightly positive: a minimal number of exons might be required for high expression levels, maybe reflecting the integration of mRNA processing and transcription (Fong and Zhou 2001). Once again, the observed pattern is significant in comparison to lowess smooths from random permutations (black curves in Fig. 3A, left panel).
As for stability, we found that highly expressed genes tend to have shorter mRNAs and shorter 3′-UTRs. In both cases, the negative association is strong. On the natural log scale, the correlation coefficients are -0.289 and -0.252, respectively (p-values 0.000). The lowess smooths are downsloping (red curves in Fig. 3B,C, left panels) and represent significant patterns in comparison to lowess smooths from random permutations (black curves in Fig. 3B,C, left panels), particularly for shorter genes.
The right panels of Figure 3, A,B,C, contain the added variable plots (see Methods) for expression on gene length after correction for exon number, mRNA length, and 3′-UTR length, respectively. In all cases, the correlation coefficients, and the lowess smooths (red curves) in comparison to lowess smooths from random permutations (black curves), establish a weakened but still significant negative association with respect to the original plot (Fig. 2, inset). Thus, splicing and mRNA stability do appear to account for a substantial portion of the negative association between expression levels and gene length, but they also leave a substantial residual effect—which may be linked to transcriptional elongation.
It must also be noted that, although we can observe individually the effects of exon number, mRNA, and 3′-UTR length, as well as their contributions to the effect of gene length, these individual effects and contributions are statistically confounded (i.e., hard to separate) on our data because of correlations existing between our splicing and stability-related parameters themselves. For instance, on the natural log scale, the correlation between exon number and mRNA length is 0.692 (p-value 0.000), and the added variable plot for expression on exon number after correcting for mRNA length (Fig. 3D) shows almost no dependence (correlation 0.043; red lowess smooth within the range of black lowess smooths from random permutations). In other words, statistically speaking, mRNA length can account for almost all the effect of exon number because of the correlation between the two variables.
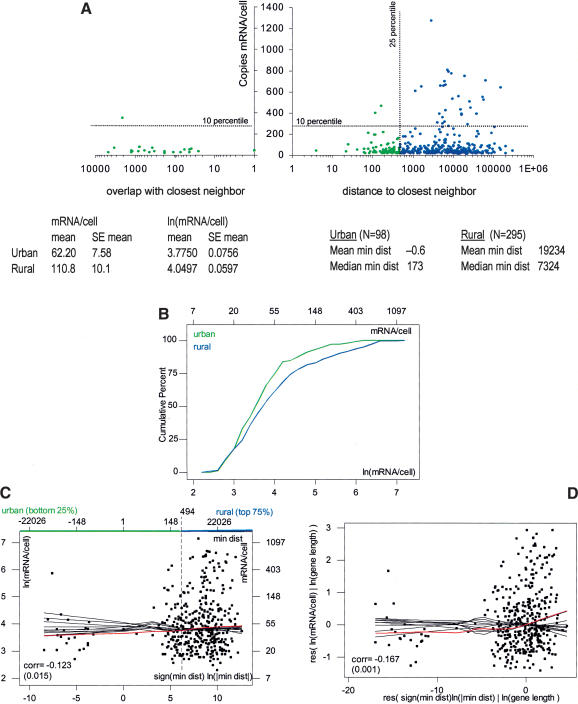
In an attempt to detect on a genome-wide scale transcriptional interference defined as in the introduction, we computed the distance between the cap sites and the poly(A) sites of each of the 393 genes and their nearest neighbors on both sides. We then tested various means of combining this distance information to partition the MHKT genes into an “urban” class (likely to be subject to interference) and a “rural” class. Remarkably, analysis of these two classes always produced similar conclusions: Genes in close proximity to other genes tend to have expression levels that are lower on average than more isolated genes. Copies of mRNA per cell versus distance to the closest neighbor (the latter on the natural log scale) are plotted in Figure 4A—for genes that overlap with their closest neighbor, the distance is negative, and the log of the size of the overlap is reported on the negative X-axis.
Figure 4.
Effects of distance to neighbors. Genes are divided into urban (green) and rural (blue) classes on the basis of their distance to the closest gene neighbor on either side (urban genes are the 25% with shortest distance). (A) A scatter plot of mRNA/cell versus distance to closest neighbor (min dist). Note that negative values correspond to sizes of overlap for genes that overlap their neighbors, and that the scale on the X-axis is logarithmic. (B) Cumulative distribution functions of ln(mRNA/cell) by class, which show depletion of highly expressed genes in the urban class. (C) A scatter plot of ln(mRNA/cell) versus sign(min_dist) ln(min_dist). This represents the log transformation for min_dist, accounting for negative values (see Methods). The plot reveals a positive association. (D) The added variable plot for ln(mRNA/cell) versus sign(min_dist) ln(min_dist) after correcting for ln(gene length). The added variable plot shows a slightly increased positive association, evidence that the relationship detected between expression and distance to closest neighbor is not a spurious byproduct of the relationship between expression and gene length (i.e., not a consequence of urban genes being longer than rural ones). Red and black curves represent lowess smooths on the actual data and on random permutations, respectively. (corr) Correlation coefficient. p-values are in parentheses. (Right table) Mean and median distances to closest neighbor for genes in the urban and rural classes. (Left table) Mean expression with its standard error, on the original, mRNA/cell, and logarithmic, ln(mRNA/cell), scale, for genes in the two classes. The mean expression level for the urban class is almost twofold lower than for the rural class.
Strikingly, all genes expressed at more than 500 copies of mRNA per cell are far from their neighbors, whereas all but two genes with a neighbor closer than 100 bp are expressed at less than 100 copies of mRNA per cell.
We adopted a simple definition of urban genes as the 25% of the MHKT genes with the shortest distance to their closest neighbor, regardless of orientation. Rural genes are defined as the remaining 75%. The tables in Figure 4 report mean and median distances to closest neighbors, as well as mean mRNA/cell and mean ln(mRNA/cell), with their standard errors, for genes in the two classes. The mean expression level for the urban class is almost twofold lower than for the rural class (62.2 ± 7.58 and 110.8 ± 10.1 mRNA/cell, respectively). Also, cumulative distribution plots of expression levels by class reveal that the urban class is depleted in highly expressed genes with respect to the rural class (green and blue curves in Fig. 4B).
Using the log scale on both axes, a scatter plot of expression versus distance to closest neighbor reveals a significant positive association: the correlation is 0.123 (p-value 0.015), and the lowess smooth is un-chance-like in comparison to lowess smooths from random permutations (red and black curves in Fig. 4C). Moreover, this positive association is not a spurious consequence of urban genes being longer than rural ones. As a matter of fact, distance to closest neighbor and gene length present a mild positive correlation of 0.10 (p-value 0.061), and the added variable plot for expression on distance after correcting for gene length (Fig. 4D) reveals a stronger positive relationship (the correlation increases to 0.163, with a p-value of 0.001, and the upward pattern in the lowess smooth is more marked).
We then examined the relationship between copies of mRNA per cell and exon number, gene length, mRNA length, and 3′-UTR length separately for the urban and rural classes (Fig. 5A,B,C,D). The lowess smooths show how the association between mRNA expression and gene length, mRNA length, 3′-UTR length, and exon number is substantially weakened in the urban but not in the rural class. In other words, transcriptional interference not only depresses expression, but also the degree to which expression is influenced by these parameters. This is evidence that transcriptional interference may dominate the effects of transcriptional elongation, mRNA stability (posttranscriptional events), and splicing proxied by these parameters, and thus occur primarily at the level of transcriptional initiation.
Figure 5.
Relationship between expression and morphological parameters within the rural and urban classes. Scatter plots of ln(mRNA/cell) versus ln(gene length), ln(mRNA length) ln(3′-UTR length), and ln(exon number), with lowess smooths for rural and urban classes (blue and green curves), and overall (red curves). Correlations by class are in the upper right corner. The relationships between expression and each of the parameters are considerably weakened for urban genes, indicating that transcriptional interference might be dominant over the other morphological parameters influencing gene expression. (corr) Correlation coefficient. p-values are in parentheses.
DISCUSSION
The most novel and striking result from our analyses is the negative association between mRNA expression and distance to the closest neighboring gene, because it indicates that transcriptional interference plays a role in determining the level of gene expression. Importantly, given that we did not take into consideration tissue-specificity and expression levels of neighboring genes, the actual effect of transcriptional interference is likely to be even stronger than what we could detect in this study.
Transcriptional interference has long been shown to affect expression of genes in tandem orientation in model systems (Proudfoot 1986). We recently extended these findings and showed that transcriptional interference has a strong negative effect on expression irrespective of the relative orientation of the transcription units (Eszterhas et al. 2002). These cell culture results are also supported by numerous reports that in vivo, selectable markers inserted in the genome by homologous recombination in ES cells have dramatic effects on gene expression (Fiering et al. 1999). At the molecular level, the mechanisms of transcriptional interference are not well understood but are likely to be complex. Steric hindrance, promoter occlusion, and RNAi are all likely contributors (Eszterhas et al. 2002).
Regardless of the mechanism, the finding that proximity of neighboring transcriptional units strongly influences mRNA expression levels in mammalian cells has implications for several cellular and evolutionary processes. For instance, mutagenesis induced by integration of man-made or naturally occurring mobile genetic elements, which is generally believed to be caused by disruption of coding or regulatory sequences (Whitelaw et al. 2001), might be greatly augmented by transcriptional interference. Reactivation of normally silenced repetitive sequence in cancer cells could depress expression of neighboring genes, and conversely, gene silencing associated with aging (Issa 2000) could activate neighboring genes. Transcriptional interference might also partially explain the recent finding that in Saccharomyces cerevisiae, adjacent pairs of genes tend to be coregulated (Cohen et al. 2000).
The associations between expression levels and gene, mRNA, and 3′-UTR lengths, as well as exon number, are more complicated to interpret, because mRNA processing is highly integrated (Proudfoot et al. 2002). Capping, splicing, and polyadenylation occur cotranscriptionally, and are all tightly linked to transcription via the interaction of various factors with the Cterminal domain of RNA polymerase II. Most of the mRNA processing mechanisms have been shown to influence and coregulate each other. In accordance with this highly interrelated picture, our splicing and stability-related parameters are strongly correlated, and their effects on expression statistically confounded. However, we can still observe these effects individually. Our results support a strong negative association between mRNA expression and mRNA stability proxied by mRNA and 3′-UTR length, and imply a complex role for splicing, with a pattern that is negative at large exon numbers, but positive at low exon numbers. This might be explained by recent findings indicating that splicing factors have a stimulatory effect on transcriptional elongation (Fong et al. 2001). For genes containing few exons, this elongation stimulus might overcome the negative effect of abortive splicing events. We also observe that splicing and mRNA stability parameters account for some but far from all of the negative association between expression and gene length, lending support to a critical role of transcriptional elongation. Long genes might be difficult to elongate because over long distances chromatin structure is a major barrier (Orphanides and Reinberg 2000) and because long genes have a higher probability of containing elongation pausing sites.
From an evolutionary point of view, as discussed in Velculescu et al. (1999), the genes of the MHKT are particularly interesting because they are likely to be among the most ancient of all genes, as they code for all the basic cellular processes. Mechanisms affecting these genes might therefore also be ancient. We propose that the effects of transcriptional interference, transcriptional elongation, and mRNA stability that we have detected are the vestiges of a simpler, primordial genomic organization in which gene expression was controlled by a minimal set of transcription factors, and modulated by the size of the DNA segments to be transcribed and the relative positions of the “genes.” In the most extreme version of this model, CG-rich primordial promoters might all have fired at the same rate and the level of expression might have been controlled entirely by interference, rate of elongation, and mRNA stability.
METHODS
Data Preparation and Preprocessing
We obtained 1183 universally expressed SAGE tags and their percell mRNA counts from http://www.sagnet.org/. We downloaded a mapping of 272,131 SAGE tags to UniGene IDs from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). The list was pared down to 410 genes by eliminating the tags that did not map to any genes, or that mapped to more than one gene, or to ribosomal RNA. When two tags mapped to the same gene, only the most highly expressed tag was retained.
We then downloaded tables containing the RefSeq annotation databases of the April 2003 genome assembly from the UC Santa Cruz genome browser Web site (http://www.genome.usc.com) and extracted or calculated values for gene length, mRNA length, 3′-UTR length, number of exons, and distance from the neighbor on each side, using MS Access 2000 and MS Excel 2000. Twelve genes were deleted from our list because they had no known neighbors, because they were duplicated, or because they had no 3′-UTR. Five single-exon genes were also eliminated because it was not clear whether they were pseudogenes. This yielded a final list of 393 genes. Gene length was defined as the distance from the cap site to the polyadenylation site. The 3′-UTR length was defined as the distance from the stop codon to the polyadenylation site.
Data Analysis
Strong concentration at low values and far-reaching “tails,” which could be observed for many of our variables (e.g., Fig. 2), were mitigated adopting natural logarithms. This simplified graphical and numerical investigation of dependence patterns.
We used Pearson correlation coefficients, which capture sign and size (min = -1, max = +1) of the linear associations between two variables.
We also used locally weighted scatter-plot smoothers (lowess; Cleveland 1979; Cook and Weisberg 1999), which capture the dependence pattern between two variables on the vertical and horizontal axes of a scatter-plot. These curves are produced by (weighted) least-square fitting of a simple line within a window sliding through the data. The size of the window controls the degree of smoothing, and is specified in terms of a certain percentage of the data points (e.g., we always used a sliding window containing 50% of the points, which represents an intermediate degree of smoothing). Moreover, these curves are made robust by iterating the fit within each window discarding outliers (we always used five iterations).
To investigate the dependence between two variables after correcting for a third quantity, we used a modified version of added variable plots (AVP; Cook 1994; Cook and Weisberg 1999). We plot residuals from the lowess smooth of the vertical axes variable on the third variable, versus residuals from the lowess smooth of the horizontal axis variable on the third variable. For example, the quantity on the vertical axis of Figure 3A, right panel, res[ln(mRNA/cell) ln(exon number)], represents vertical distances between points and the red curve in Figure 3A, left panel. The quantity on the horizontal axis of Figure 3A, right panel, res[ln(gene length) ln(exon number)], represents vertical distances from an analogous lowess for ln(gene length) on ln(exon number) (data not shown). Thus, when computing a lowess on the points of the AVP (red curve in Fig. 3A, right panel), we visualize the dependence between the component of ln(mRNA/cell) that is not explained by ln(exon number) and the component of ln(gene length) that does not “replicate” information already provided by ln(exon number) itself.
For studying the effects of transcriptional interference, we divided genes into an “urban” and a “rural” class. After computing the distance between the promoters and the poly(A) sites of each of the MHKT genes and their nearest neighbors on both sides, we considered the shortest between these two distances, min_dist. Then, we labeled “urban” the 25% of the MHKT genes with smallest min_dist, and rural the remaining 75%. Because distances can be negative (overlapping genes), when passing on the natural log scale we actually considered the logarithm of the absolute value, ln(min_dist), multiplied by the sign, sign(min_dist) (this is +1 if min_dist is positive, and -1 if it is negative).
When measuring linear association through a correlation coefficient, significance can be readily assessed through p-values. However, simple p-values cannot be provided for the significance of dependence patterns captured by lowess smooths (note that these curves, unlike typical regression functions, are nonparametric). Thus, we use random permutations of the data to compute lowess smooths that can be used as reference to gage the significance of the pattern observed on the actual data (Good 2000). In our plots, lowess smooths from random permutations are represented as thin black lines accompanying the thick red line representing the lowess computed on the actual data. For instance, in the Figure 2 inset, the values of ln(mRNA/cell) were reshuffled at random while keeping those for ln(gene length) fixed. This produces artificial data in which the marginal distributions of expression and gene length are preserved, but the dependence between them is eliminated.
Acknowledgments
E.E.B. was supported by NIH grants DK56845, HL55435, and DK061799. F.C. and W.M. were supported by grant HG-02238 from the National Human Genome Research Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1169203. Article published online before print in November 2003.
Footnotes
[Supplemental material is available online at www.genome.org. The data sets and details on data preparation and preprocessing can be found at http://bio.cse.psu.edu/dist/bouhassira/. The complete list of genes used in this study is available at http://bio.cse.psu.edu/.]
References
- Cleveland, W. 1979. Robust locally weighted regression and smoothing scatterplots. J. Am. Stat. Assoc. 74: 829-836. [Google Scholar]
- Cohen, B.A., Mitra, R.D., Hughes, J.D., and Church, G.M. 2000. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat. Genet. 26: 183-186. [DOI] [PubMed] [Google Scholar]
- Cook, R.D. 1994. On the interpretation of regression plots. J. Am. Stat. Assoc. 89: 177-189. [Google Scholar]
- Cook, R.D. and Weisberg, S. 1999. Applied regression including computing and graphics. Wiley, New York.
- Eszterhas, S.K., Bouhassira, E.E., Martin, D.I., and Fiering, S. 2002. Transcriptional interference by independently regulated genes occurs in any relative arrangement of the genes and is influenced by chromosomal integration position. Mol. Cell. Biol. 22: 469-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiering, S., Bender, M.A., and Groudine, M. 1999. Analysis of mammalian cis-regulatory DNA elements by homologous recombination. Methods Enzymol. 306: 42-66. [DOI] [PubMed] [Google Scholar]
- Fong, Y.W. and Zhou, Q. 2001. Stimulatory effect of splicing factors on transcriptional elongation. Nature 414: 929-933. [DOI] [PubMed] [Google Scholar]
- Good, P. 2000. Permutation tests:A practical guide to resampling methods for testing hypotheses. Springer Verlag, New York.
- Issa, J.P. 2000. CpG-island methylation in aging and cancer. Curr. Top. Microbiol. Immunol. 249: 101-118. [DOI] [PubMed] [Google Scholar]
- Mignone, F., Gissi, C., Liuni, S., and Pesole, G. 2002. Untranslated regions of mRNAs. Genome Biol. 3: REVIEWS0004. [DOI] [PMC free article] [PubMed]
- Mitchell, P. and Tollervey, D. 2000. mRNA stability in eukaryotes. Curr. Opin. Genet. Dev. 10: 193-198. [DOI] [PubMed] [Google Scholar]
- Moore, M.J. 2002. Nuclear RNA turnover. Cell 108: 431-434. [DOI] [PubMed] [Google Scholar]
- Orphanides, G. and Reinberg, D. 2000. RNA polymerase II elongation through chromatin. Nature 407: 471-475. [DOI] [PubMed] [Google Scholar]
- Pesole, G., Mignone, F., Gissi, C., Grillo, G., Licciulli, F., and Liuni, S. 2001. Structural and functional features of eukaryotic mRNA untranslated regions. Gene 276: 73-81. [DOI] [PubMed] [Google Scholar]
- Proudfoot, N.J. 1986. Transcriptional interference and termination between duplicated α-globin gene construct suggests a novel mechanism for gene regulation. Nature 322: 562-565. [DOI] [PubMed] [Google Scholar]
- Proudfoot, N.J., Furger, A., and Dye, M.J. 2002. Integrating mRNA processing with transcription. Cell 108: 501-512. [DOI] [PubMed] [Google Scholar]
- Velculescu, V.E., Madden, S.L., Zhang, L., Lash, A.E., Yu, J., Rago, C., Lal, A., Wang, C.J., Beaudry, G.A., Ciriello, K.M., et al. 1999. Analysis of human transcriptomes. Nat. Genet. 23: 387-388. [DOI] [PubMed] [Google Scholar]
- Vreken, P., van, d.V., de Regt, V.C., de Maat, A.L., Planta, R.J., and Raue, H.A. 1991. Turnover rate of yeast PGK mRNA can be changed by specific alterations in its trailer structure. Biochimie 73: 729-737. [DOI] [PubMed] [Google Scholar]
- Whitelaw, E. and Martin, D.I. 2001. Retrotransposons as epigenetic mediators of phenotypic variation in mammals. Nat. Genet. 27: 361-365. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://bio.cse.psu.edu/; complete list of genes used in this study.
- http://bio.cse.psu.edu/dist/bouhassira/; the data sets and details on data preparation and preprocessing.
- http://www.sagnet.org/; SAGE.
- http://www.ncbi.nlm.nih.gov; National Center for Biotechnology Informations' Web site.
- http://www.genome.usc.edu; Santa Cruz genome browser Web site.