Abstract
Developing lymphocytes must assemble antigen receptor genes encoding the B cell and T cell receptors. This process is executed by the V(D)J recombination reaction, which can be divided into DNA cleavage and DNA joining steps. The former is carried out by a lymphocyte-specific RAG endonuclease, which mediates DNA cleavage at two recombining gene segments and their flanking RAG recognition sequences. RAG cleavage generates four broken DNA ends that are repaired by non-homologous end joining forming coding and signal joints. On rare occasions, these DNA ends may join aberrantly forming chromosomal lesions such as translocations, deletions and inversions that have the potential to cause cellular transformation and lymphoid tumors. We discuss the activation of DNA damage responses by RAG-induced DSBs focusing on the component pathways that promote their normal repair and guard against their aberrant resolution. Moreover, we discuss how this DNA damage response impacts processes important for lymphocyte development.
Keywords: Antigen receptor genes, DNA damage responses, genomic stability, lymphocyte development, lymphoid tumors
Assembly of Lymphocyte Antigen Receptor Genes
The genes that encode B and T cell antigen receptors are unique in that the second exon must be assembled in developing lymphocytes from component variable (V), joining (J), and, in some cases, diversity (D) gene segments through the process of V(D)J recombination. This process of somatic DNA recombination is initiated when the RAG endonuclease introduces DNA double strand breaks (DSBs) at the border of two recombining gene segments and their flanking RAG recognition sequences, termed recombination signals (RSs) (1–3). The RAG endonuclease is composed of the RAG1 and RAG2 proteins, which are expressed primarily in developing lymphocytes (1–3). Although the catalytic nuclease activity resides in RAG1, both RAG1 and RAG2 are required for DNA cleavage (1–3). RAG activity is restricted to the G1-phase of the cell cycle because RAG2 is phosphorylated and degraded as cells enter S phase (4).
DNA cleavage by RAG leads to four broken DNA ends: a pair of coding ends and a pair of signal ends that are processed and joined by proteins of the non-homologous end joining (NHEJ) pathway of DSB repair to generate a coding joint and signal joint, respectively (5, 6). Occasionally these DSBs can be repaired aberrantly leading to the formation of chromosomal lesions such as translocations, deletions and inversions (7, 8). If the breakpoints of these chromosomal lesions lie near potential oncogenes or tumor suppressor genes, they can lead to cellular transformation and lymphoid tumors.
There are seven antigen receptor genes. The T cell receptor (TCR) α and β chain genes are completely assembled and expressed in αβ T cells, whereas the TCR γ and δ chain genes are completely assembled and expressed in γδ T cells (9, 10). The B cell receptor (BCR) is composed of a heavy (H) chain encoded by the immunoglobulin (Ig) H locus and light (L) chain encoded by either the IgLκ or IgLλ loci (9, 10). All loci contain V and J gene segments with the TCR δ, TCRβ, and IgH loci also containing D gene segments (9, 10).
The basic biochemical features of the DNA cleavage and DNA joining steps of the V(D)J recombination reaction are shared by all antigen receptor loci. However, this process is regulated in a lineage-specific manner with BCR genes completely assembled only in B cells and TCR genes completely assembled only in T cells (9, 10). Moreover, assembly is developmental stage-specific with IgH chain genes assembled in pro-B cells and IgL chain genes in pre-B cells (9, 10). Similarly, TCRβ chain genes are assembled in pro-T cells, and TCRα chain genes are assembled in pre-T cells (9, 10). Finally, assembly is regulated in the context of allelic exclusion, which ensures that a single complete productive rearrangement is generated at the TCRβ, IgH and IgL chain loci in most cells (9–11).
This regulation of antigen receptor gene assembly occurs at the DNA cleavage step of the V(D)J recombination reaction through alterations in the accessibility of antigen receptor gene segments to the endonucleolytic activity of RAG (9, 10, 12). In this regard, cis-acting transcriptional regulatory elements such as promoters and enhancers bring about alterations in DNA methylation, chromatin structure, and nuclear positioning that affect the ability of RAG to access the appropriate regions of antigen receptor loci (9, 10, 12). This modulation of accessibility is likely important not only for the regulation of antigen receptor gene assembly but also for promoting genomic stability by limiting the generation of unnecessary RAG DSBs that could be resolved aberrantly.
The V(D)J Recombination reaction
The V(D)J recombination reaction is generally divided into a DNA cleavage step that is mediated by RAG and a DNA joining step mediated by the NHEJ pathway of DNA DSB repair. The DNA cleavage step of the reaction is lymphoid-specific as RAG1 and RAG2 are expressed primarily in developing lymphocytes (1–3). In contrast, the NHEJ proteins that repair RAG DSBs are expressed in most tissues and function in general DSB repair (5, 6).
RAG-mediated DNA cleavage occurs in the setting of a synaptic complex between an appropriate pair of RSs flanking the gene segments that are undergoing recombination (1–3). RSs are composed of conserved heptamers and nonamers flanking either 12 or 23 base pair spacers that are relatively nonconserved. In order for RAG cleavage to occur, the RS pair must have dissimilar spacer length, a restriction termed the 12/23 rule. However, not all pairs of 12/23 RSs join with equal efficiency; thus, RSs can also impose restrictions on cleavage beyond the 12/23 rule referred to as B12/23 (9). DNA cleavage by the RAG proteins leads to the generation of a pair of hairpin-sealed coding ends and a pair of blunt phosphorylated signals ends. These DNA ends remain in a post-cleavage complex until they can be joined, although whether these exist in a single complex containing all four broken DNA ends or whether the two coding ends and two signal ends are maintained separately is not known.
The coding and signal end pairs generated by RAG cleavage are processed and joined by NHEJ proteins (5, 6). Prior to joining, the hairpin-sealed coding ends must be opened, which can occur asymmetrically leading to the addition of palindromic nucleotides (P nucleotides) (9). In addition, exonucleases can remove nucleotides from the coding ends, and template-independent polymerases can add non-templated nucleotides (N nucleotides) to coding ends (9). The number of nucleotides gained and lost through these processes is random, and, for that reason, only one third of completed variable region exons will be in-frame and capable of encoding an antigen receptor chain. Although this joint diversification leads to the generation of large numbers of non-functional antigen receptor genes, it is critical for the generation of a broadly active adaptive immune response. In contrast to coding ends, signal ends are joined relatively precisely with very little nucleotide addition or loss.
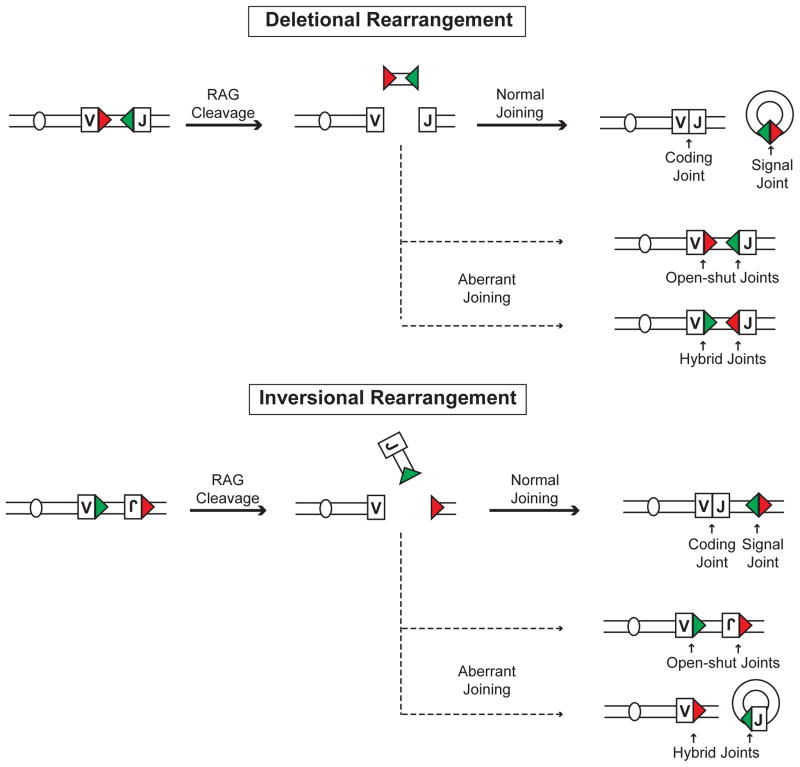
Coding joint formation is essential for antigen receptor gene assembly; however, it is also essential for maintaining the linear integrity of the chromosome, thereby preserving genomic stability. Signal joint formation is not required to complete antigen receptor gene assembly. However, it can, in some cases, be required for maintaining the linear integrity of the chromosome. If two recombining gene segments are in the same transcriptional orientation, then rearrangement leads to deletion of the intervening segment of DNA (Fig. 1). During these deletional rearrangements, the coding joint is formed in the context of the chromosome; however, the signal ends are joined on the excised DNA fragment forming an extrachromosomal circle that is lost upon cell division (Fig. 1). Thus, signal joint formation is not required for maintaining the linear integrity of the chromosome during deletional rearrangements. Alternatively, if the two recombining gene segments are in opposite transcriptional orientations, then the intervening DNA is inverted and both the signal and coding joints are formed within the chromosomal context (Fig. 1). In this situation, which is common in the IgLκ locus, both coding and signal joints are required to maintain the linear integrity of the chromosome.
Figure 1. Types of rearrangements.
Schematic of rearrangements that occur by deletion or inversion. RSs are indicated by colored triangles and the V and J gene segment by rectangles. The resulting normal signal and coding joints are depicted as are aberrant open-and-shut and hybrid joints.
There are two other types of joints that occur rarely in wild type cells and involve the inappropriate joining of coding ends to signal ends. Coding and signal ends can be rejoined in their original configuration forming an open-and-shut joint, or the coding end from one RAG DSB can be joined to the signal end at another forming a hybrid joint (Fig. 1). Although these joints would not contribute to the formation of a functional antigen receptor gene, they would maintain the linear integrity of the chromosome. How NHEJ is modulated to preferentially join the two coding ends and two signals from distinct DNA DSBs to form a functional antigen receptor gene is not known, but is likely linked to the generation of these DSBs within the context of a synaptic complex.
Signal joint formation was thought to be less efficient than coding joint formation based on the ability to readily detect un-repaired signal ends, but not un-repaired coding ends, in lymphocytes undergoing V(D)J recombination (13, 14). However, efficient signal joint formation during inversional rearrangements is required to maintain the linear integrity of the chromosome. Recent analyses have revealed that chromosomal signal joints and coding joints are formed with similar kinetics (15, 16). Moreover, signal joint formation on extrachromosomal DNA fragments during deletional rearrangement occurs with slower kinetics than chromosomal signal joint formation (16). Thus, signal joints are formed relatively efficiently within the chromosomal context where they are required to promote genomic stability.
Experimental Approaches for Studying V(D)J recombination
In vitro analyses of both DNA cleavage by the RAG proteins and coding joint formation mediated by purified NHEJ proteins have used small DNA duplex substrates to reveal many basic biochemical and enzymatic characteristics of these proteins (1–3, 17). Gellert and colleagues developed the first experimental approach for studying the V(D)J recombination reaction in cultured cells (18). Extrachromosomal plasmid substrates containing appropriate RS pairs were introduced into transformed pre-B cells that express RAG; plasmids recovered from these transfected cells were then assayed for rearrangements. This approach has also been used to assay recombination in non-lymphoid cells co-transfected with plasmids encoding RAG-1 and RAG-2 (19). Although it has proven to be a powerful tool for elucidating many important mechanistic features of both the DNA cleavage and joining steps, this approach cannot be used to delineate pathways that are specific to the generation or repair of RAG DSBs within the chromosomal context.
Rosenberg and colleagues developed a technique for studying chromosomal V(D)J recombination using Abelson murine leukemia virus transformed pre-B cell lines, hereafter referred to as abl pre-B cells (20). Abl pre-B cells harboring a temperature-sensitive Abelson murine leukemia virus, when cultured at non-permissive temperatures, undergo G1 cell cycle arrest, induce RAG expression and rearrange the endogenous IgLκ locus (20). A similar approach has also been developed that involves treating abl pre-B cells with the abl kinase inhibitor Gleevec, which also causes G1 cell cycle arrest, induction of RAG expression and robust rearrangement of the endogenous IgLκ locus as well as chromosomally integrated retroviral substrates (15, 21).
Activation of DNA Damage Responses by RAG DSBs
DNA double strand breaks are dangerous lesions that can be generated by genotoxic agents or as necessary intermediates of physiologic processes such as DNA replication and, in lymphocytes, V(D)J recombination and Ig class switch recombination (CSR). All cells mount a DNA DSB response that includes the activation of cell cycle checkpoint pathways, DNA repair pathways, and, in cells with persistent un-repaired DNA DSBs, cell death pathways (22).
DNA DSBs activate one or more PI3K-like serine/threonine kinases, which include the ataxia telangiectasia mutated (ATM) kinase, the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and the ATM and Rad3 related (ATR) kinase (22, 23). These kinases regulate the activity of target proteins through the phosphorylation of serines or threonines followed by glutamine residues (SQ/TQ). ATR is primarily activated by cells in post-replicative stages of the cell cycle. In contrast, ATM and DNA-PKcs can be activated at all stages of the cell cycle. In response to genotoxic DSBs, ATM phosphorylates hundreds of downstream proteins active in different aspects of the DNA damage response (24). Although the full extent of DNA-PKcs targets is not known, DNA-PKcs shares a subset of downstream targets with ATM (25, 26).
In response to genotoxic DNA DSBs generated in G1-phase cells, ATM and DNA-PKcs phosphorylate the Chk2 kinase, causing inactive monomers to form catalytically active dimers (25–27). Chk2 phosphorylates Cdc25a, leading to its ubiquitylation and proteosomal degradation, and, as a result, prevents G1-phase cells from transitioning into S phase (27). Transit of a cell with un-repaired DSBs into S phase would result in the formation of a replicated broken chromosome that could promote further genomic instability. In addition, ATM and DNA-PKcs both phosphorylate the p53 transcription factor, which further reinforces the G1-S checkpoint by promoting the expression of p21, a cyclin-dependent kinase-2 (CDK2) inhibitor that prevents the G1-to-S phase transition (28). In addition, p53 promotes the expression of pro-apoptotic genes, such as the Fas receptor, PUMA (p53-upregulated modulator of apoptosis) and Bax (Bcl2-associated X protein), which all promote the death of cells with persistent un-repaired DSBs (28). Finally, ATM and DNA-PKcs both participate in regulating NHEJ in G1-phase cells (22, 23). RAG DSBs also activate an ATM- and DNA-PKcs-mediated DNA damage response in G1-phase developing lymphocytes. As discussed in detail below, this response includes not only the activation of checkpoint, cell death, and repair pathways but also the activation of a cell-type-specific genetic program.
Factors Involved in RAG DSB Repair
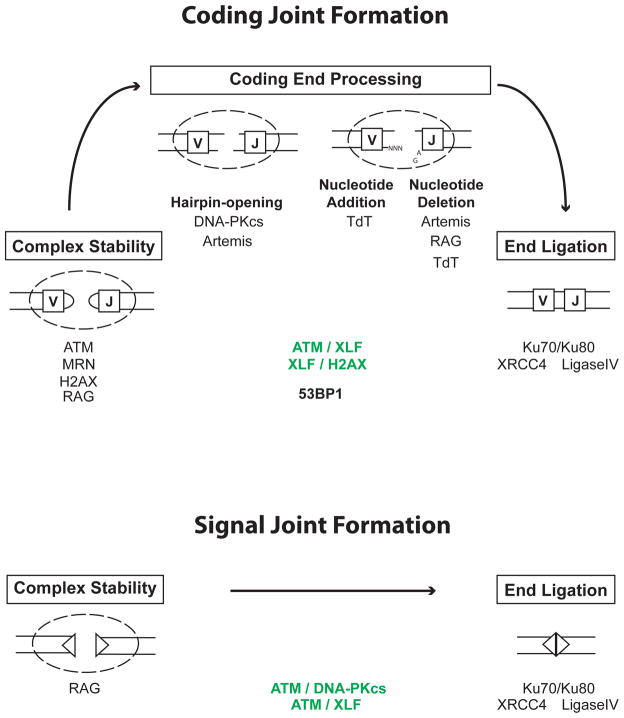
In the G1-phase of the cell cycle, DNA DSBs are predominantly repaired by NHEJ. Indeed, RAG DSBs, which are generated in G1-phase lymphocytes require NHEJ factors for efficient repair. The core NHEJ factors Ku70, Ku80, DNA Ligase IV, and XRCC4 are absolutely required for both signal and coding joint formation. Other proteins appear to have specific activity in signal joint or coding joint formation, but not both. Still other proteins may function primarily to prevent the aberrant resolution of rare un-repaired RAG DSBs and have minimal activity during normal joining. Here, we discuss the proteins that have known activities in the processing and joining of RAG-mediated DSBs (Fig. 2).
Figure 2. Factors involved in signal and coding joint formation.
RAG cleavage generates a pair of hairpin sealed coding ends (rectangles) and a pair of blunt phosphorylated signal ends (triangles). After RAG cleavage, coding ends and signal ends reside in complexes until they can be processed and joined. Coding end complexes are stabilized by ATM and MRN and may also be stabilized by H2AX and possibly RAG. ATM and MRN do not appear to be critical for signal end complex stability. However, ATM and DNA-PKcs have overlapping activities (shown in green) that are required for signal joint formation. Although the basis for this requirement is unknown, it could reflect the overlapping activity of ATM and DNA-PKcs in promoting signal end complex stability. RAG stays bound to signal ends in post-cleavage complexes generated in vitro and it has been suggested that RAG may stabilize signal ends in vivo, but a direct demonstration of this RAG function is lacking. Ku70/80 and DNA-PKcs, which are recruited early to DSBs, have been implicated in the stabilization of DNA ends prior to joining but their potential function in stabilizing RAG DSBs has not been examined.
Hairpin sealed coding ends are opened through the endonuclease activity of Artemis. Its endonuclease activity requires association with and possibly phosphorylation by activated DNA-PKcs. Nucleotides can then be added to coding ends through the activity of TdT, and nucleotides can be lost through the nuclease activity of Artemis and possibly RAG and TdT. The joining of signal and coding ends is carried out by DNA Ligase IV and XRCC4 with Ku70/Ku80 potentially being important to align DNA ends prior to joining. Although Ku function is essential for both signal and coding joint formation, its specific activities at RAG DSBs have not been directly examined. Notably, ATM and XLF have overlapping activities (shown in green) that are also important for signal and coding joint formation. In addition, XLF has overlapping activities with H2AX that are required for coding joint formation. Whether these overlapping activities are also required for signal joint formation has not been examined. The nature of these overlapping activities of XLF and ATM or H2AX remains to be determined. 53BP1 is required for optimal coding joint formation for reasons that are not clear but may involve a function in long range joining and/or preventing DNA end resection.
DNA-PK holoenzyme
The DNA-PK holoenzyme is composed of the Ku70 and Ku80 (also referred to as Ku86) heterodimer and the 460 kD DNA-PK catalytic subunit (DNA-PKcs) (29, 30). Ku70 and Ku80 are recruited rapidly to broken DNA ends after the generation of a DNA DSB, binding these ends as a heterodimeric ring structure with a central channel that accommodates the broken DNA strand (31). The orientation of the DNA strand in this channel suggests that Ku70 and Ku86 may have a role in aligning broken DNA ends prior to joining (32, 33).
The Ku heterodimer recruits DNA-PKcs to the site of a DSB. DNA-PKcs initially binds the C-terminus of Ku80, causing the translocation of the Ku heterodimer on the DNA helix and allowing DNA-PKcs to directly bind the DNA end (30). DNA-PKcs kinase activity depends on both its association with the Ku heterodimer and its interaction with the broken DNA end (30). Once activated, DNA-PKcs can phosphorylate a host of targets involved in DNA damage responses and DNA DSB repair. DNA-PKcs is also autophosphorylated at numerous SQ/TQ motifs (30, 34). This autophosphorylation may occur in trans between two DNA-PKcs proteins bound at the two broken DNA ends resulting from a single DSB (30).
DNA-PKcs can bind multiple DNA ends in a kinase-independent manner; as such, it may have activities in bridging broken DNA ends prior to their joining (35, 36). DNA-PKcs constitutively associates with XRCC4, DNA Ligase IV, and Artemis in a manner that does not depend on its kinase activity (6, 30). Thus, DNA-PKcs may have a role in recruiting NHEJ factors to DSB sites. These findings raise the possibility that DNA-PKcs may have functions in general DSB repair, and possibly in the repair of RAG DSBs, that do not depend on its catalytic activity. However, the catalytic activity of DNA-PKcs is also important as cells expressing a kinase dead mutant of DNA-PKcs exhibit defects in DSB repair (30). Many NHEJ factors have SQ/TQ motifs that serve as targets for DNA-PKcs kinase activity in vitro and in vivo (6, 30). However, mutational analyses of some of these SQ/TQ motifs have not yet revealed a function for these phosphorylation events in DSB repair.
While DNA-PKcs has important roles in the DNA end joining process, the persistence of DNA-PKcs at broken DNA ends may actually inhibit joining. In this regard, autophosphorylation of SQ/TQ motifs within the ABCDE and PQR clusters of DNA-PKcs may promote the eviction of DNA-PKcs from broken DNA ends, allowing for joining (30, 34). In addition to autophosphorylation, some SQ/TQ motifs can be phosphorylated by ATM, which may also impact DNA-PKcs activity in DSB repair (30, 34). Indeed, mice with threonine to alanine mutations of three of the TQ motifs phosphorylated by ATM exhibit defects in DNA DSB repair pathways beyond simple end joining, leading to complete stem cell failure (37).
Although DNA-PKcs, Ku70, and Ku80 function as a complex, there is a distinct difference in the phenotypes of mice deficient in DNA-PKcs and either Ku70 or Ku80 (5). Mice deficient in Ku70 or Ku80 have a complete block in lymphocyte development, are small in size, and are sterile (5). In contrast, DNA-PKcs-deficient mice develop normally and are fertile but similarly have a complete block in lymphocyte development (5). These phenotypic differences suggest that DNA-PKcs and Ku70/Ku80 may have distinct functions in DNA damage responses and DNA repair.
During V(D)J recombination, Ku70 and Ku80 are essential for both signal and coding joint formation, whereas DNA-PKcs is essential only for coding joint formation due to its requirement to promote the hairpin-opening nuclease activity of Artemis (see below) (5). Patients with mutations in Ku70 and Ku80 have not been identified. Although patients with null DNA-PKcs mutations have not been identified, some patients with radiation sensitive (RS) severe combined immunodeficiency (SCID) have a mis-sense point mutation in DNA-PKcs (L3602R) (38). SCID can be caused by mutations in many genes that encode proteins important for lymphocyte development. However, a concomitant sensitivity to ionizing radiation, RS-SCID, is indicative of potential DNA DSB repair defects and suggests that the immunodeficiency may be due to defects in the repair of RAG DSBs. Indeed, the DNA-PKcs L3602R mutation has a diminished capacity to activate Artemis (38).
Although cells with null DNA-PKcs mutations do not exhibit significant defects in the repair of signal ends, cells expressing some mutant forms of DNA-PKcs do exhibit such defects (39). In addition, cells deficient in ATM have no demonstrable defect in signal joint formation (15, 16, 40). However, cells deficient in both of these kinases have a marked defect in signal joint formation (16, 40). Thus, during V(D)J recombination, DNA-PKcs and ATM have important overlapping functions in the normal repair of signal ends. These overlapping functions depend on the proteins’ kinase activity, suggesting that they phosphorylate common downstream targets required for efficient signal joint formation (16, 40). In this regard, RAG-1 and RAG-2 each have SQ/TQ motifs that could be phosphorylated by ATM or DNA-PKcs, and it has been suggested that RAG1 and RAG2 must be displaced from signal ends prior to signal joint formation (41–44). However, cells expressing mutant forms of RAG1 or RAG2 in which all SQ/TQ motifs were mutated to AQ did not reveal defects in either signal joint or coding joint formation (43). In addition, the overlapping requirement for ATM or DNA-PKcs kinase activity during signal joining does not simply reflect the need to phosphorylate DNA-PKcs to promote its eviction from signal ends, because ATM kinase activity is required for efficient signal joint formation in DNA-PKcs-deficient cells (16, 40).
DNA Ligase IV - XRCC4
There are three mammalian ligases: DNA Ligase I, III and IV. DNA Ligase IV functions during NHEJ and V(D)J recombination (5, 45). DNA Ligase IV, XRCC4, and XLF/Cernunnos exist in a complex in the cell (46). XRCC4 promotes DNA Ligase IV stability and maintains protein levels within the cell (5, 6). In addition, XRCC4 stimulates the adenylation of lysine residues within the catalytic core of DNA Ligase IV, the first step in the formation of a phosphodiester bond (5, 6). XRCC4 may also function to align DNA ends prior to ligation (5, 6). DNA Ligase IV and XRCC4 are absolutely required for the repair of both signal and coding ends, and mice deficient in DNA Ligase IV or XRCC4 exhibit a complete block in lymphocyte development (5).
Artemis
Artemis is a member of the metallo-β-lactamase superfamily of proteins that hydrolyze covalent bonds and was first identified in a subgroup of patents with RS-SCID (47). The presence of a β-CASP domain indicates that Artemis may hydrolyze phosphodiester bonds. Artemis-deficient cells exhibit increased sensitivity to ionizing radiation (48, 49). Indeed, Artemis activity is required for the repair of a subset of genotoxic DSBs (50). From this it has been proposed that Artemis may be required to process and repair broken DNA ends with complex structures.
The coding ends generated by RAG cleavage are hairpin-sealed, and these DNA ends must be opened by hydrolysis of a phosphodiester bond prior to joining. This hairpin opening is often asymmetric from the apex leading to the generation of added palindromic (P) nucleotides at the joint. Several lines of evidence demonstrate that Artemis is required for the efficient opening of hairpin-sealed coding ends during V(D)J recombination and that this function requires DNA-PKcs. In vitro, Artemis has intrinsic 5′-to-3′ exonuclease activity (51). However, when complexed with DNA-PKcs in vitro, Artemis can open hairpin-sealed DNA ends preferentially two nucleotides 3′ of the apex, similar to what has been observed in the opening of hairpin-sealed coding ends in vivo (51). This may reflect a requirement for Artemis to associate with autophosphorylated DNA-PKcs and/or for the phosphorylation of Artemis by DNA-PKcs (17, 51, 52). Moreover, in the presence of DNA-PKcs, Artemis has endonuclease activity at 5′ and 3′ overhangs (51).
Artemis-deficient cells exhibit a block in coding joint formation and an accumulation of un-repaired hairpin-sealed coding ends, which is identical to the defect observed in DNA-PKcs-deficient cells (5, 49). As a result, Artemis-deficient mice have a severe block in B and T cell development (49). The B and T cells that do develop in these animals have antigen receptor genes with aberrant joints that exhibit an increase in the frequency and number of P nucleotides, similar to what is observed in the rare B and T cells generated in DNA-PKcs-deficient (SCID) mice (49, 53–55). These findings indicate that Artemis (and DNA-PKcs) are required for the efficient opening of hairpin-sealed coding ends during V(D)J recombination in vivo. However, in the absence of these factors, other nucleases can perform this function with far less efficiency and with important qualitative differences that could preclude the generation of a functional antigen receptor gene. These nucleases may include Mre11 and CtIP (discussed below). In addition, RAG has been reported to have hairpin opening endonuclease activity in vitro (56, 57). Although Artemis is not required for signal joint formation, signal joint fidelity is altered in Artemis-deficient cells, demonstrating that signal ends are potential substrates for Artemis activity during V(D)J recombination (58).
XLF/Cernunnos
The XRCC4-like factor (XLF), also known as Cernunnos, shares structural features with XRCC4 including an N-terminal head domain and a C-terminal coiled-coiled domain that mediates homodimerization (59, 60). XLF interacts with XRCC4 in vivo and promotes, but is not required for, the ligation activity of DNA Ligase IV (46). In this regard, XLF facilitates the joining of non-cohesive and blunt DNA ends but is dispensable for the ligation of cohesive DNA ends in vitro (61, 62).
Patients with mutations in XLF have RS-SCID, suggesting that XLF may be involved in the repair of RAG-mediated DSBs (46, 63, 64). However, the immunodeficiency observed in these patients is much less severe than that observed in Artemis-deficient patients. Additionally, mice deficient in XLF do not exhibit significant immunodeficiency (65). Moreover, although signal and coding joint formation on extrachromosomal substrates in XLF-deficient murine non-lymphoid cells is defective, XLF-deficient murine lymphocytes undergo efficient V(D)J recombination of both chromosomal and extrachromosomal substrates (63–66). Thus, it appears that XLF is not required for the repair of RAG-mediated DSBs in developing lymphocytes in mice, raising the question of why, in humans, does XLF deficiency manifest itself as RS-SCID? One possibility is that mice have compensatory mechanisms for XLF deficiency during V(D)J recombination and DNA DSB repair that may be absent in humans.
XLF and other factors may have redundant functions in the repair of RAG DSBs. This is supported by analysis of V(D)J recombination in cells deficient for XLF and the histone protein H2AX or ATM (67). Deficiency of H2AX has no demonstrable effect on the repair of RAG DSBs, and deficiency of ATM has only a mild effect (15, 68–73). However, a combined deficiency of either protein with XLF deficiency results in a severe block in lymphocyte development with a significant defect in the repair of RAG-mediated DSBs (67). This could reflect the function of XLF and H2AX (or XLF and ATM) in the same RAG DSB repair pathway. For example, H2AX prevents un-repaired RAG breaks from being aberrantly resected, and a combined deficiency of H2AX and XLF leads to more dramatic resection, suggesting that these two proteins may both modulate DNA end resection (67, 74). Alternatively, XLF and ATM or H2AX may function in unique pathways. Deficiency in any of the three pathways individually having a minimal effect on joining; however, a combined deficiency of two of these pathways leads to a block in the repair of RAG-mediated DSBs. For example, the block in coding joint formation in cells deficient in XLF and ATM could be due to a mild decrease in joining kinetics caused by XLF-deficiency coupled with the coding end instability caused by ATM deficiency (15, 61, 62, 67). Given that ATM phosphorylates H2AX, it is tempting to speculate that, during the repair of RAG DSBs, ATM and H2AX function in the same XLF-redundant pathway. However, the RAG DSB repair defects in cells deficient in XLF and H2AX are different from those observed in cells deficient in XLF and ATM, which would suggest that H2AX and ATM function in different pathways redundant with XLF in the repair of RAG DSBs (67).
TdT
Terminal deoxynucleotydltransferase (TdT) is a member of the polymerase X family of DNA polymerases that broadly functions in DNA repair (76). TdT is a template-independent DNA polymerase that adds nucleotides to 3′ DNA ends (77). TdT is expressed almost exclusively in developing lymphocytes and adds nucleotides primarily to coding ends and not signal ends (77). Although TdT is not required per se for the repair of RAG mediated DSBs, the addition of non-templated (N) nucleotides by TdT provides coding joint sequence diversity that is important for generating a varied antigen receptor repertoire.
Multiple isoforms of TdT are encoded by a single gene. In mice, the two isoforms differ only in the presence of 20 amino acids at the C-terminus in one (TdT long, TdTL) but not the other (TdT short, TdTS) (77). TdTS has polymerase activity, whereas TdTL appears to be devoid of such activity (77). Although significantly reduced, some N-region additions are found in TdT−/− mice, demonstrating that there are other template-independent polymerases, perhaps DNA polymerase μ, capable of adding N nucleotides to coding ends (78, 79). TdT may also have 3′ to 5′ exonuclease activity that could contribute to joint diversification by removing nucleotides (77). Importantly, coding joints formed in lymphocytes from TdT-deficient mice exhibit robust nucleotide loss; thus an exonuclease activity of TdTL is not essential (78, 79).
H2AX
The H2A histone variant H2AX is distributed in nucleosomes throughout the genome (80). H2AX is phosphorylated at serine 139 in its C-terminal tail by ATM, ATR and DNA-PKs, forming γ-H2AX, in chromatin across great distances flanking DNA damage (81). γ-H2AX functions, in part, through the recruitment and/or retention of DNA damage response proteins at the site of a DNA DSB (80, 82).
At RAG DSBs, γ-H2AX forms at distances of several hundred kb in flanking chromatin (83, 84). γ-H2AX may function, in part, to promote RAG DSB stability until the DNA ends can be joined (73). ATM is primarily responsible for the formation of γ-H2AX in response to RAG DSBs; however, in the absence of ATM, DNA-PKcs can also generate γ-H2AX, although at lower levels and with slower kinetics (84). H2AX-deficient mice do not have significant defects in lymphocyte development, and H2AX-deficient lymphocytes exhibit robust chromosomal V(D)J recombination, demonstrating that H2AX is not essential for the repair of RAG DSBs (71–73). However, as discussed above, H2AX and XLF have activities that are redundant in the repair of RAG DSBs (67).
The generation of γ-H2AX in chromatin flanking RAG DSBs in Artemis- or DNA Ligase IV-deficient cells prevents the aberrant resection of these persistent un-repaired DNA ends (74). The mechanism by which γ-H2AX protects RAG DSBs from resection is not known, but this DNA end protection depends on MDC-1, which binds directly to γ-H2AX (74). Interestingly, in the absence of H2AX, Artemis is not required for the efficient opening of hairpin-sealed coding ends (74). Rather, hairpin-opening depends on CtIP, a protein important for initiating DNA end resection and the generation of signal strand overhangs essential for homology-mediated DSB repair (74, 85). The DNA end resection activity of CtIP can be modulated by ATM; notably, CtIP activity in resecting RAG DSBs also depends on ATM (74, 85).
During V(D)J recombination, Artemis is thought to function primarily in opening hairpin-sealed coding ends. Thus, the hairpin-sealed coding ends opened in an Artemis-independent manner in H2AX-deficient cells should be amenable to coding joint formation. However, many of these hairpin-opened DNA ends persist un-repaired, possibly due to the presence of significant single stranded overhangs, which would inhibit NHEJ (74, 85). Broken DNA ends with significant single stranded overhangs are repaired primarily by pathways that rely on the use of homologies at the DNA ends (85). Indeed, coding joints formed in cells deficient in H2AX and Artemis demonstrate a substantial increase in the use of microhomologies (74). A requirement for microhomology during joining may lead to chromosomal deletions, which could preclude the formation of a functional antigen receptor gene or promote genomic instability by disrupting neighboring genes.
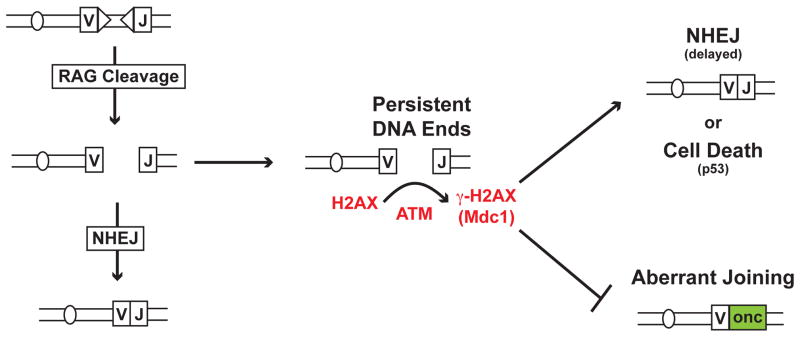
Sequence analysis of coding joints in H2AX-deficient lymphocytes does not reveal extensive deletions or a bias for microhomology use as is observed in coding joints formed in cells deficient in both Artemis and H2AX (71, 72, 74). This could be due to the compensatory function of another protein, such as XLF. Alternatively, the function of H2AX in protecting RAG DSBs from aberrant resection may not be critical at most RAG DSBs, which are rapidly repaired. Rather, H2AX may primarily be important for preventing rare un-repaired RAG DSBs from being nucleolytically processed in ways that would permit aberrant joining. Thus, γ-H2AX may maintain the structure of un-repaired broken RAG DSBs until they are either joined normally by classical NHEJ or, if they persist, activate p53 and cell death (Fig. 3).
Figure 3. Pathways that maintain genomic stability.
RAG DSBs are repaired efficiently by NHEJ. However, rare un-repaired RAG DSB must eventually be resolved by NHEJ or, if they persist un-repaired, must promote cell death to prevent their aberrant resolution as potentially dangerous chromosomal lesions. In this regard, the ATM-dependent phosphorylation of H2AX, forming γ-H2AX, and MDC1, which binds to γ-H2AX, prevents persistent un-repaired RAG DSBs from being resected in a way that allows them to access aberrant joining pathways in G1-phase lymphocytes. Thus, H2AX may maintain the structure of the DNA end in G1-phase cells such that it is either joined normally by NHEJ or activates p53-mediated cell death if it persists unjoined.
ATM
Like DNA-PKcs, ATM is a member of the PI3-kinase-like serine/threonine kinase family (23). ATM exists as an inactive homodimer that is recruited to DNA DSBs by the Mre11, Rad50, Nbs1 (MRN) complex through direct binding of ATM to Nbs1 (86–89). Once bound to MRN at the site of a DSB, ATM is activated through an autophosphorylation step that converts the inactive dimer to active monomers (86, 90, 91). However, a mutant form of ATM that cannot be phosphorylated at the canonical serine site can still initiate DNA damage responses, demonstrating that there are additional ways to activate ATM (92). Once activated, ATM phosphorylates numerous downstream targets that function in different aspects of the DNA DSB response (24).
ATM deficiency in humans leads to ataxia-telangiectasia (A–T), a disease marked by ocular telangiectasias, cerebellar ataxia, sensitivity to ionizing radiation, lymphopenia and a predisposition to lymphoid tumors with translocations involving antigen receptor genes (93). Mice deficient in ATM manifest many of these same defects (94). With respect to the lymphoid phenotypes, like A–T patients, ATM-deficient mice exhibit lymphopenia and a predisposition to the generation of thymic lymphomas with translocations involving TCR genes. Moreover, TCR locus translocations are found in a large fraction of non-transformed T cells in these mice (95). The breakpoint locations of these translocations suggest that they are derived from aberrantly repaired RAG DSBs. Indeed, mice that are deficient in ATM and RAG exhibit a significant lag in the generation of thymic lymphomas, and the lymphomas that develop do not have genomic lesions indicative of aberrantly repaired RAG DSBs (96, 97). Thus, the aberrant repair of RAG DSBs contributes to thymic lymphomas in ATM-deficient mice. However, the predisposition to thymomas in ATM-deficient mice unable to make RAG DSBs suggests that ATM has additional functions unrelated to V(D)J recombination in the prevention of thymocyte transformation.
Although mice with deficiencies in NHEJ factors have lymphopenia, they are not predisposed to lymphoid tumors, presumably due to the activation of cell cycle checkpoint and cell death pathways by un-repaired RAG DSBs. Conversely, mice deficient in p53 or Chk2 do not have defects in lymphocyte development. Although germline p53-deficient mice are prone to lymphoid tumors, most of these do not have chromosomal aberrations indicative of aberrantly repaired RAG DSBs (98, 99). However, mice with combined deficiency in NHEJ factors and p53 succumb to B cell tumors with translocations involving antigen receptor loci at an early age (100–102). These translocations are due to RAG DSBs since breeding of these mice onto a RAG-deficient background prevents tumor formation (103–105). Thus, a combined deficiency in NHEJ-mediated DSB repair and cell cycle checkpoint/death pathways closely phenocopies the lymphoid defects seen in ATM-deficient mice, suggesting that ATM functions in all of these pathways.
Indeed, ATM is activated by RAG DSBs and, in addition to initiating checkpoint and cell death pathways, ATM functions in the repair of RAG DSBs by promoting the stability of coding ends in post-cleavage complexes until they can be joined (15, 68–70, 106). In ATM-deficient pre-B cells, 10–20% of coding ends are lost from post-cleavage complexes (15). The dissociation of these DNA ends allows them to join aberrantly to other un-repaired DSBs, generating chromosomal deletions, inversions, and translocations (15, 107). The stabilizing function of ATM depends on its kinase activity. Although ATM itself can bind to DNA ends, stabilization likely occurs through the phosphorylation of downstream proteins that perform this function. These could include H2AX (as discussed above) and the MRN complex (see below), the latter of which has known activities in tethering DNA. In contrast to coding joint formation, signal joint formation is essentially normal in ATM-deficient cells due to the overlapping activity of ATM and DNA-PKcs in repairing signal ends (15, 16, 40).
Mre11/Rad50/Nbs1
The MRN complex is composed of the Mre11, Rad50 and Nbs1 proteins (88). Although Mre11, Rad50 and Nbs1 have distinct activities, they can function only in the setting of the MRN holocomplex. Soon after the generation of a DNA DSB, MRN is recruited to the site where it subsequently recruits and activates ATM (87, 88, 90, 91, 108). MRN has known functions in the repair of DNA DSBs by homologous recombination (88). Specifically, Mre11 has endonuclease and exonuclease activity, which together with CtIP initiate the resection of DNA ends required for homology-mediated DNA repair (85, 88). Mre11 may also function in the alignment of broken DNA ends prior to repair (109). Rad50 contains N-terminal Walker A and C-terminal Walker B nucleotide binding motifs and a central hinge region (88). The Walker A and B motifs form an intra-molecular dimer and bind to DNA, resulting in a conformational change at the hinge region that generates a hook domain (88). The association between two hook domains can tether two DNA strands and is thought to facilitate the association of sister chromatids during homologous recombination (88). Although the functions of Nbs1 are less clear, it may serve as a scaffold that recruits and/or retains specific DNA damage response proteins at the DSB site. In this regard, CtIP binds to the N-terminal FHA domain of Nbs1 and ATM binds to a C-terminal domain (110, 111). Mre11, Rad50 and Nbs1 can all be phosphorylated by ATM, which could alter the activity of these proteins in the repair of DNA DSBs (24, 112–114).
Humans with null Mre11, Rad50 or Nbs1 mutations have not been identified, and attempts to generate mice deficient in these proteins have failed due to early embryonic lethality, likely attributable to MRN function in homology-mediated DNA DSB repair (115–118). However, humans with hypomorphic mutations in Mre11, Rad50 and Nbs1 have been identified; these patients all exhibit hypersensitivity to ionizing radiation, but have variable defects in lymphocyte development and a variable predisposition to lymphomagenesis (88). Mice with a single hypomorphic Mre11 allele and those with two distinct Nbs1 hypomorphic alleles have been generated (119–121). As with humans, cells from all of these mice exhibit increased sensitivity to ionizing radiation; however, only one of the hypomorphic Nbs1 alleles leads to a mild immunodeficiency and a predisposition to lymphomagenesis (119–121). Together, these findings suggest that MRN functions in the repair of RAG DSBs with the observed variability in lymphoid phenotypes possibly due to the residual functions of the different hypomorphic alleles. Indeed, conditional deletion of Nbs1 in early developing thymocytes results in a near complete block in thymocyte development (122). However, this block was not rescued by expression of a TCR transgene, demonstrating that it is not primarily due to a defect in TCR gene assembly (122).
Nbs1, and presumably the entire MRN complex, is recruited to RAG DSBs generated in developing thymocytes (83). Cells expressing hypomorphic forms of Mre11 or Nbs1 have V(D)J recombination defects similar to those observed in ATM-deficient cells, although of lesser magnitude (123, 124). Thus, MRN functions in the repair of Rag-mediated DSBs. Although its specific function is not clear, it is tempting to speculate that the DNA tethering activity of Rad50 promotes the stability of coding ends in post-cleavage complexes. In this regard, phosphorylation of one or more of the MRN components by ATM may be required for this function. It remains possible that other substrates of ATM may function directly in stabilizing coding ends but may not be phosphorylated and activated in MRN-deficient cells as MRN is required to activate ATM. Multiple downstream targets of ATM were robustly phosphorylated in response to RAG DSBs in MRN-deficient abl pre-B cells demonstrating that ATM kinase activity is not globally compromised in these cells (87, 90, 91, 108, 123).
53BP1
53BP1 is a phosphorylation target of ATM recruited to DNA DSBs soon after they are generated. The localization of 53BP1 at DSBs has been linked to the formation of γ-H2AX in chromatin flanking these DSBs in addition to the presence of methylated histone H3 (K79) and histone H4 (K20) at the break site (125). 53BP1 localization also depends on the γ-H2AX- and MDC1-dependent recruitment of the RNF8 and RNF168 E3 ubiquitin ligases, which ubiquitylate histones in chromatin flanking DNA DSBs (125). 53BP1-deficient cells exhibit an increased sensitivity to ionizing radiation indicating that 53BP1 functions in DNA damage responses (126, 127). 53BP1 has been implicated in diverse activities including checkpoint activation, NHEJ-mediated DSB repair, modulation of DNA end resection and the protection and sequestration of broken DNA ends in cells passing through the cell cycle (128–133).
53BP1-deficient mice are mildly lymphopenic, indicating potential defects in V(D)J recombination (131, 134, 135). Indeed, un-repaired TCRα (Jα) coding ends are found in 53BP1-deficient thymocytes and these DNA ends are extensively resected (131). Finally, mice deficient in 53BP1 and p53 have a greater predisposition to generate lymphomas with translocations involving antigen receptor genes, which are indicative of aberrantly repaired RAG DSBs (136). Similar to V(D)J recombination, DNA DSB intermediates are generated during Ig class switch recombination (CSR). 53BP1-deficient B cells exhibit a profound defect in the repair of DNA DSBs generated during Ig CSR, leading to hyper-IgM syndrome (134, 137). Moreover, sequence analyses of joints made from these DNA ends suggest that they were also aberrantly resected prior to joining (128).
The DNA DSB repair defects observed during V(D)J recombination and Ig CSR could be due, in part, to the proposed role for 53BP1 in the joining of broken DNA ends from DSBs separated by great distances in the chromosome and the function of 53BP1 in regulating DNA end resection (128, 131, 133). H2AX and MDC1 also function in regulating DNA end resection (74). As H2AX and MDC1 are involved in the localization of 53BP1 at DNA breaks, it is possible that these three proteins function in the same pathway that preserves the integrity of DNA ends until they can be joined. Like H2AX, 53BP1 could have more substantial functions in V(D)J recombination with other proteins compensating for loss of 53BP1.
RAG1/RAG2
The RAG1 and RAG2 proteins have both been implicated in the joining step of V(D)J recombination. After cleavage in vitro, the RAG proteins remain associated with both signal end and coding end pairs, although the association with signal ends may be stronger (138, 139). The removal of RAG proteins from signal ends is required for signal joining in vitro (44). A role for RAG proteins in the joining step of the V(D)J recombination reaction has been proposed, based on the generation of RAG1 and RAG2 mutants that have normal DNA cleavage activity in vitro but are deficient in mediating V(D)J recombination of extrachromosomal substrates in vivo (140–143). The ability of these different RAG mutants to cleave chromosomal substrates was not reported. Additional evidence that the RAG proteins influence joining comes from the recent analysis of mice deficient in p53 that express a “core” RAG2 protein lacking amino acids 387–527 of its C-terminus (144). These mice develop T cell tumors with translocations involving TCR loci, a phenotype reminiscent of ATM deficient mice. This observation suggests that the full length RAG2 protein has activities in the repair of RAG DSBs, and the core RAG2 protein has defects in these activities.
Finally, RAG1 and RAG2, in vitro, have endonucleolytic activity that can open hairpin-sealed coding ends and process DNA ends (56, 57, 145). Although it is clear that the Artemis endonuclease is essential for efficient hairpin opening in vivo, it remains possible that RAG1 and RAG2 perform this function in Artemis-deficient cells. Moreover, it is possible that RAG flap endonuclease activity normally contributes to joint diversification through nucleotide loss (145).
Maintaining Genomic stability in Developing Lymphocytes
Normal individuals generate millions of RAG DSBs every hour in their developing B and T cells. The vast majority of these breaks are repaired efficiently, which both allows for the generation of functional antigen receptor genes and prevents RAG DSBs from being resolved aberrantly as potentially dangerous chromosomal translocations, deletions, or inversions. However, the presence of these chromosomal lesions in lymphoid tumors indicates that, at some level, RAG DSBs are aberrantly repaired.
Chromosomal lesions in lymphoid tumors
Chromosomal translocations are commonly found in lymphoid tumors (7, 8). Translocations are reciprocal and balanced in nature when the telomeric ends of the two participating chromosomes are exchanged. However, more complex lesions can be generated. For example, when two centromeric fragments or two telomeric fragments join, they produce dicentric or acentric chromosomes, respectively. Examples of translocations associated with lymphoid tumors that could result from aberrant RAG DSB repair include the bcl-2/IgH translocation found in follicular lymphomas and the IgH/cmyc translocations found in Burkitt’s Lymphoma (7, 8). These recurrent chromosomal lesions found in lymphoid tumors may be due to a mechanistic bias for their formation and/or to the selection of these particular lesions based on their transforming capability. Although both factors play a role, selection is likely a significant factor in the generation of tumors with recurrent translocations. In addition to translocations, DNA DSBs can be aberrantly resolved as chromosomal deletions and inversions. For example, deletion of the tumor suppressor MTS1 leads to T-ALL. Notably, the breakpoint of the deletion contains a heptamer sequence and N-nucleotides indicative of an aberrant V(D)J recombination event (146). Importantly, whereas translocations and inversions would generally require the aberrant joining of broken DNA ends from two DSBs, chromosomal deletions could form through aberrant resection and joining of broken DNA ends at a single DSB.
RAG activity in generating chromosomal lesions
The frequent occurrence of translocations involving antigen receptor loci in lymphoid tumors suggests that these lesions could result from the aberrant activity of RAG or the aberrant resolution of RAG-mediated DSBs. This is evidenced by the proximity of breakpoints to RSs and joint diversification (N and P nucleotides) indicative of RAG DSB processing (7, 8). Moreover, as discussed above, the lymphoid tumors with antigen receptor gene translocations that develop in mice deficient in ATM or in those deficient in p53 and NHEJ factors are dependent on RAG.
RAG cleavage most efficiently occurs in the setting of a synaptic complex between an appropriate (12/23 and B12/23) pair of RSs. RAG cleavage can also occur at sequences in non-antigen receptor loci that resemble RSs, referred to as cryptic RSs. Thus, reciprocal translocations could form through synapsis of an appropriate pair of RSs that lie in different antigen receptor genes (or between an RS and a cryptic RS pair or between a cryptic RS pair) on different chromosomes. For example, translocations involving the human TCRγ (Vγ) locus on chromosome 7 and the TCRδ(J δ) locus on chromosome 14 have been found in both normal and ATM-deficient lymphocytes (147, 148). Moreover, long-range synapsis between an appropriate pair of RSs in different antigen receptor genes (or between an RS and a cryptic RS pair or between a cryptic RS pair) that lie on the same chromosome would lead to large chromosomal deletions or inversions. Indeed, chromosomal inversions involving the human TCRγ (Vγ) and TCRβ (Jβ) loci, which both lie on chromosome 7 have also been found in both normal and ATM-deficient lymphocytes (147, 148). In addition, some forms of T cell lymphoma are linked to inversions occurring between the IgH and TCRα loci, which both lie on chromosome 14 (149).
All of the aforementioned lesions could form through synapsis of RSs and normal V(D)J recombination. However, analysis of a large number of unselected translocations involving the IgLκ locus and a retroviral recombination substrate in ATM-deficient abl pre-B cells revealed that many of these would violate the 12/23 rule (107). Thus, it is possible that aberrant rearrangements between distinct antigen receptor loci (or genomic regions containing cryptic RSs) may occur predominantly due to the joining of DNA ends generated at independent RAG DSBs rather than through bona fide RS synapsis. Finally, while the RAG endonuclease evolved from a transposase and truncated RAG proteins have transposase activity in vitro that could lead to translocations, full-length RAG proteins appear to catalyze these transposition events at a much lower level in vivo (150). These results suggest that the full-length RAG proteins have lost transposase activity, thereby preventing genomic instability.
Targets for aberrant RAG DSB repair
It is thought that most translocations are generated through the aberrant joining of two broken DNA ends from distinctly generated DNA DSBs (151). In lymphoid tumors with antigen receptor gene translocations, the DSB at the antigen receptor locus is frequently generated by RAG with the other DSB also being generated by RAG or by other means. In ATM-deficient pre-B cells, RAG DSBs generated at a retroviral recombination substrate frequently form translocations with the IgLκ locus, which undergoes rearrangement in these cells (107). Here the bias for the aberrant joining of two RAG DSBs may be due to the paucity of non-RAG-generated DNA DSBs in these G1-phase developing lymphocytes. Indeed, the introduction of additional genotoxic DSBs (by ionizing radiation) in these cells led to a substantial decrease in the aberrant joining of broken DNA ends generated by RAG. Translocations involving RAG DSBs in the irradiated cells targeted all chromosomes presumably at the sites of genotoxic DSBs introduced by irradiation (107).
The activation induced cytidine deaminase (AID) is expressed primarily in mature activated B cells and functions to initiate Ig class switch recombination, which involves the generation of DNA DSBs in switch regions flanking constant region exons in the IgH locus (9). DNA DSBs generated by RAG during V(D)J recombination in immature B cells and those generated by AID during CSR in mature B cells are usually temporally segregated (9). However, AID may be expressed in developing B cells in the bone marrow (152). Additionally, AID and RAG may both be expressed in transitional B cells (153). Thus, in these situations, translocations could occur between the physiologic DNA DSBs generated by AID and those generated by RAG. Moreover, the inappropriate transit of cells with un-repaired RAG DNA DSBs through the cell cycle could result in translocations between these persistent broken chromosomes and genotoxic DSBs or physiologic DSBs, for example, those generated by AID in activated mature B cells (154).
Notably, both RAG and AID can generate off-target DNA DSBs at locations outside of antigen receptor loci, and these DSBs can participate in the formation of aberrant joints. In this regard, RAG can cleave at cryptic RSs as discussed above. In addition, RAG can cut at sequences that do not resemble RSs but rather form unusual structures such as non-B DNA (155). RAG cleavage at a non-B DNA structure located at the major break point region of Bcl2 generates the IgH/Bcl2 translocations found in follicular B cell lymphomas (155). AID can have off-target effects that lead to the generation of DNA DSBs broadly throughout the genome (156–159). In addition, by altering sequences at CpG sites, AID may generate non-B DNA structures suitable for RAG cleavage (160).
Break localization as a determinant of aberrant RAG DSB repair
For DNA ends from two distinct DSBs to be joined, they must first be brought together in the nucleus. Two models have been proposed to explain how this may happen. Importantly, these two models are not mutually exclusive. In the “contact-first” model, translocations form between chromosomal regions located in close proximity to one another prior to the generation of DSBs due to a topological organization of chromosomes in the nucleus (161). On the other hand, the “breakage first” model proposes that broken DNA ends separated by great distances can migrate into close proximity and be joined (161). Broken DNA ends in mammalian cells may be relatively immobile as compared to the freely mobile broken DNA ends observed in S. cerevisiae (87, 162, 163). In studies of large numbers of translocations involving either RAG DSBs or Ig CSR DSBs, the translocation targets are distributed widely across the genome, suggesting that these DNA ends are mobile or the static topology of the genome varies significantly between individual cells (107, 164, 165). Interestingly, the majority of aberrantly resolved RAG DSBs or Ig CSR DSBs are joined to DNA ends from DSBs on the same chromosome, generating chromosomal deletions or inversions, even when these DSBs are separated by great distances (107, 164, 165). Thus, mechanisms may be in place that favor the joining of broken DNA ends from distinct DSBs on the same chromosome. Such mechanisms may explain how two independently generated broken DNA ends in different switch regions can be efficiently joined during Ig CSR (166).
Pathways used for the aberrant repair of RAG DSBs
RAG DSBs can be resolved aberrantly in wild type cells with intact NHEJ. However, these lesions also form in NHEJ-deficient cells, demonstrating that they can be generated through alternate end joining (AEJ) pathways. AEJ is probably best defined as joining that occurs in the absence of core NHEJ factors (XRCC4, DNA Ligase IV and Ku70/80) and may reflect the activity of one or more distinct DNA end joining pathways (6–8). The use of microhomologies during AEJ is more common than during NHEJ but is not an absolute requirement (6–8). Little is currently known about the components of AEJ pathway(s). However, since these joints form in DNA Ligase IV-deficient cells demonstrates, they must use a different mammalian ligase -- either DNA Ligase I or DNA Ligase III. Recent studies suggest that DNA Ligase III is a component of AEJ, but, in some circumstances, DNA Ligase I can also function in this regard (167, 168). In addition, MRN, CtIP and PARP1 have been implicated as functioning in the AEJ pathway (124, 169–174).
Analysis of breakpoint junctions of the translocations involving I-SceI-generated DSBs revealed that the frequency and extent of microhomology usage and deletions was not substantially different between wild-type and NHEJ-deficient cells (175). Thus, translocations may preferentially use AEJ even in wild-type cells where NHEJ is intact. Why would aberrant joining of two DNA ends favor utilization of AEJ over NHEJ in wild-type cells? One possibility is that persistent un-repaired DNA DSBs may be vulnerable to a mode of processing that prohibits efficient NHEJ. In this regard, resection of DNA ends may generate significant single strand overhangs that inhibit NHEJ (85). However, these ends may be joined efficiently by AEJ pathways, which can utilize microhomologies exposed by the resection process. Consistent with this notion, persistent RAG DSBs in H2AX-deficient cells are resected in a CtIP-dependent manner. Although many of these ends persist un-repaired, the joints that do form exhibit chromosomal deletions and an increased usage of microhomologies as compared to joints formed in wild-type cells (74). Moreover, the chromosomal translocations involving I-SceI DSBs are reduced in number in CtIP-deficient cells (172).
Mature B cells deficient in core NHEJ factors exhibit only a 50% reduction in Ig CSR (7). Moreover, most of the joints that form in the mutant cells have microhomologies, suggesting that they are products of AEJ pathways (7). Thus, Ig CSR proceeds efficiently by AEJ in NHEJ-deficient cells. In contrast, RAG DSB repair is almost completely abolished in NHEJ-deficient lymphocytes. Why is it that RAG DSBs are not efficiently joined by AEJ? In this regard, the RAG-2 protein may function to prevent access of RAG DSBs to AEJ pathways, although how this occurs is not currently known (176).
Perspectives on maintaining genomic stability in developing lymphocytes
The major function of V(D)J recombination is to assemble the genes encoding lymphocyte antigen receptors. However, additional pathways must be in place to prevent rare un-repaired RAG DSBs from being aberrantly resolved and disrupting genomic stability. In this regard, the resolution of RAG DSBs as potentially dangerous chromosomal lesions requires failures in multiple components of the DNA damage response. First, RAG DSBs must persist or escape efficient repair. Second, the un-repaired RAG DSBs must avoid the activation of p53 and cell death pathways. Third, un-repaired RAG breaks must dissociate from their post-cleavage complex and associate with other cellular DSBs to which they will ultimately join. Fourth, un-repaired DSBs must not be processed in a way that prevents joining by NHEJ or AEJ.
As tumor formation requires the inactivation of multiple component pathways of the DNA damage response, only combined deficiencies in factors affecting single component pathways would lead to the generation of lymphoid tumors. In this regard, mice with combined deficiencies of p53 and NHEJ factors exhibit a high frequency of lymphoid tumors with translocations involving antigen receptor genes, whereas mice with singular deficiencies of p53 or NHEJ factors do not. Alternatively, the inactivation of a single protein that regulates multiple component pathways, such as ATM, could lead to lymphoid tumors. In the case of ATM-deficiency, these pathways include maintaining coding ends in post-cleavage complexes and activating p53 in response to un-repaired RAG DSBs.
The requirement to generate complete antigen receptor genes while maintaining genomic stability may be achieved primarily through pathways that promote the efficient NHEJ-mediated repair of RAG DSBs. Given the sheer number of RAG DSBs generated during lymphocyte development, it is likely that not all of these DSBs are repaired with the same efficiency. Even a small fraction of these breaks that persist unrepaired could have a significant impact. Indeed, while conditional deletion of p53 at the pro-B cell stage in mice does not perturb B cell development, these mice readily develop B cell lymphomas with translocations involving the IgH locus (177). These data suggest that, even in cells with proficient NHEJ pathways, rare RAG DSBs may persist un-repaired and may be capable of aberrant repair if these DSBs are unable to activate cell death pathways.
Moreover, there may be additional pathways that do not have a significant role in NHEJ; rather they have evolved primarily to deal with rare persistent DSBs, ensuring that such recalcitrant DSBs remain suitable substrates for NHEJ and, if not appropriately joined, result in cell death rather than aberrant repair (Fig. 3). In this regard, H2AX-deficient mice exhibit near normal lymphocyte development, and V(D)J recombination proceeds with normal kinetics in H2AX-deficient lymphocytes (71–73). However, H2AX-deficient mice are predisposed to lymphoid tumors that can have translocations involving antigen receptor loci (71, 98, 178). Together, these data suggest that H2AX may be required to prevent rare un-repaired RAG DSBs from accessing aberrant repair pathways.
Genetic program activated by RAG DSBs
Once activated by DSBs, ATM can regulate gene expression by modulating transcription factor activity. Moreover, ATM also has post-transcriptional regulatory activity altering miRNA processing (179). ATM directly phosphorylates and regulates the activity of the transcription factor p53 (22). In response to genotoxic DNA DSBs, ATM-mediated phosphorylation stabilizes the p53 transcription factor, enhancing its transcriptional activity (28). p53 regulates many genes, such as Bax, PUMA and the Fas receptor, that promote cell death in response to DNA damage (28). Persistent un-repaired RAG DSBs in developing lymphocytes activate ATM and p53 (106, 180, 181). However, the unopposed activation of p53 by RAG DSBs in wild-type developing lymphocytes could lead to their premature death rather than repair of RAG DSBs and successful completion of antigen receptor gene assembly. Conceivably, the activation of p53 could be delayed and only occur in the presence of persistent un-repaired RAG DSBs. Alternatively, RAG DSBs may also activate pro-survival pathways that, when integrated with pro-apoptotic pathways, ultimately determine cell fate. Pro-survival pathways may predominate early after DSB generation allowing the cell time to repair the DSB, whereas pro-apoptotic pathways may predominate at later time points, eliminating cells with persistent un-repaired DSBs.
In addition to p53, RAG DSBs activate the classical (p50/RelA) NFκB pathway, which, in other settings, is known to enhance the expression of pro-survival genes (182). Activation of the classical NFκB pathway by RAG DSBs depends on ATM and NEMO (Ikkγ) similar to its activation by genotoxic DSBs (182, 183). ATM directly phosphorylates NEMO in the nucleus, which results in its translocation to the cytoplasm and activation of the classical NFκB pathway (183).
In pre-B cells, RAG DSBs activate a broad genetic program involving over 250 genes that rely on transcriptional pathways in addition to p53 and NFκB (182). Approximately half of these break-dependent gene expression changes require ATM, suggesting that other kinases, including perhaps DNA-PKcs, are also capable of regulating gene expression in response to RAG DSBs (182). As expected, the cohort of genes regulated by RAG DSBs comprise those that function in canonical DNA damage responses such as cell survival and cell death. For example, Pim2 and Bcl3, which are both up-regulated by RAG DSBs, have known activities in promoting cell survival (182, 184, 185).
Remarkably, many of the genes regulated by ATM in response to RAG DSBs are not directly involved in canonical DNA damage responses (182). Rather, these genes have known activities in lymphocyte development or function. For example, the expression of CD62L, CD69 and SWAP70 are up-regulated by RAG DSBs in pre-B cells; these proteins all have activities in lymphocyte migration and homing (see refs in182). In this regard, the repositioning of pre-B cells actively assembling IgL chain genes to specific bone marrow niches may be important for the appropriate stepwise maturation of these lymphocyte precursors (186).
Several of the genes that are responsive to RAG DSBs in pre-B cells are also regulated by genotoxic DSBs (182). This finding suggests that at least some portion of this genetic program is not specific to RAG DSBs. Rather, developing G1-phase lymphocytes may be poised to activate a specific genetic program when any DSB is sensed. As RAG is the most common source of DSBs in these G1-phase cells, their generation and the resulting activation of DNA damage responses could serve as a cellular sentinel for the initiation of antigen receptor gene assembly. Importantly, these gene expression changes are activated by transient RAG DSBs, which are normally repaired with rapid kinetics in wild-type cells (182). RAG-dependent gene expression changes were observed in both developing B and T cells (182). Moreover, recent studies have shown that DNA DSBs generated in mature B cells during Ig CSR also activate ATM and a genetic program important for plasma cell differentiation (187). Together, these findings suggest that signals emanating from DNA DSB intermediates during physiologic processes may regulate cell-type specific functions (182, 187, 188). Moreover, the inappropriate activation of these genetic programs by genotoxic DSBs (e.g., those generated by ionizing radiation or chemotherapeutic agents) could disrupt normal cellular development and/or function.
The notion that RAG DSBs activate a genetic program important for developing lymphocytes may seem at odds with the observation that TCR and BCR transgenes drive T and B cell development, respectively, in RAG-deficient mice. However, the genetic program promoted by RAG DSBs may be important mainly for developing lymphocytes actively rearranging antigen receptor genes. Forcing the expression of TCR and BCR transgenes in RAG-deficient mice would bypass these requirements just as it bypasses the important requirement for RAG gene expression.
Finally, as discussed above, antigen receptor assembly is regulated in several different contexts (9–11). In this regard, it is possible that the generation of a RAG DSB provides signals through the DNA damage response that regulates subsequent rearrangement steps. This could, for example, prevent the generation of additional RAG DSBs until the initial DSB has been resolved and the rearrangement product tested for functionality. This level of regulation may be important for allelic exclusion, for example, by enabling interallelic ordering of V(D)J recombination (9–11). Indeed, recent studies have suggested that the activation of ATM via RAG DSBs at the IgH locus prevents the generation of a RAG DSB at the other IgH allele, possibly by repositioning it to peri-centric heterochromatin (189).
Conclusions and future directions
All developing lymphocytes must generate and repair several DNA DSBs in order to complete antigen receptor gene assembly. These physiologic DNA DSBs activate DNA damage responses just as those breaks initiated by genotoxic agents such as ionizing radiation or chemotherapeutic agents. The RAG DSB-initiated DNA damage response promotes the efficient repair of these breaks, prevents their aberrant repair and activates cell death pathways in the event the break cannot be repaired. Many of the proteins and processes required for the NHEJ-mediated repair of RAG DSBs have been characterized. However, the findings reported in recent studies emphasize that there are likely additional important undefined pathways of the DNA damage response in developing lymphocytes that have remained ill-defined due to functional redundancies. Identifying these pathways and defining their function will be an important priority. Moreover, whether joining pathways distinct from NHEJ are primarily responsible for the aberrant resolution of RAG DSBs and if so, how these pathways are normally held in check will need to be determined. Finally, the full breadth of processes regulated by signals emanating from RAG DSBs that directly or indirectly regulate antigen receptor gene assembly and other broader developmental processes remains to be determined.
Acknowledgments
We thank Drs. Andre Nussenzweig, Eugene Oltz, Craig Bassing and Jacqueline Payton for critical review of the manuscript. This work is supported by the National Institutes of Health grants AI074953 (B.P.S.), AI47829 (B.P.S.) and CA136470 (B.P.S.).
References
- 1.Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 2.Oettinger MA. V(D)J recombination: on the cutting edge. Curr Opin Cell Biol. 1999;11:325–9. doi: 10.1016/S0955-0674(99)80044-1. [DOI] [PubMed] [Google Scholar]
- 3.Gellert M. V(D)J recombination: rag proteins, repair factors, and regulation. Annu Rev Biochem. 2002;71:101–32. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 4.Desiderio S, Lin WC, Li Z. The cell cycle and V(D)J recombination. Curr Top Microbiol Immunol. 1996;217:45–59. doi: 10.1007/978-3-642-50140-1_4. [DOI] [PubMed] [Google Scholar]
- 5.Rooney S, Chaudhuri J, Alt FW. The role of the non-homologous end-joining pathway in lymphocyte development. Immunol Rev. 2004;200:115–31. doi: 10.1111/j.0105-2896.2004.00165.x. [DOI] [PubMed] [Google Scholar]
- 6.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gostissa M, Alt FW, Chiarle R. Mechanisms that promote and suppress chromosomal translocations in lymphocytes. Annu Rev Immunol. 2011;29:319–50. doi: 10.1146/annurev-immunol-031210-101329. [DOI] [PubMed] [Google Scholar]
- 8.Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell. 2010;141:27–38. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dudley DD, Chaudhuri J, Bassing CH, Alt FW. Mechanism and control of V(D)J recombination versus class switch recombination: similarities and differences. Adv Immunol. 2005;86:43–112. doi: 10.1016/S0065-2776(04)86002-4. [DOI] [PubMed] [Google Scholar]
- 10.Cobb RM, Oestreich KJ, Osipovich OA, Oltz EM. Accessibility control of V(D)J recombination. Adv Immunol. 2006;91:45–109. doi: 10.1016/S0065-2776(06)91002-5. [DOI] [PubMed] [Google Scholar]
- 11.Brady BL, Steinel NC, Bassing CH. Antigen receptor allelic exclusion: an update and reappraisal. J Immunol. 2010;185:3801–8. doi: 10.4049/jimmunol.1001158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat Rev Immunol. 2011;11:251–63. doi: 10.1038/nri2941. [DOI] [PubMed] [Google Scholar]
- 13.Roth DB, Nakajima PB, Menetski JP, Bosma MJ, Gellert M. V(D)J recombination in mouse thymocytes: double-strand breaks near T cell receptor delta rearrangement signals. Cell. 1992;69:41–53. doi: 10.1016/0092-8674(92)90117-u. [DOI] [PubMed] [Google Scholar]
- 14.Ramsden DA, Gellert M. Formation and resolution of double-strand break intermediates in V(D)J rearrangement. Genes Dev. 1995;9:2409–20. doi: 10.1101/gad.9.19.2409. [DOI] [PubMed] [Google Scholar]
- 15.Bredemeyer AL, Sharma GG, Huang CY, Helmink BA, Walker LM, Khor KC, Nuskey B, Sullivan KE, Pandita TK, Bassing CH, Sleckman BP. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 2006;442:466–70. doi: 10.1038/nature04866. [DOI] [PubMed] [Google Scholar]
- 16.Gapud EJ, Dorsett Y, Yin B, Callen E, Bredemeyer A, Mahowald GK, Omi KQ, Walker LM, Bednarski JJ, McKinnon PJ, Bassing CH, Nussenzweig A, Sleckman BP. Ataxia telangiectasia mutated (Atm) and DNA-PKcs kinases have overlapping activities during chromosomal signal joint formation. Proc Natl Acad Sci U S A. 2011;108:2022–7. doi: 10.1073/pnas.1013295108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu H, Shimazaki N, Raval P, Gu J, Watanabe G, Schwarz K, Swanson PC, Lieber MR. A biochemically defined system for coding joint formation in V(D)J recombination. Mol Cell. 2008;31:485–97. doi: 10.1016/j.molcel.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hesse JE, Lieber MR, Gellert M, Mizuuchi K. Extrachromosomal DNA substrates in pre-B cells undergo inversion or deletion at immunoglobulin V-(D)-J joining signals. Cell. 1987;49:775–83. doi: 10.1016/0092-8674(87)90615-5. [DOI] [PubMed] [Google Scholar]
- 19.Sadofsky MJ, Hesse JE, McBlane JF, Gellert M. Expression and V(D)J recombination activity of mutated RAG-1 proteins. Nucleic Acids Res. 1993;21:5644–50. doi: 10.1093/nar/21.24.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen YY, Wang LC, Huang MS, Rosenberg N. An active v-abl protein tyrosine kinase blocks immunoglobulin light-chain gene rearrangement. Genes Dev. 1994;8:688–97. doi: 10.1101/gad.8.6.688. [DOI] [PubMed] [Google Scholar]
- 21.Muljo SA, Schlissel MS. A small molecule Abl kinase inhibitor induces differentiation of Abelson virus-transformed pre-B cell lines. Nat Immunol. 2003;4:31–7. doi: 10.1038/ni870. [DOI] [PubMed] [Google Scholar]
- 22.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–68. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 24.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–6. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 25.Callen E, Jankovic M, Wong N, Zha S, Chen HT, Difilippantonio S, Di Virgilio M, Heidkamp G, Alt FW, Nussenzweig A, Nussenzweig M. Essential role for DNA-PKcs in DNA double-strand break repair and apoptosis in ATM-deficient lymphocytes. Mol Cell. 2009;34:285–97. doi: 10.1016/j.molcel.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomimatsu N, Mukherjee B, Burma S. Distinct roles of ATR and DNA-PKcs in triggering DNA damage responses in ATM-deficient cells. EMBO Rep. 2009;10:629–35. doi: 10.1038/embor.2009.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartek J, Falck J, Lukas J. CHK2 kinase--a busy messenger. Nat Rev Mol Cell Biol. 2001;2:877–86. doi: 10.1038/35103059. [DOI] [PubMed] [Google Scholar]
- 28.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–31. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 29.Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–42. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 30.Meek K, Dang V, Lees-Miller SP. DNA-PK: the means to justify the ends? Adv Immunol. 2008;99:33–58. doi: 10.1016/S0065-2776(08)00602-0. [DOI] [PubMed] [Google Scholar]
- 31.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–14. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 32.Feldmann E, Schmiemann V, Goedecke W, Reichenberger S, Pfeiffer P. DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res. 2000;28:2585–96. doi: 10.1093/nar/28.13.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabotyanski EB, Gomelsky L, Han JO, Stamato TD, Roth DB. Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res. 1998;26:5333–42. doi: 10.1093/nar/26.23.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weterings E, Chen DJ. DNA-dependent protein kinase in nonhomologous end joining: a lock with multiple keys? J Cell Biol. 2007;179:183–6. doi: 10.1083/jcb.200705106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeFazio LG, Stansel RM, Griffith JD, Chu G. Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J. 2002;21:3192–200. doi: 10.1093/emboj/cdf299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merkle D, Douglas P, Moorhead GB, Leonenko Z, Yu Y, Cramb D, Bazett-Jones DP, Lees-Miller SP. The DNA-dependent protein kinase interacts with DNA to form a protein-DNA complex that is disrupted by phosphorylation. Biochemistry. 2002;41:12706–14. doi: 10.1021/bi0263558. [DOI] [PubMed] [Google Scholar]
- 37.Zhang S, Yajima H, Huynh H, Zheng J, Callen E, Chen HT, Wong N, Bunting S, Lin YF, Li M, Lee KJ, Story M, Gapud E, Sleckman BP, Nussenzweig A, Zhang CC, Chen DJ, Chen BP. Congenital bone marrow failure in DNA-PKcs mutant mice associated with deficiencies in DNA repair. J Cell Biol. 2011;193:295–305. doi: 10.1083/jcb.201009074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Burg M, Ijspeert H, Verkaik NS, Turul T, Wiegant WW, Morotomi-Yano K, Mari PO, Tezcan I, Chen DJ, Zdzienicka MZ, van Dongen JJ, van Gent DC. A DNA-PKcs mutation in a radiosensitive T-B- SCID patient inhibits Artemis activation and nonhomologous end-joining. J Clin Invest. 2009;119:91–8. doi: 10.1172/JCI37141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gapud EJ, Sleckman BP. Unique and redundant functions of ATM and DNA-PKcs during V(D)J recombination. Cell Cycle. 2011;10:1928–35. doi: 10.4161/cc.10.12.16011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zha S, Jiang W, Fujiwara Y, Patel H, Goff PH, Brush JW, Dubois RL, Alt FW. Ataxia telangiectasia-mutated protein and DNA-dependent protein kinase have complementary V(D)J recombination functions. Proc Natl Acad Sci U S A. 2011;108:2028–33. doi: 10.1073/pnas.1019293108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin JM, Landree MA, Roth DB. V(D)J recombination catalyzed by mutant RAG proteins lacking consensus DNA-PK phosphorylation sites. Mol Immunol. 1999;36:1263–9. doi: 10.1016/s0161-5890(99)00099-1. [DOI] [PubMed] [Google Scholar]
- 42.Hah YS, Lee JH, Kim DR. DNA-dependent protein kinase mediates V(D)J recombination via RAG2 phosphorylation. J Biochem Mol Biol. 2007;40:432–8. doi: 10.5483/bmbrep.2007.40.3.432. [DOI] [PubMed] [Google Scholar]
- 43.Gapud EJ, Lee BS, Mahowald GK, Bassing CH, Sleckman BP. Repair of Chromosomal RAG-Mediated DNA Breaks by Mutant RAG Proteins Lacking Phosphatidylinositol 3-Like Kinase Consensus Phosphorylation Sites. J Immunol. 2011;187:1826–34. doi: 10.4049/jimmunol.1101388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones JM, Gellert M. Intermediates in V(D)J recombination: a stable RAG1/2 complex sequesters cleaved RSS ends. Proc Natl Acad Sci U S A. 2001;98:12926–31. doi: 10.1073/pnas.221471198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4:712–20. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 46.Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–13. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 47.Moshous D, Callebaut I, de Chasseval R, Corneo B, Cavazzana-Calvo M, Le Deist F, Tezcan I, Sanal O, Bertrand Y, Philippe N, Fischer A, de Villartay JP. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–86. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 48.Rooney S, Alt FW, Lombard D, Whitlow S, Eckersdorff M, Fleming J, Fugmann S, Ferguson DO, Schatz DG, Sekiguchi J. Defective DNA repair and increased genomic instability in Artemis-deficient murine cells. J Exp Med. 2003;197:553–65. doi: 10.1084/jem.20021891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rooney S, Sekiguchi J, Zhu C, Cheng HL, Manis J, Whitlow S, DeVido J, Foy D, Chaudhuri J, Lombard D, Alt FW. Leaky Scid phenotype associated with defective V(D)J coding end processing in Artemis-deficient mice. Mol Cell. 2002;10:1379–90. doi: 10.1016/s1097-2765(02)00755-4. [DOI] [PubMed] [Google Scholar]
- 50.Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, Parker AR, Jackson SP, Gennery A, Jeggo PA, Lobrich M. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol Cell. 2004;16:715–24. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 51.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–94. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 52.Goodarzi AA, Yu Y, Riballo E, Douglas P, Walker SA, Ye R, Harer C, Marchetti C, Morrice N, Jeggo PA, Lees-Miller SP. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. Embo J. 2006;25:3880–9. doi: 10.1038/sj.emboj.7601255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bosma GC, Fried M, Custer RP, Carroll A, Gibson DM, Bosma MJ. Evidence of functional lymphocytes in some (leaky) scid mice. J Exp Med. 1988;167:1016–33. doi: 10.1084/jem.167.3.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schuler W, Ruetsch NR, Amsler M, Bosma MJ. Coding joint formation of endogenous T cell receptor genes in lymphoid cells from scid mice: unusual P-nucleotide additions in VJ-coding joints. Eur J Immunol. 1991;21:589–96. doi: 10.1002/eji.1830210309. [DOI] [PubMed] [Google Scholar]
- 55.Kienker LJ, Kuziel WA, Tucker PW. T cell receptor gamma and delta gene junctional sequences in SCID mice: excessive P nucleotide insertion. J Exp Med. 1991;174:769–73. doi: 10.1084/jem.174.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Besmer E, Mansilla-Soto J, Cassard S, Sawchuk DJ, Brown G, Sadofsky M, Lewis SM, Nussenzweig MC, Cortes P. Hairpin coding end opening is mediated by RAG1 and RAG2 proteins. Mol Cell. 1998;2:817–28. doi: 10.1016/s1097-2765(00)80296-8. [DOI] [PubMed] [Google Scholar]
- 57.Shockett PE, Schatz DG. DNA hairpin opening mediated by the RAG1 and RAG2 proteins. Mol Cell Biol. 1999;19:4159–66. doi: 10.1128/mcb.19.6.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Touvrey C, Couedel C, Soulas P, Couderc R, Jasin M, de Villartay JP, Marche PN, Jouvin-Marche E, Candeias SM. Distinct effects of DNA-PKcs and Artemis inactivation on signal joint formation in vivo. Mol Immunol. 2008;45:3383–91. doi: 10.1016/j.molimm.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 59.Li Y, Chirgadze DY, Bolanos-Garcia VM, Sibanda BL, Davies OR, Ahnesorg P, Jackson SP, Blundell TL. Crystal structure of human XLF/Cernunnos reveals unexpected differences from XRCC4 with implications for NHEJ. EMBO J. 2008;27:290–300. doi: 10.1038/sj.emboj.7601942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andres SN, Modesti M, Tsai CJ, Chu G, Junop MS. Crystal structure of human XLF: a twist in nonhomologous DNA end-joining. Mol Cell. 2007;28:1093–101. doi: 10.1016/j.molcel.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 61.Gu J, Lu H, Tsai AG, Schwarz K, Lieber MR. Single-stranded DNA ligation and XLF-stimulated incompatible DNA end ligation by the XRCC4-DNA ligase IV complex: influence of terminal DNA sequence. Nucleic Acids Res. 2007;35:5755–62. doi: 10.1093/nar/gkm579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsai CJ, Kim SA, Chu G. Cernunnos/XLF promotes the ligation of mismatched and noncohesive DNA ends. Proc Natl Acad Sci U S A. 2007;104:7851–6. doi: 10.1073/pnas.0702620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche MC, Sanal O, Plebani A, Stephan JL, Hufnagel M, le Deist F, Fischer A, Durandy A, de Villartay JP, Revy P. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–99. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 64.Dai Y, Kysela B, Hanakahi LA, Manolis K, Riballo E, Stumm M, Harville TO, West SC, Oettinger MA, Jeggo PA. Nonhomologous end joining and V(D)J recombination require an additional factor. Proc Natl Acad Sci U S A. 2003;100:2462–7. doi: 10.1073/pnas.0437964100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li G, Alt FW, Cheng HL, Brush JW, Goff PH, Murphy MM, Franco S, Zhang Y, Zha S. Lymphocyte-specific compensation for XLF/cernunnos end-joining functions in V(D)J recombination. Mol Cell. 2008;31:631–40. doi: 10.1016/j.molcel.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zha S, Alt FW, Cheng HL, Brush JW, Li G. Defective DNA repair and increased genomic instability in Cernunnos-XLF-deficient murine ES cells. Proc Natl Acad Sci U S A. 2007;104:4518–23. doi: 10.1073/pnas.0611734104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zha S, Guo C, Boboila C, Oksenych V, Cheng HL, Zhang Y, Wesemann DR, Yuen G, Patel H, Goff PH, Dubois RL, Alt FW. ATM damage response and XLF repair factor are functionally redundant in joining DNA breaks. Nature. 2011;469:250–4. doi: 10.1038/nature09604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang CY, Sharma GG, Walker LM, Bassing CH, Pandita TK, Sleckman BP. Defects in coding joint formation in vivo in developing ATM-deficient B and T lymphocytes. J Exp Med. 2007;204:1371–81. doi: 10.1084/jem.20061460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matei IR, Gladdy RA, Nutter LM, Canty A, Guidos CJ, Danska JS. ATM deficiency disrupts TCR{alpha} locus integrity and the maturation of CD4+CD8+ thymocytes. Blood. 2006 doi: 10.1182/blood-2006-05-020917. [DOI] [PubMed] [Google Scholar]
- 70.Vacchio MS, Olaru A, Livak F, Hodes RJ. ATM deficiency impairs thymocyte maturation because of defective resolution of T cell receptor {alpha} locus coding end breaks. Proc Natl Acad Sci U S A. 2007;104:6323–8. doi: 10.1073/pnas.0611222104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ, Redon C, Pilch DR, Olaru A, Eckhaus M, Camerini-Otero RD, Tessarollo L, Livak F, Manova K, Bonner WM, Nussenzweig MC, Nussenzweig A. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–7. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bassing CH, Chua KF, Sekiguchi J, Suh H, Whitlow SR, Fleming JC, Monroe BC, Ciccone DN, Yan C, Vlasakova K, Livingston DM, Ferguson DO, Scully R, Alt FW. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc Natl Acad Sci U S A. 2002;99:8173–8. doi: 10.1073/pnas.122228699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yin B, Savic V, Juntilla MM, Bredemeyer AL, Yang-Iott KS, Helmink BA, Koretzky GA, Sleckman BP, Bassing CH. Histone H2AX stabilizes broken DNA strands to suppress chromosome breaks and translocations during V(D)J recombination. J Exp Med. 2009;206:2625–39. doi: 10.1084/jem.20091320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Helmink BA, Tubbs AT, Dorsett Y, Bednarski JJ, Walker LM, Feng Z, Sharma GG, McKinnon PJ, Zhang J, Bassing CH, Sleckman BP. H2AX prevents CtIP-mediated DNA end resection and aberrant repair in G1-phase lymphocytes. Nature. 2011;469:245–9. doi: 10.1038/nature09585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zha S, Sekiguchi J, Brush JW, Bassing CH, Alt FW. Complementary functions of ATM and H2AX in development and suppression of genomic instability. Proc Natl Acad Sci U S A. 2008;105:9302–6. doi: 10.1073/pnas.0803520105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ramadan K, Shevelev I, Hubscher U. The DNA-polymerase-X family: controllers of DNA quality? Nat Rev Mol Cell Biol. 2004;5:1038–43. doi: 10.1038/nrm1530. [DOI] [PubMed] [Google Scholar]
- 77.Motea EA, Berdis AJ. Terminal deoxynucleotidyl transferase: the story of a misguided DNA polymerase. Biochim Biophys Acta. 2009;1804:1151–66. doi: 10.1016/j.bbapap.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Komori T, Okada A, Stewart V, Alt FW. Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science. 1993;261:1171–5. doi: 10.1126/science.8356451. [DOI] [PubMed] [Google Scholar]
- 79.Gilfillan S, Dierich A, Lemeur M, Benoist C, Mathis D. Mice lacking TdT: mature animals with an immature lymphocyte repertoire. Science. 1993;261:1175–8. doi: 10.1126/science.8356452. [DOI] [PubMed] [Google Scholar]
- 80.Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A. H2AX: the histone guardian of the genome. DNA Repair (Amst) 2004;3:959–67. doi: 10.1016/j.dnarep.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 81.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–68. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 82.Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003;5:675–9. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 83.Chen HT, Bhandoola A, Difilippantonio MJ, Zhu J, Brown MJ, Tai X, Rogakou EP, Brotz TM, Bonner WM, Ried T, Nussenzweig A. Response to RAG-mediated VDJ cleavage by NBS1 and gamma-H2AX. Science. 2000;290:1962–5. doi: 10.1126/science.290.5498.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Savic V, Yin B, Maas NL, Bredemeyer AL, Carpenter AC, Helmink BA, Yang-Iott KS, Sleckman BP, Bassing CH. Formation of dynamic gamma-H2AX domains along broken DNA strands is distinctly regulated by ATM and MDC1 and dependent upon H2AX densities in chromatin. Mol Cell. 2009;34:298–310. doi: 10.1016/j.molcel.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nat Struct Mol Biol. 2010;17:11–6. doi: 10.1038/nsmb.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 87.Nelms BE, Maser RS, MacKay JF, Lagally MG, Petrini JH. In situ visualization of DNA double-strand break repair in human fibroblasts. Science. 1998;280:590–2. doi: 10.1126/science.280.5363.590. [DOI] [PubMed] [Google Scholar]
- 88.Stracker TH, Theunissen JW, Morales M, Petrini JH. The Mre11 complex and the metabolism of chromosome breaks: the importance of communicating and holding things together. DNA Repair (Amst) 2004;3:845–54. doi: 10.1016/j.dnarep.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 89.D’Amours D, Jackson SP. The Mre11 complex: at the crossroads of dna repair and checkpoint signalling. Nat Rev Mol Cell Biol. 2002;3:317–27. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- 90.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–4. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 91.Lee JH, Paull TT. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–6. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- 92.Pellegrini M, Celeste A, Difilippantonio S, Guo R, Wang W, Feigenbaum L, Nussenzweig A. Autophosphorylation at serine 1987 is dispensable for murine Atm activation in vivo. Nature. 2006;443:222–5. doi: 10.1038/nature05112. [DOI] [PubMed] [Google Scholar]
- 93.Lavin MF, Shiloh Y. The genetic defect in ataxia-telangiectasia. Annu Rev Immunol. 1997;15:177–202. doi: 10.1146/annurev.immunol.15.1.177. [DOI] [PubMed] [Google Scholar]
- 94.Matei IR, Guidos CJ, Danska JS. ATM-dependent DNA damage surveillance in T-cell development and leukemogenesis: the DSB connection. Immunol Rev. 2006;209:142–58. doi: 10.1111/j.0105-2896.2006.00361.x. [DOI] [PubMed] [Google Scholar]
- 95.Liyanage M, Weaver Z, Barlow C, Coleman A, Pankratz DG, Anderson S, Wynshaw-Boris A, Ried T. Abnormal rearrangement within the alpha/delta T-cell receptor locus in lymphomas from Atm-deficient mice. Blood. 2000;96:1940–6. [PubMed] [Google Scholar]
- 96.Liao MJ, Van Dyke T. Critical role for Atm in suppressing V(D)J recombination-driven thymic lymphoma. Genes Dev. 1999;13:1246–50. doi: 10.1101/gad.13.10.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Petiniot LK, Weaver Z, Barlow C, Shen R, Eckhaus M, Steinberg SM, Ried T, Wynshaw-Boris A, Hodes RJ. Recombinase-activating gene (RAG) 2-mediated V(D)J recombination is not essential for tumorigenesis in Atm-deficient mice. Proc Natl Acad Sci U S A. 2000;97:6664–9. doi: 10.1073/pnas.97.12.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bassing CH, Suh H, Ferguson DO, Chua KF, Manis J, Eckersdorff M, Gleason M, Bronson R, Lee C, Alt FW. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003;114:359–70. doi: 10.1016/s0092-8674(03)00566-x. [DOI] [PubMed] [Google Scholar]
- 99.Hirao A, Cheung A, Duncan G, Girard PM, Elia AJ, Wakeham A, Okada H, Sarkissian T, Wong JA, Sakai T, De Stanchina E, Bristow RG, Suda T, Lowe SW, Jeggo PA, Elledge SJ, Mak TW. Chk2 is a tumor suppressor that regulates apoptosis in both an ataxia telangiectasia mutated (ATM)-dependent and an ATM-independent manner. Mol Cell Biol. 2002;22:6521–32. doi: 10.1128/MCB.22.18.6521-6532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Frank KM, Sharpless NE, Gao Y, Sekiguchi JM, Ferguson DO, Zhu C, Manis JP, Horner J, DePinho RA, Alt FW. DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol Cell. 2000;5:993–1002. doi: 10.1016/s1097-2765(00)80264-6. [DOI] [PubMed] [Google Scholar]
- 101.Difilippantonio MJ, Zhu J, Chen HT, Meffre E, Nussenzweig MC, Max EE, Ried T, Nussenzweig A. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature. 2000;404:510–4. doi: 10.1038/35006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gao Y, Ferguson DO, Xie W, Manis JP, Sekiguchi J, Frank KM, Chaudhuri J, Horner J, DePinho RA, Alt FW. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature. 2000;404:897–900. doi: 10.1038/35009138. [DOI] [PubMed] [Google Scholar]
- 103.Zhu C, Mills KD, Ferguson DO, Lee C, Manis J, Fleming J, Gao Y, Morton CC, Alt FW. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–21. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
- 104.Difilippantonio MJ, Petersen S, Chen HT, Johnson R, Jasin M, Kanaar R, Ried T, Nussenzweig A. Evidence for replicative repair of DNA double-strand breaks leading to oncogenic translocation and gene amplification. J Exp Med. 2002;196:469–80. doi: 10.1084/jem.20020851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gladdy RA, Taylor MD, Williams CJ, Grandal I, Karaskova J, Squire JA, Rutka JT, Guidos CJ, Danska JS. The RAG-1/2 endonuclease causes genomic instability and controls CNS complications of lymphoblastic leukemia in p53/Prkdc-deficient mice. Cancer Cell. 2003;3:37–50. doi: 10.1016/s1535-6108(02)00236-2. [DOI] [PubMed] [Google Scholar]
- 106.Perkins EJ, Nair A, Cowley DO, Van Dyke T, Chang Y, Ramsden DA. Sensing of intermediates in V(D)J recombination by ATM. Genes Dev. 2002;16:159–64. doi: 10.1101/gad.956902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mahowald GK, Baron JM, Mahowald MA, Kulkarni S, Bredemeyer AL, Bassing CH, Sleckman BP. Aberrantly resolved RAG-mediated DNA breaks in Atm-deficient lymphocytes target chromosomal breakpoints in cis. Proc Natl Acad Sci U S A. 2009;106:18339–44. doi: 10.1073/pnas.0902545106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y. Requirement of the MRN complex for ATM activation by DNA damage. Embo J. 2003;22:5612–21. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, Moiani D, Carney JP, Russell P, Tainer JA. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, Classen S, Glover JN, Iwasaki H, Russell P, Tainer JA. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chen L, Nievera CJ, Lee AY, Wu X. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J Biol Chem. 2008;283:7713–20. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- 112.Lim DS, Kim ST, Xu B, Maser RS, Lin J, Petrini JH, Kastan MB. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature. 2000;404:613–7. doi: 10.1038/35007091. [DOI] [PubMed] [Google Scholar]
- 113.Wu X, Ranganathan V, Weisman DS, Heine WF, Ciccone DN, O’Neill TB, Crick KE, Pierce KA, Lane WS, Rathbun G, Livingston DM, Weaver DT. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature. 2000;405:477–82. doi: 10.1038/35013089. [DOI] [PubMed] [Google Scholar]
- 114.Gatei M, Young D, Cerosaletti KM, Desai-Mehta A, Spring K, Kozlov S, Lavin MF, Gatti RA, Concannon P, Khanna K. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat Genet. 2000;25:115–9. doi: 10.1038/75508. [DOI] [PubMed] [Google Scholar]
- 115.Xiao Y, Weaver DT. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 1997;25:2985–91. doi: 10.1093/nar/25.15.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Luo G, Yao MS, Bender CF, Mills M, Bladl AR, Bradley A, Petrini JH. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc Natl Acad Sci U S A. 1999;96:7376–81. doi: 10.1073/pnas.96.13.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhu J, Petersen S, Tessarollo L, Nussenzweig A. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr Biol. 2001;11:105–9. doi: 10.1016/s0960-9822(01)00019-7. [DOI] [PubMed] [Google Scholar]
- 118.Difilippantonio S, Celeste A, Fernandez-Capetillo O, Chen HT, Reina San Martin B, Van Laethem F, Yang YP, Petukhova GV, Eckhaus M, Feigenbaum L, Manova K, Kruhlak M, Camerini-Otero RD, Sharan S, Nussenzweig M, Nussenzweig A. Role of Nbs1 in the activation of the Atm kinase revealed in humanized mouse models. Nat Cell Biol. 2005;7:675–85. doi: 10.1038/ncb1270. [DOI] [PubMed] [Google Scholar]
- 119.Kang J, Bronson RT, Xu Y. Targeted disruption of NBS1 reveals its roles in mouse development and DNA repair. Embo J. 2002;21:1447–55. doi: 10.1093/emboj/21.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Williams BR, Mirzoeva OK, Morgan WF, Lin J, Dunnick W, Petrini JH. A murine model of Nijmegen breakage syndrome. Curr Biol. 2002;12:648–53. doi: 10.1016/s0960-9822(02)00763-7. [DOI] [PubMed] [Google Scholar]
- 121.Theunissen JW, Kaplan MI, Hunt PA, Williams BR, Ferguson DO, Alt FW, Petrini JH. Checkpoint failure and chromosomal instability without lymphomagenesis in Mre11(ATLD1/ATLD1) mice. Mol Cell. 2003;12:1511–23. doi: 10.1016/s1097-2765(03)00455-6. [DOI] [PubMed] [Google Scholar]
- 122.Saidi A, Li T, Weih F, Concannon P, Wang ZQ. Dual functions of Nbs1 in the repair of DNA breaks and proliferation ensure proper V(D)J recombination and T-cell development. Mol Cell Biol. 2010;30:5572–81. doi: 10.1128/MCB.00917-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Helmink BA, Bredemeyer AL, Lee BS, Huang CY, Sharma GG, Walker LM, Bednarski JJ, Lee WL, Pandita TK, Bassing CH, Sleckman BP. MRN complex function in the repair of chromosomal Rag-mediated DNA double-strand breaks. J Exp Med. 2009;206:669–79. doi: 10.1084/jem.20081326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Deriano L, Stracker TH, Baker A, Petrini JH, Roth DB. Roles for NBS1 in alternative nonhomologous end-joining of V(D)J recombination intermediates. Mol Cell. 2009;34:13–25. doi: 10.1016/j.molcel.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.FitzGerald JE, Grenon M, Lowndes NF. 53BP1: function and mechanisms of focal recruitment. Biochem Soc Trans. 2009;37:897–904. doi: 10.1042/BST0370897. [DOI] [PubMed] [Google Scholar]
- 126.Morales JC, Xia Z, Lu T, Aldrich MB, Wang B, Rosales C, Kellems RE, Hittelman WN, Elledge SJ, Carpenter PB. Role for the BRCA1 C-terminal repeats (BRCT) protein 53BP1 in maintaining genomic stability. J Biol Chem. 2003;278:14971–7. doi: 10.1074/jbc.M212484200. [DOI] [PubMed] [Google Scholar]
- 127.Ward IM, Minn K, van Deursen J, Chen J. p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol Cell Biol. 2003;23:2556–63. doi: 10.1128/MCB.23.7.2556-2563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bothmer A, Robbiani DF, Feldhahn N, Gazumyan A, Nussenzweig A, Nussenzweig MC. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J Exp Med. 2010;207:855–65. doi: 10.1084/jem.20100244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS, Grofte M, Chan KL, Hickson ID, Bartek J, Lukas J. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol. 2011;13:243–53. doi: 10.1038/ncb2201. [DOI] [PubMed] [Google Scholar]
- 130.Mochan TA, Venere M, DiTullio RA, Jr, Halazonetis TD. 53BP1, an activator of ATM in response to DNA damage. DNA Repair (Amst) 2004;3:945–52. doi: 10.1016/j.dnarep.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 131.Difilippantonio S, Gapud E, Wong N, Huang CY, Mahowald G, Chen HT, Kruhlak MJ, Callen E, Livak F, Nussenzweig MC, Sleckman BP, Nussenzweig A. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008;456:529–33. doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Xie A, Hartlerode A, Stucki M, Odate S, Puget N, Kwok A, Nagaraju G, Yan C, Alt FW, Chen J, Jackson SP, Scully R. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol Cell. 2007;28:1045–57. doi: 10.1016/j.molcel.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, Xu X, Deng CX, Finkel T, Nussenzweig M, Stark JM, Nussenzweig A. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–54. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Manis JP, Morales JC, Xia Z, Kutok JL, Alt FW, Carpenter PB. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat Immunol. 2004;5:481–7. doi: 10.1038/ni1067. [DOI] [PubMed] [Google Scholar]
- 135.Ward IM, Minn K, Jorda KG, Chen J. Accumulation of checkpoint protein 53BP1 at DNA breaks involves its binding to phosphorylated histone H2AX. J Biol Chem. 2003;278:19579–82. doi: 10.1074/jbc.C300117200. [DOI] [PubMed] [Google Scholar]
- 136.Ward IM, Difilippantonio S, Minn K, Mueller MD, Molina JR, Yu X, Frisk CS, Ried T, Nussenzweig A, Chen J. 53BP1 cooperates with p53 and functions as a haploinsufficient tumor suppressor in mice. Mol Cell Biol. 2005;25:10079–86. doi: 10.1128/MCB.25.22.10079-10086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Reina-San-Martin B, Chen J, Nussenzweig A, Nussenzweig MC. Enhanced intra-switch region recombination during immunoglobulin class switch recombination in 53BP1 −/− B cells. Eur J Immunol. 2007;37:235–9. doi: 10.1002/eji.200636789. [DOI] [PubMed] [Google Scholar]
- 138.Agrawal A, Schatz DG. RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell. 1997;89:43–53. doi: 10.1016/s0092-8674(00)80181-6. [DOI] [PubMed] [Google Scholar]
- 139.Hiom K, Gellert M. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol Cell. 1998;1:1011–9. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- 140.Yarnell Schultz H, Landree MA, Qiu JX, Kale SB, Roth DB. Joining-deficient RAG1 mutants block V(D)J recombination in vivo and hairpin opening in vitro. Mol Cell. 2001;7:65–75. doi: 10.1016/s1097-2765(01)00155-1. [DOI] [PubMed] [Google Scholar]
- 141.Qiu JX, Kale SB, Yarnell Schultz H, Roth DB. Separation-of-function mutants reveal critical roles for RAG2 in both the cleavage and joining steps of V(D)J recombination. Mol Cell. 2001;7:77–87. doi: 10.1016/s1097-2765(01)00156-3. [DOI] [PubMed] [Google Scholar]
- 142.Tsai CL, Drejer AH, Schatz DG. Evidence of a critical architectural function for the RAG proteins in end processing, protection, and joining in V(D)J recombination. Genes Dev. 2002;16:1934–49. doi: 10.1101/gad.984502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fugmann SD, Schatz DG. Identification of basic residues in RAG2 critical for DNA binding by the RAG1-RAG2 complex. Mol Cell. 2001;8:899–910. doi: 10.1016/s1097-2765(01)00352-5. [DOI] [PubMed] [Google Scholar]
- 144.Deriano L, Chaumeil J, Coussens M, Multani A, Chou Y, Alekseyenko AV, Chang S, Skok JA, Roth DB. The RAG2 C terminus suppresses genomic instability and lymphomagenesis. Nature. 2011;471:119–23. doi: 10.1038/nature09755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Santagata S, Besmer E, Villa A, Bozzi F, Allingham JS, Sobacchi C, Haniford DB, Vezzoni P, Nussenzweig MC, Pan ZQ, Cortes P. The RAG1/RAG2 complex constitutes a 3′ flap endonuclease: implications for junctional diversity in V(D)J and transpositional recombination. Mol Cell. 1999;4:935–47. doi: 10.1016/s1097-2765(00)80223-3. [DOI] [PubMed] [Google Scholar]
- 146.Cayuela JM, Gardie B, Sigaux F. Disruption of the multiple tumor suppressor gene MTS1/p16(INK4a)/CDKN2 by illegitimate V(D)J recombinase activity in T-cell acute lymphoblastic leukemias. Blood. 1997;90:3720–6. [PubMed] [Google Scholar]
- 147.Lipkowitz S, Stern MH, Kirsch IR. Hybrid T cell receptor genes formed by interlocus recombination in normal and ataxia-telangiectasis lymphocytes. J Exp Med. 1990;172:409–18. doi: 10.1084/jem.172.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kobayashi Y, Tycko B, Soreng AL, Sklar J. Transrearrangements between antigen receptor genes in normal human lymphoid tissues and in ataxia telangiectasia. J Immunol. 1991;147:3201–9. [PubMed] [Google Scholar]
- 149.Baer R, Chen KC, Smith SD, Rabbitts TH. Fusion of an immunoglobulin variable gene and a T cell receptor constant gene in the chromosome 14 inversion associated with T cell tumors. Cell. 1985;43:705–13. doi: 10.1016/0092-8674(85)90243-0. [DOI] [PubMed] [Google Scholar]
- 150.Matthews AG, Oettinger MA. Regulation of RAG transposition. Adv Exp Med Biol. 2009;650:16–31. doi: 10.1007/978-1-4419-0296-2_2. [DOI] [PubMed] [Google Scholar]
- 151.Richardson C, Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature. 2000;405:697–700. doi: 10.1038/35015097. [DOI] [PubMed] [Google Scholar]
- 152.Han JH, Akira S, Calame K, Beutler B, Selsing E, Imanishi-Kari T. Class switch recombination and somatic hypermutation in early mouse B cells are mediated by B cell and Toll-like receptors. Immunity. 2007;27:64–75. doi: 10.1016/j.immuni.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wang JH, Gostissa M, Yan CT, Goff P, Hickernell T, Hansen E, Difilippantonio S, Wesemann DR, Zarrin AA, Rajewsky K, Nussenzweig A, Alt FW. Mechanisms promoting translocations in editing and switching peripheral B cells. Nature. 2009;460:231–6. doi: 10.1038/nature08159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Callen E, Jankovic M, Difilippantonio S, Daniel JA, Chen HT, Celeste A, Pellegrini M, McBride K, Wangsa D, Bredemeyer AL, Sleckman BP, Ried T, Nussenzweig M, Nussenzweig A. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 2007;130:63–75. doi: 10.1016/j.cell.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 155.Raghavan SC, Lieber MR. DNA structures at chromosomal translocation sites. Bioessays. 2006;28:480–94. doi: 10.1002/bies.20353. [DOI] [PubMed] [Google Scholar]
- 156.Yamane A, Resch W, Kuo N, Kuchen S, Li Z, Sun HW, Robbiani DF, McBride K, Nussenzweig MC, Casellas R. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat Immunol. 2010;12:62–9. doi: 10.1038/ni.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Robbiani DF, Bunting S, Feldhahn N, Bothmer A, Camps J, Deroubaix S, McBride KM, Klein IA, Stone G, Eisenreich TR, Ried T, Nussenzweig A, Nussenzweig MC. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol Cell. 2009;36:631–41. doi: 10.1016/j.molcel.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Shen HM, Peters A, Baron B, Zhu X, Storb U. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science. 1998;280:1750–2. doi: 10.1126/science.280.5370.1750. [DOI] [PubMed] [Google Scholar]
- 159.Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–5. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 160.Tsai AG, Lu H, Raghavan SC, Muschen M, Hsieh CL, Lieber MR. Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell. 2008;135:1130–42. doi: 10.1016/j.cell.2008.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Misteli T, Soutoglou E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol. 2009;10:243–54. doi: 10.1038/nrm2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Haber JE, Leung WY. Lack of chromosome territoriality in yeast: promiscuous rejoining of broken chromosome ends. Proc Natl Acad Sci U S A. 1996;93:13949–54. doi: 10.1073/pnas.93.24.13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol. 2007;9:675–82. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Klein IA, Resch W, Jankovic M, Oliveira T, Yamane A, Nakahashi H, Di Virgilio M, Bothmer A, Nussenzweig A, Robbiani DF, Casellas R, Nussenzweig M. Translocation-Capture Sequencing Reveals the Extent and Nature of Chromosomal Rearrangements in B Lymphocytes. Cell. 2011 doi: 10.1016/j.cell.2011.07.048. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chiarle R, Zhang Y, Frock RL, Lewis SM, Molinie B, Ho YJ, Myers DR, Choi VW, Compagno M, Malkin DJ, Neuberg D, Monti S, Giallourakis CG, Gostissa M, Alt FW. Genome-Wide Translocation Sequencing Reveals Mechanisms of Chromosome Breaks and Rearrangements in B Cells. Cell. 2011 doi: 10.1016/j.cell.2011.07.049. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zarrin AA, Del Vecchio C, Tseng E, Gleason M, Zarin P, Tian M, Alt FW. Antibody class switching mediated by yeast endonuclease-generated DNA breaks. Science. 2007;315:377–81. doi: 10.1126/science.1136386. [DOI] [PubMed] [Google Scholar]
- 167.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem. 2004;279:55117–26. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 168.Simsek D, Brunet E, Wong SY, Katyal S, Gao Y, McKinnon PJ, Lou J, Zhang L, Li J, Rebar EJ, Gregory PD, Holmes MC, Jasin M. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 2011;7:e1002080. doi: 10.1371/journal.pgen.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol. 2009;16:814–8. doi: 10.1038/nsmb.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, Lopez BS. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol. 2009;16:819–24. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- 171.Lee-Theilen M, Matthews AJ, Kelly D, Zheng S, Chaudhuri J. CtIP promotes microhomology-mediated alternative end joining during class-switch recombination. Nat Struct Mol Biol. 2011;18:75–9. doi: 10.1038/nsmb.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhang Y, Jasin M. An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat Struct Mol Biol. 2011;18:80–4. doi: 10.1038/nsmb.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wang M, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–82. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Robert I, Dantzer F, Reina-San-Martin B. Parp1 facilitates alternative NHEJ, whereas Parp2 suppresses IgH/c-myc translocations during immunoglobulin class switch recombination. J Exp Med. 2009;206:1047–56. doi: 10.1084/jem.20082468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat Struct Mol Biol. 2010;17:410–6. doi: 10.1038/nsmb.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong SY, Arnal S, Holub AJ, Weller GR, Pancake BA, Shah S, Brandt VL, Meek K, Roth DB. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–6. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 177.Rowh MA, Demicco A, Horowitz JE, Yin B, Yang-Iott KS, Fusello AM, Hobeika E, Reth M, Bassing CH. Tp53 deletion in B lineage cells predisposes mice to lymphomas with oncogenic translocations. Oncogene. 2010 doi: 10.1038/onc.2011.191. [DOI] [PubMed] [Google Scholar]
- 178.Celeste A, Difilippantonio S, Difilippantonio MJ, Fernandez-Capetillo O, Pilch DR, Sedelnikova OA, Eckhaus M, Ried T, Bonner WM, Nussenzweig A. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 2003;114:371–83. doi: 10.1016/s0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zhang X, Wan G, Berger FG, He X, Lu X. The ATM kinase induces microRNA biogenesis in the DNA damage response. Mol Cell. 2011;41:371–83. doi: 10.1016/j.molcel.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Guidos CJ, Williams CJ, Grandal I, Knowles G, Huang MT, Danska JS. V(D)J recombination activates a p53-dependent DNA damage checkpoint in scid lymphocyte precursors. Genes Dev. 1996;10:2038–54. doi: 10.1101/gad.10.16.2038. [DOI] [PubMed] [Google Scholar]
- 181.Nacht M, Strasser A, Chan YR, Harris AW, Schlissel M, Bronson RT, Jacks T. Mutations in the p53 and SCID genes cooperate in tumorigenesis. Genes Dev. 1996;10:2055–66. doi: 10.1101/gad.10.16.2055. [DOI] [PubMed] [Google Scholar]
- 182.Bredemeyer AL, Helmink BA, Innes CL, Calderon B, McGinnis LM, Mahowald GK, Gapud EJ, Walker LM, Collins JB, Weaver BK, Mandik-Nayak L, Schreiber RD, Allen PM, May MJ, Paules RS, Bassing CH, Sleckman BP. DNA double-strand breaks activate a multi-functional genetic program in developing lymphocytes. Nature. 2008;456:819–23. doi: 10.1038/nature07392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wu ZH, Shi Y, Tibbetts RS, Miyamoto S. Molecular linkage between the kinase ATM and NF-kappaB signaling in response to genotoxic stimuli. Science. 2006;311:1141–6. doi: 10.1126/science.1121513. [DOI] [PubMed] [Google Scholar]
- 184.Kashatus D, Cogswell P, Baldwin AS. Expression of the Bcl-3 proto-oncogene suppresses p53 activation. Genes Dev. 2006;20:225–35. doi: 10.1101/gad.1352206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Amaravadi R, Thompson CB. The survival kinases Akt and Pim as potential pharmacological targets. J Clin Invest. 2005;115:2618–24. doi: 10.1172/JCI26273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Johnson K, Hashimshony T, Sawai CM, Pongubala JM, Skok JA, Aifantis I, Singh H. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity. 2008;28:335–45. doi: 10.1016/j.immuni.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 187.Sherman MH, Kuraishy AI, Deshpande C, Hong JS, Cacalano NA, Gatti RA, Manis JP, Damore MA, Pellegrini M, Teitell MA. AID-induced genotoxic stress promotes B cell differentiation in the germinal center via ATM and LKB1 signaling. Mol Cell. 2010;39:873–85. doi: 10.1016/j.molcel.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Sherman MH, Bassing CH, Teitell MA. Regulation of cell differentiation by the DNA damage response. Trends Cell Biol. 2011;21:312–9. doi: 10.1016/j.tcb.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hewitt SL, Yin B, Ji Y, Chaumeil J, Marszalek K, Tenthorey J, Salvagiotto G, Steinel N, Ramsey LB, Ghysdael J, Farrar MA, Sleckman BP, Schatz DG, Busslinger M, Bassing CH, Skok JA. RAG-1 and ATM coordinate monoallelic recombination and nuclear positioning of immunoglobulin loci. Nat Immunol. 2009;10:655–64. doi: 10.1038/ni.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]