Abstract
Yeast surface display is being employed to engineer desirable properties into proteins for a broad variety of applications. Labeling with soluble ligands enables rapid and quantitative analysis of yeast-displayed libraries by flow cytometry, while cell-surface selections allow screening of libraries with insoluble or even as-yet-uncharacterized binding targets. In parallel, the utilization of yeast surface display for protein characterization, including in particular the mapping of functional epitopes mediating protein–protein interactions, represents a significant recent advance.
Introduction
Frequently, improvements in protein function are sought with respect to binding affinity, catalytic activity, and/or structural properties. Given our limited, albeit expanding, understanding of protein sequence/function relationships, achieving the desired improvements through rational protein design is still difficult. However, approaches involving random mutagenesis and directed evolution have been applied with great success for obtaining proteins with defined characteristics.
Yeast surface display is a particularly powerful platform for engineering proteins by directed evolution. Since its introduction 10 years ago [1], yeast surface display has been used to engineer a variety of proteins for improved affinity, specificity, expression, stability, and catalytic activity. A significant feature of the yeast surface display system is its employment of a eukaryotic host possessing the secretory biosynthetic apparatus for promoting efficient oxidative protein folding and N-linked glycosylation. As such, a diverse assortment of proteins has been successfully displayed on the surface of yeast, enabling their subsequent engineering by yeast surface display. As shown in Figure 1 , these include growth factors, antibody fragments, and complex cell-surface receptors such as epidermal growth factor receptor (EGFR), demonstrating the complexity of proteins amenable to engineering by yeast display. However, although well suited to biosynthesis of secreted eukaryotic proteins, the yeast secretory pathway is likely to inefficiently express some cytoplasmic or nuclear proteins due to the presence of multiple reduced cysteines (e.g. zinc finger proteins). Other key beneficial attributes of yeast display include: rapid and quantitative library screening by fluorescence-activated cell sorting; minimization of artifacts due to host-expression-bias through concurrent expression labeling; and convenient evaluation of mutant characteristics (e.g. affinity, stability) in surface-displayed format without soluble expression and purification of each individual clone [2•].
Figure 1.
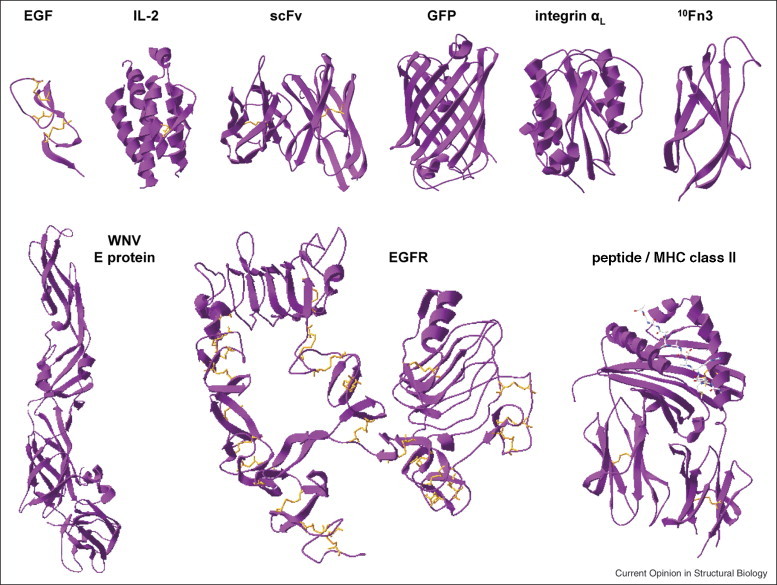
A selection of proteins successfully displayed as Aga2p fusions on the surface of yeast. Top row, left to right: human epidermal growth factor [31••] (1JL9), human interleukin-2 [61] (2B51), single-chain antibody fragment 4m5.3 [62] (1X9Q), green fluorescent protein [63] (1EMA), human αL integrin inserted domain [19] (1LFA), and human fibronectin [18] (1FNA). Bottom row, left to right: West Nile Virus envelope protein [32•] (2I69), human EGF receptor ectodomain [27] (1NQL), and human MHC class II HLA-DR4αβ in complex with peptide [64] (2SEB). The PDB IDs for the structures shown are noted in parentheses. This figure was generated using Swiss-Pdb Viewer [65].
Equipped with these features, yeast surface display is now a well-established method for protein directed evolution. As illustrated in Figure 2 , the number of published studies employing yeast surface display is currently in an exponential growth phase. A recent direct comparison of the yeast and phage display systems, using identical immune antibody libraries and target antigens, also found yeast display to sample the library repertoire ‘considerably more fully’ while being ‘less labor-intensive’ [3••]. Notably, of the 12 novel clones identified by yeast display, only five were functional when subcloned and displayed on phage, although 11 could be expressed solubly in functional form in Escherichia coli [3••].
Figure 2.
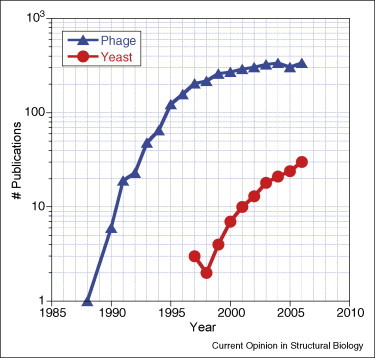
Comparison of numbers of studies employing phage or yeast display.
Here we review recent applications of yeast surface display, highlighting its role in both protein engineering and characterization. We also discuss recent methodological developments, including new techniques for library screening, that have further expanded the utility of this display platform.
Identifying protein–protein interactions
Recently, several groups used yeast surface display to identify natural protein–protein interactions. For instance, yeast surface display was employed for a proteome-wide search of proteins that interact with either EGFR or focal adhesion kinase, in a tyrosine phosphorylation-dependent manner [4]. By displaying a human cDNA library on the surface of yeast and screening with synthetic phosphopeptides, the authors identified several interactions previously unreported [4]. Renner and colleagues recently used yeast-displayed tumor antigens to assess tumor-specific antibody responses in cancer patients [5, 6] and have also screened a yeast-displayed cancer-patient cDNA library for novel tumor antigens [7]. Notably, the screening of yeast-displayed libraries revealed many tumor antigens not previously detected from prokaryote-displayed libraries [7]. However, given the formal possibility that mimotopes could be selected due to expression of frameshifted peptides, confirmation of antisera binding to expressed gene products is necessary following such screens. In addition, several groups have isolated novel lead antibodies binding to a variety of targets, from immune [8] or nonimmune [9, 10, 11, 12] human single-chain variable fragment (scFv) libraries.
Improving affinity and engineering specificity
Affinity and specificity are key parameters governing a protein's function as a diagnostic or therapeutic agent, and yeast surface display has been widely applied for improving or altering these binding properties. Recently, the affinity maturation of several scFvs [13, 14] was reported. More notably, yeast surface display has been employed to selectively expand or restrict binding specificity. For example, after affinity maturing several scFvs against botulinum neurotoxin type A1 [15], Marks and colleagues were able to broaden the specificity of the most potent clone, using a dual-selection strategy, to achieve high-affinity binding to type A2 while retaining high-affinity binding to type A1 [16••]. Conversely, Weaver-Feldhaus et al. engineered conformational specificity into calmodulin-binding scFvs, obtaining clones that recognize only one of the Ca2+-free or Ca2+-bound calmodulin forms [10].
In addition to antibody fragments, yeast surface display has been used to affinity mature proteins with a variety of other folds. Two groups recently reported engineering the 10th human fibronectin type III domain (10Fn3) alternative scaffold, for high-affinity binding to model antigens such as maltose-binding protein [17••] and lysozyme [18]. Jin et al. have also applied yeast surface display to evolve the αL integrin inserted domain, achieving a 200,000-fold increase in ligand-binding affinity [19]. Aside from reporting a remarkable affinity improvement, this study demonstrates the utility of yeast surface display for probing protein allostery, as the mutations responsible for improving affinity locate to a region proposed to control protein conformation [19]. Other molecules recently engineered for improved affinity by yeast display include EGF [20] and single-chain T-cell receptors (scTCRs) [21, 22••, 23•].
A notable advantage to engineering affinity by yeast surface display is the ability to characterize isolated mutants directly in display format. This feature eliminates the need for soluble expression and purification of individual clones and becomes especially significant when many clones require characterization. Importantly, as shown in Figure 3 , the equilibrium dissociation constants measured through titration of proteins on yeast have, to date, shown consistency with those measured by a variety of other methods.
Figure 3.
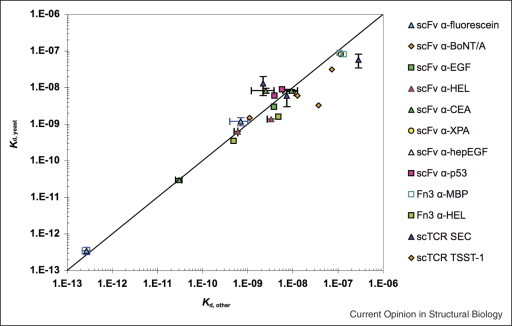
Comparison of dissociation constants determined by yeast surface display, Kd YSD, and other methods, Kd other. scFv binding to fluorescein; Kd other determined by fluorescence quenching. scFv binding botulinum neurotoxin type A; Kd other determined by surface plasmon resonance (SPR) [15]. scFv binding to carcinoembryonic antigen (CEA); Kd other determined by titration of soluble scFv against mammalian-cell-surface-expressed CEA (Michael M Schmidt, unpublished results). scFv binding to hen egg white lysozyme (HEL); Kd other determined by fluorescence quench titration. scFv binding to p53 peptides; Kd other determined by SPR. scFv binding to EGF; Kd other determined by SPR. scFv binding to heparin-binding EGF; Kd other determined by SPR. scFv binding to xeroderma pigmentosum-complementing protein group A; Kd other determined by SPR. Fibronectin binding to HEL; Kd other determined by equilibrium competition titration using purified fibronectin mutants [18]. Fibronection binding to maltose-binding protein; Kd other determined by SPR [17••]. scTCR binding staphylococcal enterotoxin C3; Kd other determined by SPR. scTCR binding toxic shock syndrome toxin-1; Kd other determined by SPR [21]. Figure modified from Figure 8 of Lipovsek et al. [18].
Increasing stability and expression
Thermal stability and soluble expression level are often critical parameters determining a protein's practical utility. Protein stability affects shelf life and suitability for applications at elevated temperatures. Concurrently, a protein's expression level strongly influences its cost of production.
The display level of a protein on the surface of yeast, as a fusion to the yeast agglutinin protein Aga2p, has been shown to correlate with both thermal stability as well as soluble expression level [24], enabling engineering of protein stability and expression by yeast surface display. Several groups have employed the procedure of random mutagenesis, yeast display induction, and high-display screening to improve the stability and expression of proteins, including scTCRs [21, 22••], major histocompatability complex (MHC) class I molecule H-2Ld [25], tumor antigen NY-ESO-1 [26], and the ectodomain of EGFR [27]. In all cases, stability engineering enabled subsequent soluble expression in bacteria [21, 22••, 25] or yeast [26, 27], where efforts for the wild-type version had been unsuccessful. However, this stability engineering approach may not be suitable in all cases, as display levels did not correlate with stability for artificial proteins of particularly high thermal stability [28].
Improvement of soluble expression levels can also be achieved through engineering the expression host instead of the protein itself. Here, yeast display allows the establishment of the critical link between the desirable high-secretion phenotype and its causative genotype. Wentz and Shusta recently employed yeast surface display to screen a yeast cDNA library for yeast genes whose over-expression improved the yield of scFvs and scTCRs [29]. The authors found Aga2p fusion to alter the secretory processing of the heterologous protein, necessitating the performance of screens under conditions where the dominant determinant of display level is the heterologous protein and not Aga2p [29]. Nonetheless, they identified several genes whose over-expression enhanced secretion even in the absence of Aga2p. An approach that eliminates the potential artifacts resulting from Aga2p fusion but that still maintains the link between secretory phenotype and genotype has been reported by Rakestraw et al. [30]. Here, yeast are first tagged with the target protein's binding partner, which subsequently captures the protein of interest as it is secreted by the cell [30]. Mathematical modeling and experimental observations indicate that this selection method is capable of distinguishing subtle differences in secretion level and also possesses a sizable time window for library screening [30]. In addition, since the binding partner is used as bait, only clones secreting well-folded proteins are selected.
Mapping functional protein epitopes
Identifying the key residues that mediate protein–protein interactions provides insight into biological processes and can also facilitate protein design. Recently, the yeast surface display platform was adapted for identifying such residues in a systematic and high-throughput manner. Specifically, as demonstrated by Chao et al. for EGFR and several anti-EGFR antibodies, screening yeast-displayed libraries of the antigen yields epitope maps with residue-level resolution [31••]. Notably, this technique enables the identification of discontinuous and conformational epitopes. Also, it interrogates protein–protein interactions more comprehensively than alanine scanning. Indeed, Chao et al. noted several energetically important residues for which alanine scanning would have yielded false negative results [31••].
Several other groups have applied this technique for the characterization of antibody–antigen interactions. For instance, Diamond and colleagues used this approach to determine the antigenic epitopes recognized by various antibodies and scFvs capable of neutralizing West Nile Virus [32•, 33, 34, 35, 36]. Importantly, for antibody E16, whose structure in complex with its ligand was determined, the results of yeast-display epitope mapping studies [32•, 33] were validated by crystallographic data [33]. Other antibodies recently analyzed by this method include those against botulinum neurotoxin [37], the B and T lymphocyte attenuator [38], the nucleocapsid protein of severe acute respiratory syndrome coronavirus [39], and, in a case of serendipity, NY-ESO-1 [26]. While the examples cited here all represent antibody–antigen interactions, this technique should be applicable for dissecting protein–protein interactions in general. However, a potential limitation is its requirement for an independent means of verifying proper antigen folding.
Engineering proteins against insoluble or unknown targets
One of the key advantages to engineering and characterizing proteins by yeast surface display is the ability to analyze large populations rapidly and quantitatively by flow cytometry. However, this approach requires a soluble ligand that is not always available. Recently, this limitation was addressed by two new screening methods, both employing intact mammalian cells [40••, 41••].
First, Wang and Shusta reported screening yeast-displayed libraries against integral membrane targets by monolayer panning [40••]. Here, desirable clones become selectively enriched by virtue of their affinity for the cell-surface target, while low-affinity clones are washed away from the monolayer [40••]. Several selection conditions, such as wash stringency and ligand density, can affect the success of this approach and were thoroughly investigated for a model system [40••]. This approach enabled the isolation from a nonimmune human scFv library of clones that recognize antigens expressed on the surface of brain endothelial cells [11]. Notably, the identities of the cognate antigens were not defined or known at the outset of selection. However, the isolated scFvs, in yeast-display format, allowed these antigens to be immunoprecipitated from cell lysates for further characterization [11]. Belcher and colleagues have employed an analogous panning approach to screen libraries of yeast-displayed scFvs and peptides for ones possessing high affinity and specificity for inorganic materials such as cadmium sulfide [12, 42] and sapphire [43].
Independently, Kranz and colleagues have developed a cell-surface screening method using density centrifugation [41••]. This approach takes advantage of the differential sedimentation characteristics of yeast and mammalian cells, which, upon centrifugation, sediment through or settle above the density medium Ficoll-Paque, respectively. As yeast expressing high-affinity variants can form conjugates with mammalian cells, they are selectively enriched through retention in the upper layer [41••]. This method has enabled the isolation of high-affinity TCR mutants specific for either class I or class II MHC, which are particularly difficult to solubilize [41••].
Immobilized proteins and enzymes
The recent literature also includes reports of protein immobilization by yeast surface display. Analogous to covalent linkage of proteins to solid beads, such immobilization offers proteins a physical support that often improves stability and facilitates reusability. However, unlike bead conjugation, yeast display does not require additional steps of protein purification and immobilization.
Recent examples of yeast surface immobilized proteins include metal-binding metallothioneins, which sequester toxic cadmium ions [44], as well as streptavidin [45] and protein A [46], which enable capture of desired soluble proteins. Mutants of the integrin αL inserted domain have also been displayed on the surface of yeast, for investigating the effects of ligand-binding affinity on cell adhesion and rolling [47]. In addition, several groups have proposed using antigen-displaying yeast as preventative or therapeutic vaccines [48, 49, 50].
A host of enzymes have been functionally displayed on the surface of yeast, including lipase [51, 52], biotin ligase [53], organophosphorous hydrolase [54], carboxylesterase [55], epimerase [56], cyclodextrin glucanotransferase [57], and neurolysin [58]. While efforts to engineer mutants with improved catalytic activity have been reported [51, 52], systematic and high-throughput examples of catalysis engineering have yet to be described. The use of yeast surface display for engineering enzymes and their substrates will be an interesting direction for the future.
Conclusion
In summary, yeast surface display facilitates efforts to engineer proteins with defined characteristics, and can also provide valuable quantitative information regarding protein–protein interactions. The eukaryotic nature of the yeast secretory pathway has enabled the study and manipulation of even complex proteins by this method. However, yeast and mammalian glycosylation structures differ, prompting development of human cell display [59]. Nonetheless, yeast display remains suitable for most proteins of interest; for situations where glycosylation differences matter, yeast strains possessing the human glycosylation pathway may also serve as an alternative [60].
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
SAG was supported by AI065824 and CA101830 from the National Institutes of Health and a National Science Foundation Graduate Fellowship.
References
- 1.Boder E.T., Wittrup K.D. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 2•.Chao G., Lau W.L., Hackel B.J., Sazinsky S.L., Lippow S.M., Wittrup K.D. Isolating and engineering human antibodies using yeast surface display. Nat Protoc. 2006;1:755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]; This paper provides a detailed and comprehensive protocol for engineering proteins by yeast surface display. While the focus is on isolating and engineering scFvs, the procedures are applicable for engineering any protein that can be displayed by yeast and whose binding target is available in soluble form.
- 3••.Bowley D.R., Labrijn A.F., Zwick M.B., Burton D.R. Antigen selection from an HIV-1 immune antibody library displayed on yeast yields many novel antibodies compared to selection from the same library displayed on phage. Protein Eng Des Sel. 2007;20:81–90. doi: 10.1093/protein/gzl057. [DOI] [PubMed] [Google Scholar]; This study offers the first direct comparison of yeast surface display and phage display. From identical libraries, screened with the same antigen, yeast display was found to identify many more high-affinity clones and also required less effort. These two significant advantages of yeast display were attributed to, respectively, eukaryotic processing and flow-cytometry-based library screening and clone analysis.
- 4.Bidlingmaier S., Liu B. Construction and application of a yeast surface-displayed human cDNA library to identify post-translational modification-dependent protein–protein interactions. Mol Cell Proteomics. 2006;5:533–540. doi: 10.1074/mcp.M500309-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Mischo A., Kubuschok B., Ertan K., Preuss K.-D., Romeike B., Regitz E., Schormann C., de Bruijn D., Wadle A., Neumann F. Prospective study on the expression of cancer testis genes and antibody responses in 100 consecutive patients with primary breast cancer. Int J Cancer. 2006;118:696–703. doi: 10.1002/ijc.21352. [DOI] [PubMed] [Google Scholar]
- 6.Wadle A., Kubuschok B., Imig J., Wüllner B., Wittig C., Zwick C., Mischo A., Wätzig K., Romeike B.F.M., Lindemann W. Serological immune response to cancer testis antigens in patients with pancreatic cancer. Int J Cancer. 2006;119:117–125. doi: 10.1002/ijc.21744. [DOI] [PubMed] [Google Scholar]
- 7.Wadle A., Mischo A., Imig J., Wüllner B., Hensel D., Wätzig K., Neumann F., Kubuschok B., Schmidt W., Old L.J. Serological identification of breast cancer-related antigens from a Saccharomyces cerevisiae surface display library. Int J Cancer. 2005;117:104–113. doi: 10.1002/ijc.21147. [DOI] [PubMed] [Google Scholar]
- 8.Lee H.-W., Lee S.-H., Park K.-J., Kim J.-S., Kwon M.-H., Kim Y.-S. Construction and characterization of a pseudo-immune human antibody library using yeast surface display. Biochem Biophys Res Commun. 2006;346:896–903. doi: 10.1016/j.bbrc.2006.05.202. [DOI] [PubMed] [Google Scholar]
- 9.Scholler N., Garvik B., Quarles T., Jiang S., Urban N. Method for generation of in vivo biotinylated recombinant antibodies by yeast mating. J Immunol Methods. 2006;317:132–143. doi: 10.1016/j.jim.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weaver-Feldhaus J.M., Miller K.D., Feldhaus M.J., Siegel R.W. Directed evolution for the development of conformation-specific affinity reagents using yeast display. Protein Eng Des Sel. 2005;18:527–536. doi: 10.1093/protein/gzi060. [DOI] [PubMed] [Google Scholar]
- 11.Wang X.X., Cho Y.K., Shusta E.V. Mining a yeast library for brain endothelial cell-binding antibodies. Nat Methods. 2007;4:143–145. doi: 10.1038/nmeth993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peelle B.R., Krauland E.M., Wittrup K.D., Belcher A.M. Probing the interface between biomolecules and inorganic materials using yeast surface display and genetic engineering. Acta Biomater. 2005;1:145–154. doi: 10.1016/j.actbio.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 13.Rajpal A., Beyaz N., Haber L., Cappuccilli G., Yee H., Bhatt R.R., Takeuchi T., Lerner R.A., Crea R. A general method for greatly improving the affinity of antibodies by using combinatorial libraries. Proc Natl Acad Sci USA. 2005;102:8466–8471. doi: 10.1073/pnas.0503543102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z., Kim G.-B., Woo J.-H., Liu Y.Y., Mathias A., Stavrou S., Neville D.M. Improvement of a recombinant anti-monkey anti-CD3 diphtheria toxin based immunotoxin by yeast display affinity maturation of the scFv. Bioconjugate Chem. 2007;18:947–955. doi: 10.1021/bc0603438. [DOI] [PubMed] [Google Scholar]
- 15.Razai A., Garcia-Rodriguez C., Lou J., Geren I.N., Forsyth C.M., Robles Y., Tsai R., Smith T.J., Smith L.A., Siegel R.W. Molecular evolution of antibody affinity for sensitive detection of botulinum neurotoxin type A. J Mol Biol. 2005;351:158–169. doi: 10.1016/j.jmb.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 16••.Garcia-Rodriguez C., Levy R., Arndt J.W., Forsyth C.M., Razai A., Lou J., Geren I., Stevens R.C., Marks J.D. Molecular evolution of antibody cross-reactivity for two subtypes of type A botulinum neurotoxin. Nat Biotechnol. 2007;25:107–116. doi: 10.1038/nbt1269. [DOI] [PubMed] [Google Scholar]; This study illustrates the use of yeast surface display for selectively expanding antibody reactivity. Starting from a scFv that recognizes botulinum neurotoxin type A1 with high affinity, the authors identified a mutant that recognizes type A2 with high affinity as well. Notably, the selection process required independent and simultaneous quantification of binding to both target antigens, which was enabled by cell-surface display and flow cytometry.
- 17••.Koide A., Gilbreth R.N., Esaki K., Tereshko V., Koide S. High-affinity single-domain binding proteins with a binary-code interface. Proc Natl Acad Sci USA. 2007;104:6632–6637. doi: 10.1073/pnas.0700149104. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this work, fibronectin scaffold binders with only tyrosine and serine in their variable loops were identified by first screening a phage-displayed library, followed by a yeast-displayed library for fine affinity discrimination.
- 18.Lipovsek D., Lippow S.M., Hackel B.J., Gregson M.W., Cheng P., Kapila A., Wittrup K.D. Evolution of an interloop disulfide bond in high-affinity antibody mimics based on fibronectin type III domain and selected by yeast surface display: molecular convergence with single-domain camelid and shark antibodies. J Mol Biol. 2007;368:1024–1041. doi: 10.1016/j.jmb.2007.02.029. [DOI] [PubMed] [Google Scholar]
- 19.Jin M., Song G., Carman C.V., Kim Y.-S., Astrof N.S., Shimaoka M., Wittrup D.K., Springer T.A. Directed evolution to probe protein allostery and integrin I domains of 200,000-fold higher affinity. Proc Natl Acad Sci USA. 2006;103:5758–5763. doi: 10.1073/pnas.0601164103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cochran J.R., Kim Y.-S., Lippow S.M., Rao B., Wittrup K.D. Improved mutants from directed evolution are biased to orthologous substitutions. Protein Eng Des Sel. 2006;19:245–253. doi: 10.1093/protein/gzl006. [DOI] [PubMed] [Google Scholar]
- 21.Buonpane R.A., Moza B., Sundberg E.J., Kranz D.M. Characterization of T cell receptors engineered for high affinity against toxic shock syndrome toxin-1. J Mol Biol. 2005;353:308–321. doi: 10.1016/j.jmb.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 22••.Weber K.S., Donermeyer D.L., Allen P.M., Kranz D.M. Class II-restricted T cell receptor engineered in vitro for higher affinity retains peptide specificity and function. Proc Natl Acad Sci USA. 2005;102:19033–19038. doi: 10.1073/pnas.0507554102. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study, a panel of higher-affinity mutants of a class II-restricted TCR was generated, found to retain exquisite peptide specificity, and used to address the functional consequences of TCR affinity on T cell activation.
- 23•.Buonpane R.A., Churchill H.R.O., Moza B., Sundberg E.J., Peterson M.L., Schlievert P.M., Kranz D.M. Neutralization of staphylococcal enterotoxin B by soluble, high-affinity receptor antagonists. Nat Med. 2007;13:725–729. doi: 10.1038/nm1584. [DOI] [PubMed] [Google Scholar]; This study describes the affinity maturation of a TCR V domain against the superantigen staphylococcal enterotoxin B (SEB). A mutant possessing a three-million-fold increase in binding affinity was isolated and found to potently antagonize lethal SEB activity in animal models.
- 24.Shusta E.V., Kieke M.C., Parke E., Kranz D.M., Wittrup K.D. Yeast polypeptide fusion surface display levels predict thermal stability and soluble secretion efficiency. J Mol Biol. 1999;292:949–956. doi: 10.1006/jmbi.1999.3130. [DOI] [PubMed] [Google Scholar]
- 25.Jones L.L., Brophy S.E., Bankovich A.J., Colf L.A., Hanick N.A., Garcia K.C., Kranz D.M. Engineering and characterization of a stabilized α1/α2 module of the class I major histocompatibility complex product Ld. J Biol Chem. 2006;281:25734–25744. doi: 10.1074/jbc.M604343200. [DOI] [PubMed] [Google Scholar]
- 26.Piatesi A., Howland S.W., Rakestraw J.A., Renner C., Robson N., Cebon J., Maraskovsky E., Ritter G., Old L., Wittrup K.D. Directed evolution for improved secretion of cancer-testis antigen NY-ESO-1 from yeast. Protein Expr Purif. 2006;48:232–242. doi: 10.1016/j.pep.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 27.Kim Y.-S., Bhandari R., Cochran J.R., Kuriyan J., Wittrup K.D. Directed evolution of the epidermal growth factor receptor extracellular domain for expression in yeast. Proteins Struct Funct Bioinformatics. 2006;62:1026–1035. doi: 10.1002/prot.20618. [DOI] [PubMed] [Google Scholar]
- 28.Park S., Xu Y., Stowell X.F., Gai F., Saven J.G., Boder E.T. Limitations of yeast surface display in engineering proteins of high thermostability. Protein Eng Des Sel. 2006;19:211–217. doi: 10.1093/protein/gzl003. [DOI] [PubMed] [Google Scholar]
- 29.Wentz A.E., Shusta E.V. A novel high-throughput screen reveals yeast genes that increase secretion of heterologous proteins. Appl Environ Microbiol. 2007;73:1189–1198. doi: 10.1128/AEM.02427-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rakestraw J.A., Baskaran A.R., Wittrup K.D. A flow cytometric assay for screening improved heterologous protein secretion in yeast. Biotechnol Prog. 2006;22:1200–1208. doi: 10.1021/bp0600233. [DOI] [PubMed] [Google Scholar]
- 31••.Chao G., Cochran J.R., Dane Wittrup K. Fine epitope mapping of anti-epidermal growth factor receptor antibodies through random mutagenesis and yeast surface display. J Mol Biol. 2004;342:539–550. doi: 10.1016/j.jmb.2004.07.053. [DOI] [PubMed] [Google Scholar]; This study details the application of yeast surface display for identifying key residues mediating protein–protein interactions. By constructing a library of randomly mutagenized EGFR variants and screening with anti-EGFR antibodies, the authors mapped the binding epitopes of these antibodies with residue resolution. Significantly, this technique identifies discontinuous and heat-denaturable epitopes, samples substitutions to amino acids other than alanine, and requires no soluble expression and purification of mutants.
- 32•.Oliphant T., Engle M., Nybakken G.E., Doane C., Johnson S., Huang L., Gorlatov S., Mehlhop E., Marri A., Chung K.M. Development of a humanized monoclonal antibody with therapeutic potential against West Nile Virus. Nat Med. 2005;11:522–530. doi: 10.1038/nm1240. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study applies yeast surface display for determining the antigenic epitopes recognized by a panel of monoclonal antibodies specific for the envelope protein of West Nile Virus. Significantly, the epitope map of antibody E16 has been validated by crystallographic data, presented in Nybakken et al.
- 33.Nybakken G.E., Oliphant T., Johnson S., Burke S., Diamond M.S., Fremont D.H. Structural basis of West Nile Virus neutralization by a therapeutic antibody. Nature. 2005;437:764–769. doi: 10.1038/nature03956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gould L.H., Sui J., Foellmer H., Oliphant T., Wang T., Ledizet M., Murakami A., Noonan K., Lambeth C., Kar K. Protective and therapeutic capacity of human single-chain Fv–Fc fusion proteins against West Nile Virus. J Virol. 2005;79:14606–14613. doi: 10.1128/JVI.79.23.14606-14613.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oliphant T., Nybakken G.E., Engle M., Xu Q., Nelson C.A., Sukupolvi-Petty S., Marri A., Lachmi B.-E., Olshevsky U., Fremont D.H. Antibody recognition and neutralization determinants on domains I and II of West Nile Virus envelope protein. J Virol. 2006;80:12149–12159. doi: 10.1128/JVI.01732-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chung K.M., Nybakken G.E., Thompson B.S., Engle M.J., Marri A., Fremont D.H., Diamond M.S. Antibodies against West Nile Virus nonstructural protein NS1 prevent lethal infection through Fc γ receptor-dependent and -independent mechanisms. J Virol. 2006;80:1340–1351. doi: 10.1128/JVI.80.3.1340-1351.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levy R., Forsyth C.M., LaPorte S.L., Geren I.N., Smith L.A., Marks J.D. Fine and domain-level epitope mapping of botulinum neurotoxin type A neutralizing antibodies by yeast surface display. J Mol Biol. 2007;365:196–210. doi: 10.1016/j.jmb.2006.09.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hurchla M.A., Sedy J.R., Gavrielli M., Drake C.G., Murphy T.L., Murphy K.M. B and T lymphocyte attenuator exhibits structural and expression polymorphisms and is highly induced in anergic CD4+ T cells. J Immunol. 2005;174:3377–3385. doi: 10.4049/jimmunol.174.6.3377. [DOI] [PubMed] [Google Scholar]
- 39.Liang Y., Wan Y., Qiu L.-W., Zhou J., Ni B., Guo B., Zou Q., Zou L., Zhou W., Jia Z. Comprehensive antibody epitope mapping of the nucleocapsid protein of severe acute respiratory syndrome (SARS) coronavirus: insight into the humoral immunity of SARS. Clin Chem. 2005;51:1382–1396. doi: 10.1373/clinchem.2005.051045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40••.Wang X.X., Shusta E.V. The use of scFv-displaying yeast in mammalian cell surface selections. J Immunol Methods. 2005;304:30–42. doi: 10.1016/j.jim.2005.05.006. [DOI] [PubMed] [Google Scholar]; This study describes a method for screening yeast-displayed libraries by monolayer panning, which is well suited for engineering of proteins whose binding targets are not readily solubilized and expressed by adherent cells.
- 41••.Richman S.A., Healan S.J., Weber K.S., Donermeyer D.L., Dossett M.L., Greenberg P.D., Allen P.M., Kranz D.M. Development of a novel strategy for engineering high-affinity proteins by yeast display. Protein Eng Des Sel. 2006;19:255–264. doi: 10.1093/protein/gzl008. [DOI] [PubMed] [Google Scholar]; This study describes a method for screening yeast-displayed libraries by density centrifugation, which is well suited for engineering proteins whose binding targets are not readily solubilized and expressed by cells grown in suspension.
- 42.Peelle B.R., Krauland E.M., Wittrup K.D., Belcher A.M. Design criteria for engineering inorganic material-specific peptides. Langmuir. 2005;21:6929–6933. doi: 10.1021/la050261s. [DOI] [PubMed] [Google Scholar]
- 43.Krauland EM, Peelle BR, Wittrup KD, Belcher AM: Peptide tags for enhanced cellular and protein adhesion to single-crystalline sapphire. Biotechnol Bioeng 2007:n/a. [DOI] [PubMed]
- 44.Kuroda K., Ueda M. Effective display of metallothionein tandem repeats on the bioadsorption of cadmium ion. Appl Microbiol Biotechnol. 2006;70:458–463. doi: 10.1007/s00253-005-0093-8. [DOI] [PubMed] [Google Scholar]
- 45.Furukawa H., Tanino T., Fukuda H., Kondo A. Development of novel yeast cell surface display system for homo-oligomeric protein by coexpression of native and anchored subunits. Biotechnol Prog. 2006;22:994–997. doi: 10.1021/bp0601342. [DOI] [PubMed] [Google Scholar]
- 46.Shibasaki S., Kawabata A., Ishii J., Yagi S., Kadonosono T., Kato M., Fukuda N., Kondo A., Ueda M. Construction of a novel synergistic system for production and recovery of secreted recombinant proteins by the cell surface engineering. Appl Microbiol Biotechnol. 2007;75:821–828. doi: 10.1007/s00253-007-0868-1. [DOI] [PubMed] [Google Scholar]
- 47.Pepper L.R., Hammer D.A., Boder E.T. Rolling adhesion of αl I domain mutants decorrelated from binding affinity. J Mol Biol. 2006;360:37–44. doi: 10.1016/j.jmb.2006.04.049. [DOI] [PubMed] [Google Scholar]
- 48.Zhu K., Chi Z., Li J., Zhang F., Li M., Yasoda H.N., Wu L. The surface display of haemolysin from vibrio harveyi on yeast cells and their potential applications as live vaccine in marine fish. Vaccine. 2006;24:6046–6052. doi: 10.1016/j.vaccine.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 49.Tamaru Y., Ohtsuka M., Kato K., Manabe S., Kuroda K., Sanada M., Ueda M. Application of the arming system for the expression of the 380R antigen from Red Sea Bream Iridovirus (RSIV) on the surface of yeast cells: a first step for the development of an oral vaccine. Biotechnol Prog. 2006;22:949–953. doi: 10.1021/bp060130x. [DOI] [PubMed] [Google Scholar]
- 50.Wadle A., Held G., Neumann F., Kleber S., Wuellner B., Asemissen A.M., Kubuschok B., Scheibenbogen C., Breinig T., Meyerhans A. Cross-presentation of HLA Class I epitopes from influenza matrix protein produced in Saccharomyces cerevisiae. Vaccine. 2006;24:6272–6281. doi: 10.1016/j.vaccine.2006.05.096. [DOI] [PubMed] [Google Scholar]
- 51.Shiraga S., Kawakami M., Ishiguro M., Ueda M. Enhanced reactivity of Rhizopus oryzae lipase displayed on yeast cell surfaces in organic solvents: potential as a whole-cell biocatalyst in organic solvents. Appl Environ Microbiol. 2005;71:4335–4338. doi: 10.1128/AEM.71.8.4335-4338.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shiraga S., Ishiguro M., Fukami H., Nakao M., Ueda M. Creation of Rhizopus oryzae lipase having a unique oxyanion hole by combinatorial mutagenesis in the lid domain. Appl Microbiol Biotechnol. 2005;68:779–785. doi: 10.1007/s00253-005-1935-0. [DOI] [PubMed] [Google Scholar]
- 53.Parthasarathy R., Bajaj J., Boder E.T. An immobilized biotin ligase: surface display of Escherichia coli BirA on Saccharomyces cerevisiae. Biotechnol Prog. 2005;21:1627–1631. doi: 10.1021/bp050279t. [DOI] [PubMed] [Google Scholar]
- 54.Takayama K., Suye S.-I., Kuroda K., Ueda M., Kitaguchi T., Tsuchiyama K., Fukuda T., Chen W., Mulchandani A. Surface display of organophosphorus hydrolase on Saccharomyces cerevisiae. Biotechnol Prog. 2006;22:939–943. doi: 10.1021/bp060107b. [DOI] [PubMed] [Google Scholar]
- 55.Breinig F., Diehl B., Rau S., Zimmer C., Schwab H., Schmitt M.J. Cell surface expression of bacterial esterase a by Saccharomyces cerevisiae and its enhancement by constitutive activation of the cellular unfolded protein response. Appl Environ Microbiol. 2006;72:7140–7147. doi: 10.1128/AEM.00503-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang H.-C., Bi J.-Y., Chen C., Huang G.-L., Qi Q.-S., Xiao M., Wang P.G. Immobilization of UDP-galactose 4-epimerase from Escherichia coli on the yeast cell surface. Biosci Biotechnol Biochem. 2006;70:2302–2306. doi: 10.1271/bbb.60134. [DOI] [PubMed] [Google Scholar]
- 57.Shim J.-H., Seo N.-S., Roh S.-A., Kim J.-W., Cha H., Park K.-H. Improved bread-baking process using Saccharomyces cerevisiae displayed with engineered cyclodextrin glucanotransferase. J Agric Food Chem. 2007;55:4735–4740. doi: 10.1021/jf070217d. [DOI] [PubMed] [Google Scholar]
- 58.Kadonosono T., Kato M., Ueda M. Metallopeptidase, neurolysin, as a novel molecular tool for analysis of properties of cancer-producing matrix metalloproteinases-2 and -9. Appl Microbiol Biotechnol. 2007;75:1285–1291. doi: 10.1007/s00253-007-0952-6. [DOI] [PubMed] [Google Scholar]
- 59.Ho M., Nagata S., Pastan I. Isolation of anti-CD22 Fv with high affinity by Fv display on human cells. Proc Natl Acad Sci USA. 2006;103:9637–9642. doi: 10.1073/pnas.0603653103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hamilton S.R., Bobrowicz P., Bobrowicz B., Davidson R.C., Li H., Mitchell T., Nett J.H., Rausch S., Stadheim T.A., Wischnewski H. Production of complex human glycoproteins in yeast. Science. 2003;301:1244–1246. doi: 10.1126/science.1088166. [DOI] [PubMed] [Google Scholar]
- 61.Rao B.M., Driver I., Lauffenburger D.A., Wittrup K.D. High-affinity CD25-binding IL-2 mutants potently stimulate persistent T cell growth. Biochemistry. 2005;44:10696–10701. doi: 10.1021/bi050436x. [DOI] [PubMed] [Google Scholar]
- 62.Boder E.T., Midelfort K.S., Wittrup K.D. Directed evolution of antibody fragments with monovalent femtomolar antigen-binding affinity. Proc Natl Acad Sci USA. 2000;97:10701–10705. doi: 10.1073/pnas.170297297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang D., Shusta E.V. Secretion and surface display of green fluorescent protein using the yeast Saccharomyces cerevisiae. Biotechnol Prog. 2005;21:349–357. doi: 10.1021/bp0497482. [DOI] [PubMed] [Google Scholar]
- 64.Boder E.T., Bill J.R., Nields A.W., Marrack P.C., Kappler J.W. Yeast surface display of a noncovalent MHC class II heterodimer complexed with antigenic peptide. Biotechnol Bioeng. 2005;92:485–491. doi: 10.1002/bit.20616. [DOI] [PubMed] [Google Scholar]
- 65.Guex N., Peitsch M.C. Swiss-model and the swiss-pdbviewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]


