Abstract
This article investigates the expression patterns of 160 genes that are expressed during early mouse development. The cDNAs were isolated from 7.5 d postcoitum (dpc) endoderm, a region that comprises visceral endoderm (VE), definitive endoderm, and the node–tissues that are required for the initial steps of axial specification and tissue patterning in the mouse. To avoid examining the same gene more than once, and to exclude potentially ubiquitously expressed housekeeping genes, cDNA sequence was derived from 1978 clones of the Endoderm library. These yielded 1440 distinct cDNAs, of which 123 proved to be novel in the mouse. In situ hybridization analysis was carried out on 160 of the cDNAs, and of these, 29 (18%) proved to have restricted expression patterns.
The genomic sequences of many animals are now known, including C. elegans, human, mouse, and Drosophila (The C. elegans genome consortium 1998; Adams et al. 2000; Lander et al. 2001; Venter et al. 2001; Aparicio et al. 2002; Carlton et al. 2002; Dehal et al. 2002; Gardner et al. 2002; Waterston et al. 2002), and the sequences of others will be available very soon. The task now facing biologists is to discover the functions of the genes that have been identified through these sequencing projects. For some organisms, such as C. elegans, it is possible to adopt a systematic approach to ablating gene function (Fraser et al. 2000; Kamath et al. 2003). For vertebrates, and especially mammals, a systematic approach of this sort is a daunting prospect, but a widespread analysis of gene function is nevertheless essential for a proper understanding of development and disease.
The most tractable mammalian species for such an analysis is the mouse, in which it is possible to mutate gene function randomly, by using γ-irradiation, chemical mutagenesis or gene traps (Stanford et al. 2001), or a directed fashion by means of homologous recombination in embryonic stem cells (Doetschman et al. 1987; Thomas and Capecchi 1987). Mutagenesis has proved a very useful approach, but it is limited in some respects because redundancy or compensation may mask functional requirements and because early lethality may conceal later roles of some genes. The necessity to maintain large numbers of mutant strains also presents practical difficulties.
Homologous recombination overcomes these problems by allowing the ablation of specific genes at particular times in development and in a tissue-specific manner. It is not yet feasible, however, to contemplate targeting the entire proteome in this way, so it is necessary to decide which genes to target first. Work from several species indicates that one criterion might be based on gene expression patterns. In situ hybridization analyses of random clones from unmodified, normalized, or subtracted cDNA libraries has identified many genes with restricted expression patterns that hint at particular embryonic functions (Gawantka et al. 1998; Neidhardt et al. 2000; Christiansen et al. 2001; Kudoh et al. 2001). In addition, the results have allowed the definition of “synexpression groups,” the members of which are expressed in similar patterns and may be regulated in similar ways and act in the same molecular pathways (Gawantka et al. 1998; Niehrs and Pollet 1999).
In this article we refine this approach by using sequence comparisons to reduce cDNA library complexity and to remove unwanted molecules (see below). We use a cDNA library constructed from 7.5 d postcoitum (dpc) endoderm (Harrison et al. 1995), a region that comprises VE, definitive endoderm, and the node–tissues that are required for the initial steps of axial specification and tissue patterning in the mouse embryo (Beddington and Robertson 1999; Lu et al. 2001; Hamada et al. 2002). This Endoderm library, together with four others (whole Embryonic Region, Ectoderm, Mesoderm, and Primitive Streak), has already proved its worth in subtractive and differential hybridization experiments that have identified regionally expressed genes that are required for normal development (Harrison et al. 1995; Dunwoodie et al. 1997, 1998, 2002; Dunwoodie and Beddington 2002; Martinez Barbera et al. 2002).
Analysis of 1978 sequences derived from the endoderm library identified 1440 different cDNAs, of which 123 proved to be novel in the mouse. In situ hybridization analysis was carried out on 160of the cDNAs, and of these, 18% proved to have restricted expression. This work provides valuable information about the repertoire of gene expression in the endoderm of the mouse embryo and may supply pointers as to which genes merit further investigation concerning their roles in development and disease (Anderson and Beddington 1997).
RESULTS
Sequence Analyses
cDNA clones (3072) were selected at random from the Endoderm library, and 2635 sequence tags were generated by single-pass 3′ sequencing (Avner et al. 2001). Repetitive and poor-quality sequence was masked, and any sequence tag of <199 nucleotides after masking was discarded. Analysis of the remaining 1978 sequences is presented in Table 1. Each sequence was compared by using BLASTN with mouse expressed sequence tag (EST) clusters (TIGR Tentative Consensus sequences or TCs version 8.0, June 1, 2002; http://www.tigr.org/tdb/tgi/mgi) and with predicted mouse transcripts in ENSEMBL (version 8.3c.1, July 12, 2002; http://www.ensembl.org/Mus_musculus/). Sequence matches were considered significant if alignment of >50nucleotides was observed and the significance value was less than e-30. All remaining sequences were considered novel.
Table 1.
Summary of Endoderm Sequence Analysis
| cDNA clones selected for sequencing | 3072 |
| Successful single pass sequence | 2635 |
| Masked sequences >199 base pairs in length | 1978 |
| Sequences matching ENSEMBL genes | 1355 |
| Sequences matching TIGR-TC EST clusters only | 496 |
| Novel sequences | 127 |
| Nonredundant clones matching mouse ESTs | 1317 |
| Nonredundant novel clones | 123 |
Of the 1978 sequences, 1851 clones matched a defined EST (TIGR-TC) cluster, an ENSEMBL gene or transcript, or both. The remaining 127 clones matched neither data set and are classified as novel. Clustering of the 1851 sequences that matched the TIGR-TC or EMSEMBL databases generated a non-redundant set of 1317 known cDNAs. The 127 novel sequences were compared with each other by using BLASTN, using significance limits similar to those described above. This procedure reduced the number of novel cDNAs to 123. All sequences described in this article are available in GenBank, and cDNAs can be obtained from the UK Human Genome Mapping Project Resource Centre (http://www.hgmp.mrc.ac.uk/geneservice/reagents/products/cdna_resources/index.shtml).
Expression Analysis
Of the 1978 cDNAs described above, 160were chosen for expression analysis. Clones were selected so as to exclude housekeeping genes and genes previously studied in a developmental context, but to include completely novel sequences, previously unknown sequences that had also been identified in other organisms, cDNAs encoding putative transcriptional regulators, splicing factors, signaling molecules, cell-cycle regulators, cytoskeletal proteins, and cDNAs encoding homologs of proteins implicated in human disease (for examples, see Table 2).
Table 2.
Sequence Analysis of cDNA Clones With Restricted Expression
| Sequence ID | Frequency | Representative ID | Description |
|---|---|---|---|
| t8219b01 | 1 | ||
| t7822b10 | 2 | ENSMUSG00000013236 | Protein-tyrosine phosphatase, receptor-type (Ptpt9; EC 3.1.3.48) |
| r8220b29 | 1 | TC469486 | |
| s8609b60 | 2 | ENSMUSG00000019970 | Serum and glucocorticoid-regulated kinase (Sgk; EC 2.7.1) |
| m8708a09 | 4 | ENSMUSG00000021728 | Embigin precursor, also known as Teratocarcinoma glycoprotein 70 (GP-70) |
| v8130b53 | 9 | TC461859 | Solute carrier family 2 (facilitated glucose transporter) member 3 (Slc2a3) |
| t7825b42 | 3 | TC511260 | Sp120 (Hnrpu) |
| s8609b24 | 1 | ENSMUSG00000039878 | Similar to LIV-1, estrogen-regulated |
| r8316a33 | 1 | ENSMUSG00000024253 | Dynein 2 light intermediate chain (mD2LIC) |
| v8130b25 | 1 | ENSMUSG00000028162 | |
| r8707a53 | 3 | ENSMUSG00000023906 | Claudin-6 |
| m8708a22 | 1 | ENSMUSG00000039676 | Calcyphosine |
| p7822b53 | 1 | ENSMUSG00000005505 | Weakly similar to ring canal protein; contains BTB/POZ domain |
| t8130b59 | 1 | TC503400 | |
| t8417b56 | 1 | ENSMUSG0000002764 | Neuronatin, also known as Peg5 (isoform 2) |
| t8219b25 | 1 | TC488224 | Similar (16%) to KIAA0802 protein (Homo sapiens) |
| w8609b57 | 1 | ENSMUSG00000029032 | Neuroblastoma; similar to Rho GEF 16; |
| t7822b19 | 1 | ENSMUSG00000021681 | Paternally expressed gene 3 (Peg3) |
| k8709a24 | 1 | ENSMUSG00000031665 | Sal-like 1 (Sall1) |
| r8220b09 | 1 | TC501397 | Silica-induced gene 41 (Silg41); similar to arg/ser-rich splicing factor (transformer2) |
| t8130b26 | 1 | ENSMUSG00000042142 | Rb-binding protein 2 (Rb-BP2); also known as Plu-1 |
| m8708a39 | 1 | ENSMUSG00000022761 | Leucine-zipper-like transcriptional regulator 1 (Lztr1) |
| s8129b58 | 1 | ENSMUSG00000026833 | Pancortins 1 and/or 3 |
| k8220b03 | 1 | ENSMUSG00000029381 | Shroom (actin binding protein) (Shrm) |
| r8220b57 | 1 | ENSMUSG00000005566 | Transcription intermediary factor 1-β (Tif1-β) |
| r8319a44 | 2 | 14-3-3 protein σ | |
| k8709a20 | 2 | ENSMUSG00000020849 | 14-3-3 protein ε (protein kinase C inhibitor protein-1) |
| k8710a07 | 1 | ENSMUSG00000021667 | Nop seven associated protein 2 (Nsa2p); also known as Lnr42, TINP1 or HCLG1 |
| t8219b26 | 1 | ENSMUSG00000032376 | Ubiquitin-specific protease 3 (Ubp7) |
The table shows 29 cDNAs with restricted patterns of expression. The clones are shown in the same order as in Figure 1, with the first three being members of the visceral endoderm synexpression group. Headings represent (1) the sequence identification number, (2) the number of times the sequence was isolated, (3) the ENSEMBL gene number of the cDNA or its TIGR Cluster number, and (4) gene name or family. Genes for which no description is appended bear no resemblance to any other in the databases.
Expression patterns were categorized subjectively as “ubiquitous” (64; 40%) if similar levels of expression were observed in all tissues, as “widespread” (57; 36%) if expression was observed in several but not all tissues (frequently with different levels in different tissues), as “restricted” (29; 18%) if transcripts were localized to just a few regions in at least one of the stages examined, and as “undetectable” (10; 6%). The expression patterns of all the restricted cDNAs and of one ubiquitous and two widespread clones are illustrated in Figure 1 and described in the Appendix. Details of the restricted cDNAs are summarized in Table 2, which lists the clones in the same order as in Figure 1, with the first three being members of the visceral endoderm synexpression group (see below). A Supplement to Table 2 (available online at www.genome.org) lists the cDNAs with widespread and ubiquitous expression.
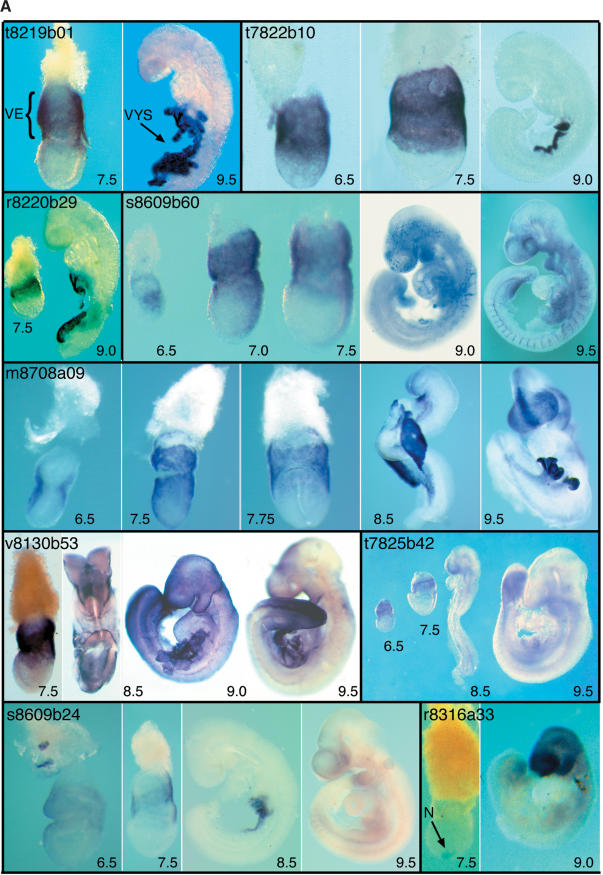
Figure 1.
Images of the expression patterns of all the “restricted” genes (beginning with the three genes in the synexpression group), two of the widespread cDNAs, and one ubiquitously expressed sequence. Images representing individual clones are enclosed by black lines; sequence identifiers and other information are indicated on the figures. A more detailed description of the expression patterns, together with explanations of the annotations, is provided in the Appendix. The restricted genes are listed in the same order as in Table 2.
Of the 29 restricted expression patterns identified, 22 are expressed in the tissues from which the library was made, of which three (t8219b01, t7822b10, and r8220b29) are exclusively expressed in these tissues. Seven genes were not expressed at detectable levels in the source tissues (w8609b57, r8220b09, t8130b26, m8708a39, r8220b57, r8319a44, t8219b26). Examination of the restricted expression patterns revealed just one group of genes with a similar expression pattern at all stages examined (6.5–9.5 dpc). This synexpression group (Niehrs and Pollet 1999) comprises the three clones, t8219b01, t7822b10, and r8220b29, that are expressed exclusively in the tissues from which the Endoderm library was constructed. All three are expressed in VE at 6.5 and 7.5 dpc and in the yolk sac at 8.5 and 9.5 dpc (Fig. 1). Of the three, only t7822b10 has been described previously. It encodes a receptor-type protein tyrosine phosphatase termed Ptpt9, the loss of function of which causes abnormalities of the central and peripheral nervous systems and of the neuroendocrine system (Elchebly et al. 1999; Wallace et al. 1999; Batt et al. 2002). We do not know whether the three genes have related functions, because no known motifs have been identified in t8219b01 or r8220b29. Ptpt9 maps to chromosome 17 (54.5Mb), whereas t8219b01 maps to chromosome 8 (60.6Mb) and r8220b29 to chromosome 5 (127.4Mb). The coordinated expression of the three genes is therefore unlikely to be a consequence of their genomic organization.
In addition to this single synexpression group, we have also identified three “coexpression groups,” all members of which are expressed in the same tissue at a particular stage of development and therefore may cooperate in the specification of that tissue in which they are expressed. Members of a coexpression group may also be expressed in other regions, and their expression patterns at earlier and later stages may also diverge. In defining these groups, we omit the ubiquitously expressed and widespread clones (which are likely to have housekeeping functions), and focus particularly on the signaling centers in the 7.5-dpc embryo from which the Endoderm library was derived. Thus, Table 3 lists the clones expressed in the VE coexpression group (the largest) and the node and definitive endoderm coexpression groups.
Table 3.
Coexpression Groups
| Coexpression group | Clones |
|---|---|
| Visceral endoderm | s8609b60, m8708a09, v8130b53, t7825b42, s8609b24, v8130b25 |
| Node | v8130b25, r8316a33, m8708a22, p7822b53, t8130b59 |
| Gut endoderm | v8130b25, r8707a53 |
The three coexpression groups are based on the constituent tissues of the 7.5-dpc mouse embryo. Only genes with restricted expression pattens are included.
DISCUSSION
Endoderm cDNA Sequence Analysis
At 7.5 dpc, the endoderm that surrounds the embryonic region of the mouse conceptus, from which the endoderm library is derived, is a single layer of ∼700 cells (Snow 1977). This tissue comprises the node (which is required to establish the anteriorposterior, dorso-ventral, and left-right axes of the embryo), VE (which is important for nutrient exchange and for initiating anterior patterning), and the definitive endoderm (which is also involved in anterior patterning; Anderson and Beddington 1997; Beddington and Robertson 1999; Bielinska et al. 1999; Lu et al. 2001; Hamada et al. 2002). The node, VE, and definitive endoderm go on to form the notochord and floor plate of the neural tube, yolk sac endoderm, and gut endoderm (GE) respectively.
Although the mouse genome has been almost completely sequenced (Waterston et al. 2002), our data indicate that transcript identification is incomplete. Indeed, sequencing of just 1978 clones of the Endoderm library has identified no fewer than 123 novel cDNAs. Therefore, our work provides a valuable source of ESTs, which will be useful in functional genomic projects and expression profiling. Further sequencing of the library will be required to draw conclusions about the complexity of gene expression in the endoderm, but we note that two-thirds of the 1978 sequences analyzed were represented only once, indicating that many more transcripts remain to be isolated from the original 5.8 × 105 independent clones (Harrison et al. 1995).
Endoderm cDNA Expression Analysis
In this article we have studied the expression patterns of 160 cDNAs derived from a mouse endoderm cDNA library. Our screen differs from related screens (Gawantka et al. 1998; Neidhardt et al. 2000; Christiansen et al. 2001; Kudoh et al. 2001) because cDNAs were sequenced and clustered before carrying out expression analyses. This ensured that each transcript was studied only once, an important issue when analyzing mouse development because obtaining mouse embryos at the appropriate stages is more costly and time-consuming than doing the same in chicken, frog, or fish.
Many transcripts proved to have ubiquitous or widespread expression patterns, but the expression of 29 (18%) was restricted to particular tissues at least in one of the time points examined. Such cDNAs are of interest because they may provide useful molecular markers for those tissues and because their expression patterns may provide hints as to their developmental functions.
A sequence-based approach such as that taken here may assist in the identification of cDNAs with restricted expression patterns. In addressing this point, it is difficult to make direct comparisons with other screens because definitions of “restricted” may vary, because other screens have used different species at different stages, and because of the way in which cDNA clones were selected. Nevertheless, screens that have selected cDNAs at random, whether using parent libraries or even subtracted cDNA libraries, have tended to obtain lower proportions of restricted expression patterns than those described in this article (Table 4; Neidhardt et al. 2000; Christiansen et al. 2001; Kudoh et al. 2001). In contrast, a screen making use of a library normalized by colony hybridization rather than by sequence analysis (Neidhardt et al. 2000) obtained a very similar proportion to that reported here, emphasizing the importance of normalization in screens of this sort, especially when material might be limiting. We opted to use of the parent cDNA library rather than a subtracted version so as to avoid the loss of rare clones. Interestingly, a similar percentage of restricted patterns at 9.5 dpc was obtained in an expression analysis of murine orthologs of all genes on human chromosome 21 (Gitton et al. 2002; Reymond et al. 2002).
Table 4.
Frequency of Selection of Restricted cDNAs in Different Expression Screens
| Reference | Species | Stages screened | cDNA library | Library type | Number of clones screened | Restricted cDNAs |
|---|---|---|---|---|---|---|
| This study | Mouse | 6.5—9.5 dpc | 7.5-dpc endoderm | Parent | 160 | 18% |
| Neidhardt et al. 2000 | Mouse | 9.5 dpc | 9.5-dpc embryo | Parent | 989 | 6% |
| Neidhardt et al. 2000 | Mouse | 9.5 dpc | 9.5-dpc embryo | Subtracted | 3737 | 7% |
| Neidhardt et al. 2000 | Mouse | 9.5 dpc | 9.5-dpc embryo | Normalized | 622 | 18% |
| Reymond et al. 2002 | Mouse | 9.5 dpc | Orthologues of human chromosome 21 genes | — | 158 | 21% |
| Reymond et al. 2002 | Mouse | 10.5 dpc | idem | — | 158 | 28% |
| Reymond et al. 2002 | Mouse | 14.5 dpc (sections) | idem | — | 158 | 42% |
| Gitton et al. 2002 | Mouse | 9.5 dpc | idem | — | 158 | 21% |
| Christiansen et al. 2001 | Chick | HH* 9—12 | Hindbrain HH* 10—11a | Subtracted | 445 | 8% |
| Kudoh et al. 2001 | Zebrafish | Shield, 3 somites, 15 somites, 24 hpf | Early somitogenesis embryo | Normalized | 2765 | 13% |
| Gawantka et al. 1998 | X. laevis | Stages 10+, 13, 30 | Neurula stage embryo | Parent | 1765 | 25%b |
Stage according to Hamburger and Hamilton (1951)
This figure is reduced to 16% if one considers only unique cDNAs with a restricted expression pattern
One benefit of a screen such as this is that it enables the definition of sets of coregulated genes, or “synexpression groups” (Niehrs and Pollet 1999) as well as coexpression groups. In defining such groups, we omit widespread and ubiquitous clones so as to exclude “housekeeping” genes. As described above, we found a single synexpression group, which comprises genes expressed in the VE at 6.5 and 7.5 dpc and in the yolk sac at 8.5 and 9.5 dpc. In addition, we defined coexpression groups for VE, node, and GE. Each coexpression group contains the genes that are expressed in the tissue in question at 7.5 dpc (Table 3), with the VE group containing six clones; the node group, five clones; and the definitive gut group, two clones. Members of a coexpression group may cooperate in the specification or function of the tissue in question.
METHODS
Endoderm cDNA Sequence Analysis
Clones from the Endoderm library were randomly picked and gridded into 384-well plates (Genetix Ltd) using an automated colony picker (Meier-Ewert et al. 1993). They were sequenced from the 3′ end, vector sequence was removed, and repeats and regions of poor quality were masked by using PHRED (http://www.phrap.org/phrap.docs/phred.html). Sequences containing <200 nucleotides were not analyzed further. Sequence data have been submitted to the EMBL database.
BLASTN (NCBI: ftp://ftp.ncbi.nih.gov/blast/executables/) was used to compare each sequence with two publicly available mouse gene data sets: the ENSEMBL gene predictions for mouse (version 8.3c.1, July 12, 2002; http://www.ensembl.org/Mus_musculus) and the TIGR Gene Index (TIGR-Tentative Consensus sequences or TCs version 8.0, June 1, 2002; http://www.tigr.org/tdb/tgi/mgi/). Alignments were inspected manually, and possible homology or novelty was further investigated by using BLASTP (NCBI: ftp://ftp.ncbi.nih.gov/blast/executables/). Sequences that failed to match an ENSEMBL gene or a TIGR TC were considered as potentially novel. To determine redundancy within the clone set, sequences that mapped to the same ENSEMBL predicted gene were considered redundant. Similarly, sequences that lacked an ENSEMBL mapping but shared the same TIGR TC were considered redundant. Sequences that failed to map to an ENSEMBL prediction or a TIGR TC were considered nonredundant.
RNA In Situ Hybridization
Mouse embryos were collected from CBA/Ca × C57Bl10or C57BL6 × C57BL6 matings at 6.5, 7.5, 8.5, and 9.5 dpc. Extra-embryonic membranes were removed in M2 medium (Hogan et al. 1994) containing 10% fetal calf serum. Embryos were fixed overnight in 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS) at 4°C, after which they were dehydrated in increasing concentrations of methanol in PBS and stored in 100% methanol at -20°C until use. Antisense RNA probes were generated as described (Harrison et al. 1995) and whole-mount RNA in situ hybridization (WISH) was performed according to the method of Wilkinson (1992). Hybridization conditions were those of Rosen and Beddington (1993), except that embryo powder was omitted from the procedure, and treatment with 10mg/mL proteinase K was 5 min for embryos at 6.5 to 7.5 dpc and 12 min for embryos at 8.5 to 9.5 dpc. Embryos were processed in 12-well plates (Costar) in 12-μm mesh nets for embryos at ≤7.5 dpc, and 74-μm mesh nets for embryos at ≥8.5 dpc. At least three embryos of each stage were examined for each probe, and restricted expression patterns were confirmed by an independent set of hybridizations. After stopping the staining reaction, embryos were postfixed in 4% PFA, 0.1% glutaraldehyde in PBS for 1 h at room temperature and stored in 0.4% PFA at 4°C. Photographs were taken by using a dissecting microscope (Nikon) and tungsten film (Kodak 64T). Images were digitized by using a Polaroid SprintScan 35 scanner.
Acknowledgments
Dedicated to the memory of Rosa Beddington (March 23, 1956 to May 18, 2001).
This work was supported by an MRC Special Project Grant (G9118913) and EEC Contract PL 962414. S.G. is an NHMRC RD Wright Fellow. R.S.-N. is a Gulbenkian PhD Program in Biology and Medicine student and was funded by the Portuguese Foundation for Science and Technology. J.M.B. was supported by a Human Frontier Science Programme Long Term Fellowship. S.L.D. is a Pharmacia Foundation of Australia Senior Research Fellow. We are grateful to B. Gorick and the Human Genome Mapping Project at Hinxton UK for help with replication of the 7.5-dpc mouse endoderm library, and to Michael Wiles and Patricia Ruiz for initial sequence analysis. We also thank Simon Bullock, Juan Pedro Martinez Barbera, and Tristan Rodriguez for their help and advice throughout the course of this work and their comments on the manuscript. “Restricted” expression patterns have been submitted to the Mouse Gene Expression database (GXD) http://www.informatics.jax.org/mgihome/GXD/aboutGXD.shtml.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
APPENDIX
Expression Patterns of “Restricted”cDNAs
t8219b01
At the mid streak stage, expression of clone t8219b01 is detected in the VE. Expression at later stages is restricted to the visceral yolk sac (VYS).
t7822b10
At 6.0dpc, Ptpt9 expression is restricted to the VE and later to the VYS. Loss of function of this gene has been reported to cause abnormalities of the central and peripheral nervous systems and of the neuroendocrine system (Elchebly et al. 1999; Wallace et al. 1999; Batt et al. 2002).
r8220b29
At the mid streak stage, clone r8220b29 expression is detected in the VE (7.5 dpc). Expression at later stages (9.0dpc) is restricted to the VYS.
s8609b60
At the onset of gastrulation, Sgk is strongly expressed in the VE overlying the nascent mesodermal wings and, more weakly, in the mesoderm itself. Transcripts are also detected in the VE overlying the extra-embryonic ectoderm. As gastrulation proceeds, the latter domain of expression becomes more robust, and in the embryo proper, it is strongest in the regions juxtaposing the primitive streak. At 8.5 and 9.5 dpc, Sgk transcripts are found in the vasculature as well as in the eye and branchial arches. Loss of function of this gene reduces the ability of mice to reduce Na+ excretion when subjected to dietary NaCl restriction (Wulff et al. 2002). The expression pattern of Sgk has been described by Lee and colleagues (2001).
m8708a09
At 6.5 dpc, Embigin is strongly expressed in the VE at the junction between extra-embryonic and embryonic portions of the conceptus. By 7.5 dpc, expression occurs throughout the VE and, more weakly, in the definitive endoderm. At head-fold stages, Embigin transcription occurs in anterior definitive endoderm, with strong expression also detectable in the VE. At 8.5 dpc, transcripts are present in the forebrain neuroepithelium, the foregut diverticulum, and the yolk sac. By 9.5 dpc, expression is strong in forebrain neuroepithelium (especially in the dorsal midline) and also occurs in the mid- and hindbrain. Transcripts are also detectable in branchial arches and the nephrogenic cord. The early expression pattern of this gene has been described by Shimono and Behringer (1999); later stages, by Fan and colleagues (1998).
v8130b53
At the late gastrula stage, strong expression of Slc2a3 is detected in the VE (7.5 dpc). Later, expression is seen in the surface ectoderm (8.5 and 9.0dpc) and the VYS. As development proceeds, expression in surface ectoderm persists but decreases anteriorly. Expression in the yolk sac is still detectable at 9.5 and 10.5 dpc (data not shown).
t7825b42
At egg cylinder stages, mouse Sp120 is most strongly expressed in the extra-embryonic half of the conceptus, with only weak expression in the embryonic half, mostly in the primitive streak. At 8.5 and 9.5 dpc, robust expression is seen in the tailbud and presomitic mesoderm, when transcripts are also present in ventral forebrain, branchial arches, and the limb buds.
s8609b24
At egg cylinder stages, expression of s8609b24 occurs in the VE overlying the extra-embryonic portion of the conceptus and the most proximal region of the epiblast. At 6.5 dpc, VE expression covers most of the conceptus, although it is weaker distally and completely absent from the most proximal region. At 7.5 dpc, expression persists in the progeny of the VE cells, coming to lie over the extra-embryonic ectoderm; transcripts are still absent from the most proximal VE. By 8.5 dpc, expression is confined to the yolk sac, but at 9.5 dpc, there is widespread, albeit weak, expression in the embryo proper, particularly in the forebrain, anterior midbrain, branchial arches, and gut.
r8316a33
mD2LIC expression is first detected in the node at the mid to late streak stage. Expression persists in the node at the late neural plate/early head-fold stage, but is reduced by the eight-somite stage when widespread expression is detectable throughout the embryo (data not shown). This widespread expression persists and becomes stronger in the 25-somite stage embryo. By 11 dpc, expression is detected in GE and the heart (data not shown).
v8130b25
At 6.5 dpc, expression of v8130b25 is restricted to the VE overlying the embryonic and extra-embryonic ectoderm. By 7.5 dpc, expression is observed in the node, and at 8.5 dpc, this gene is strongly expressed in the VYS, GE, and developing blood cells. By 9.5 dpc, expression is associated with the vasculature, heart, branchial arch, and brain.
r8707a53
Expression of Claudin-6 is detectable in the forebrain, in the VYS, and throughout the GE from 9.0dpc. At 9.0dpc, expression in the forebrain is predominantly ventral, whereas at 9.5 dpc, it is mainly dorsal.
m8708a22
Calcyphosine is weakly expressed in extra-embryonic ectoderm at 6.5 dpc (data not shown). At 7.0dpc, expression occurs throughout the extra-embryonic ectoderm and the epiblast, with maximal expression in the node. During elongation of the streak, highest expression is seen in the node. At 8.5 and 9.5 dpc, expression is ubiquitous.
p7822b53
Expression of p7822b53 is restricted to the node of the gastrulating embryo.
t8130b59
Expression of clone t8130b59 is detectable in the node at 7.5 dpc and in the branchial arches and otic vesicles at 9.5 dpc.
t8417b56
At 6.5 dpc, Neuronatin is expressed weakly in the embryonic half of the conceptus. By 7.0dpc, transcripts are present throughout the mesoderm and ectoderm, and maximal expression is then seen in the posterior head-folds (arrows). At 8.5 to 9.5 dpc, Neuronatin expression is detectable in the ventral forebrain, branchial arches, and foregut diverticulum. Forebrain expression is more widespread at this time, and expression also occurs throughout the trunk mesoderm. Expression of neuronatin at 8.5 and 9.5 dpc has also been described by Wijnholds et al. (1995), who detected expression in rhombomeres 3 and 5 of the hindbrain.
t8219b25
Before gastrulation, weak transcription of t8219b25 occurs throughout the epiblast, and this widespread embryonic expression persists until the late head-fold stage (8.0dpc). By 8.5 to 9.0 dpc, expression is detected in the diencephalon and midbrain, with weaker expression in the hindbrain and spinal cord. At 9.5 dpc, expression occurs in the roofplate and first branchial arch, with elevated expression detected in the hindbrain and anterior spinal cord.
w8609b57
At the onset of gastrulation, Neuroblastoma is strongly expressed in a single domain comprising the most proximal region of the egg cylinder and a proximo-distal stripe within the ectoplacental cone (arrow). This domain persists during head-fold stages, when the gene becomes weakly expressed throughout the VE and more strongly in the head-fold pocket, and notochord (7.5 dpc; arrow). At 8.5 dpc, expression is strong in notochord and ventral forebrain, with weak activation in the foregut diverticulum. By 9.5 dpc, epithelial expression extends from the ventral forebrain to the fourth branchial arch, with transcription also occurring in the otic vesicle.
t7822b19
At 6.5 dpc, Peg3 expression occurs in the anterior VE (long arrow) and the primitive streak (short arrow). By 7.5 dpc, expression is widespread in embryonic mesoderm and allantois. Loss of function of Peg3 causes growth retardation and an impairment of maternal behavior that frequently results in death of the off-spring (Li et al. 1999).
k8709a24
At egg cylinder stages, Sall1 is expressed in the anterior and, more weakly, in the posterior epiblast. At head-fold stages, transcripts become restricted to anterior neural folds, and at 8.5 dpc, this expression resolves into ventral neural plate and neural groove. Weak expression is also seen in the branchial arch region and posterior trunk. At 9.5 dpc, Sall1 is expressed in the ventral forebrain, anterior midbrain, the midbrain/hindbrain boundary, branchial arch ectoderm, posterior trunk, and, most prominently, mesonephros and presomitic mesoderm and somites. SALL1 is implicated in Townes-Brocks syndrome (Kohlhase et al. 1998), and loss of function of Sall1 indicates that the gene is required for ureteric bud invasion during kidney development (Nishinakamura et al. 2001). Expression of Sall1 at 7.5, 8.5, and 9.5 dpc has been reported by Buck and colleagues (2001).
r8220b09
Expression of Silg41 occurs in the extra-embryonic ectoderm before and at the onset of gastrulation, at 6.0to 6.5 dpc.
t8130b26
Rb-BP2expression is restricted to the embryonic ectoderm from 6.0–7.5 dpc. By 7.75 dpc, transcripts are strongly detectable in the anterior definitive endoderm as well as in the chorion and allantois. By 8.5 dpc, expression is restricted to the forebrain.
m8708a39
At 6.5 dpc, Lztr-1 is expressed in the epiblast and in extraembryonic ectoderm and/or endoderm adjacent to the ectoplacental cone (arrow). At 7.5 dpc, although expression is widespread in the embryonic region, it is stronger posteriorly and downregulated in the node. At head-fold stages, Lztr-1 expression is most prominent in the neural folds and nascent neural tube. At 9.5 dpc, expression is high in the forebrain, branchial arches, and limb buds.
s8129b58
At the onset of gastrulation, Pancortin-1 and/or -3 is expressed at the junction between embryonic and extra-embryonic portions of the conceptus, with higher levels anteriorly. As gastrulation proceeds, expression occurs in the amnion and chorion and becomes widespread within the embryo proper. During somatogenesis (8.5 dpc), expression becomes restricted to rhombomere 4 (arrow), to the junction between the diencephalon and mesencephalon, and to anterior and posterior portions of trunk mesenchyme. At 9.5 dpc, spotty expression is detectable in the midbrain in the earliest differentiating neurons. Expression also occurs in the olfactory placodes and in some cranial ganglia. Expression in the limb buds is initially widespread but becomes restricted to posterior regions as development proceeds. Expression of the closely related genes Noelin 1 and 2 at 10.5 dpc has been described by Moreno and Bronner-Fraser (2002). The expression pattern they describe is similar, although not identical, to that described in this article at 9.5 dpc.
k8220b03
At 7.5 dpc, Shrm expression is detected throughout all embryonic tissues. Particularly strong expression occurs in the rostral region of presomitic mesoderm and later in the most posterior somites. Weaker expression is detected in the neural epithelium at 8.5 dpc. Somitic expression persists in older embryos, particularly in cells giving rise to ventral sclerotome. At 9.5 dpc, there is weak expression in the brain. Loss-of-function experiments indicate that Shroom, an actin-binding protein, is required for neural tube morphogenesis (Hildebrand and Soriano 1999).
r8220b57
Expression of Tif-1 β is restricted to the advancing primitive streak at 7.5 dpc, and later at 9.0dpc, it is strongest in the tailbud, presomitic mesoderm, nascent somites, branchial arches, and limb buds.
r8319a44
Expression of 14-3-3 σ is detected at the onset of gastrulation (6.5 dpc) and up to late streak stages (7.5 dpc) in the extra-embryonic ectoderm and ectoplacental cone. At the onset of gastrulation, transcripts are localized to the apical surface of cells (arrow). At somites stages (8.5, 9.5 dpc), expression occurs in surface ectoderm precursors along the distal edges of the neural folds and then, briefly, in a thin line above the neural tube. Expression is observed in branchial arches.
k8709a20
14-3-3 ε is ubiquitously expressed at 6.5 dpc but is then downregulated such that by 7.5 dpc, transcripts are barely detectable. At 8.5 dpc, weak expression occurs in the forebrain and heart. At 9.5 dpc, forebrain expression is prominent, together with strong expression in the midbrain and branchial arches. These observations complement work by McConnell and colleagues (1995), which has analyzed expression of 14-3-3 ε from 8.5 dpc and found that expression is high in neural tissue by 12.5 dpc.
k8710a07
Nsa2p is expressed throughout the epiblast and extraembryonic ectoderm at 6.5 dpc. At 7.5 dpc, it continues to be expressed in all internal cell layers of the conceptus. By 8.5 and 9.5 dpc, expression is strongest in the branchial arches, neural tube, and, particularly, the forebrain. Low-level expression also occurs throughout the lateral mesoderm.
t8219b26
Expression of Ubp7 is detected in the extra-embryonic ectoderm at the onset of gastrulation (6.5 dpc) and in the primitive streak and emerging mesoderm during gastrulation (7.5 dpc). At 7.75 and 9.5 dpc, widespread expression occurs in some mesodermal derivatives.
p8224a43
An example of a “widespread” cDNA. Expression is ubiquitous but occurs at different levels in different tissues.
t8130b25
An example of a “widespread” cDNA. Expression is ubiquitous but occurs at different levels in different tissues.
k8311b01
An example of a “ubiquitous” cDNA Expression is completely ubiquitous in both embryonic and extra-embryonic tissues.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1362303. Article published online before print in November 2003.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Adams, M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185-2195. [DOI] [PubMed] [Google Scholar]
- Anderson, K. and Beddington, R. 1997. Pattern formation and developmental mechanisms. Curr. Opin. Genet. Dev. 7: 455-458. [DOI] [PubMed] [Google Scholar]
- Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J.M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. 2002. A radiation hybrid transcript map of the mouse genome. Nat. Genet. 29: 194-200. [DOI] [PubMed] [Google Scholar]
- Avner, P., Bruls, T., Poras, I., Eley, L., Gas, S., Ruiz, P., Wiles, M.V., Sousa-Nunes, R., Kettleborough, R., Rana, A. 2001. A radiation hybrid transcript map of the mouse genome. Nat. Genet. 29: 194-200. [DOI] [PubMed] [Google Scholar]
- Batt, J., Asa, S., Fladd, C., and Rotin, D. 2002. Pituitary, pancreatic and gut neuroendocrine defects in protein tyrosine phosphatase-sigma–deficient mice. Mol. Endocrinol. 16: 155-169. [DOI] [PubMed] [Google Scholar]
- Beddington, R.S.P. and Robertson, E.J. 1999. Axis development and early asymmetry in mammals. Cell 96: 195-209. [DOI] [PubMed] [Google Scholar]
- Bielinska, M., Narita, N., and Wilson, D.B. 1999. Distinct roles for visceral endoderm during embryonic mouse development. Int. J. Dev. Biol. 43: 183-205. [PubMed] [Google Scholar]
- Buck, A., Kispert, A., and Kohlhase, J. 2001. Embryonic expression of the murine homologue of SALL1, the gene mutated in Townes-Brocks syndrome. Mech. Dev. 104: 143-146. [DOI] [PubMed] [Google Scholar]
- The C. elegans genome consortium. 1998. Genome sequence of the nematode C. elegans: A platform for investigating biology. The C. elegans Sequencing Consortium. Science 282: 2012-2018. [DOI] [PubMed] [Google Scholar]
- Carlton, J.M., Angiuoli, S.V., Suh, B.B., Kooij, T.W., Pertea, M., Silva, J.C., Ermolaeva, M.D., Allen, J.E., Selengut, J.D., Koo, H.L., et al. 2002. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature 419: 512-519. [DOI] [PubMed] [Google Scholar]
- Christiansen, J.H., Coles, E.G., Robinson, V., Pasini, A., and Wilkinson, D.G. 2001. Screening from a subtracted embryonic chick hindbrain cDNA library: Identification of genes expressed during hindbrain, midbrain and cranial neural crest development. Mech. Dev. 102: 119-133. [DOI] [PubMed] [Google Scholar]
- Dehal, P., Satou, Y., Campbell, R.K., Chapman, J., Degnan, B., De Tomaso, A., Davidson, B., Di Gregorio, A., Gelpke, M., Goodstein, D.M., et al. 2002. The draft genome of Ciona intestinalis: Insights into chordate and vertebrate origins. Science 298: 2157-2167. [DOI] [PubMed] [Google Scholar]
- Doetschman, T., Gregg, R.G., Maeda, N., Hooper, M.L., Melton, D.W., Thompson, S., and Smithies, O. 1987. Targetted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature 330: 576-578. [DOI] [PubMed] [Google Scholar]
- Dunwoodie, S.L. and Beddington, R.S.P. 2002. The expression of the imprinted gene lpl is restricted to extra-embryonic tissues and embryonic lateral mesoderm during early mouse development. Int. J. Dev. Biol. 46: 459-466. [PubMed] [Google Scholar]
- Dunwoodie, S.L., Henrique, D., Harrison, S.M., and Beddington, R.S.P. 1997. Mouse Dll3: A novel divergent Δ gene which may complement the function of other Δ homologues during early pattern formation in the mouse embryo. Development 124: 3065-3076. [DOI] [PubMed] [Google Scholar]
- Dunwoodie, S.L., Rodriguez, T.A., and Beddington, R.S.P. 1998. Msg1 and Mrg1, founding members of a gene family, show distinct patterns of gene expression during mouse embryogenesis. Mech. Dev. 72: 27-40. [DOI] [PubMed] [Google Scholar]
- Dunwoodie, S.L., Clements, M., Sparrow, D.B., Sa, X., Conlon, R.A., and Beddington, R.S.P. 2002. Axial skeletal defects caused by mutation in the spondylocostal dysplasia/pudgy gene Dll3 are associated with disruption of the segmentation clock within the presomitic mesoderm. Development 129: 1795-1806. [DOI] [PubMed] [Google Scholar]
- Elchebly, M., Wagner, J., Kennedy, T.E., Lanctot, C., Michaliszyn, E., Itie, A., Drouin, J., and Tremblay, M.L. 1999. Neuroendocrine dysplasia in mice lacking protein tyrosine phosphatase sigma. Nat. Genet. 21: 330-333. [DOI] [PubMed] [Google Scholar]
- Fan, Q.W., Kadomatsu, K., Uchimura, K., and Muramatsu, T. 1998. Embigin/basigin subgroup of the immunoglobulin superfamily: Different modes of expression during mouse embryogenesis and correlated expression with carbohydrate antigenic markers. Dev. Growth Diff. 40: 277-286. [DOI] [PubMed] [Google Scholar]
- Fraser, A.G., Kamath, R.S., Zipperlen, P., Martinez-Campos, M., Sohrmann, M., and Ahringer, J. 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408: 325-330. [DOI] [PubMed] [Google Scholar]
- Gardner, M.J., Hall, N., Fung, E., White, O., Berriman, M., Hyman, R.W., Carlton, J.M., Pain, A., Nelson, K.E., Bowman, S., et al. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419: 498-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawantka, V., Pollet, N., Delius, H., Vingron, M., Pfister, R., Nitsch, R., Blumenstock, C., and Niehrs, C. 1998. Gene expression screening in Xenopus identifies molecular pathways, predicts gene function and provides a global view of embryonic patterning. Mech. Dev. 77: 95-141. [DOI] [PubMed] [Google Scholar]
- Gitton, Y., Dahmane, N., Baik, S., Ruiz i Altaba, A., Neidhardt, L., Scholze, M., Herrmann, B.G., Kahlem, P., Benkahla, A., Schrinner, S., et al. 2002. A gene expression map of human chromosome 21 orthologues in the mouse. Nature 420: 586-590. [DOI] [PubMed] [Google Scholar]
- Hamada, H., Meno, C., Watanabe, D., and Saijoh, Y. 2002. Establishment of vertebrate left-right asymmetry. Nat. Rev. Genet. 3: 103-113. [DOI] [PubMed] [Google Scholar]
- Hamburger, V. and Hamilton, H.L. 1951. A series of normal stages in the development of the chick embryo. J. Morphol. 88: 49-92. [PubMed] [Google Scholar]
- Harrison, S.M., Dunwoodie, S.L., Arkell, R.M., Lehrach, H., and Beddington, R.S.P. 1995. Isolation of novel tissue-specific genes from cDNA libraries representing the individual tissue constituents of the gastrulating mouse embryo. Development 121: 2479-2489. [DOI] [PubMed] [Google Scholar]
- Harrison, S.M., Houzelstein, D., Dunwoodie, S.L., and Beddington, R.S.P. 2000. Sp5, a new member of the Sp1 family, is dynamically expressed during development and genetically interacts with Brachyury. Dev. Biol. 227: 358-372. [DOI] [PubMed] [Google Scholar]
- Hildebrand, J.D. and Soriano, P. 1999. Shroom, a PDZ domain-containing actin-binding protein, is required for neural tube morphogenesis in mice. Cell 99: 485-497. [DOI] [PubMed] [Google Scholar]
- Hogan, B., Beddington, R., Costantini, F., and Lacy, E. 1994. Manipulating the mouse embryo: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Kamath, R.S., Fraser, A.G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohrmann, M., et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231-237. [DOI] [PubMed] [Google Scholar]
- Kohlhase, J., Wischermann, A., Reichenbach, H., Froster, U., and Engel, W. 1998. Mutations in the SALL1 putative transcription factor gene cause Townes-Brocks syndrome. Nat. Genet. 18: 81-83. [DOI] [PubMed] [Google Scholar]
- Kudoh, T., Tsang, M., Hukriede, N.A., Chen, X., Dedekian, M., Clarke, C.J., Kiang, A., Schultz, S., Epstein, J.A., Toyama, R., et al. 2001. A gene expression screen in zebrafish embryogenesis. Genome Res. 11: 1979-1987. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Lee, E., Lein, E.S., and Firestone, G.L. 2001. Tissue-specific expression of the transcriptionally regulated serum and glucocorticoid-inducible protein kinase (Sgk) during mouse embryogenesis. Mech. Dev. 103: 177-181. [DOI] [PubMed] [Google Scholar]
- Li, L., Keverne, E.B., Aparicio, S.A., Ishino, F., Barton, S.C., and Surani, M.A. 1999. Regulation of maternal behavior and offspring growth by paternally expressed Peg3. Science 284: 330-333. [DOI] [PubMed] [Google Scholar]
- Lu, C.C., Brennan, J., and Robertson, E.J. 2001. From fertilization to gastrulation: Axis formation in the mouse embryo. Curr. Opin. Genet. Dev. 11: 384-392. [DOI] [PubMed] [Google Scholar]
- Martinez Barbera, J.P., Rodriguez, T.A., Greene, N.D., Weninger, W.J., Simeone, A., Copp, A.J., Beddington, R.S.P., and Dunwoodie, S. 2002. Folic acid prevents exencephaly in Cited2 deficient mice. Hum. Mol. Genet. 11: 283-293. [DOI] [PubMed] [Google Scholar]
- McConnell, J.E., Armstrong, J.F., Hodges, P.E., and Bard, J.B. 1995. The mouse 14-3-3 epsilon isoform, a kinase regulator whose expression pattern is modulated in mesenchyme and neuronal differentiation. Dev. Biol. 169: 218-228. [DOI] [PubMed] [Google Scholar]
- Meier-Ewert, S., Maier, E., Ahmadi, A., Curtis, J., and Lehrach, H. 1993. An automated approach to generating expressed sequence catalogues. Nature 361: 375-376. [DOI] [PubMed] [Google Scholar]
- Moreno, T.A. and Bronner-Fraser, M. 2002. Neural expression of mouse Noelin-1/2 and comparison with other vertebrates. Mech. Dev. 119: 121. [DOI] [PubMed] [Google Scholar]
- Neidhardt, L., Gasca, S., Wertz, K., Obermayr, F., Worpenberg, S., Lehrach, H., and Herrmann, B.G. 2000. Large-scale screen for genes controlling mammalian embryogenesis, using high-throughput gene expression analysis in mouse embryos. Mech. Dev. 98: 77-94. [DOI] [PubMed] [Google Scholar]
- Niehrs, C. and Pollet, N. 1999. Synexpression groups in eukaryotes. Nature 402: 483-487. [DOI] [PubMed] [Google Scholar]
- Nishinakamura, R., Matsumoto, Y., Nakao, K., Nakamura, K., Sato, A., Copeland, N.G., Gilbert, D.J., Jenkins, N.A., Scully, S., Lacey, D.L., et al. 2001. Murine homolog of SALL1 is essential for ureteric bud invasion in kidney development. Development 128: 3105-3115. [DOI] [PubMed] [Google Scholar]
- Reymond, A., Marigo, V., Yaylaoglu, M.B., Leoni, A., Ucla, C., Scamuffa, N., Caccioppoli, C., Dermitzakis, E.T., Lyle, R., Banfi, S., et al. 2002. Human chromosome 21 gene expression atlas in the mouse. Nature 420: 582-586. [DOI] [PubMed] [Google Scholar]
- Rosen, B. and Beddington, R.S.P. 1993. Whole-mount in situ hybridization in the mouse embryo: Gene expression in three dimensions. Trends Genet. 9: 162-167. [DOI] [PubMed] [Google Scholar]
- Shimono, A. and Behringer, R.R. 1999. Isolation of novel cDNAs by subtractions between the anterior mesendoderm of single mouse gastrula stage embryos. Dev. Biol. 209: 369-380. [DOI] [PubMed] [Google Scholar]
- Snow, M.H.L. 1977. Gastrulation in the mouse: Growth and regionalization of the epiblast. J. Embryol. Exp. Morph. 42: 293-303. [Google Scholar]
- Stanford, W.L., Cohn, J.B., and Cordes, S.P. 2001. Gene-trap mutagenesis: Past, present and beyond. Nat. Rev. Genet. 2: 756-768. [DOI] [PubMed] [Google Scholar]
- Thomas, K.R. and Capecchi, M.R. 1987. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell 51: 503-512. [DOI] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- Wallace, M.J., Batt, J., Fladd, C.A., Henderson, J.T., Skarnes, W., and Rotin, D. 1999. Neuronal defects and posterior pituitary hypoplasia in mice lacking the receptor tyrosine phosphatase PTPσ. Nat. Genet. 21: 334-338. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Wijnholds, J., Chowdhury, K., Wehr, R., and Gruss, P. 1995. Segment-specific expression of the neuronatin gene during early hindbrain development. Dev. Biol. 171: 73-84. [DOI] [PubMed] [Google Scholar]
- Wilkinson, D.G. 1992. Whole mount in situ hybridisation of vertebrate embryos. In In situ hybridisation, pp. 75-83. IRL Press, Oxford, UK.
- Wulff, P., Vallon, V., Huang, D.Y., Volkl, H., Yu, F., Richter, K., Jansen, M., Schlunz, M., Klingel, K., Loffing, J., et al. 2002. Impaired renal Na+ retention in the sgk1-knockout mouse. J. Clin. Invest. 110: 1263-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.tigr.org/tdb/tgi/mgi; TIGR Tentative Consensus sequences, version 8.0.
- http://www.ensembl.org/Mus_musculus/; predicted mouse transcripts in ENSEMBL.
- http://www.hgmp.mrc.ac.uk/geneservice/reagents/products/cdna_resources/index.shtml; the UK Human Genome Mapping Project Resource Centre.
- http://www.phrap.org/phrap.docs/phred.html; PHRED.
- ftp://ftp.ncbi.nih.gov/blast/executables/; BLASTN and BLASTP.
- http://www.tigr.org/tdb/tgi/mgi/; TIGR Gene Index.