Abstract
Autoimmune diseases are thought to be initiated by exposures to foreign antigens that cross-react with endogenous molecules. Scleroderma is an autoimmune connective tissue disease in which patients make antibodies to a limited group of autoantigens, including RPC1, encoded by the POLR3A gene. As patients with scleroderma and antibodies against RPC1 are at increased risk for cancer, we hypothesized that the “foreign” antigens in this autoimmune disease are encoded by somatically mutated genes in the patients’ incipient cancers. Studying cancers from scleroderma patients, we found genetic alterations of the POLR3A locus in six of eight patients with antibodies to RPC1 but not in eight patients without antibodies to RPC1. Analyses of peripheral blood lymphocytes and serum suggested that POLR3A mutations triggered cellular immunity and cross-reactive humoral immune responses. These results offer insight into the pathogenesis of scleroderma and provide support for the idea that acquired immunity helps to control naturally occurring cancers.
Systemic sclerosis (scleroderma) is a chronic autoimmune rheumatic disease associated with widespread obliterative vasculopathy and tissue fibrosis (1, 2). A striking feature of this disease is the temporal clustering of scleroderma and cancer that has been observed in patients with autoantibodies to RNA polymerase III subunit (RPC1) but not in patients with autoantibodies to topoisomerase 1 (TOP1) or centromere protein B (CENPB) (3). A variety of potential mechanisms could explain the occurrence of cancers in scleroderma patients with autoantibodies to RPC1 (4). For example, it is possible that a defective immune system responsible for the autoimmune disease predisposes to neoplasia, and that this effect is more prominent in patients with antibodies to RPC1 than in the other subgroups. Alternatively, it is possible that the cytotoxic, mutagenic therapies used to treat scleroderma patients with more fulminant disease leads to cancer in these individuals; patients with antibodies to RPC1 tend to have more severe disease than those with other antibodies. Finally, the reverse scenario is possible: Cancer might trigger scleroderma in patients with antibodies to RPC1. In particular, we considered whether occasional cancers might harbor missense mutations in the polymerase III polypeptide A (POLR3A) gene. If the altered protein encoded by the mutant POLR3A gene were recognized by the patient’s immune system, an immune response against the tumor could theoretically be generated. If cross-reactive with the normal RPC1 protein, this immune response could in turn injure selected tissues, thereby inducing scleroderma. Experiments to test this hypothesis were performed, as described below.
Genetic Analysis
We began by searching for missense mutations in the POLR3A gene in tumors from scleroderma patients. We collected tumor and normal tissue samples from eight scleroderma patients who had autoantibodies to RPC1. We also evaluated eight scleroderma patients who had autoantibodies to TOP1 or to CENPB and developed cancers (Table 1). Five of the patients with antibodies to RPC1 developed cancer before scleroderma (median of 0.4 years before scleroderma onset), whereas the remaining three developed cancer 0.3 to 2.5 years after the onset of scleroderma (Table 1). In contrast, patients with autoantibodies to CENPB or TOP1 who developed cancers only did so a median of 14.2 years after the onset of their scleroderma (Table 1). The characteristics of the 16 scleroderma patients, including tumor type, age of diagnosis of cancer, cancer-scleroderma interval, and autoantibody status, are listed in Table 1; additional clinical information is provided in table S1 and (5).
Table 1. Selected clinical and genetic characteristics of the scleroderma patients evaluated in this study.
NA, not applicable.
| Patient no. | Scleroderma duration at diagnosis of cancer (years) | Auto- antibodies to: | Age at diagnosis of cancer | Cancer type | Cancer subtype | Cancer stage | POLR3A mutation (% mutant alleles) | POLR3A mutation (genomic position on chr. 10) | POLR3A mutation (amino acid change) | POLR3A loss of heterozygosity (LOH) |
|---|---|---|---|---|---|---|---|---|---|---|
| SCL-1 | − 0.2 | RPC1 | 51 | Breast cancer | Invasive ductal | IA | ND† | NA | NA | LOH |
| SCL-2 | − 0.1 | RPC1 | 42.3 | Lung cancer | Small cell carcinoma | I or II* | 26% | 79414962C>G | p.E1072Q | LOH |
| SCL-4 | − 0.4 | RPC1 | 44 | Ovarian cancer | Adenocarcinoma | IIIC | 4.3% | 79407320C>G | p.K1365N | No LOH |
| SCL-13 | 0.3 | RPC1 | 51.1 | Breast cancer | Invasive ductal | IIB | No mutation detected | NA | NA | LOH |
| SCL-35 | −2 | RPC1 | 50.9 | Breast cancer | Ductal carcinoma in situ | 0 | No mutation detected | NA | NA | No LOH |
| SCL-42 | 1.5 | RPC1 | 47.5 | Breast cancer | Invasive ductal | IIA | 31% | 79455393A>G | p.I104T | LOH |
| SCL-81 | − 4.2 | RPC1 | 54.6 | Colorectal cancer | Adenocarcinoma | III | No mutation detected | NA | NA | LOH |
| SCL-82 | 2.5 | RPC1 | 51.1 | Breast cancer | Ductal carcinoma in situ | 0 | No mutation detected | NA | NA | No LOH |
| SCL-5 | 9.2 | TOP1 | 74.6 | Lung cancer | Adenocarcinoma | IB | No mutation detected | NA | NA | No LOH |
| SCL-8 | 0.4 | TOP1 | 65.1 | Breast cancer | Infiltrating lobular | IIIA | No mutation detected | NA | NA | No LOH |
| SCL-11 | 13.4 | TOP1 | 55.7 | Breast cancer | Infiltrating lobular | IIIC | No mutation detected | NA | NA | No LOH |
| SCL-12 | 34 | CENPB | 68.6 | Anal cancer | Squamous cell carcinoma | I | No mutation detected | NA | NA | Uninformative‡ |
| SCL-19 | 34 | TOP1 | 74.1 | Breast cancer | Ductal carcinoma in situ | 0 | No mutation detected | NA | NA | No LOH |
| SCL-24 | 36.9 | CENPB | 64.2 | B cell lymphoma | Extranodal, mantle cell | IV | No mutation detected | NA | NA | No LOH |
| SCL-32 | −2.5 | CENPB | 43.1 | Breast cancer | Invasive ductal | I | No mutation detected | NA | NA | Uninformative‡ |
| SCL-85 | 15 | TOP1 | 52.1 | Breast cancer | Invasive ductal | IIA | No mutation detected | NA | NA | No LOH |
Patient records indicate only that the disease was localized.
No mutation detected.
“Uninformative” indicates that none of the evaluated SNPs were heterozygous in the normal cells of the patient.
Formalin-fixed, paraffin-embedded tumors from each of the 16 patients were microdissected to enrich for neoplastic cell content, and DNA was purified, blunt-ended, and ligated to adapters suitable for library preparation (5). Libraries from peripheral blood cells of each patient were similarly prepared. After amplification of the 32 libraries (16 tumor, 16 matched normal), the polymerase chain reaction (PCR) products were captured by using PCR-generated fragments containing all coding sequences of the POLR3A, TOP1, and CENPB genes (5). The captured fragments were evaluated by sequencing on an Illumina instrument, achieving an average coverage of 516 reads per base of the 53 coding exons of the three genes (range: 95- to 2011-fold).
This sequence revealed three somatic, mis-sense variants in POLR3A and none in TOP1 or CENPB (Table 1). All three variants were in the patients with autoantibodies to RPC1. The three somatic mutations were each validated by massively parallel sequencing of PCR products generated from the regions surrounding the mutations (5). Notably, both the capture approach and the direct-PCR sequencing approach showed that one of the three somatic mutations was decidedly subclonal, that is, was present in only a subset of the neoplastic cells: The fraction of mutant alleles in the lung cancer from patient SCL-2 was only 4.3%, far less than the estimated fraction of neoplastic cells in the microdissected sample used for DNA purification (Table 1) (5).
Given the subclonal nature of one of these mutations, we considered whether cells containing these mutations were selected against during tumor growth, perhaps even disappearing as a result of an immune response. The most frequent way to lose a mutant allele in human cancers is through a gross chromosomal event that results in loss of the entire gene and the surrounding chromosomal region (loss of heterozygosity, LOH) (6). To search for evidence of such losses, we designed 19 primer pairs that could each amplify a small fragment containing at least one common single-nucleotide polymorphism (SNP) within or surrounding the POLR3A gene (table S2). These primer pairs were used in a multiplexed protocol to evaluate all 16 tumors (5). Five of the eight tumors from scleroderma patients with autoanti-bodies to RPC1 exhibited LOH (Table 2). These five tumors included three that did not contain a detectable somatic mutation of POLR3A (Table 1). The fraction of neoplastic cells that had undergone LOH could be estimated from the allelic ratios of the SNP data, and in four of the five cases, was subclonal (Table 2). Notably, none of the tumors from patients with antibodies to TOP1 or CENPB exhibited LOH of the region containing POLR3A (Table 2). As an additional control, we evaluated 21 SNPs within or surrounding the TOP1 locus on chromosome 20 (table S2) and found that none of the 16 tumors from scleroderma patients, regardless of autoantibody status, had undergone LOH of this region (table S3).
Table 2. Allelic ratios of SNP loci within and closely surrounding the POLR3A gene.
NI: Noninformative; i.e., the SNP was not heterozygous in the normal cells of the patient (see supplementary materials and methods).
| Chr. 10 position | SNP ID | Patients with RPC-1 antibodies
|
Patients without RPC-1 antibodies
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SCL-1 | SCL-2 | SCL-4 | SCL-13 | SCL-35 | SCL-42 | SCL-81 | SCL-82 | SCL-5 | SCL-8 | SCL-11 | SCL-12 | SCL-19 | SCL-24 | SCL-32 | SCL-85 | ||
| 79,213,314 | rs1054608 | 55% | 88% | 102% | NI | NI | 68% | 83% | NI | NI | NI | 99% | NI | 99% | 103% | NI | NI |
| 79,222,098 | rs2165046 | 52% | 88% | 99% | NI | NI | 69% | 76% | NI | NI | NI | 99% | NI | 94% | 102% | NI | NI |
| 79,222,113 | rs1058203 | NI | NI | NI | NI | 100% | NI | 78% | NI | NI | 102% | NI | NI | NI | 99% | NI | 99% |
| 79,222,157 | rs1058202 | 50% | NI | 102% | NI | NI | NI | NI | NI | NI | 102% | NI | NI | NI | NI | NI | 97% |
| 79,230,809 | rs10762763 | 53% | 86% | 103% | NI | NI | 69% | 85% | NI | NI | NI | 102% | NI | 102% | 104% | NI | NI |
| 79,235,661 | rs2289311 | 55% | 91% | 103% | NI | NI | 66% | 88% | NI | 97% | NI | 104% | NI | 100% | 103% | NI | NI |
| 79,260,691 | rs10824579 | NI | 93% | 97% | NI | NI | 70% | 73% | NI | NI | NI | 96% | NI | 97% | 103% | NI | NI |
| 79,323,400 | rs1248888 | 80% | NI | NI | NI | 102% | NI | 87% | NI | 101% | 102% | NI | NI | NI | 101% | NI | 96% |
| 79,406,970 | rs2241547 | 78% | NI | NI | NI | 104% | NI | NI | NI | NI | NI | NI | NI | NI | NI | NI | 102% |
| 79,415,741 | rs12241228 | NI | NI | NI | NI | NI | NI | NI | NI | 100% | NI | NI | NI | NI | NI | NI | NI |
| 79,415,795 | rs3815891 | 70% | NI | NI | NI | NI | NI | NI | NI | 103% | NI | NI | NI | NI | NI | NI | 102% |
| 79,419,810 | rs7094028 | 79% | NI | NI | NI | NI | NI | NI | NI | 98% | NI | NI | NI | NI | NI | NI | 104% |
| 79,424,105 | rs2818827 | 75% | NI | 104% | NI | 100% | NI | NI | NI | NI | 102% | NI | NI | NI | NI | NI | 102% |
| 79,424,140 | rs12267816 | NI | NI | NI | NI | NI | NI | NI | NI | 104% | NI | NI | NI | NI | NI | NI | NI |
| 79,442,860 | rs2493568 | 76% | NI | 96% | 44% | 101% | NI | 91% | NI | NI | 103% | NI | NI | NI | NI | NI | 101% |
| 79,514,037 | rs67287610 | NI | NI | NI | 45% | 100% | NI | 95% | NI | 99% | NI | NI | NI | NI | NI | NI | NI |
| 79,514,072 | rs4979801 | 85% | NI | NI | 46% | 101% | NI | 95% | NI | NI | 96% | NI | NI | NI | NI | NI | 104% |
| 79,546,360 | rs2253909 | NI | NI | 94% | 40% | 102% | NI | 76% | NI | 102% | 99% | NI | NI | NI | NI | NI | NI |
| 79,549,686 | rs2253513 | 78% | NI | 102% | 41% | 102% | NI | 88% | NI | NI | 100% | NI | NI | NI | NI | NI | 90% |
| 79,573,735 | rs2114907 | 82% | NI | 103% | 43% | 104% | NI | 78% | 96% | NI | 104% | 100% | NI | NI | NI | NI | NI |
| 79,615,946 | rs1249134 | 86% | 90% | 98% | 44% | 102% | NI | 85% | NI | NI | NI | 99% | NI | 104% | NI | NI | NI |
| 79,618,728 | rs1249126 | 75% | 91% | 103% | 46% | 101% | NI | 74% | NI | NI | NI | NI | NI | 98% | NI | NI | NI |
| 79,654,802 | rs2434123 | NI | NI | NI | 46% | NI | 67% | NI | 102% | NI | 104% | NI | NI | NI | NI | NI | NI |
| Allelic ratio average* (%) | 71% | 88% | 99% | 44% | 101% | 69% | 83% | 99% | 99% | 101% | 100% | NI | 99% | 102% | NI | 100% | |
| Allelic ratio SD (%) | 13% | 4% | 6% | 2% | 4% | 3% | 7% | 4% | 5% | 3% | 2% | NI | 3% | 2% | NI | 4% | |
Entries represent the allelic ratios of the indicated SNPs. The values in normal individuals were 100% ± 3.10%. Allelic ratios less than 2 SDs from the mean (i.e., <94%) are highlighted in bold. A tumor was considered to exhibit LOH if more than 3/4 of the informative SNPs exhibited alleic ratios <94%.
In summary, six of eight tumors from scleroderma patients with autoantibodies to RPC1 harbored genetic alterations affecting the POLR3A locus compared to zero of eight tumors from scleroderma patients without anti-RPC1 antibodies (P < 0.01, Fisher exact probability test, two-tailed).
Immunological Analysis
We began the immunological analysis of these patients by addressing whether RPC1 autoantibodies recognized the mutated protein differently from the wild type (WT) form of the protein. Each of the three abnormal forms of the protein found in scleroderma patients was synthesized by in vitro transcription-translation (IVTT) (5). Wild-type and patient-matched mutant RPC1 were then subjected to immunoprecipitation analysis with sera from patients or from normal individuals (control sera). In each case, mutant and WT proteins were precipitated similarly by patient serum, but not precipitated by control sera (fig. S1), demonstrating that the autoantibodies do not discriminate between WT and mutant versions of the antigen.
We next constructed a custom peptide micro-array to comprehensively identify linear antigenic regions of the RPC1 protein. We synthesized 276 overlapping peptides of 15 amino acids in length, each offset by five amino acids from the previous peptide and covering the entire length of RPC1 (table S4). Peptides that contained each of the three somatic mutations described above were also synthesized (three peptides for each mutant; table S4). These peptides were printed on microarrays and used to assess serum from the three patients with antibodies to RPC1 (SCL-02, SCL-04, and SCL-42) whose cancers harbored POLR3A mutations, and four control patients (SCL-200, SCL-201, SCL-202, SCL-203) who had scleroderma and antibodies to RPC1 but who did not have cancers. Each of the seven serum samples displayed reactivity with at least two of the peptides on the array (fig. S2). Notably, there was no reactivity to the mutant peptides or their wild type counterparts (i.e., WT amino acids in place of mutant amino acids) in sera from the patients whose cancers harbored these mutations (or in the other patients).
Having shown that there was no demonstrable humoral immune response specific to the mutant RPC1 proteins, we sought to determine whether there was a cellular immune response directed against the mutants. We first performed high-resolution class I and II human leukocyte antigen (HLA) typing on the three scleroderma patients in whom somatically mutated POLR3A genes were identified (table S5). IEDB analysis resource Consensus tools (7–9) were then used to determine whether peptides containing the specific mutations in individual patients were likely to bind with high affinity to that patient’s HLA alleles. In patient SCL-42, both WT and mutant epitopes were predicted to bind with high affinity to both alleles of the patient’s class II DR HLA (table S6). This was particularly pronounced for HLA-DR*0701, where the predicted median inhibitory concentration (IC50) was <1 nM for both the mutant (FHVGYFRAVIGTLQMI) and WT peptides (FHVGYFRAVIGILQMI; table S6). High-affinity binding of the WT and mutant peptides to this patient’s other DR allele (HLA-DR*1001) was also predicted (table S6). In patient SCL-4, the mutant peptide was predicted to bind to this patient’s HLA-DR*0101 allele with an affinity of 4 nM, 18-fold higher than the affinity of the WT peptide (table S6). The WT peptide in this region was also predicted to bind, albeit less strongly, to this patient’s second allele (26 nM to HLA-DR*1101). Neither WT nor mutant peptides were predicted to bind with high affinity to the class II molecules of patient SCL-2 (table S6). The algorithms also predicted binding of patient-matched WT and mutant peptides to a single HLA class I allele in each patient, though the binding affinities were only moderate (27 to 78 nM, table S6).
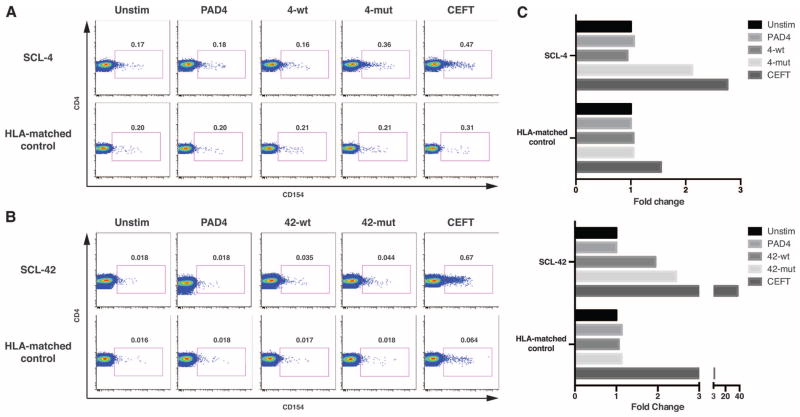
CD4 cells are known to recognize peptides presented by HLA class II alleles and play central roles in both tumor immunity and auto-immunity (10, 11). Given this knowledge and our finding that the predicted affinities for class II peptides were much higher than for class I peptides, we searched for CD4 T cells recognizing the predicted peptides in peripheral blood mono-nuclear cells (PBMCs) from patients whose tumors contained POLR3A mutations. CD154 expression at 18 hours after peptide stimulation was used to identify peptide-activated CD4+ T cells (12, 13). In patient SCL-4, CD4 T cell activation was observed in response to the patient-matched mutant peptide but not to the WT peptide (Fig. 1, A and C). Moreover, no CD4 T cell responses to these peptides were observed in T cells from a healthy control matched with SCL-4 at HLA-DR*1101 (Fig. 1A). Thus, the experimental data confirmed the in silico predictions.
Fig. 1. Mutant and WT peptide-specific CD4+ T cells in patients SCL-4 and SCL-42.
CD154 expression on CD4+ T cells was assayed after stimulation (18 hours) with patient-specific WT or mutant RPC1 peptides, PAD4 peptide (negative control), or a pool of peptides from infectious agent antigens (CEFT, positive control). Healthy donors matched for one HLA-DR allele were used as controls. Experiments on SCL-4 (A) and SCL-42 (B) were repeated on separate blood draws, three and two times, respectively, with similar results. Gate frequencies are expressed as percentage of CD4+ T cells. (C) Frequency of peptide-reactive CD4 T cells expressed as fold change over CD154+ CD4 T cells in the unstimulated negative control.
The experimental data also confirmed the predicted reactivity of T cells from patient SCL-42, with a twofold increase in the number of CD4+ CD154+ T cells in response to both the WT and mutant SCL-42 peptides over control conditions. The frequency of responding cells was about a log lower in SCL-42 compared to SCL-4, with ~1:5000 CD4 Tcells responding (Fig. 1, B and C). The CD4 T cell responses to WT and mutant SCL-42 peptides were abolished by treatment with anti–HLA-DR antibodies but not by an isotype control (fig. S3). As in patient SCL-4, no response to RPC1 peptides was observed in T cells from a healthy control matched with SCL-42 at HLA-DR*0701 (Fig. 1B). As predicted by the in silico binding algorithms (table S6), patient SCL-2 did not respond to either WT or mutant peptides, but did express CD154 in response to the positive control stimulus, demonstrating that her cells were immune competent (fig. S5).
These data document the existence of CD4 Tcells reactive with peptides containing the RPC1 mutations in two of the three patients studied. The reactivity was patient, peptide, and HLA-type specific. The frequencies of mutant peptide–reactive CD4 T cells observed in these scleroderma patients (~1:600 to ~1:5000, Fig. 1) were in the range observed for antigen-specific CD4+ T cells observed in other autoimmune processes (14).
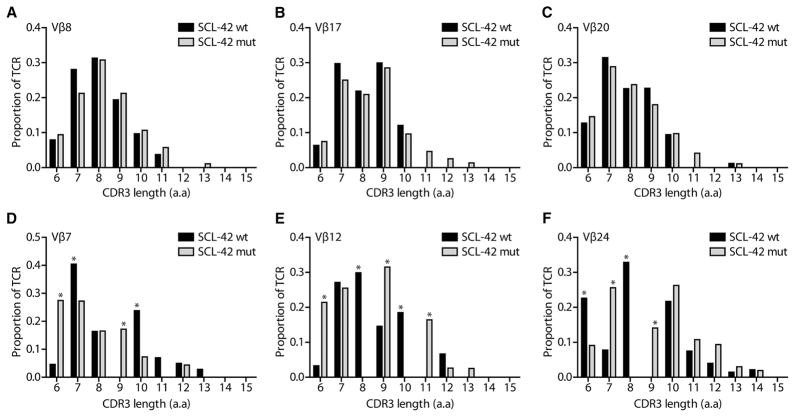
SCL-4 responded only to the mutant peptide, whereas patient SCL-42 responded to the mutant as well as to the WT peptides (Fig. 1C). It was possible that the CD4 T cells that were activated in response to the mutant peptide in SCL-42 were the same as those responding to the WT peptide. To evaluate this issue, we performed T cell receptor (TCR) spectratyping of T cells stimulated by either WT or mutant peptides. Out of the 22 Vβ families analyzed, 12 displayed a similar distribution of their CDR3 lengths in response to WT and mutant peptides, including Vβ8, Vβ17, and Vβ20 (Fig. 2, A to C). In contrast, significant differences in the distribution of CDR3 lengths were observed for several other Vβs (Vβ3, Vβ5, Vβ7, Vβ12, Vβ16, and Vβ24) (Fig. 2, D to F). For some Vβs, marked skewing in CDR3 lengths was observed, with >25% of TCRs from cells treated with either the mutant or the WT form of the peptide represented by a single CDR3 length. These data suggested that the T cells responding to the mutant peptides were not, in general, those responding to the WT peptides.
Fig. 2. Vβ-family usage and CDR3 length in patient SCL-42 PBMCs stimulated with WT or mutant peptides.
SCL-42 PBMCs were stimulated for 6 days with patient-specific mutant (gray bars) and corresponding WT (black bars) RPC1 peptides. No appreciable differences in TCR diversity were observed in Vβ8 (A), Vβ17 (B), and Vβ20 (C) TCR families. Skewing of the CDR3 length distribution in Vβ7 (D), Vβ12 (E), and Vβ24 (F) TCR families was observed, and CDR3 lengths that differed by >15% between WT and mutant stimulated PBMCs are indicated (*). CDR3 length is expressed in amino acids (a.a).
To characterize the TCRs in more detail, we determined the sequence of the CDR3 regions in the Vβ7, Vβ12, and Vβ24 PCR products (5). Two notable findings were revealed by massively parallel sequencing of these regions. First, the sequences of the dominant TCRs generated from T cells stimulated with the WT peptide were completely distinct from those stimulated by the mutant peptide (Table 3). In five of six dominant TCRs identified by sequencing, the WT- and mutant-specific CDR3 sequences were precisely the lengths predicted by the spectratype analysis (Table 3). The sequencing results therefore strongly supported the conclusion from spectra-typing that the mutant and WT peptides had stimulated many distinct T cell clones. Second, there was a high degree of redundancy among the amino acid sequences—but not the nucleotide sequences—of the TCRs identified in this experiment. For example, we identified 17 different nucleotide sequences (represented by 2066 clusters on the sequencing instrument) that encoded the identical CDR3 amino acid sequence in Tcells stimulated by the mutant peptide (Table 3). As T cells, unlike B cells, do not undergo continued evolution once a successful V(D)J rearrangement has occurred (15, 16), these data document the existence of multiple, independent T cell clones responding, and presumably binding, to the same mutant peptide–HLA complex.
Table 3. Dominant TCR sequences identified by massively parallel sequencing after stimulation with WT or mutant peptides.
Abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
| Vβ | CDR3 | Jβ | CDR3 length (amino acids) | Total no. of sequences | No. of unique sequences | ||
|---|---|---|---|---|---|---|---|
| SCL-42 WT-stimulated TCRs | |||||||
| Vβ7 | LCASS | PNELGQGSAF | FGQGT | Jβ1.1 | 10* | 160 | 1 |
| Vβ12 | FCASS | DEVNTEAF | FGQGT | Jβ1.1 | 8* | 18 | 1 |
| Vβ24 | LCATS | TGTVMNTEAF | FGQGT | Jβ1.1 | 10 | 1332 | 6 |
| SCL-42 mutant-stimulated TCRs | |||||||
| Vβ7 | LCASS | ESWTDYGYT | FGSGT | Jβ1.2 | 9* | 97 | 1 |
| Vβ12 | FCASS | DGGTRHEQF | FGPGT | Jβ2.1 | 9* | 160 | 2 |
| Vβ24 | LCATS | RDTVNQPQH | FGDGT | Jβ1.5 | 9* | 2066 | 17 |
Corresponds to the expected length based on TCR spectratyping.
Finally, we developed CDR3-specific Taqman assays to verify that distinct populations of WT and mutant-specific T cells were present in the peripheral blood of SCL-42 before the short-term cultures used in the experiments described above. The Vβ24 TCRs were chosen for this experiment because their CDR3 sequences were the most abundant in the sequencing analysis and were each encoded by multiple distinct nucleotide sequences (Table 3). The TCRs expected to bind the mutant and WT peptides were detected in uncultured SCL-42 PBMCs (fig. S4). Neither TCR was detectable in the PMBCs of patient SCL-4, used as a control.
Discussion
A subset of patients with scleroderma and other autoimmune rheumatic diseases manifest cancer around the time of autoimmune disease diagnosis, suggesting that the two processes might be linked mechanistically (3, 4, 17). In scleroderma, this temporal clustering of scleroderma and cancer appears limited to the subgroup of patients with antibodies to RPC1 (3). In the current work, we demonstrated that the POLR3A locus is genetically altered (by somatic mutation or LOH) in six of eight cancers of patients with antibodies to RPC1, but not in cancers from scleroderma patients with other autoantibody specificities. Moreover, T cells reactive with the mutant forms of RPC1 could be identified in the peripheral blood of two of the three patients tested. These T cells did not simply cross-react with the WT form of the peptides, because T cells from patient SCL-4 were not stimulated by the WT form, and the sequences of the TCRs conferring responsiveness to the WT and mutant peptides in SCL-42 were largely unrelated.
These genetic and immunologic findings suggest mutation in POLR3A as the initiator of the immune response to RPC1 in an important subset of scleroderma patients. The alternative to this conclusion—that the onset of scleroderma and the cancer genomes of these patients were unrelated and that the missense mutations and Tcell responses directed against the same mutations were coincidental—is unlikely given the rarity of POLR3A mutations in cancer in general (0.7%, P < 10−20) (18) and the absence of alterations at this locus in scleroderma patients without antibodies to RPC (P < 0.01). Additionally, in patient SCL-42, there were multiple different nucleotide sequences encoding TCRs with the identical amino acid sequence in T cells stimulated by the mutant peptide (Table 3). This provides strong support for the conclusion that the mutant POLR3A gene product acted as an immunogen initiating the anti-RPC1 immune response in vivo.
Antibodies from all patients with POLR3A mutations recognized WT and mutant versions of RPC1 to a similar extent, and no antibodies directed specifically against the WT versus mutant peptides could be demonstrated. This suggests that the humoral response does not directly target the area of the mutation or discriminate between mutant and WT versions of RPC1. The inability of autoantibodies to discriminate between the mutant and WT forms of the antigen is consistent with previous studies showing that a cross-reactive humoral response is typical when a novel form of an antigen initially stimulates Tcells that specifically recognize the modified antigen (19, 20). The antibody cross-reactivity might contribute to B cell–mediated diversification of autoimmunity, spreading T cell responses to the WT autoantigen (21, 22).
Our data therefore suggest that the “foreign” antigen triggering the autoimmune response in scleroderma patients is actually a tumor antigen. This complements previous observations indicating that cancers can elicit immune responses. Some cases of paraneoplastic syndrome are caused by autoimmunity to proteins expressed in tumors (23); these responses are directed exclusively to the normal protein, and there is no evidence that the gene(s) are mutated in the tumors. Conversely, mutant genes in human tumors can elicit an immune response against the mutant gene product (24–26); these immune responses have not been shown to elicit a cross-reactive response to the normal gene product that could result in autoimmunity. Finally, an in vitro–generated protein containing multiple (but not single) mutations, when injected into mice, can elicit a broad, cross-reactive immune response against the normal protein that results in auto-immunity (27). In these mice, tumor cells expressing only the WT protein can also be targeted by the subsequent immune response. Our results show that an analogous situation appears to occur in humans when a single, strongly immunogenic epitope is created by somatic mutation in a patient with an appropriate MHC type. However, the generation of an autoreactive immune response alone may not be sufficient to generate the self-sustaining tissue injury seen in scleroderma, and additional factors (genetic, environmental, or target tissue–specific) may be required (28).
Our cohort included cancer patients without anti-RPC1 antibodies (Table 1). Although the interval between scleroderma and cancer onset in these patients was long (median of 14.2 years), two patients (SCL-8 and SCL-32) had relatively short intervals. We did not identify genetic alterations of TOPO1 or CENPB in these two patients. Whether their cancers were adventitious, related to therapy, or due to mutations in genes encoding homologs of TOPO1 or CENPB or proteins that interact with them is unknown but are intriguing hypotheses for future study. Similar factors could also explain the absence of genetic alterations of POLR3A in two of the eight patients with antibodies to RPC1.
The relatively low fraction of neoplastic cells with genetic alterations in the cancers from some of these patients (Tables 1 and 2) suggests that immunoediting of the cancer had occurred, with cells containing these mutations selected against during tumor growth (29). The emergence of cancer in RPC1-positive scleroderma patients may thereby represent escape of the tumor from immune pressure. We speculate that cancers harboring POLR3A mutations had stimulated scleroderma in most patients with the RPC1 form of the disease. However, in the majority of these patients, the immune response had eradicated the cancer by the time scleroderma developed. Patients with a short cancer–autoimmune disease interval have also been described for other autoimmune rheumatic disease phenotypes (e.g., myositis, vasculitis, systemic lupus erythematosus), and similar mechanisms may be operative in these diseases (17, 30, 31). Given the ubiquitous presence of somatic mutations in solid tumors (32), these new data add credence to the idea that immunoediting could play a major role in limiting the incidence of human cancer—an old hypothesis (33, 34) that has recently garnered more attention (35–37). The data also suggest that this family of autoantigens might be used to generate biologically effective antitumor immunity.
Supplementary Material
Acknowledgments
We thank J. Ptak, N. Silliman. L. Dobbyn, J. Schaeffer, and M. Sampedro for expert technical assistance and J. Wu for advice on capturing. These studies were supported by The Virginia and D. K. Ludwig Fund for Cancer Research, The Donald B. and Dorothy L. Stabler Foundation, the Scleroderma Research Foundation, the Rheumatology Research Foundation Career Development Bridge Funding Award, and NIH grants K23 AR061439, P30-AR053503, CA43460, CA 57345, and CA 62924.
Footnotes
References and Notes
- 1.Gabrielli A, Avvedimento EV, Krieg T. N Engl J Med. 2009;360:1989–2003. doi: 10.1056/NEJMra0806188. [DOI] [PubMed] [Google Scholar]
- 2.Harris ML, Rosen A. Curr Opin Rheumatol. 2003;15:778–784. doi: 10.1097/00002281-200311000-00016. [DOI] [PubMed] [Google Scholar]
- 3.Shah AA, Rosen A, Hummers L, Wigley F, Casciola-Rosen L. Arthritis Rheum. 2010;62:2787–2795. doi: 10.1002/art.27549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shah AA, Rosen A. Curr Opin Rheumatol. 2011;23:530–535. doi: 10.1097/BOR.0b013e32834a5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Materials and methods are available as supplementary materials on Science Online.
- 6.Knudson AG. J Cancer Res Clin Oncol. 1996;122:135–140. doi: 10.1007/BF01366952. [DOI] [PubMed] [Google Scholar]
- 7.Kim Y, et al. Nucleic Acids Res. 2012;40(W1):W525–W530. [Google Scholar]
- 8.Wang P, et al. PLOS Comput Biol. 2008;4:e1000048. doi: 10.1371/journal.pcbi.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang P, et al. BMC Bioinformatics. 2010;11:568. doi: 10.1186/1471-2105-11-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding ZC, Zhou G. Clin Dev Immunol. 2012;2012:890178. doi: 10.1155/2012/890178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mellman I, Coukos G, Dranoff G. Nature. 2011;480:480–489. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chattopadhyay PK, Yu J, Roederer M. Nat Med. 2005;11:1113–1117. doi: 10.1038/nm1293. [DOI] [PubMed] [Google Scholar]
- 13.Frentsch M, et al. Nat Med. 2005;11:1118–1124. doi: 10.1038/nm1292. [DOI] [PubMed] [Google Scholar]
- 14.Nepom GT, et al. Arthritis Rheum. 2002;46:5–12. doi: 10.1002/1529-0131(200201)46:1<5::AID-ART10063>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 15.Jackson KJ, Kidd MJ, Wang Y, Collins AM. Front Immunol. 2013;4:263. doi: 10.3389/fimmu.2013.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McHeyzer-Williams MG, Davis MM. Science. 1995;268:106–111. doi: 10.1126/science.7535476. [DOI] [PubMed] [Google Scholar]
- 17.Buchbinder R, Forbes A, Hall S, Dennett X, Giles G. Ann Intern Med. 2001;134:1087–1095. doi: 10.7326/0003-4819-134-12-200106190-00008. [DOI] [PubMed] [Google Scholar]
- 18.Forbes SA, et al. Nucleic Acids Res. 2011;39 :D945–D950. doi: 10.1093/nar/gkq929. (Database) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doyle HA, Mamula MJ. Ann N Y Acad Sci. 2005;1050:1–9. doi: 10.1196/annals.1313.001. [DOI] [PubMed] [Google Scholar]
- 20.Mamula MJ, et al. J Biol Chem. 1999;274:22321–22327. doi: 10.1074/jbc.274.32.22321. [DOI] [PubMed] [Google Scholar]
- 21.Lin RH, Mamula MJ, Hardin JA, Janeway CA., Jr J Exp Med. 1991;173:1433–1439. doi: 10.1084/jem.173.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mamula MJ, Fatenejad S, Craft J. J Immunol. 1994;152:1453–1461. [PubMed] [Google Scholar]
- 23.Albert ML, Darnell RB. Nat Rev Cancer. 2004;4:36–44. doi: 10.1038/nrc1255. [DOI] [PubMed] [Google Scholar]
- 24.Gaudin C, Kremer F, Angevin E, Scott V, Triebel F. J Immunol. 1999;162:1730–1738. [PubMed] [Google Scholar]
- 25.Wang RF, Wang X, Atwood AC, Topalian SL, Rosenberg SA. Science. 1999;284:1351–1354. doi: 10.1126/science.284.5418.1351. [DOI] [PubMed] [Google Scholar]
- 26.Wölfel T, et al. Science. 1995;269:1281–1284. doi: 10.1126/science.7652577. [DOI] [PubMed] [Google Scholar]
- 27.Engelhorn ME, et al. Nat Med. 2006;12:198–206. doi: 10.1038/nm1363. [DOI] [PubMed] [Google Scholar]
- 28.Casciola-Rosen L, et al. J Exp Med. 2005;201:591–601. doi: 10.1084/jem.20041367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schreiber RD, Old LJ, Smyth MJ. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 30.Bernatsky S, et al. Arthritis Rheum. 2005;52:1481–1490. doi: 10.1002/art.21029. [DOI] [PubMed] [Google Scholar]
- 31.Tatsis E, Reinhold-Keller E, Steindorf K, Feller AC, Gross WL. Arthritis Rheum. 1999;42:751–756. doi: 10.1002/1529-0131(199904)42:4<751::AID-ANR19>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 32.Vogelstein B, et al. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burnet M. BMJ. 1957;1:841–847. doi: 10.1136/bmj.1.5023.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burnet FM. Prog Exp Tumor Res. 1970;13:1–27. doi: 10.1159/000386035. [DOI] [PubMed] [Google Scholar]
- 35.Quezada SA, Peggs KS. Br J Cancer. 2013;108:1560–1565. doi: 10.1038/bjc.2013.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DuPage M, Mazumdar C, Schmidt LM, Cheung AF, Jacks T. Nature. 2012;482:405–409. doi: 10.1038/nature10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsushita H, et al. Nature. 2012;482:400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.