Abstract
Understanding genome-wide links between genotype and phenotype has generally been difficult due to both the complexity of phenotypes, and until recently, inaccessibility to large numbers of genes that might underlie a trait. To address this issue, we establish the association between particular RNAi phenotypes in Caenorhabditis elegans and sequence characteristics of the corresponding proteins and DNA. We find that genes showing RNAi phenotypes are long and highly expressed with little noncoding DNA and high rates of synonymous site substitution (KS). In addition, genes conferring RNAi phenotypes have significantly lower rates of nonsynonymous site substitution (KA). Collectively, these sequence features explain nearly 20% of the difference between the sets of loci that display or lack a RNAi-mediated effect, and reflect aspects both of the RNAi mechanism and the biological function of the genes. For example, the particularly low rate of evolution of genes in the sterility RNAi phenotype class suggests a role of C. elegans life history in shaping these patterns of sequence and expression characteristics on phenotypes. This approach also allows prediction of a set of heretofore-uncharacterized loci for which we expect future RNAi studies to reveal phenotypic effects (i.e., false negatives in present screens).
Despite the availability of complete genome sequences and advances in methodologies for functional screens, the molecular bases of most phenotypes remain unknown, even in genetic model organisms. Recently, large-scale functional studies have assayed phenotypes for the majority of genes in the nematode Caenorhabditis elegans (Kamath et al. 2003) and in the budding yeast Saccharomyces cerevisiae (Winzeler et al. 1999), placing genes into broad functional classes (e.g., fecundity, viability, morphology). These resources for functional information make it feasible to couple phenotypic categorizations with the molecular features of all of the genes associated with a particular phenotype. In this study, we link compositional features of proteins and DNA, as well as rates of molecular evolution, to phenotypic traits for the C. elegans genome.
Because proteins are the active, functional forms of most genes, and amino acid properties are likely to influence phenotypes by contributing to protein structure (Torshin 2001), the amino acid composition of proteins has provided a starting point for connecting sequence characteristics to organismal phenotypes. For example, one pattern that has emerged from analyses of protein composition is that natural selection acts on translational processes to minimize the costs of protein synthesis (Barrai et al. 1995; Dufton 1997), favoring both translational efficiency (Moriyama and Powell 1998; Marais and Duret 2001) and the use of energetically inexpensive amino acids (Akashi and Gojobori 2002; Seligmann 2003). Consequently, it is possible to address the question of whether the energy devoted to translation varies among genes contributing to different phenotypes.
In addition to insights from the compositional features of DNA and proteins, rates of molecular evolution can portray overall responses to natural selection and thus bear on the relationship between genotype and phenotype. Evolutionary rates have long been considered in the context of gene function; for example, many individual structural proteins essential to central cellular processes (housekeeping genes), such as histones and actin, experience strong functional constraints and evolve slowly (Wilson et al. 1977). In contrast with housekeeping genes, many genes involved in reproduction and immunity show evidence for rapid rates of evolution across a broad range of taxa (Coulthart and Singh 1988; Lee et al. 1995; Ferris et al. 1997; Civetta and Singh 1998; Wyckoff et al. 2000; Swanson et al. 2001; Swanson and Vacquier 2002; Waterston et al. 2002). Although genes associated with particular phenotypes may be predicted to evolve at different rates, an elucidation of general patterns relating phenotypic traits to coding sequence evolution awaits analysis at the genomic scale.
In C. elegans, it is now possible to assess the genomic distribution of protein and DNA compositional profiles and evolutionary rates among loci that are involved in a variety of known phenotypes as assayed by RNAi (Fire et al. 1998; Timmons and Fire 1998). A recent study inhibited 86% of C. elegans' predicted genes by RNAi and identified 1528 loci that demonstrated phenotypic effects (Kamath et al. 2003). In addition, large-scale studies of gene-expression levels from different stages of development (Hill et al. 2000) provide another category of functional information on these genes. Here, we address the question of whether sets of genes characterized by different RNAi phenotypes vary in their compositional features, levels of expression, and evolutionary rates. We find that loci with scoreable RNAi phenotypes evolve more slowly (but likely experience higher mutation rates), are longer, and exhibit higher levels of expression than do loci lacking an RNAi phenotype. We also find that the degree of sterility conferred by RNAi is negatively associated with evolutionary rate and that, unexpectedly, loci involved in sterility tend to evolve more slowly than loci that influence embryonic survival or other phenotypes. From our results, we generate a predictive model of protein function as related to its sequence features and identify a large set of loci that have features typical of loci with an RNAi phenotype, but for which large-scale screens have not identified an RNAi-mediated effect.
RESULTS
Molecular Correlates of RNAi Phenotype
On the basis of 14 attributes of protein composition and the frequencies of individual amino acids for each of nearly 22,000 predicted transcripts in C. elegans (Table 1), we found that virtually all features differed significantly between the sets of loci with an observed or with an unobserved/untested RNAi phenotype (Kamath et al. 2003) in univariate analyses (Wilcoxon rank sum test, all P ≤ 0.0014; Table 1; Fig. 1), with the only exceptions being four amino acid characteristics (the frequencies of histidine, methionine, proline, and of tiny residues). For these same genes, we also estimated codon bias (Fop) and base composition, and for a subset of 7267 loci, we calculated rates of synonymous and nonsynonymous site substitution on the basis of C. elegans- C. briggsae comparisons. Loci with an observed RNAi phenotype differed from remaining loci for all of these features as well (Table 1); those exhibiting RNAi phenotypes demonstrate strong codon bias, high G+C composition, and low substitution rates. Partitioning mRNA expression levels (Hill et al. 2000) between the RNAi classes revealed that loci with an observed RNAi phenotype have significantly higher levels of expression than do other loci (Table 1). This pattern of higher expression for loci with RNAi phenotypes persists even when sterility-related loci (which contain a disproportionate representation of highly expressed protein synthesis genes) are excluded from the analysis (Wilcoxon P < 0.0001).
Table 1.
Features of Genes That Differ Between Those That Exhibit or Lack an Observed RNAi Phenotype
|
Occurrence in loci with RNAi phenotypesa
|
||||||
|---|---|---|---|---|---|---|
| Higher | Lower | |||||
| Amino Acid Characteristics | ||||||
| Number of Residuesb** | **** | |||||
| Total Molecular Weightd | **** | |||||
| Average Residue Weight | **** | |||||
| Total Costd | **** | |||||
| Cost per Residue | **** | |||||
| Essential Residues (%) | **** | |||||
| Total Protein Charge | **** | |||||
| Charged Residues (%) | **** | |||||
| Isoelectric Point | **** | |||||
| Tiny Residues (%) | ns | |||||
| Small Residues (%) | *** | |||||
| Aliphatic Residues (%) | **** | |||||
| Aromatic Residues (%) | **** | |||||
| Polar Residues (%) | **** | |||||
| Basic Residues (%) | **** | |||||
| Acidic Residues (%) | **** | |||||
| Amino Acid Composition (%) | ||||||
| Alanine | **** | |||||
| Arginine | **** | |||||
| Asparagine | **** | |||||
| Aspartate | **** | |||||
| Cysteine | **** | |||||
| Glutamic Acid | **** | |||||
| Glutamine | **** | |||||
| Glycine | **** | |||||
| Histidine | ns | |||||
| Isoleucine | **** | |||||
| Leucine | **** | |||||
| Lysine | **** | |||||
| Methionine | ns | |||||
| Phenylalanine | **** | |||||
| Proline | ns | |||||
| Serine | **** | |||||
| Threonine | **** | |||||
| Tryptophan | **** | |||||
| Tyrosine | **** | |||||
| Valine | ** | |||||
| Expression Levels | ||||||
| Average across developmentb**** | **** | |||||
| Sequence Characteristics | ||||||
| Fop | **** | |||||
| G+C Content | **** | |||||
| Fraction Codingb**** | **** | |||||
| KA/KSd | **** | |||||
| KSb****c | **** | |||||
| KAb**** | **** | |||||
Significance levels from Wilcoxon rank sum test indicated by ****P < 0.0001, ***P < 0.001 **P < 0.01, *P < 0.05, (ns) no significant difference (P > 0.05)
Significance in logistic multiple regression analysis with asterisk P-value cutoffs as in a
KS is significantly higher among loci with RNAi phenotypes in the multivariate analysis
Not included in multivariate analyses
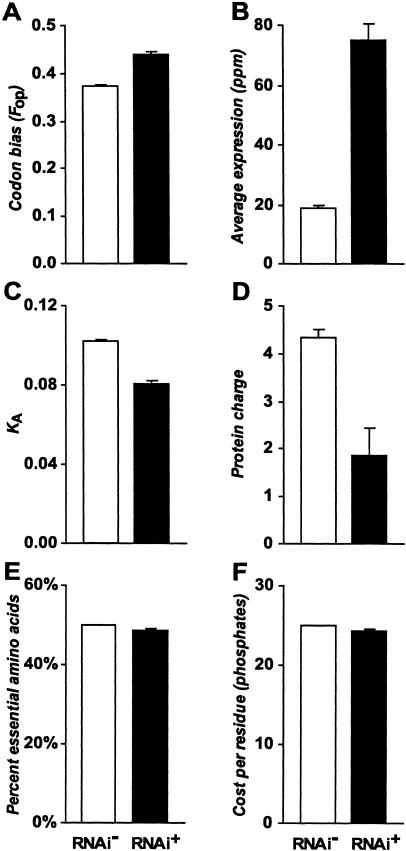
Figure 1.
Comparison of features between genes with no observed phenotype (RNAi-) and those with an RNAi phenotype (RNAi+). Codon bias (A), average gene expression (B), rate of nonsynonymous site substitution (C), total protein charge (D), percent essential amino acids (E), and the per-residue cost (number of high-energy phosphate bonds) of proteins (F) each differed significantly between the two phenotype classes in univariate analyses (all P < 0.0001). Error bars, ±1 SE.
Because many of these variables covary, we applied multivariate approaches to explore the independent effects of the different compositional features, thus excluding some features due to extreme covariation (e.g., molecular weight, total protein cost, KA/KS). In a logistic multiple regression, KA, KS, expression level, number of residues, and the fraction of coding sequence each contributed significantly to differences between the groups of loci that display and do not display observed RNAi phenotypes (all P < 0.002). Overall, this model explains 19.7% of the difference between the two groups of loci (χ2 = 964.5, df = 40, P < 0.0001), and a model restricted to the five independently significant factors alone accounts for 14.3% of the difference (χ2 = 696.2, df = 5, P < 0.0001).
Patterns of Molecular Evolution Within and Among RNAi Phenotypes
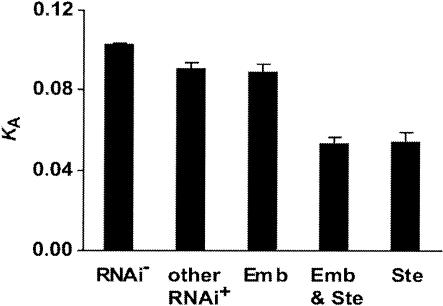
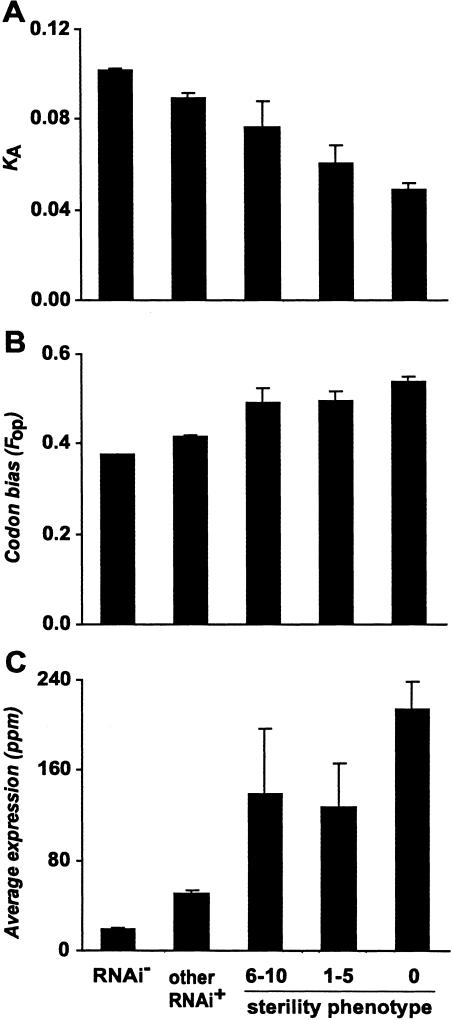
Loci with a known RNAi phenotype show a significantly lower mean rate of protein evolution—by more than 20%—than do loci without an RNAi phenotype (Wilcoxon P < 0.0001; Fig. 1). Much of this difference derives from the very low mean KA of sterility-related genes (KA = 0.053 ± 0.003 SE; Fig. 2); however, loci with RNAi phenotypes not associated with sterility still exhibit reduced rates of protein evolution (KA = 0.09 ± 0.002 SE) relative to those with no observed RNAi phenotype (KA = 0.10 ± 0.0007 SE; Wilcoxon P < 0.0001). The subset of 52 sterility-related loci with functional categorizations that are more comparable to rapidly evolving genes in other species (transcription factors, small molecule transporters, signaling molecules, genes of unknown function; Kamath et al. 2003) also demonstrate slow rates of evolution (KA = 0.072 ± 0.0072SE cf. Fig. 2; Wilcoxon P < 0.0001). Furthermore, proteins with sterility-related phenotypes evolve more slowly than do those that impact embryonic viability (Wilcoxon P < 0.0001; Fig. 2). Among loci showing a sterility phenotype, those exhibiting a stronger influence on fertility (i.e., those resulting in fewer progeny) tend to evolve slower (Spearman's ρ = -0.32, P < 0.0001; Fig. 3). Such loci also exhibit more extreme codon usage bias (Spearman's ρ = 0.31, P < 0.0001) and a tendency toward higher levels of expression throughout development (Spearman's ρ = 0.25, P < 0.0001; Fig. 3). In addition, loci exhibiting greater numbers of different RNAi phenotypes evolve more slowly (Spearman's ρ = -0.14, P < 0.0001).
Figure 2.
Mean nonsynonymous substitution rate (KA) in mutually exclusive groupings of genes with RNAi phenotypes of sterility (Ste), embryonic lethality (Emb), sterility and embryonic lethality, some other observed phenotype (other RNAi+), and no observed phenotype (RNAi-). Ste and Emb loci include loci with both partial and complete sterility or lethality, respectively. Error bars, ±1 SE.
Figure 3.
Nonsynonymous substitution rate (KA), codon usage bias (Fop), and average expression in genes assigned a sterility phenotype. Mean KA (A), Fop (B), and average expression (C) differ among genes with no observed phenotype (RNAi-), a nonsterile phenotype (other RNAi+), or a sterility phenotype characterized by 6-10, 1-5, or 0 offspring. Error bars, ±1 SE.
How Good Is RNAi at Producing Phenotypes?
Having determined the characteristics of genes that result in observable RNAi phenotypes, it is possible to estimate the proportion of loci that display these compositional features, but did not produce a phenotype experimentally (potential false negatives), as well as those loci that have features atypical of loci with observable RNAi phenotypes (potential false positives). Applying discriminant function analysis to a model with 40 compositional, sequence, expression, and evolutionary features indicates that, of the 5888 loci included, 229 show an RNAi phenotype, but have compositional characteristics that are not typical of loci with an RNAi-mediated effect. An additional 1253 loci that were not shown to have an RNAi phenotype experimentally display features that are expected of loci showing RNAi effects. The most extreme cases form reasonable candidates from which to expect future RNAi studies to identify phenotypic effects, and include, for example, the ribosomal protein genes T24B8.1 (rpl-32), B0513.3 (rpl-29), and M01F1.2(rpl-16) (Supplemental Table 1 available online at www.genome.org). By having restricted this analysis to loci with divergence information, in which genes with paralogs were filtered out, this list should be further enriched for genes expected to yield RNAi effects.
DISCUSSION
We show that genes with observable RNAi phenotypes possess identifying features that can be inferred from their sequences and expression levels. In general, these loci are long, expressed at high levels, and experience slow rates of evolution. Why do these molecular characteristics correlate with the incidence of an RNAi phenotype? Virtually every measured compositional feature was found to differ between those loci that display an RNAi-mediated effect and those loci that lack a detectable effect (Table 1). Biological processes are likely to underlie these differences, although some differences probably also reflect features associated with the effectiveness of RNAi. For example, the significantly lower incidence of RNAi phenotypes among loci with a small percentage of coding sequence probably reflects the use of genomic DNA templates to generate dsRNA by Kamath et al. (2003), which can include introns and thus reduce the efficacy of RNAi. If in general, however, RNAi methods identify those genes whose functions are most sensitive to mutation in natural populations (such that more mutations cause phenotypic variants among different alleles), then such loci would be more visible to selection in nature. This suggests that loci identified by RNAi would experience stronger purifying selection than other loci, because most non-neutral mutations are deleterious (Kimura 1983), consistent with our finding that loci exhibiting an RNAi phenotype in C. elegans have a lower KA (Fig. 1).
Expression and Translational Efficiency
Loci with observed RNAi phenotypes exhibit high-expression levels and strong codon biases (Table 1; Fig. 1). This pattern could be explained by highly expressed genes exerting more pleiotropic effects or encoding proteins that have more interactions with other gene products. Consistent with this hypothesis, in a limited set of 21 loci with interaction information from C. elegans (Walhout et al. 2002), expression level shows a positive trend with the number of interacting partners (Spearman's ρ = 0.43, P = 0.055). In general, expression level increases as the number of observed RNAi phenotypic classes increases for a given locus (Spearman's ρ = 0.26, P < 0.0001); because genes with dsRNA predicted to target multiple loci (e.g., in a multi-gene family) were excluded from the list of RNAi genes (Kamath et al. 2003), these observations are probably not a consequence of sequence similarity within gene families. High pleiotropy and many interacting partners might be typical of proteins involved in major cellular processes common to many tissue types. Cumulatively, this suggests that, despite individual examples of critical genes with low-copy transcripts (Koopman et al. 1990; Carmi and Meyer 1999), genes with clear ties to phenotype and fitness tend to be highly expressed.
Because RNAi efficiency is reduced with low doses of dsRNA (Gonczy et al. 2000), one might expect an inherent RNAi bias to disproportionately affect loci with low levels of expression—this pattern is not observed; in fact, we find that RNAi effects are more common among genes with high levels of expression. We suspect that the positive association between expression level and the incidence of an RNAi effect might also relate to the hypothesized amplification mechanism for triggering an RNAi response (Fire et al. 1998), because the presence of highly abundant mRNAs may result in a more dramatic, nonlinear amplification of interfering RNAs than for rare mRNAs, which prompts a more complete block to translation. Previous work has described the strong positive association between codon bias and amount of expression in C. elegans due to selection for translational efficiency (Marais and Duret 2001; Castillo-Davis and Hartl 2002), an association that we also recover. In addition, we find a negative association between expression level and the per-residue cost of amino acids (Spearman's ρ = -0.25, P < 0.0001), mirroring results from bacteria (Akashi and Gojobori 2002) and suggesting a potential tradeoff between the costs related to transcription/translation and the energy required to synthesize proteins. However, interpretations regarding per-residue cost in C. elegans should be made cautiously because the metric of cost is based on bacterial energetic expenditure values that likely differ from those in animals (e.g., half of the amino acids are not synthesized in worms, some essential amino acids may be limiting in nature or subject to different costs of metabolism). Regardless, the relative contributions of codon bias and per-residue protein cost indicate that selection for translational efficiency is likely to be stronger than selection for per-residue cost minimization in C. elegans. This observation is not unexpected, given that the amino acids synthesized by C. elegans are primarily the energetically least expensive (Sayre et al. 1963; Vanfleteren 1973).
Compositional Features of Proteins
Genes that show an RNAi-mediated phenotype possess unique compositional features. The significance of most individual effects disappears in multivariate analyses, indicating that many of these compositional features covary. However, protein length (hence, total protein cost) remains a significant factor distinguishing between the occurrence of and lack of an RNAi phenotype after accounting for covariation (Table 1; Fig. 1). In line with the evidence that proteins with many phenotypes evolve slower, an association between the number of protein interactions and the number of protein domains could explain the pattern of longer proteins among loci with an observable RNAi phenotype. Although genes with C. briggsae orthologs more often have an RNAi phenotype than genes that lack an ortholog (Fisher's exact test, P < 0.0001), a lower accuracy of gene annotation among loci without RNAi phenotypes should not adversely affect our conclusions, because multivariate analyses were limited to genes with identified orthologs in C. briggsae. We also expect that a significant fraction of the remaining variation between loci that display and do not display an RNAi effect can be explained by additional features of sequences, such as components of DNA and gene structure, regulatory sequence elements, and information about protein domains.
Our analyses also identified loci that display attributes typical of loci that exhibit an RNAi phenotype, but for which Kamath et al. (2003) identified no effect of RNAi. Because genes expressed in some cell types (e.g., sperm, neurons) are not efficiently blocked by RNAi (Fraser et al. 2000), and genes with subtle phenotypes were not scored in the genomic screen (Kamath et al. 2003), all genes with functional consequences cannot be discriminated in this way. Nevertheless, we hypothesize that future functional studies will identify RNAi-mediated phenotypic effects for many of the genes in this list (Supplemental Table 1). The efficacy and specificity of RNAi on these loci also will depend on which portion of the sequence is targeted with dsRNA; inclusion of noncoding intron sequences in dsRNA is likely to reduce the efficacy of RNAi and local sequence similarity, despite a lack of overall paralogy, may destroy RNAi specificity. In addition to these factors and mRNA stability, protein stability will also contribute to the incidence of a robust RNAi effect, which may be reflected by the additional ∼5% variance explained by the inclusion of protein compositional features in the multiple regression analysis, despite their being individually nonsignificant. A remaining challenge for our understanding of gene function and the action of interfering dsRNA is to characterize the factors that render some cell and tissue types unsusceptible to RNAi.
Molecular Evolution Among RNAi Phenotypes
Among the RNAi phenotypes, loci related to fecundity show particularly pronounced deviations in their rates of molecular evolution. Proteins implicated in fecundity by RNAi evolve slower than do those proteins that influence viability or other organismal phenotypes (Fig. 2). Moreover, loci that confer greater sterility through RNAi evolve more slowly than do those with only a partially sterilizing effect (Fig. 3; Wilcoxon P = 0.026). Such observations contrast with the findings in many other organisms that loci involved in reproduction evolve rapidly (Coulthart and Singh 1988; Lee et al. 1995; Ferris et al. 1997; Civetta and Singh 1998; Wyckoff et al. 2000; Swanson et al. 2001; Swanson and Vacquier 2002; Waterston et al. 2002).
Because C. elegans populations are composed of self-fertilizing hermaphrodites and rare males, there is likely to be little sexual selection, which is thought to drive the rapid evolution of reproduction-related loci in obligately outcrossing species (Andersson 1994; Rice and Holland 1997). Consequently, the slow rate of molecular evolution among fecundity-related loci might reflect purifying selection on genes involved in basic components of reproduction, such as oocyte provisioning and germline development. Consistent with this idea, protein synthesis (goodness of fit χ2 = 562.1, P < 0.0001) and cell architecture (goodness of fit χ2 = 229.3, P < 0.0001) genes are represented disproportionately among those loci that cause sterility when subjected to RNAi. Among fecundity-related loci that may be more analogous to rapidly evolving genes in other species (transcription factors, signaling molecules, small molecule transporters, and genes of unknown function), strong purifying selection (low KA) is still observed. However, because RNAi does not efficiently block the action of genes expressed in sperm (Fraser et al. 2000), our analyses cannot speak to any patterns for genes underlying sperm competition. The greater evolutionary constraint on fecundity-related proteins than on proteins that influence viability is surprising and may relate to the fact that proteins involved in fecundity show much higher average levels of expression and codon bias. The combined action of selection at the levels of both protein function and translational efficiency, in the absence of a strong effect of potentially countervailing sexual selection, likely causes the dramatic conservation of fecundity-related loci.
Genes with an observed RNAi phenotype exhibit higher substitution rates at synonymous codon sites (KS) than loci without an observed RNAi phenotype, once corrected for other variables. This independent association with KS—an estimator of mutation rate—is unexpected, given that it is opposite the univariate pattern (Table 1); mutation rate differences in mammals depend, in part, on methylated dinucleotide composition (Subramanian and Kumar 2003), and some similar mechanism of DNA modification could also underlie differences in mutation rate in C. elegans. However, the mammalian mechanism cannot be the operative force in C. elegans, because its genome lacks both a typical DNA methyltransferase and any evidence of methylated 5-positioned carbons of cytosine (Bird 2002). Transcription-mediated mutation (i.e., highly transcribed regions more susceptible to mutation by being single-stranded more often) cannot be ruled out, although the negative association between expression level and KS suggests that it is unlikely to play a role. Furthermore, because (1) genes that show an RNAi phenotype are over-represented in the central portions of chromosomes (Kamath et al. 2003), (2) rates of recombination are lower in chromosomal centers (Barnes et al. 1995), and (3) recombination might be mutagenic in C. elegans (Marais et al. 2001; Cutter and Payseur 2003), this pattern of higher KS among loci with an RNAi phenotype remains enigmatic.
METHODS
Data Acquisition
We retrieved all predicted transcripts of C. elegans (21,198; The C. elegans Sequencing Consortium 1998) and C. briggsae (14,713) from the March 4, 2003 release of the Ensembl genome browser (http://www.ensembl.org). Phenotypes for each of 19,213 loci were based on the RNAi screen of Kamath et al. (2003), 1,528 of which included one or more of the following phenotypic designations: embryonic lethality (100%, 50%-80%, 20%-40%, 10%, no lethality detected), sterility (0 progeny, 1-5 progeny, 6-10 progeny, no sterility detected), developmental (slow growth, larval arrest), and >20 post-embryonic phenotypes. Microarray expression levels for 18,588 genes at eight timepoints across C. elegans' development (oocyte, 0 h, 12h, 24 h, 36 h, 48 h, 60 h, 2 wk) expressed in parts per million transcripts (ppm) were obtained from Hill et al. (2000). Expression levels for loci reported as absent at a particular timepoint were considered as having a value of 0 ppm, and average expression values across timepoints were used as proxy for overall expression level.
Sequence Composition
We calculated the compositional properties of the conceptual amino acid translations of the 21,198 C. elegans transcripts using the Emboss Pepstats routine (http://www.emboss.org). These properties include the frequency of each amino acid and of functionally defined residues (size, aromaticity, aliphaticity, polarity, charge, and acidity), length, charge, molecular weight, and isoelectric point. The execution of Pepstats and the extraction of output from the program were automated using a Perl script developed by the authors. We estimated costs of protein metabolism as the number of high-energy phosphate bonds used for a particular protein according to the estimates of Akashi and Gojobori (2002). Applying this approach, we calculated the biosynthetic cost for each protein based on total amino acid content. We also computed the fraction of essential amino acids (Sayre et al. 1963; Vanfleteren 1973) and the fraction coding of total predicted gene length. For estimates of codon composition, we used the program CodonW (http://www.molbiol.ox.ac.uk/cu/codonW.html) to calculate base composition and the Fop index of codon usage bias (Ikemura 1985; Stenico et al. 1994; Sharp and Bradnam 1997).
Evolutionary Rates
We used a reciprocal best-hit BLAST approach to identify orthologs (Rivera et al. 1998) among the predicted transcripts from all genes of C. elegans and C. briggsae applying an E-value maximum of 10-6. Potential paralogs, defined as those genes with better within- than between-genome bitscore hit values, were discarded. Sequences were aligned on the basis of conceptual amino acid translations, and corresponding nucleotide sequence alignments were evaluated with Diverge (Genetics Computer Group [GCG], Madison, WI) to calculate rates of synonymous (KS) and nonsynonymous (KA) site substitution with the method of Li (Li et al. 1985; Pamilo and Bianchi 1993; Li 1993). We subsequently removed as potential false positive matches all gene pairs for which KA or KS could not be computed, and those that fell within the upper decile of the KA distribution (KA ≥ 0.25). This procedure yielded a final set of 7267 putatively orthologous gene pairs used in subsequent analyses.
Statistical Procedures
To evaluate associations between protein characteristics, evolutionary rate, and phenotype, we used Wilcoxon rank sum tests for phenotypes with two categories and analysis of variance (ANOVA) for phenotypes with more than two categories. The number of residues in a protein, KA, Fop, and expression levels (after 1 ppm replaced 0 ppm values) were log10 transformed and fraction coding was arcsin-square-root transformed to restore normality in parametric analyses. Logistic multiple regression and discriminant function analyses were implemented in JMP v5.0 to create profiles of amino acid properties for the different phenotypic classes (data for all variables for 5888 loci). Standard errors (SE) are used to indicate dispersion about mean values.
Acknowledgments
We are grateful for the efforts of the Sanger Institute and the Washington University Genome Sequencing Center in making C. briggsae sequences publicly available. We are indebted to H. Ochman for his encouragement and discussion throughout the development of this work. We also thank N. Merchant, G. Nelson, and S. Miller of the University of Arizona Biotechnology Computing Facility for computational advice and assistance. This work benefited from the input of the IGERT fellows and the manuscript was improved by the comments of N. Moran, M. Nodine, H. Ochman, and two anonymous reviewers. This work was conducted as part of the University of Arizona NSF Integrative Graduate Education Research Training (IGERT) grant Genomics Initiative (DGE-0114420).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1659203.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Akashi, H. and Gojobori, T. 2002. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc. Natl. Acad. Sci. 99: 3695-3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, M. 1994. Sexual selection. Princeton University Press, Princeton, NJ.
- Barnes, T.M., Kohara, Y., Coulson, A., and Hekimi, S. 1995. Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics 141: 159-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrai, I., Volinia, S., and Scapoli, C. 1995. The usage of oligopeptides in proteins correlates negatively with molecular-weight. Int. J. Peptide Prot. Res. 45: 326-331. [DOI] [PubMed] [Google Scholar]
- Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes & Dev. 16: 6-21. [DOI] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science 282: 2012-2018. [DOI] [PubMed] [Google Scholar]
- Carmi, I. and Meyer, B.J. 1999. The primary sex determination signal of Caenorhabditis elegans. Genetics 152: 999-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Davis, C.I. and Hartl, D.L. 2002. Genome evolution and developmental constraint in Caenorhabditis elegans. Mol. Biol. Evol. 19: 728-735. [DOI] [PubMed] [Google Scholar]
- Civetta, A. and Singh, R.S. 1998. Sex-related genes, directional sexual selection, and speciation. Mol. Biol. Evol. 15: 901-909. [DOI] [PubMed] [Google Scholar]
- Coulthart, M.B. and Singh, R.S. 1988. High-level of divergence of male-reproductive-tract proteins, between Drosophila melanogaster and its sibling species, Drosophila simulans. Mol. Biol. Evol. 5: 182-191. [DOI] [PubMed] [Google Scholar]
- Cutter, A.D. and Payseur, B.A. 2003. Selection at linked sites in the partial selfer Caenorhabditis elegans. Mol. Biol. Evol. 20: 665-673. [DOI] [PubMed] [Google Scholar]
- Dufton, M.J. 1997. Genetic code synonym quotas and amino acid complexity: Cutting the cost of proteins? J. Theor. Biol. 187: 165-173. [DOI] [PubMed] [Google Scholar]
- Ferris, P.J., Pavlovic, C., Fabry, S., and Goodenough, U.W. 1997. Rapid evolution of sex-related genes in Chlamydomonas. Proc. Natl. Acad. Sci. 94: 8634-8639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire, A., Xu, S.Q., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806-811. [DOI] [PubMed] [Google Scholar]
- Fraser, A.G., Kamath, R.S., Zipperlen, P., Martinez-Campos, M., Sohrmann, M., and Ahringer, J. 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408: 325-330. [DOI] [PubMed] [Google Scholar]
- Gonczy, P., Echeverri, C., Oegema, K., Coulson, A., Jones, S.J.M., Copley, R.R., Duperon, J., Oegema, J., Brehm, M., Cassin, E., et al. 2000. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 408: 331-336. [DOI] [PubMed] [Google Scholar]
- Hill, A.A., Hunter, C.P., Tsung, B.T., Tucker-Kellogg, G., and Brown, E.L. 2000. Genomic analysis of gene expression in C. elegans. Science 290: 809-812. [DOI] [PubMed] [Google Scholar]
- Ikemura, T. 1985. Codon usage and transfer-RNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 2: 13-34. [DOI] [PubMed] [Google Scholar]
- Kamath, R.S., Fraser, A.G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohrmann, M., et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231-237. [DOI] [PubMed] [Google Scholar]
- Kimura, M. 1983. Rare variant alleles in the light of the neutral theory. Mol. Biol. Evol. 1: 84-93. [DOI] [PubMed] [Google Scholar]
- Koopman, P., Munsterberg, A., Capel, B., Vivian, N., and Lovellbadge, R. 1990. Expression of a candidate sex-determining gene during mouse testis differentiation. Nature 348: 450-452. [DOI] [PubMed] [Google Scholar]
- Lee, Y.H., Ota, T., and Vacquier, V.D. 1995. Positive selection is a general phenomenon in the evolution of abalone sperm lysin. Mol. Biol. Evol. 12: 231-238. [DOI] [PubMed] [Google Scholar]
- Li, W.-H. 1993. Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J. Mol. Evol. 36: 96-99. [DOI] [PubMed] [Google Scholar]
- Li, W.-H., Wu, C.I., and Luo, C.C. 1985. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol. Biol. Evol. 2: 150-174. [DOI] [PubMed] [Google Scholar]
- Marais, G. and Duret, L. 2001. Synonymous codon usage, accuracy of translation, and gene length in Caenorhabditis elegans. J. Mol. Evol. 52: 275-280. [DOI] [PubMed] [Google Scholar]
- Marais, G., Mouchiroud, D., and Duret, L. 2001. Does recombination improve selection on codon usage? Lessons from nematode and fly complete genomes. Proc. Natl. Acad. Sci. 98: 5688-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama, E.N. and Powell, J.R. 1998. Gene length and codon usage bias in Drosophila melanogaster, Saccharomyces cerevisiae and Escherichia coli. Nucleic Acids Res. 26: 3188-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamilo, P. and Bianchi, N.O. 1993. Evolution of the zfx and zfy genes—Rates and interdependence between the genes. Mol. Biol. Evol. 10: 271-281. [DOI] [PubMed] [Google Scholar]
- Rice, W.R. and Holland, B. 1997. The enemies within: Intergenomic conflict, interlocus contest evolution (ICE), and the intraspecific Red Queen. Behav. Ecol. Sociobiol. 41: 1-10. [Google Scholar]
- Rivera, M.C., Jain, R., Moore, J.E., and Lake, J.A. 1998. Genomic evidence for two functionally distinct gene classes. Proc. Natl. Acad. Sci. 95: 6239-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayre, F.W., Hansen, E.L., and Yarwood, E.A. 1963. Biochemical aspects of nutrition of Caenorhabditis briggsae. Exp. Parasitol. 13: 98-107. [DOI] [PubMed] [Google Scholar]
- Seligmann, H. 2003. Cost-minimization of amino acid usage. J. Mol. Evol. 56: 151-161. [DOI] [PubMed] [Google Scholar]
- Sharp, P.M. and Bradnam, K.R. 1997. Codon usage in C. elegans. In C. elegans II (ed. D.L. Riddle, T. Blumenthal, B.J. Meyer, and J.R. Priess), pp. 1053-1057. Cold Spring Harbor Laboratory Press, New York.
- Stenico, M., Lloyd, A.T., and Sharp, P.M. 1994. Codon usage in Caenorhabditis elegans—Delineation of translational selection and mutational biases. Nucleic Acids Res. 22: 2437-2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, S. and Kumar, S. 2003. Neutral substitutions occur at a faster rate in exons than in noncoding DNA in primate genomes. Genome Res. 13: 838-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, W.J. and Vacquier, V.D. 2002. The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3: 137-144. [DOI] [PubMed] [Google Scholar]
- Swanson, W.J., Zhang, Z.H., Wolfner, M.F., and Aquadro, C.F. 2001. Positive Darwinian selection drives the evolution of several female reproductive proteins in mammals. Proc. Natl. Acad. Sci. 98: 2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons, L. and Fire, A. 1998. Specific interference by ingested dsRNA. Nature 395: 854. [DOI] [PubMed] [Google Scholar]
- Torshin, I.Y. 2001. Clustering amino acid contents of protein domains: Biochemical functions of proteins and implications for origin of biological macromolecules. Front. Biosci. 6: A1-A12. [DOI] [PubMed] [Google Scholar]
- Vanfleteren, J.R. 1973. Amino-acid requirements of free-living nematode Caenorhabditis briggsae. Nematologica 19: 93-99. [Google Scholar]
- Walhout, A.J.M., Reboul, J., Shtanko, O., Bertin, N., Vaglio, P., Ge, H., Lee, H., Doucette-Stamm, L., Gunsalus, K.C., Schetter, A.J., et al. 2002. Integrating interactome, phenome, and transcriptome mapping data for the C. elegans germline. Curr. Biol. 12: 1952-1958. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Wilson, A.C., Carlson, S.S., and White, T.J. 1977. Biochemical evolution. Annu. Rev. Biochem. 46: 573-639. [DOI] [PubMed] [Google Scholar]
- Winzeler, E.A., Shoemaker, D.D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J.D., Bussey, H., et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901-906. [DOI] [PubMed] [Google Scholar]
- Wyckoff, G.J., Wang, W., and Wu, C.I. 2000. Rapid evolution of male reproductive genes in the descent of man. Nature 403: 304-309. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.ensembl.org; source for C. elegans and C. briggsae predicted transcripts.
- http://www.emboss.org; source for Pepstats routine.
- http://www.molbiol.ox.ac.uk/cu/codonW.html; codonW program for calculating codon bias.