Abstract
Enriching the surface density of immobilized capture antibodies enhances the detection signal of antibody sandwich microarrays. In this study, we improved the detection sensitivity of our previously developed P-Si (porous silicon) antibody microarray by optimizing concentrations of the capturing antibody. We investigated immunoassays using a P-Si microarray at three different capture antibody (PSA - prostate specific antigen) concentrations, analyzing the influence of the antibody density on the assay detection sensitivity. The LOD (limit of detection) for PSA was 2.5ngmL−1, 80pgmL−1, and 800fgmL−1 when arraying the PSA antibody, H117 at the concentration 15µgmL−1, 35µgmL−1 and 154µgmL−1, respectively. We further investigated PSA spiked into human female serum in the range of 800fgmL−1 to 500ngmL−1. The microarray showed a LOD of 800fgmL−1 and a dynamic range of 800 fgmL−1 to 80ngmL−1 in serum spiked samples.
Keywords: Antibody microarray, Porous silicon, Sandwich immunoassay, Prostate specific antigen
1.Introduction
Microarray based immunoassays are currently undergoing intense developments for the detection of low abundant protein biomarkers in human biofluids such as serum, urine and CSF (cerebrospinal fluid). The microarray format can ultimately offer advantages in terms of a low amount antibody consumption, high sensitive readout, and multiplex performance. Such developments could hold promise of earlier diagnosis of disease, reducing the need for biopsy and providing post therapy monitoring of patients for recurrence [1, 2]. There are mainly two types of microarray-based immunoassays when analyzing a biofluid without performing any chemical modification or labeling of the sample:
-
The sandwich microarray antibodies are spotted on solid surfaces. The biofluid with the target analyte is subsequently incubated on the array for specific binding to the primary antibody. After addition of a secondary antibody that is allowed to bind the target, the sandwich complex is formed. Sandwich assays are widely used for diagnostics, frequently in 96-well formats. The translation of these assays into a miniaturized format is an attractive approach to minimize the consumption of sample and analyte. The most difficult step is to obtain a matched sandwich antibody pair [3, 4].
As an alternative, an array of spotted biofluids (sample) can be probed with individual antibodies named reverse phase type immunoassay [5].
In reverse phase assay, many different samples (cell or tissue lysates) are immobilized in a microarray format and simultaneously analyzed for the presence of a single protein using a target-specific antibody. This enables label-free analysis of biological samples by simply arraying the biofluid and detecting the biomarkers with an antibody, e.g. by fluorescent labeling of the antibody or by catalyzed signal amplification and colorimetric readout [5, 6]. Such an assay has a potential for detection of autoantibodies in the classifications of different autoimmune disease [7], but due to its inherent properties it can not be used for analysis of low abundantly expressed biomarkers [8]
Sandwich immunoassays have become a major work horse in clinical diagnostics since they offer high detection sensitivity gained by the enrichment of the target proteins to the capturing antibody [9]. The assay specificity is greatly increased by using matched antibody pairs.
To increase the assay sensitivity, several amplification methods have been proposed that are linked to modifying the detection antibodies by e.g. dendritic amplification [10], catalyzed signal amplification with colorimetric readout [11,12] or detection with rolling-circle amplification [13]. On the other hand, enriching the concentration of the capture antibody may also enhance sensitivity yet maintaining a simple assay protocol. Increased density of the immobilized antibody on the each microarray spot can offer improved capturing capacity of target antigen, which in turn increases the number of antigen bound to the primary antibody and consequently more completed sandwich pairs are achieved at the end of the assay leading to increased detection signals [4,14].
To enrich a capture antibody on the surface, the substrate or chip is of utmost importance. The substrate used in our current study is an in house developed three- dimensional porous silicon surface. It has proven to be highly compatible with protein microarray technology based on its spot quality, spot density and sensitivity [14]. As a model biomarker we used PSA (prostate specific antigen), which is the most commonly used biomarker for prostate disease, e.g. prostate cancer. PSA, a kallikrein-related peptidase, occurs in free (unbound) and bound (complex) forms secreted from the epithelial cell in the prostate gland [15]. Although PSA has its limitations to distinguish between malignant and benign prostate diseases, it still remains as a valuable biomarker capable of discriminating different prostate cancer stages and potential indicators of recurrence in patient after radical prostatectomy (RP) [16,17].
The most commonly used diagnostic cut-off value for PSA in plasma is 2–3 ngmL−1, higher values often merit further investigation, e.g. a prostate biopsy [15]. In order to detect prostate cancer (PCa) recurrence after radical prostatectomy by measuring PSA in plasma a three order of magnitude lower limit of detection is required [18]. To achieving such a demand, much higher sensitive detection methods are required. D. Liu et al. [19] used gold nanoparticle -based probe to increase detection sensitivity of PSA in serum. They selected five prostate cancer patients’ serum and diluted until sub pictogram per milliliter using PBST buffer. LOD could be told sub to few pgmL−1 in the paper. Another nanoparticle based PSA immunoassay (so called bio barcode system) was published by C.S. Thaxton et al. [20] to monitor prostate cancer recurrence. LOD of this system push down to 330 fg/mL and showed possibility of early detection of prostate cancer recurrence. Single molecular based digital ELISA systems are one of important direction toward ultra sensitive assay development. D.M Rissin et. al. realized the femtoliter chamber array platform, which occupies single bead with one-target molecules in one chamber [21,22]. This assay platform detected PSA in sera under 14 fgmL−1 and also shown reliability of the assay for post-RP. Similar approach but increasing number of chambers enormously could be limit of detection less than 2 aM [22].
Although above methods are impressive in its low LOD, our developed P-Si based microarray platform has great advantage of its simplicity and robustness. P-Si surface does not require any chemical treatment such as amine, or epoxy coating for immobilization of antibody. Since physical adsorption is main motif to bind antibody on the surface, it does not require any laborious procedure such as incubation, humidity control and temperature control. It only takes few minutes to finish up antibody immobilization on the surface so that it is possible to reduce total assay time [23,24]. The P-Si microarray platform also dealt with 80 clinical plasma samples to quantify total and free PSA (duplex assay) [25]. The samples analyzed by this microarray obtained 0.14ngmL−1 of LOD with dynamic range from 0.4 to 74.9 ngmL−1 at total PSA and 0.76 ngmL−1 of LOD with dynamic range from 0.87 to 295 ngmL−1 in case of freePSA.
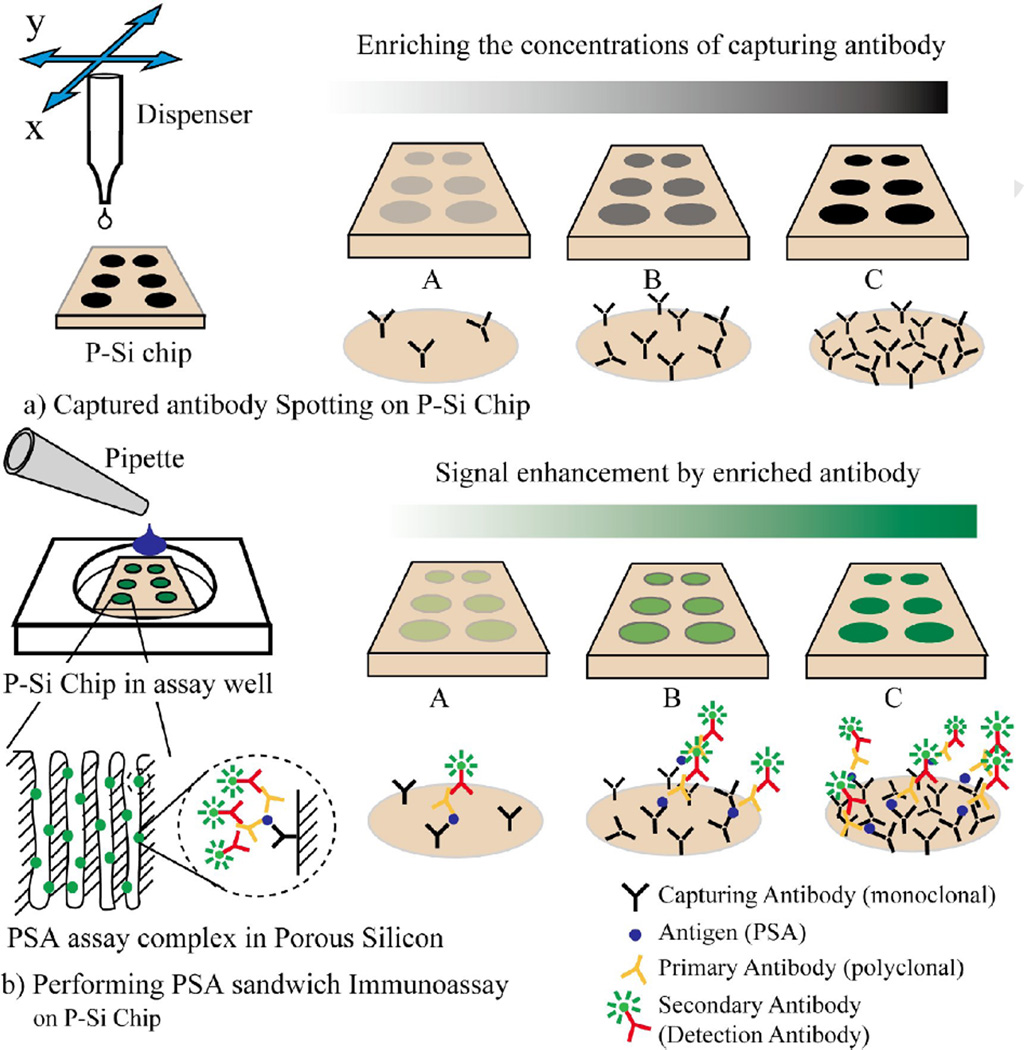
In this study, we improved detection sensitivity of our previously developed P-Si based antibody microarray by changing the concentration of the capturing antibody when spotting the microarray. Figure 1 schematically shows the effect of enriching the capture antibody in the immunoassay. Three different concentrations of capturing antibody were arrayed and evaluated in detecting PSA in buffer (PBS). The P-Si microarray, with optimized primary antibody concentration, was subsequently evaluated in detecting PSA spiked in human female serum. The reported data shows that the PSA detection sensitivity was improved about three orders of magnitude by increasing the concentration of capturing antibody in the P-Si microarray, with a LOD of 800fgmL−1.
Figure 1.
Capturing antibody was spotted on the solid support (P-Si chip). (a) By changing the capture antibody concentration the surface density of the spotted antibody increased. (b) Enhanced microarray signal readouts were obtained at higher density of capturing antibody microarray.
2. Materials and Methods
2.1 Porous silicon fabrication
Morphology and geometry of pores layers are affect physical property and characteristics of P-Si chip and they are strongly governed by a large number of etching parameters such as HF concentration, current density, anodization time, illumination, crystal orientation, silicon type, doping levels [23]. Generally good uniformity and mechanical stability of porous layer, low intrinsic fluorescent, and low wetting ability, i.e., a high liquid contact angle are demanded for porous support for protein immobilization [23,26]. The fabrication procedure of porous silicon in this paper was followed to optimum condition for immobilization of antibody as described previously [23]. In brief, silicon, 6–8 ohm·cm resistivity (boron doped p-type), <100> orientation, was purchased from Addision Engineering (San Jose, CA, USA). The wafer was placed in middle of an electrochemical-etching cell. The electrolyte solution consisted of 3.6 % hydrofluoric acid and 90.7 % dimethylformamide (Merck, Darmstad, Germany). The silicon was anodized for 70 min with backside illumination. Current density during anodization was 90mA/m2 after which the silicon was washed in ethanol three times and diced into 3x3 mm pieces to fit a microtiter plate format (Corning Costar Corporation, Cambridge, MA, USA).
2.2 Proteins and Reagents
The monoclonal mouse antibody against PSA (H117) was produced as previously described [18]. The polyclonal sheep anti–PSA antibody and Alexa Fluor 488 labeled donkey anti–sheep antibody were purchased at Abcam, Cambridge, UK, (ab35355) and Jackson ImmunoReaserch, West Grove, PA, USA, (713-545-003), respectively. Prostate specific antigen (PSA) from human semen was obtained from Sigma-Aldrich, St. Louis, MO, USA.
2.3 Analytical samples
Human female serum was obtained from a healthy blood donor, aliquoted, and stored −80 °C. The serum was spiked with PSA in a titration series ranging from 800 fgmL−1 to 80 ngmL−1.
2.4 Sandwich immunoassay
The porous silicon wafer was diced into 3×3 mm chips to fit into microtiter platewells. Droplets of approximately 300 pl monoclonal mouse antibody (H117) were dispensed in a 9×9 array format onto a porous silicon chip at a spot to spot distance of 250µm using an in-house developed piezoelectric micro-dispenser [24,27]. Three different concentrations of capturing antibodies (15µgmL−1, 35µgmL−1, and 154µgmL−1) were arrayed on each P-Si chip and PSA was assayed in PBS buffer to evaluate the influence of antibody concentration. The corresponding assays were subsequently performed on PSA spiked human female serum samples. The mean spot intensities, calculated from 12 spots for each sample, were used to derive a calibration curve for PSA in the concentration range 800 pgmL−1 to 80ngmL−1.
The antibody-activated chips were loaded in 96 well plates for the immunoassay. To remove loosely bound antibody, the chips were washed three times by 10 mM PBS. After blocking in 100µl 5% (w/v) non-fat dry milk in PBS (Bio-Rad, Hercules, CA, USA) for 1 hour to prevent non-specific binding, the chips were washed 3 times in PBS-Tween solution (0.05% Tween 20 in 10mM PBS) and 100 µl of PSA spiked human female serum pipetted into each well and incubated for 1 hour. After an additional washing step, the chips were incubated with 100µl of polyclonal sheep anti-PSA antibody (1/1000 dilution of its original concentration) as the secondary antibody. Following a washing step, 1µgmL−1 of 100 µl Alexa Fluor 488 labelled anti- sheep polyclonal detector antibody was added to the chips and incubated for 1 hour. Finally, the chips were washed 3 times and dried in room temperature. Fluorescence readout was performed by a BX51WI microscope with laser confocal unit (Olympus, Japan). For reproducibility, all assay were performed in two independence chips.
2.5 Morphology analysis by FESEM
The morphology of the porous silicon surface was analysed by a field emission scanning electron microscope (FESEM; JEOL JSM-6700F), Figure 2. Before SEM analysis, a 10-nm layer of platinum was deposited on the samples.
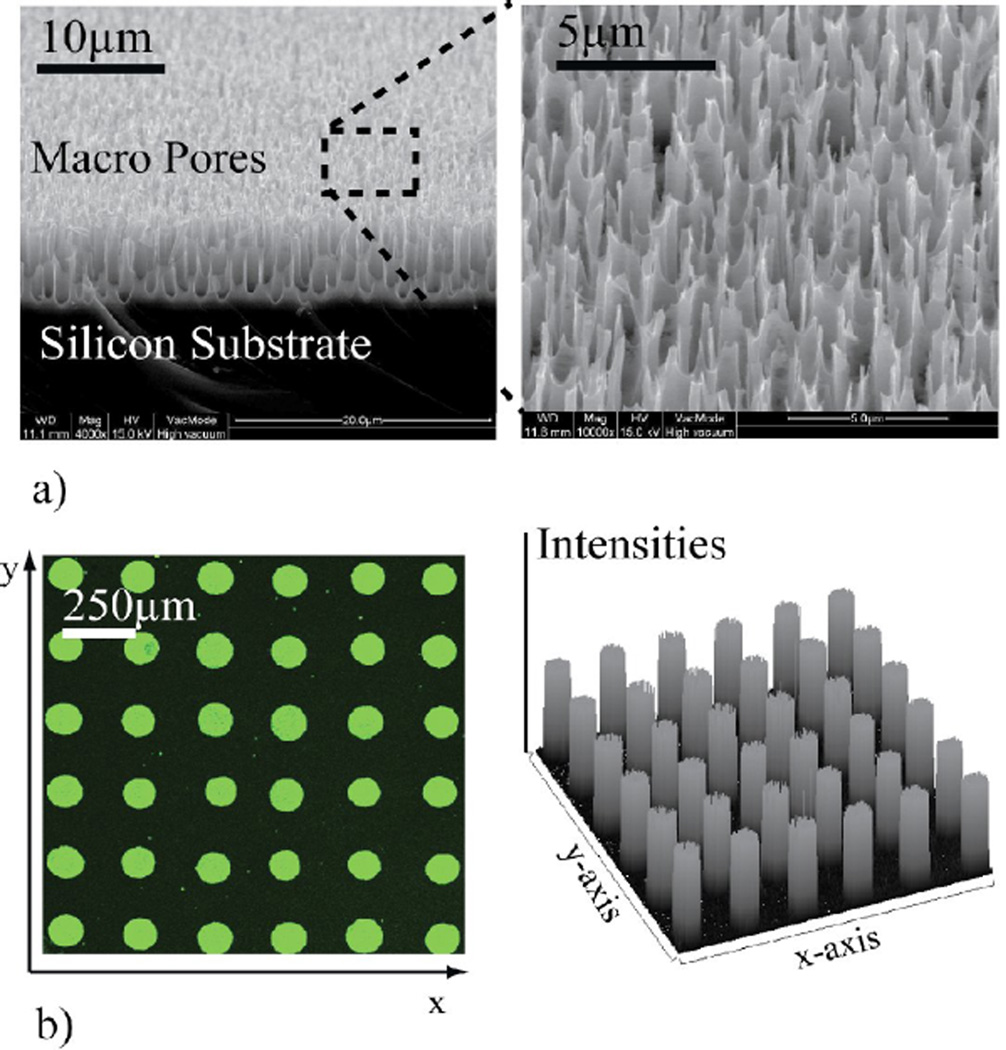
Figure 2.
a) Field emission scanning electron microscope image of the porous silicon surface. Macro pore structure is shown in the zoom insert of the surface. b) Part of spot images and its profile scanning from one of sandwich assay.
2.6 Measurement
The intensity of each spot was quantified by an open source image processing tool kit, ImageJ (http://rsbweb.nih.gov/ij/). 9 spots of each P-Si chip were chosen to quantification of data, which means totally 18 spots were used for data analysis since all experiment were performed within two independent chips. The intensity of each spot image was measured and averaged across its circular area. Local background signal was collected the same way and subtracted from the spot signals, generating mean spot intensities as presented in the graphs. The LOD (limit of detection) was defined as the lowest detectable PSA concentration corresponding to at least two standard deviations above the mean spot intensities of the negative control (N) signals.
3. Results and Discussion
3.1 Surface morphology of porous silicon (P-Si) and microarray format
The 3-D morphology of the micro/nano porous silicon surface layer offers a high capacity of antibody immobilization. Figure 2-a shows macroporous layers with a characteristic size of sub micro −1µm on the silicon surface by FESEM images. Captured antibody (H117) was microarrayed onto the 3-D porous silicon surface using an in-house developed piezoelectric microdispenser. Figure 2-b shows an example of spots and the homogenous intensity profile of a microarray obtained by the micro-/nanoporous surface morphology.
3.2 Assay performance against concentrations of capturing antibody
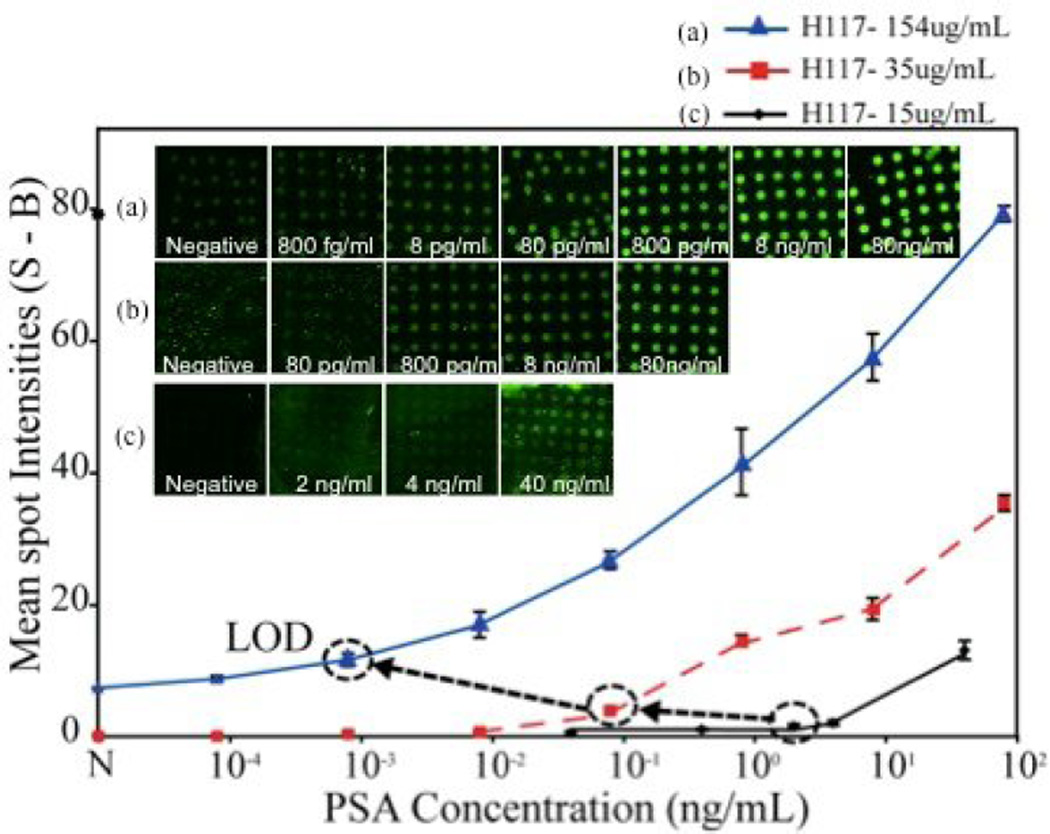
In general, a surface bound immunoassay becomes more sensitive, if the affinity of the capture antibody increases and/or if the surface density of capture antibody molecules increases [4,14]. To optimize our P-Si microarray we evaluated the assay sensitivity by arraying the PSA capture antibody, H117, at three different concentrations and performed the PSA immunoassays in buffer solution. In Figure 3 the microarray fluorescence readout versus the PSA concentration is presented for three different antibody concentrations (15, 35 and 154 µgmL−1). The increased concentration of the capture antibody assay yielded a lowered PSA LOD from 2.5 ngmL−1 at an antibody concentration of 15 µgmL−1 to a LOD of 800 fgmL−1 at an antibody concentration of 154 µgmL−1. It was however noted that at an antibody concentration of 154 ugmL−1 the negative control (N) displayed an elevated signal above zero caused by unspecific binding between capturing antibody and its binding partner (such as secondary or the detector antibody). It was also noted that the dynamic range was extended from 100–102 ngmL−1 to 10−3–102 ngmL−1 as the concentration of the antibody was increased. The signal intensities were enhanced in proportion to the concentration of the capturing antibody, which followed the expectations of improved LOD with increased surface density of the capture antibody. Table 1 shows coefficient of variance and mean spot intensities of the figure 3. In most case, spots reproducibility within the chips has good reliability (CVs ~ 10–20%). However, three cases such as two negative controls in capturing antibody concentration of 15µgmL−1 and 35µgmL−1, and 8 pgmL−1 of PSA when the capturing antibody was 35 µgmL−1 are significantly high CV (~ 70 to 85 %). That is caused by no or less signal difference between spot images and background. In comparison to our earlier study using a nanoparticle enhanced microarray assay [24], the P-Si microassay`s reported herein displayed an improved LOD of two orders of magnitude (800 fgmL−1 when arraying the capture antibody, H117, at a concentration of 154 µgmL−1 as compared to 0.07 – 0.14 ngmL−1 of the earlier nanoparticle assay)
Figure 3.
Titration series of PSA in buffer (PBS) solution at three different concentrations of the capturing antibody H117 (15 µgmL−1, µgmL−1 and 154 µgmL−1). The LOD was found to be 2.5 ngmL−1 when the capturing antibody was 15 µgmL−1 and was reduced to 80 pgmL−1 and 800 fgmL−1 when the capturing antibody concentrations were 35 µgmL−1 and 154 µgmL−1, respectively. Signal of negative control (N) increases significantly at the higher concentration of H117 (154µgmL−1).
Table 1.
Mean spot intensities and its coefficient of variance (CV) of antibody microarrays
| PSA concenentration | Mean spot intensities |
CV(%) n=18 |
|
|---|---|---|---|
| H117 Conc. (15µgmL) | Negative | 0.41 | 80 |
| 2 ngmL−1 | 1.45 | 20 | |
| 4 ngmL−1 | 1.92 | 19 | |
| 20 ngmL−1 | 13.0 | 10 | |
| H117 Conc. (35µgmL) | Negative | 0.34 | 85 |
| 8 pgmL−1 | 0.41 | 75 | |
| 80 pgmL−1 | 3.85 | 6 | |
| 800 pgmL−1 | 14.56 | 5 | |
| 8 ngmL−1 | 19.23 | 8 | |
| 80 ngmL−1 | 35.28 | 3 | |
| H117 Conc. (154µgmL) | Negative | 7.22 | 10 |
| 80 fgmL−1 | 8.63 | 5 | |
| 800 fgmL−1 | 11.5 | 8.3 | |
| 8 pgmL−1 | 19.42 | 6.8 | |
| 80 pgmL−1 | 26.65 | 5.1 | |
| 800 pgmL−1 | 41.48 | 3.0 | |
| 8 ngmL−1 | 57.39 | 6.1 | |
| 80 ngmL−1 | 79.10 | 2 | |
3.3 PSA microarray assays in female human serum
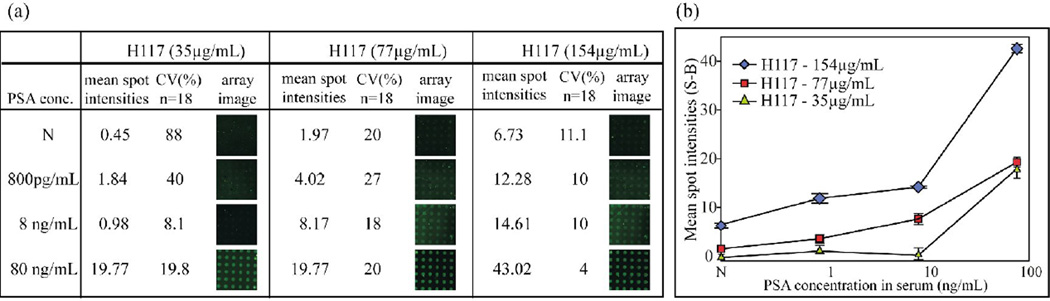
The P-Si microarray assay was subsequently evaluated in human serum. Three P-Si microarrays with antibody concentrations of 35 µgmL−1, 77 µgmL−1 and 154 µgmL−1 were primarily tested in PSA-spiked human female serum at three different PSA concentrations; 800pgmL−1, 8ngmL−1 and 80ngmL−1. The choice of antibody concentration was based on the data in Figure 3. The corresponding titration series of PSA in female serum was recorded for microarrays with three different antibody concentrations: 35, 77 and 154 µgmL−1, Figure 4. In case of the 35 µgmL−1 antibody concentration, spots signals could not be clearly distinguished against the negative controls (N) at a PSA level of 8 ngmL−1. CVs are varied 10 to 20 % that is similar to that of assay in PBS buffer (figure 3). Over 80 % CV value appears in negative control when capturing antibody concentration is 15µgmL−1 that is caused by less resolution between signal and background. At a H117 concentration of 154 ugmL−1 the negative control also increased significantly in accordance with observations in Figure 4. Therefore, 77 ugmL−1 of H117 was selected as an optimal capture antibody concentration for PSA assay in serum. It was also noted that the signal intensities of PSA-spiked serum samples were significantly lower (about 50%) than those obtained with PSA in PBS buffer. This result agrees with the 30–40 % loss of immunoaccessible PSA seen in earlier studies using purified PSA added to serum. [18,27,28]
Figure 4.
PSA-spiked human female serum analyzed with the sandwich microarray at three different capturing antibody concentrations (H117: 35 µgmL−1, 77 µgmL−1 and 154 µgmL−1). Increased assay sensitivity was observed with the elevated concentration of the capturing antibody (H117). The negative signal also increased at the higher concentrations of H117.
3.4 Improvement of the detection limit for determination of PSA
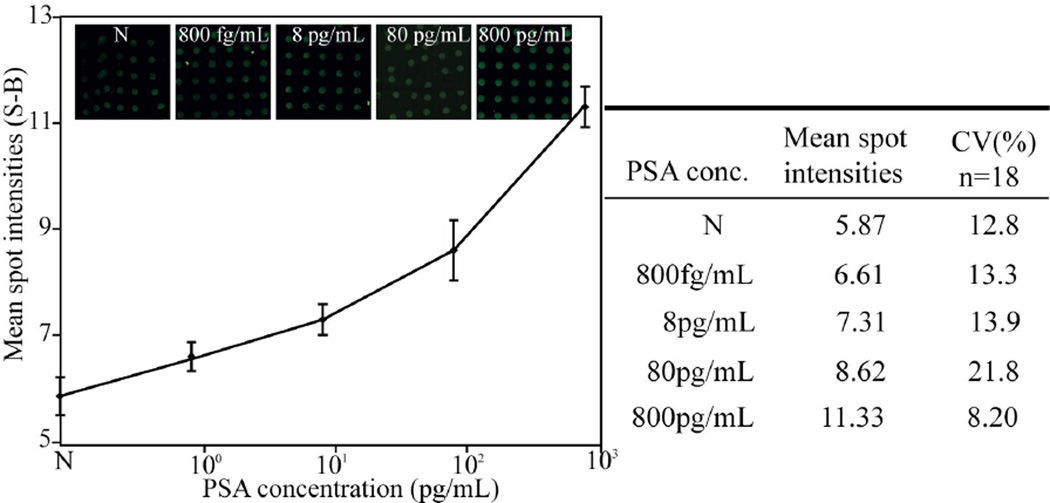
To evaluate the performance of the optimized PSA assay, titration of PSA spiked serum was conducted at a PSA concentration range from 800fgmL−1 to 800 pgmL−1. We chose an antibody concentration of 77 µgmL−1 since it showed good assay readout with a minimum of negative (N) control, Figure 4. Figure 5 shows the titrations of PSA in serum. Limit of detection, LOD, was found to be around 800 fgmL−1 based on a criterion of two standard deviations above the negative control (N). The optimized microarray PSA assay displays a broad dynamic range of five orders of magnitude from 800fgmL−1 to 80 ngmL−1, which now opens for detection of patients with early tumor relapse after radical prostate ectomy.
Figure 5.
Titration of PSA spiked in female serum as monitored with the sandwich antibody microarray. The assay was performed using a capturing antibody concentration of 77µgmL−1. Mean spot intensities and standard deviations were calculated from the spots by imaging via a 10x lens. The size of each spot was around 120 µm and spot to spot distance about 250 µm. Each mean spot intensity (y-axis) was calculated from 12 microarray spots (S) subtracted by background signal (B). The LOD was defined as the lowest detectable signal compared to the false positive (N) and was found to be 800 fgmL−1.
4. Conclusions
In this work, we demonstrate the importance in optimizing the capture antibody concentration in surface bound immunoassays. An increased capture antibody concentration enables our P-Si microarray to detect sub/low pico-gram per milliliter of PSA in undiluted human serum with a broad dynamic range of five orders of magnitude. The sensitivity of our protein microarray for PSA detection covers relevant diagnostic cut off points (from 0.6 ngmL−1 to 4ngmL−1) generally used as indicators of potential malignant prostate disease. The microarray format, an improved LOD (800fgmL−1) and a wide dynamic range (800 fgmL−1 to 80 ngmL−1) allows for earlier detection of recurrent disease in prostate cancer patients.
Highlights.
We develop Porous Silicon based microarray for PSA detection in human serum
We optimize density of capturing antibody for improving sandwich immunoassay of PSA
Porous silicon has high capacity of capturing of antibody
The microarray offers 800fgmL−1(0.6amol) of PSA in human serum
It also has broad detection dynamic ranges ((800 fgmL−1 to 80 ngmL−1)
Acknowledgement
This study was supported by grants Swedish Research Council (Grant no. 621-2009-5361) and STINT Institutional Grant IG2010 2068. We also greatly acknowledged Swedish Cancer Society funding no. to 11-0624, National Cancer Institute (grant numbers no. R33 CA 127768-02, P50-CA92629, R01 CA160816) and the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre based at Oxford University Hospitals NHS Trust and University of Oxford, and Fundación Federico SA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jacobs JM, Adkins JN, Qian W, Liu T, Shen Y, Camp DG, II, Smith RD, et al. J. Proteome Res. 2005;4:1073–1085. doi: 10.1021/pr0500657. [DOI] [PubMed] [Google Scholar]
- 2.Drukier AK, Ossetrova N, Schors E, Krasik G, Grigoriev I, Koenig C, Sulkowski M, Holcman J, Brown LR, Tomaszewski JE, Schnall MD, Sainsbury R, Lokshin AE, Godovac-Zimmermann J. J. Proteome Res. 2006;5:1906–1915. doi: 10.1021/pr0600834. [DOI] [PubMed] [Google Scholar]
- 3.Gembitsky DS, Lawlor K, Jacovina A, Yaneva M, Tempst P. Mol. Cell Proteomics. 2004;3:1102–1118. doi: 10.1074/mcp.M400075-MCP200. [DOI] [PubMed] [Google Scholar]
- 4.Saviranta P, Okon R, Brinker A, Warashina M, Eppinger J, Geierstanger BH. Clin. Chem. 2004;50:1907–1920. doi: 10.1373/clinchem.2004.037929. [DOI] [PubMed] [Google Scholar]
- 5.Tibes R, Qiu Y, Lu Y, Hennessy B, Andreeff M, Mills GB, Kornblau SM. Mol. Cancer Ther. 2006;5:2512–2521. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 6.Wulfkuhle JD, Aquino JA, Calvert VS, Fishman DA, Coukos G, Liotta LA, Petricoin EF., III Proteomics. 2003;3:2085–2090. doi: 10.1002/pmic.200300591. [DOI] [PubMed] [Google Scholar]
- 7.Sheehan KM, Calvert VS, Kay EW, Lu Y, Fishman D, Espina V, Aquino J, Speer R, Araujo R, Mills GB, Liotta LA, Petricoin EF, III, Wulfkuhle JD. Molecular & Cellular Proteomics. 2005;4:346–355. doi: 10.1074/mcp.T500003-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Järås K, Ressine A, Nilsson E, Malm J, Marko-Varga G, Lilja H, Laurell T. Anal. Chem. 2007;79:5817–5825. doi: 10.1021/ac0709955. [DOI] [PubMed] [Google Scholar]
- 9.Poetz O, Schwenk JM, Kramer S, Stoll D, Templin MF, Joos TO. Mech. Ageing Dev. 2005;126:161–170. doi: 10.1016/j.mad.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 10.Hosokawa K, Omata M, Maeda M. Anal. Chem. 2007;79:6000–6004. doi: 10.1021/ac070659o. [DOI] [PubMed] [Google Scholar]
- 11.Rapkiewicz A, Espina V, Zujewski JA, Lebowitz PF, Filie A, Wulfkuhle J, Camphausen K, Petricoin EF, Liotta LA, Abati A. Cancer. 2007;111:173–184. doi: 10.1002/cncr.22686. [DOI] [PubMed] [Google Scholar]
- 12.Nishizuka S, Charboneau L, Young L, Major S, Reinhold WC, Waltham M, Kouros-Mehr H, Bussey KJ, Lee JK, Espina V, Munson PJ, Petricoin E, III, Liotta LA, WeinsteinS JN. Proc. Natl. Acad. Sci. 2003;100:14229–14234. doi: 10.1073/pnas.2331323100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schweitzer B, Wiltshire S, Lambert J, O'Malley S, Kukanskis K, Zhu Z, Kingsmore SF, Lizardi PM, Ward DC. Proc. Natl. Acad. Sci. 2000;97:10113–10119. doi: 10.1073/pnas.170237197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Templin MF, Stoll D, Schrenk M, Traub PC, Vöhringer CF, Joos TO. Trends in biotechnology. 2002;20:160–166. doi: 10.1016/s0167-7799(01)01910-2. [DOI] [PubMed] [Google Scholar]
- 15.Lilja H, Ulmert D, Vickers AJ. Nat. Rev. Cancer. 2008;8:268–278. doi: 10.1038/nrc2351. [DOI] [PubMed] [Google Scholar]
- 16.Stephenson AJ, Kattan MW, Eastham JA, Dotan ZA, Bianco FJ, Lilja H, Scardino PT. Journal of clinical oncology. 2006;24:3973–3978. doi: 10.1200/JCO.2005.04.0756. [DOI] [PubMed] [Google Scholar]
- 17.Steuber T, Vickers AJ, Haese A, Becker C, Pettersson K, Chun F, Kattan MW, Eastham JA, Scardino PT, Huland H, Lilja H. Int. J. Cancer. 2006;118:1234–1240. doi: 10.1002/ijc.21474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lilja H, Christensson A, Dahlén U, Matikainen MT, Nilsson O, Pettersson K, Lövgren T. Clin. Chem. 1991;37:1618–1625. [PubMed] [Google Scholar]
- 19.Liu D, huang X, Wang Z, Jin A, Sun X, Zhu L, Wang F, Ma Y, Niu G, Hight Walker AR, Chen X. ACSNano [Google Scholar]
- 20.Thaxton CS, Elghanian R, Thomas AD, Stoeva SI, Lee JS, Smith ND, Schaeffer AJ, Klocker H, Horninger W, Bartsch G, Mirkin CA. PNAS. 2009;106:18437–18442. doi: 10.1073/pnas.0904719106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rissin DM, Kan CW, Campbell TG, Howes SC, Fournier DR, Song L, Piech T, Patel PP, Chang L, Rivnak AJ, Ferrell EP, Randall JD, Provuncher GK, Walt DR, Duffy DC. Nature Biotechnology. 2010;28:595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson DH, Hanlon DW, Provuncher GK, Chang L, Song L, Patel PP, Ferrell EP, Lepor H, Partin AW, Chan DW, Sokoll LJ, Cheli CD, Thiel RP, Fournier DR, Duffy DC. Clinical Chemistry. 2011;57(12):1712–1721. doi: 10.1373/clinchem.2011.169540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ressine A, Ekström S, Marko-Varga G, Laurell T, Ressine A. Anal. Chem. 2003;75:6968–6974. doi: 10.1021/ac034425q. [DOI] [PubMed] [Google Scholar]
- 24.Järås K, Tajudin AA, Ressine A, Soukka T, Marko-Varga G, Bjartell A, Malm J, Laurell T, Lilja H. J. Proteome Research. 2008;7:1308–1314. doi: 10.1021/pr700591j. [DOI] [PubMed] [Google Scholar]
- 25.Järås K, Adler B, Tojo A, Malm Johan, Marko-Varga G, Lilja H, Laurell T. Clinica Chimica Acta. 2012;414:76–84. doi: 10.1016/j.cca.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foll H, Christophersen M, Carstensen J, Hasse G. Sci. Eng. R. 2002;39:93–141. [Google Scholar]
- 27.Laurell T, Wallman L, Nilsson J. J. Micromech. Microeng. 1999;9:369–376. [Google Scholar]
- 28.Christensson A, Laurell C, Lilja H. Eur. J. Biochem. 1990;194:755–763. doi: 10.1111/j.1432-1033.1990.tb19466.x. [DOI] [PubMed] [Google Scholar]