Abstract
DNA methylation is found in many eukaryotes, but its function is still controversial. We have studied the methylation of plant and animal genomes using a PCR-based technique amenable for high throughput. Repetitive elements are methylated in both organisms, but whereas most mammalian exons are methylated, plant exons are not. Thus, targeting of methylation specifically to transposons appears to be restricted to plants. We propose that the mechanistic basis of this difference may involve RNA interference. Sequencing strategies that depend on differential methylation are predicted to have different outcomes in plant and mammalian genomes.
The pattern of cytosine methylation in eukaryotic genomes has been investigated in a number of organisms, and differs substantially (Bird 2002). In plants, for example, methylation is found at symmetric CpXpG and asymmetric CpXpX sites, as well as the canonical CpG sites found in most mammalian genomes (Gruenbaum et al. 1981; Meyer et al. 1994). DNA Methyltransferases present both in plants and animals (i.e., MET1 in Arabidopsis, ZMET1 in maize, and DNMT1 in mammals) are involved in maintenance of methylation of CpG dinucleotides (Bestor 1992; Kishimoto et al. 2001). Methylation of CpXpG and asymmetric sites is carried out by chromomethylases (i.e., CMT3, and ZMET2), and the domains rearranged methylases (DRM1 and DRM2), respectively, which are encoded by plant-specific gene families (Cao et al. 2000; Papa et al. 2001; Cao and Jacobsen 2002). In addition, chromatin-remodeling proteins also affect DNA methylation. Arabidopsis decrease in DNA methylation 1 (DDM1) is a SWI2/SNF2 protein that is responsible for genome-wide methylation (Vongs et al. 1993), and KRYPTONITE is a histone lysine 9 methylase that directs methylation of some CpXpG sequences (Jackson et al. 2002; Johnson et al. 2002).
The level of 5-methylcytosine (5mC) varies substantially among plants, but accounts for up to 25% of all cytosines in maize (Papa et al. 2001). In mammals, the distribution of 5mC undergoes radical changes during early development (Monk et al. 1987), but it is prominent in repetitive sequences and absent from CpG islands (Yoder et al. 1997; Bird 2002). In plants, silent transposable elements are methylated (Bennetzen et al. 1994; Flavell 1994; Martienssen 1998), and can be reactivated in methylation-defective mutants (Miura et al. 2001; Singer et al. 2001). However, methylation is unusual in plant genes, and is restricted to the 5′- and 3′-flanking regions in the few cases in which it was studied in detail (Walbot and Warren 1990; Patterson et al. 1993).
In animals, there are two alternative views concerning the role of DNA methylation. One view is that DNA methylation silences transposons as it does in plants (Yoder et al. 1997). Supporting this hypothesis, it has been shown that retrotransposon transcripts accumulate in methylation-defective mutant mice (Walsh et al. 1998) and in interspecific mammalian hybrids that have hypomethylated genomes (O'Neill et al. 1998). The second view is that methylation targets both genes and repetitive DNA in order to decrease transcriptional noise. Only CpG islands are protected from methylation to permit access to promoters (Bird 1995; Tornaletti and Pfeifer 1995; Tada et al. 1997).
Methylation has also been found associated with cancer. A reduction in overall levels of DNA methylation as well as hypermethylation of CpG islands have been observed in cancerous cells (Feinberg et al. 1988; Jones and Baylin 2002). It is not clear whether this global demethylation can reactivate silent transposons leading to chromosome instability (Robertson 2001), although, recently, it has been reported that mice with decreased DNA methylation levels due to low DNMT1 expression show increased genomic rearrangements and develop tumors at high frequency (Eden et al. 2003; Gaudet et al. 2003).
Using a modified technique (Chotai and Payne 1998), which we refer to as McrPCR, we have studied DNA methylation in plants and mammals. With this unbiased approach, amenable for high throughput, we show that exon methylation is extensive in animals, but not in plants, indicating that whereas transposon silencing may be an important role for DNA methylation in plants, it's role is more complicated in mammalian genomes. Our results have implications for the application of methylation-based enrichment strategies in plant and animal genomic sequencing.
RESULTS AND DISCUSSION
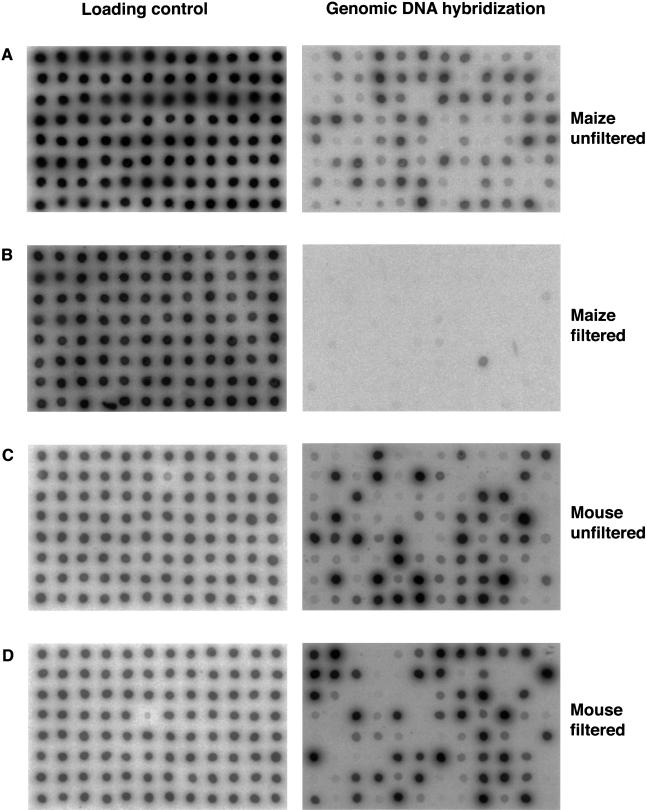
We showed previously that most transposons are methylated in maize, using a gene-enrichment sequencing strategy known as Methylation Filtration (Rabinowicz et al. 1999; Rabinowicz 2003). That is, maize genomic libraries constructed using a methylation restrictive Escherichia coli host (i.e., mcrBC+) substantially exclude highly repetitive sequences, and are enriched for genes. Similar enrichment is observed for several other plant species (P. Rabinowicz, A. Budiman, J. Bedell, N. Lakey, W.R. McCombie, and R.Martienssen, unpubl.). To investigate the distribution of DNA methylation in the mouse genome, we applied Methylation Filtration by constructing a short insert genome shotgun library in the McrBC+ strain JM107 using mouse DNA. McrBC requires two [A/G]mC half-sites to restrict DNA (Sutherland et al. 1992). Figure 1 shows 96 clones from a mouse-filtered library spotted onto a nylon membrane hybridized with labeled mouse genomic DNA. The hybridizing clones contain high-copy DNA. In contrast to maize, there is no enrichment for mouse low-copy sequences in McrBC+ as opposed to McrBC- strains. One explanation could be that highly repetitive sequences have suffered CG suppression and have few CG-methylation sites. Alternatively, the lack of differential enrichment could mean that genes and transposons are not differentially methylated in mouse.
Figure 1.
Dot blot hybridizations from maize unfiltered (A) and filtered (B) and mouse unfiltered (C) and filtered (D) libraries. A total of 96 clones were spotted in each membrane and hybridized against total genomic DNA to identify repetitive clones (right) or against vector DNA as loading control (left). Maize data (A,B) reproduced with permission from Nature Genetics (Rabinowicz et al. 1999).
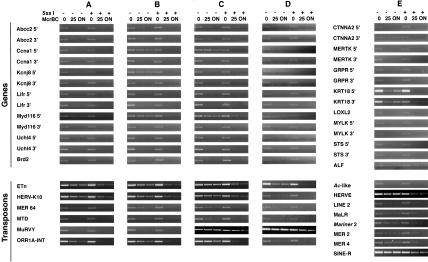
To distinguish these possibilities, we examined DNA methylation in plant and animal genes and transposons using McrPCR. Genomic DNA was first digested with McrBC in vitro. Specific primers were then used to amplify transposon and exon sequences from randomly selected genes (see Supplemental information available at www.genome.org). As a control, the experiment was repeated using genomic DNA pretreated with the CpG methylase Sss I (Fig. 2). We applied McrPCR to maize-leaf DNA and mouse-spleen DNA. A total of 95% of plant exons can be quantitatively amplified after complete digestion with McrBC, indicating that they are unmethylated, whereas they cannot be amplified if pretreated with Sss I (Fig. 3). In contrast, only 15% of mouse exons could be amplified following McrBC digestion, indicating that most mouse exons were methylated (Fig. 4). Transposons gave similar patterns in each organism, indicating that they are methylated so long as they contain CpG sites. We confirmed the unmethylated status of the maize exons by comparing their sequences with the Methylation Filtered (undermethylated) sequences present in GenBank, using BLASTN (Altschul et al. 1997). More than 60% of the exons (or 70% of the genes) had at least one almost perfect match with a Methylation Filtered sequence (data not shown). Consistently, there are currently ∼200 Mb of the undermethylated fraction of the maize genome in GenBank, which correspond to approximately half of the gene space (L. Palmer, P. Rabinowicz, V. Balija, A. O'Shaughnessy, L. Nascimento, S. Dike, M. de la Bastide, R. Martienssen, and W. McCombie, in prep.).
Figure 2.
McrPCR. Equal samples of genomic DNA are either pretreated with CpG methylase (Sss I methylase), or not, and both samples are digested in a time course with McrBC (only time zero and complete digestion are shown). PCR is then performed using specific primers (arrows). If the target sequence (gray box) is methylated in the genome, there will be a decrease in the amount of PCR product following McrBC digestion with or without Sss I methylase pretreatment. If the target is not methylated, a decrease in PCR yield will only be evident following Sss I methylase pretreatment.
Figure 3.
Transposons, but not genes, are methylated in maize. Genomic DNA was digested with McrBC during 0 min, 25 min, or 8 h, with or without Sss I methylase pretreatment. In genes for which two exons were tested, their relative location (5′ or 3′) is indicated. Genes can be found in the Maize-Targeted Mutagenesis (MTM, see Supplemental information, May et al. 2003)
Figure 4.
Genes and transposons are methylated in mouse and human. In wild-type (A) or transgenic control (B) mouse spleen, both genes and transposons are methylated. In mouse lymphoma cells (C), mouse cells in culture (D), or human cells in culture (E), genes remain methylated, whereas some transposons become demethylated.
Mouse and human fibroblast cells had levels of exon methylation comparable with those seen in mouse spleen (Fig. 4). The mouse retrovirus MuRVY, however, was de-methylated in cultured cells. In mouse lymphoma DNA, exon methylation was not affected, although transposon methylation was reduced as in cultured cells (Fig. 4). Of six repetitive elements, three retroelements (MuRVY, ETn, and ORR1A-INT) lost some methylation in lymphoma cells compared with wild-type spleen. Levels of 5mC are reduced in cancer cells (Goelz et al. 1985; Feinberg et al. 1988; Robertson 2001), and we are currently investigating whether this demethylation results in retrotransposition, which has been shown to cause large genomic rearrangements in human cells (Gilbert et al. 2002; Symer et al. 2002). Methylation also represses recombination that may contribute to genome instability in cancer (Colot and Rossignol 1999).
McrPCR is very sensitive and straightforward to apply in high throughput. In contrast, Southern analysis using methylation-sensitive restriction enzymes usually monitors methylation at sites near, rather than within genes, whereas higher-resolution techniques, such as bisulphite sequencing, can be difficult to quantify in high throughput (Adorjan et al. 2002). We have used McrPCR to show that most coding sequences are methylated in mammals (Bird 1999; Simmen et al. 1999), whereas plant genes are mostly unmethylated under identical conditions. Transposons and other repeats are methylated in both plants and animals. Our results suggest that almost all of the exons, and all of the corresponding genes, will be represented in Methylation Filtration shotgun libraries of plant genomes (Rabinowicz et al. 2003). Analysis of a large number of maize Methylation Filtration reads shows comprehensive genome coverage, further supporting this prediction (L. Palmer, P. Rabinowicz, V. Balija, A. O'Shaughnessy, L. Nascimento, S. Dike, M. de la Bastide, R. Martienssen, and W. McCombie, in prep.). Furthermore, the 5′ exon of the heme oxidase (Heme Ox) gene, which shows methylation by McrPCR analysis, was still found among the Methylation Filtered reads in GenBank. This supports the notion that even variably methylated genes can be recovered if a few different tissues or developmental stages are used to construct Methylation Filtered libraries.
The striking difference in methylation patterns between plant and mammalian genes suggests that either methylation plays different roles in these organisms, or that targeting mechanisms differ, or both. All plants studied so far have high levels of genomic methylation limited to repeats and transposable elements. However, whereas DNA methylation certainly helps to silence plant transposons, active transposons have been found in a very wide range of plants, including maize (McClintock 1951). Further, some animals and fungi lack methylation completely, and transposons are specifically unmethylated in a primitive chordate (Simmen et al. 1999).
In mammals, transposons are methylated, but this does not distinguish them from genes. Further, a sharp reduction in methylation in preimplantation embryos does not appear to lead to transposon activation, although mutants in DNA methylation do (Monk et al. 1987). One possibility is that exon methylation is due to a spreading from nearby transposons. In human, however, we observed that unmethylated exons are, on average, closer to a transposon than methylated exons, and that there is no bias against methylation of the first exon, even when they are close to CpG islands (data not shown).
What might be the explanation for exon methylation in mammals? Mammalian genes are more than 10 times larger than maize genes, and may use methylation to reduce spurious transcription. Recently, it has been shown that histone H3 lysine-9 methylation can be triggered by RNA interference in yeast and tetrahymena (Taverna et al. 2002; Volpe et al. 2002), and that histone H3 lysine-9 methylation can guide DNA methylation in fungi and plants (Tamaru and Selker 2001; Jackson et al. 2002; Johnson et al. 2002). This may be the mechanism underlying transposon methylation (Martienssen and Colot 2001; Hamilton et al. 2002; Zilberman et al. 2003). Whereas mammalian cells support RNA interference, they lack the RNA-dependent RNA polymerase (RdRP) found in plants and fungi. In fission yeast, RdRP binds chromatin and is a key component in targeting histone modification (Volpe et al. 2002). Perhaps, in its absence, mammalian genomes cannot differentially target DNA methylation to genes and transposons, resulting in a general methylation that decreases a potentially deleterious transcriptional background (Bird 1995).
METHODS
Strains
Maize DNA was prepared from inbred B73. Mouse genomic DNA used for library construction and hybridizations was prepared from day 13.5 C57BL/6 embryos (Jackson Labs), and mouse-cell culture DNA was prepared from primary C57BL/6 MEFs. Human-cell DNA was extracted from IMR90 embryonic lung fibroblasts. Mouse spleen DNA was prepared from adult C57BL/6 mice and from Eμ-Myc transgenic mice (Adams et al. 1985) without any detectable tumor burden. Lymphoma DNA was extracted from lymph nodes of Eμ-Myc mice harboring palpable B-cell lymphomas. These lymph nodes comprise >95% tumor cells (Schmitt et al. 2002).
McrPCR and Methylation Filtration
Genomic DNA was prepared by phenol-chloroform extraction and ethanol precipitation, and was then nebulized as described (Wilson and Mardis 1997). Libraries and dot-blot hybridizations were prepared as described previously (Rabinowicz et al. 1999). Sss I pre-treatment and McrBC digestion was performed as recommended by the supplier (New England Biolabs). Amplification was carried out using 25–50 ng of template DNA, 0.5 mM primers (see Supplemental information), 0.5 mM dNTPs, 1 U of Taq polymerase (QIAGEN) in 20 μL of 1× reaction buffer. Reactions were in 96-well PCR plates heated to 95°C for 5 min, followed by 26 cycles of 30 sec at 95°C, 45 sec at 58°C, and 45 sec at 72°C. An extension of 10 min at 72°C followed the last cycle.
Acknowledgments
This work was supported by grants from USDA (97-35300-4564) and NSF (DBI-0110143) Plant Genome Programs.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1784803.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Adams, J.M., Harris, A.W., Pinkert, C.A., Corcoran, L.M., Alexander, W.S., Cory, S., Palmiter, R.D., and Brinster, R.L. 1985. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 318: 533-538. [DOI] [PubMed] [Google Scholar]
- Adorjan, P., Distler, J., Lipscher, E., Model, F., Muller, J., Pelet, C., Braun, A., Florl, A.R., Gutig, D., Grabs, G., et al. 2002. Tumour class prediction and discovery by microarray-based DNA methylation analysis. Nucleic Acids Res. 30: e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen, J.L., Schrick, K., Springer, P.S., Brown, W.E., and SanMiguel, P. 1994. Active maize genes are unmodified and flanked by diverse classes of modified, highly repetitive DNA. Genome 37: 565-576. [DOI] [PubMed] [Google Scholar]
- Bestor, T.H. 1992. Activation of mammalian DNA methyltransferase by cleavage of a Zn binding regulatory domain. EMBO J. 11: 2611-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird, A. 1999. DNA methylation de novo. Science 286: 2287-2288. [DOI] [PubMed] [Google Scholar]
- Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes & Dev. 16: 6-21. [DOI] [PubMed] [Google Scholar]
- Bird, A.P. 1995. Gene number, noise reduction and biological complexity. Trends Genet. 11: 94-100. [DOI] [PubMed] [Google Scholar]
- Cao, X. and Jacobsen, S.E. 2002. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc. Natl. Acad. Sci. 99: 16491-16498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, X., Springer, N.M., Muszynski, M.G., Phillips, R.L., Kaeppler, S., and Jacobsen, S.E. 2000. Conserved plant genes with similarity to mammalian de novo DNA methyltransferases. Proc. Natl. Acad. Sci. 97: 4979-4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotai, K.A. and Payne, S.J. 1998. A rapid, PCR based test for differential molecular diagnosis of Prader-Willi and Angelman syndromes. J. Med. Genet. 35: 472-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot, V. and Rossignol, J.L. 1999. Eukaryotic DNA methylation as an evolutionary device. Bioessays 21: 402-411. [DOI] [PubMed] [Google Scholar]
- Eden, A., Gaudet, F., Waghmare, A., and Jaenisch, R. 2003. Chromosomal instability and tumors promoted by DNA hypomethylation. Science 300: 455. [DOI] [PubMed] [Google Scholar]
- Feinberg, A.P., Gehrke, C.W., Kuo, K.C., and Ehrlich, M. 1988. Reduced genomic 5-methylcytosine content in human colonic neoplasia. Cancer Res. 48: 1159-1161. [PubMed] [Google Scholar]
- Flavell, R.B. 1994. Inactivation of gene expression in plants as a consequence of specific sequence duplication. Proc. Natl. Acad. Sci. 91: 3490-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet, F., Hodgson, J.G., Eden, A., Jackson-Grusby, L., Dausman, J., Gray, J.W., Leonhardt, H., and Jaenisch, R. 2003. Induction of tumors in mice by genomic hypomethylation. Science 300: 489-492. [DOI] [PubMed] [Google Scholar]
- Gilbert, N., Lutz-Prigge, S., and Moran, J.V. 2002. Genomic deletions created upon LINE-1 retrotransposition. Cell 110: 315-325. [DOI] [PubMed] [Google Scholar]
- Goelz, S.E., Vogelstein, B., Hamilton, S.R., and Feinberg, A.P. 1985. Hypomethylation of DNA from benign and malignant human colon neoplasms. Science 228: 187-190. [DOI] [PubMed] [Google Scholar]
- Gruenbaum, Y., Naveh-Many, T., Cedar, H., and Razin, A. 1981. Sequence specificity of methylation in higher plant DNA. Nature 292: 860-862. [DOI] [PubMed] [Google Scholar]
- Hamilton, A., Voinnet, O., Chappell, L., and Baulcombe, D. 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21: 4671-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, J.P., Lindroth, A.M., Cao, X., and Jacobsen, S.E. 2002. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416: 556-560. [DOI] [PubMed] [Google Scholar]
- Johnson, L., Cao, X., and Jacobsen, S. 2002. Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 12: 1360-1367. [DOI] [PubMed] [Google Scholar]
- Jones, P.A. and Baylin, S.B. 2002. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 3: 415-428. [DOI] [PubMed] [Google Scholar]
- Kishimoto, N., Sakai, H., Jackson, J., Jacobsen, S.E., Meyerowitz, E.M., Dennis, E.S., and Finnegan, E.J. 2001. Site specificity of the Arabidopsis METI DNA methyltransferase demonstrated through hypermethylation of the superman locus. Plant Mol. Biol. 46: 171-183. [DOI] [PubMed] [Google Scholar]
- Martienssen, R. 1998. Transposons, DNA methylation and gene control. Trends Genet. 14: 263-264. [DOI] [PubMed] [Google Scholar]
- Martienssen, R.A. and Colot, V. 2001. DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science 293: 1070-1074. [DOI] [PubMed] [Google Scholar]
- May, B.P., Liu, H., Vollbrecht, E., Senior, L., Rabinowicz, P.D., Roh, D., Pan, X., Stein, L., Freeling, M., Alexander, D., et al. 2003. Maize-targeted mutagenesis: A knockout resource for maize. Proc. Natl. Acad. Sci. 100: 11541-11546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B. 1951. Chromosome organization and genic expression. Cold Spring Harb. Symp. Quant. Biol. 16: 13-47. [DOI] [PubMed] [Google Scholar]
- Meyer, P., Niedenhof, I., and ten Lohuis, M. 1994. Evidence for cytosine methylation of non-symmetrical sequences in transgenic Petunia hybrida. EMBO J. 13: 2084-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, A., Yonebayashi, S., Watanabe, K., Toyama, T., Shimada, H., and Kakutani, T. 2001. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411: 212-214. [DOI] [PubMed] [Google Scholar]
- Monk, M., Boubelik, M., and Lehnert, S. 1987. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development 99: 371-382. [DOI] [PubMed] [Google Scholar]
- O'Neill, R.J., O'Neill, M.J., and Graves, J.A. 1998. Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature 393: 68-72. [DOI] [PubMed] [Google Scholar]
- Papa, C.M., Springer, N.M., Muszynski, M.G., Meeley, R., and Kaeppler, S.M. 2001. Maize chromomethylase Zea methyltransferase2 is required for CpNpG methylation. Plant Cell 13: 1919-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, G.I., Thorpe, C.J., and Chandler, V.L. 1993. Paramutation, an allelic interaction, is associated with a stable and heritable reduction of transcription of the maize b regulatory gene. Genetics 135: 881-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowicz, P.D. 2003. Constructing gene enriched genomic libraries using methylation filtration technology. Methods Mol. Biol. 236: 21-36. [DOI] [PubMed] [Google Scholar]
- Rabinowicz, P.D., Schutz, K., Dedhia, N., Yordan, C., Parnell, L.D., Stein, L., McCombie, W.R., and Martienssen, R.A. 1999. Differential methylation of genes and retrotransposons facilitates shotgun sequencing of the maize genome. Nat. Genet. 23: 305-308. [DOI] [PubMed] [Google Scholar]
- Rabinowicz, P.D., McCombie, W.R., and Martienssen, R.A. 2003. Gene enrichment in plant genomic shotgun libraries. Curr. Opin. Plant Biol. 6: 150-156. [DOI] [PubMed] [Google Scholar]
- Robertson, K.D. 2001. DNA methylation, methyltransferases, and cancer. Oncogene 20: 3139-3155. [DOI] [PubMed] [Google Scholar]
- Schmitt, C.A., Fridman, J.S., Yang, M., Baranov, E., Hoffman, R.M., and Lowe, S.W. 2002. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell 1: 289-298. [DOI] [PubMed] [Google Scholar]
- Simmen, M.W., Leitgeb, S., Charlton, J., Jones, S.J., Harris, B.R., Clark, V.H., and Bird, A. 1999. Nonmethylated transposable elements and methylated genes in a chordate genome. Science 283: 1164-1167. [DOI] [PubMed] [Google Scholar]
- Singer, T., Yordan, C., and Martienssen, R.A. 2001. Robertson's Mutator transposons in A. thaliana are regulated by the chromatin-remodeling gene Decrease in DNA Methylation (DDM1). Genes & Dev. 15: 591-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland, E., Coe, L., and Raleigh, E.A. 1992. McrBC: A multisubunit GTP-dependent restriction endonuclease. J. Mol. Biol. 225: 327-348. [DOI] [PubMed] [Google Scholar]
- Symer, D.E., Connelly, C., Szak, S.T., Caputo, E.M., Cost, G.J., Parmigiani, G., and Boeke, J.D. 2002. Human l1 retrotransposition is associated with genetic instability in vivo. Cell 110: 327-338. [DOI] [PubMed] [Google Scholar]
- Tada, M., Tada, T., Lefebvre, L., Barton, S.C., and Surani, M.A. 1997. Embryonic germ cells induce epigenetic reprogramming of somatic nucleus in hybrid cells. EMBO J. 16: 6510-6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru, H. and Selker, E.U. 2001. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414: 277-283. [DOI] [PubMed] [Google Scholar]
- Taverna, S.D., Coyne, R.S., and Allis, C.D. 2002. Methylation of histone h3 at lysine 9 targets programmed DNA elimination in tetrahymena. Cell 110: 701-711. [DOI] [PubMed] [Google Scholar]
- Tornaletti, S. and Pfeifer, G.P. 1995. Complete and tissue-independent methylation of CpG sites in the p53 gene: Implications for mutations in human cancers. Oncogene 10: 1493-1499. [PubMed] [Google Scholar]
- Volpe, T.A., Kidner, C., Hall, I.M., Teng, G., Grewal, S.I., and Martienssen, R.A. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833-1837. [DOI] [PubMed] [Google Scholar]
- Vongs, A., Kakutani, T., Martienssen, R.A., and Richards, E.J. 1993. Arabidopsis thaliana DNA methylation mutants. Science 260: 1926-1928. [DOI] [PubMed] [Google Scholar]
- Walbot, V. and Warren, C. 1990. DNA methylation in the Alcohol dehydrogenase-1 gene of maize. Plant Mol. Biol. 15: 121-125. [DOI] [PubMed] [Google Scholar]
- Walsh, C.P., Chaillet, J.R., and Bestor, T.H. 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20: 116-117. [DOI] [PubMed] [Google Scholar]
- Wilson, R.K. and Mardis, E.R. 1997. Shotgun sequencing. In Genome analysis. A laboratory manual (eds. B. Birren, E.D. Green, S. Klapholz, R.M. Myers, and J. Roskams), pp. 397-454. Cold Spring Harbor Laboratory Press, Plainview, NY.
- Yoder, J.A., Walsh, C.P., and Bestor, T.H. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13: 335-340. [DOI] [PubMed] [Google Scholar]
- Zilberman, D., Cao, X., and Jacobsen, S.E. 2003. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716-719. [DOI] [PubMed] [Google Scholar]