Abstract
Protein expression in the cell is affected by various sequence-dependent features. Several such sequence-dependent features have been individually studied,yet they have not been compared quantitatively in terms of their relative influence on protein expression,and a hierarchy of these elements has not been determined. Here we present a quantitative analysis examining sequence-dependent features involved in prokaryotic translation,namely,the base-pairing potential between the mRNA Shine-Dalgarno sequence and the ribosomal RNA,codon bias,and the identity of the stop codon. We analyzed these features both at intra- and intergenomic levels using the Escherichia coli and Haemophilus influenzae genomes. Within each genome,we examined the relationship between each feature and protein expression levels determined by 2D-gel analyses. At the intergenomic level,comparative genomic principles were applied to study the relative preservation of the different sequence-dependent properties between orthologs. From these analyses,we determined that biased codon usage is the property that is most highly associated with protein expression and that is most conserved. The identity of the stop codon and the base-pairing potential of the mRNA Shine-Dalgarno sequence and the rRNA seem to have less of an effect on protein expression.
Protein levels in the cell are affected by many factors that operate at the various stages of gene expression including, primarily, transcription and translation. Many of the recent studies have focused on the stage of transcription and on features that affect mRNA abundance (e.g., Spellman et al. 1998; Selinger et al. 2000; Lee et al. 2002; Tjaden et al. 2002), while translation has been somewhat neglected. This is for two main reasons: (1) The present wide use of DNA microarray technology has led to the accumulation of vast amounts of mRNA expression data that motivate numerous studies on transcriptional regulation. In contrast, protein expression data are still quite scarce. (2) It has conventionally been held that protein levels depend, to a large extent, on corresponding mRNA levels, and that it is therefore sufficient to study the transcription stage comprehensively. Recent studies, however, show that the latter statement is not necessarily true. At least in eukaryotes, there are several studies that measured the levels of corresponding mRNAs and proteins under the same conditions and showed that the changes in protein levels cannot be explained by the mRNA changes per se (Futcher et al. 1999; Gygi et al. 1999; Ideker et al. 2001; Washburn et al. 2003). In prokaryotes, as transcription and translation are tightly coupled, it has been widely accepted that cellular protein levels depend directly on mRNA abundance. Still, even in prokaryotes, it is not clear whether mRNA levels alone can account for the variation in protein levels, and if they can be used as predictive indicators of protein levels. Indeed, data on protein levels and mRNA levels in prokaryotes start to accumulate, but there still is a lack of consistent data of the two quantities measured under the same conditions that will enable direct evaluation of this questioned relationship. In parallel, it is important to examine whether there are other attributes, beyond those associated with transcription, that show an association with protein expression and may be used as predictive indicators of protein levels. In the present study, we analyze features associated with translation in prokaryotes and their relative influence on protein levels.
Translation in prokaryotes involves three major steps: initiation, elongation, and termination. Translation efficiency, therefore, may be influenced by each of these steps, which, in turn, are affected by sequence-dependent features.
Initiation of translation involves the binding of the ribosome to the mRNA. The 3′-end of the 16S RNA component of the ribosome binds, via base-pairing, to a short sequence upstream of the start codon, the Shine-Dalgarno (SD) sequence (Shine and Dalgarno 1974). The free energy gained by this interaction can be used as a parameter that reflects the efficiency of translation initiation (Schurr et al. 1993; Osada et al. 1999). Differences in the mRNA SD sequence binding potential to the rRNA, as indicated by the computed free energy values, may explain differences in protein expression. (Note that in this paper we refer to proteins and to the genes encoding them interchangeably. When referring to highly expressed genes, we refer to the genes encoding highly expressed proteins, whose expression was measured by protein abundance.)
The rate of translational elongation has been suggested to be influenced by codon bias. This conjecture is based on two findings: (1) The levels of the different tRNA species in the cell have been shown to correlate with the abundance of their corresponding codons (Ikemura 1985). (2) Highly expressed genes have been shown to have a strong bias toward the more frequent codons (Kanaya et al. 1999; Karlin et al. 2001). Thus, highly expressed genes are expected to use those codons that have higher levels of corresponding tRNAs, enhancing the rate at which translation progresses.
The codons preceding the stop codon (Mottagui-Tabar et al. 1994; Bjornsson et al. 1996), as well as the identity of the stop codon and the base following it (Poole et al. 1995), have been shown to influence the efficiency of translation termination. It has been shown that inefficient stop codons lead to a lower amount of protein (Jin et al. 2002). However, the stop codon has been shown not to be very conserved among Escherichia coli and Bacillus subtilis homologs (Rocha et al. 1999).
Several statistical studies have been performed to examine the prevalence of the different sequence features in the genes encoding highly expressed proteins, and to estimate the correlation between various sequence-dependent features. Karlin and colleagues (2001) showed for several bacteria that proteins documented as highly expressed use the most frequent codons preferentially. Rocha et al. (1999) analyzed the stop and start signals in B. subtilis in relation to G + C content and putative secondary structures. Two other studies have pointed out a correlation between the mRNA SD sequence and codon bias (Sakai et al. 2001; Ma et al. 2002). However, there has not been an assessment of the relative influence of different features on protein expression using measured protein levels in the cell.
Using genomic data and experimental data on protein levels in both E. coli (VanBogelen et al. 1996; Blattner et al. 1997; Link et al. 1997) and Haemophilus influenzae (Fleischmann et al. 1995; Langen et al. 2000), we assessed the relative contribution of the various sequence features to the determination of protein levels. These include the features described above, namely, the binding potential of the mRNA SD sequence and the ribosomal RNA, codon bias, and the identity of the stop codon. First, we assessed the relative influence of each feature on protein expression within each genome by comparing highly and not highly expressed genes. Secondly, we assessed the relative importance of the sequence-dependent features through a comparative genomics approach. A commonly accepted principle of comparative genomics is that sequences with a crucial function are conserved among different, related organisms. Consequently, conserved sequences are implied to have an important function. Here we apply this notion in a broader manner, to assess the relative importance of the examined features for the determination of protein levels. Although different organisms live under different conditions and have different requirements, it seems a reasonable assumption that most principles of regulation would remain conserved in organisms that are not too diverged. In our analysis we do not compare the sequences per se, but their quantitative measures. Thus, by our approach the two studied organisms may have different rRNA SD sequences and completely different codon usages. What we test is the correlation between the quantitative measures representing these features in orthologous genes.
Both the intra- and intergenomic analyses provide quantitative estimates as to the relative influence that the various sequence features related to translation have on protein expression. We show that biased codon usage highly correlates with protein levels, whereas the stop codon and the base-pairing potential between the mRNA SD sequence and the rRNA seem to have less of an effect on protein expression.
RESULTS
General Overview of the Analysis
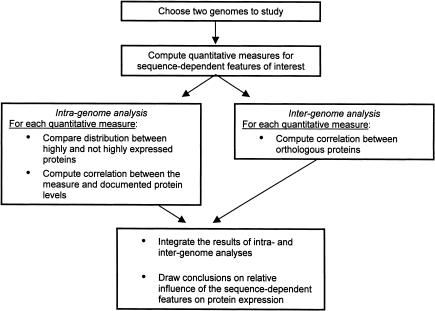
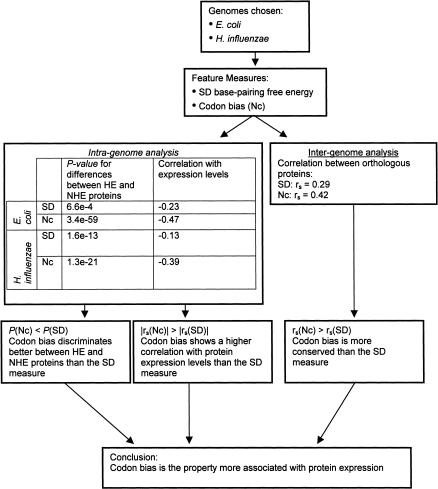
We assess the relative influence of the different sequence features on protein levels by performing both “intragenome” and “intergenome” analyses (Fig. 1). To this end, we need to choose genomes for which protein expression data are available, and to represent the sequence features by quantitative measures (see Methods section). In the intragenome analysis, we compare for each measure of a sequence feature its distribution between highly expressed (HE) and not highly expressed (NHE) proteins encoded in the genome. Additionally, we compute the correlation coefficient between the measure and protein expression levels. The sequence feature showing a more significant distinction between HE and NHE proteins and a higher correlation coefficient is considered as having a greater effect on protein expression. In the intergenome analysis, we compare for each sequence feature its values between orthologous genes in the two genomes by computing a correlation coefficient. The property that shows a significantly higher correlation between orthologs is considered as having a greater effect on protein levels. Consistent results in the intra- and intergenome analyses should provide further support to our conclusions.
Figure 1.
Overview of the analysis.
Data Organization
Because an important aspect of our analysis regards conservation of features that are sequence-dependent, it was necessary to select a pair of organisms that have sufficiently diverged in their genome sequences, to avoid conclusions based on sequence conservation per se owing to insufficient evolutionary divergence. Yet it was also necessary to ensure that the two organisms are close enough to expect similarity in their molecular mechanisms and underlying molecular principles. Two organisms that were found to comply with these requirements were E. coli and H. influenzae, both with fully sequenced genomes (Fleischmann et al. 1995; Tatusov et al. 1996; Blattner et al. 1997). Both organisms belong to the γ subdivision of proteobacteria; yet they are also sufficiently diverged. Their GC content is very different (50.7% for E. coli and 38% for H. influenzae), and the distributions of their codon frequencies (Nakamura et al. 2000) are significantly different (P ≪ 1e-50 by a χ2 test). In addition, for both, large-scale 2D-gel analyses have been carried out (e.g., VanBogelen et al. 1996; Link et al. 1997; Langen et al. 2000), providing a set of experimentally determined highly expressed proteins in both organisms. For E. coli, there were several sets of highly expressed proteins, extracted in different conditions (VanBogelen et al. 1996; Link et al. 1997). All these proteins were included in the HE group and were compared with NHE proteins. For the correlation analysis of the features with the expression levels, we included only the proteins isolated from bacteria that were grown on minimal media and harvested during growth phase (Link et al. 1997). As to H. influenzae, the study included various gels and fractions from a variety of chromatography steps performed to enrich for low-abundance proteins (Langen et al. 2000). However, only levels of proteins from the primary gel were measured in that study, and these were used in our analysis as HE proteins. Of the 3938 genes from GenBank predicted by GeneMarkS (Besemer et al. 2001) in E. coli, 424 are HE, and 163 of these are from growth phase. Of the 1683 genes from H. influenzae, 297 are HE.
The COG database (Clusters of Orthologous Groups of proteins; Tatusov et al. 2001) was used both to filter out false predictions from the NHE set, as well as to find pairs of orthologs within the two organisms. This database contains groups of orthologous proteins from 43 complete genomes, most of which are bacterial. For E. coli, 2953 of the NHE proteins are in COGs. For H. influenzae, 1243 of the NHE proteins are in COGs. Pairs of proteins from E. coli and H. influenzae, within COGs, which were each the best hit of the other, were taken as orthologs. This, when intersected with the GeneMarkS predictions, resulted in a set of 1271 pairs of orthologs. Out of the 1271 orthologs, 129 were HE in both genomes, 153 were HE in E. coli but NHE in H. influenzae, 141 were NHE in E. coli but HE in H. influenzae, and 848 were NHE in both genomes. Thus, the property of high/low expression is highly conserved (P = 4e-30 by a χ2 test).
Analysis of Sequence-Dependent Features
The SD Sequence Upstream of the Translation Initiation Site
The ribosome binding site in prokaryotes is characterized by an mRNA sequence upstream of the translation initiation site that is capable of base-pairing with the 3′-end of the 16S rRNA (Shine and Dalgarno 1974). It is possible that sequences that better fit the 3′ 16S rRNA SD sequence will make more efficient translation initiation sites. To examine whether there is a correlation between protein levels and the potential of the mRNA SD sequence to bind to the 16S rRNA, we computed the free energy gained by the binding of these two sequences (Schurr et al. 1993; Osada et al. 1999). The more compatible rRNA–mRNA SD sequences are expected to have lower free energy values.
We first compared the SD free energy values between HE and NHE proteins in each of the two genomes. The HE proteins were those detected in the 2D-gel experiments (VanBogelen et al. 1996; Link et al. 1997; Langen et al. 2000). The NHE proteins were all other proteins in our data sets that were also in the COG database. The SD free energy values were found to be lower in HE proteins compared with other proteins.
This result was found to be slightly significant for E. coli (P = 6.63e-4 for the Wilcoxon rank sum test, n1 = 424 proteins, n2 = 2953 proteins), and very significant for H. influenzae (P = 1.62e-13 for the Wilcoxon rank sum test, n1 = 297 proteins, n2 = 1243 proteins).
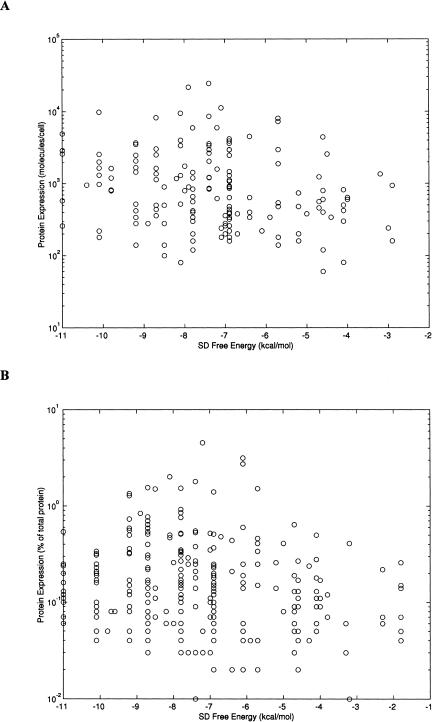
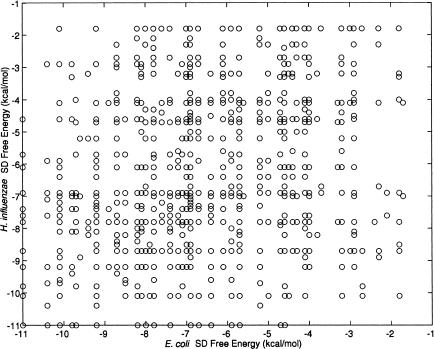
To further investigate the influence of the mRNA–rRNA binding on translation efficiency, the correlation between the free energy and the protein expression levels was computed. As shown in Figure 2, the correlation between these two properties is very low, with rs = -0.23 for E. coli (P < 0.005, n = 163) and rs = -0.13 for H. influenzae (P < 0.05, n = 297). The intergenomic analysis showed a low correlation between the SD free energy values of orthologous proteins (n = 1271, rs = 0.29, P < 0.001; Fig. 3).
Figure 2.
Shine-Dalgarno (SD) free energy values versus protein abundance. The quantitative measure of the SD mRNA sequence is the free energy gain upon its base-pairing with the 16S rRNA. Scatter plots of the free energy values versus protein abundance are shown for 163 highly expressed E. coli proteins (A) and for 297 highly expressed H. influenzae proteins (B). Protein abundance is shown on a log scale.
Figure 3.
Correlation between SD free energy values of orthologs. A total of 1271 orthologs were included in this analysis.
Codon Bias
Proteins that use more frequent codons have been shown to be highly expressed (Ikemura 1985). To examine quantitatively the relationship between codon bias and protein levels, two measures of the codon bias were used: the commonly used CAI (Sharp and Li 1987) and the effective number of codons (Nc; Wright 1990; see Methods section). Comparison by the Wilcoxon rank sum test of both the CAI values and the Nc values between HE and NHE proteins in each of the two genomes revealed a significant difference (for E. coli, P < 1e-100 for CAI and P = 3.35e-59 for Nc; and for H. influenzae, P < 1e-100 for CAI and P = 1.26e-21 for Nc). This indicates that the HE protein group has a strongly biased use of codons.
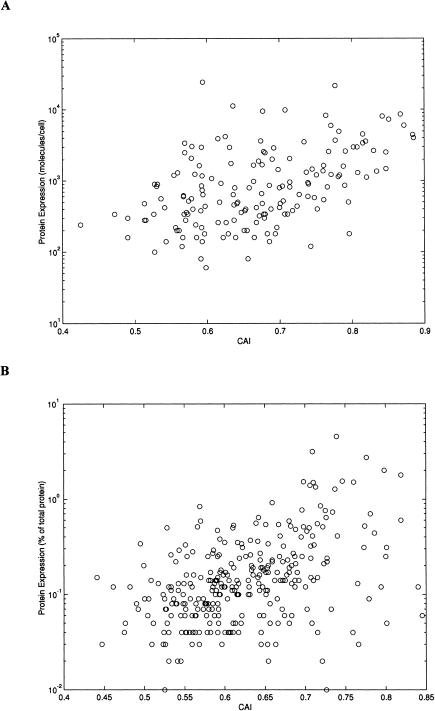
The analysis of correlation between the protein levels and the codon measures pointed to the same phenomenon: For CAI values, the Spearman correlation coefficients were 0.47 (P < 0.001, n = 163 proteins) and 0.44 (P < 0.001, n = 297 proteins) for E. coli and H. influenzae, respectively (Fig. 4). Likewise, the correlation coefficients between the protein levels and Nc values were -0.47 for E. coli and -0.39 for H. influenzae (P < 0.001 for both).
Figure 4.
Relationship between codon bias and protein abundance. The bias in codon usage is expressed by the CAI measure. Scatter plots of CAI versus relative protein abundance are shown for 163 highly expressed E. coli proteins (A) and for 297 highly expressed H. influenzae proteins (B). Protein abundance is shown on a log scale.
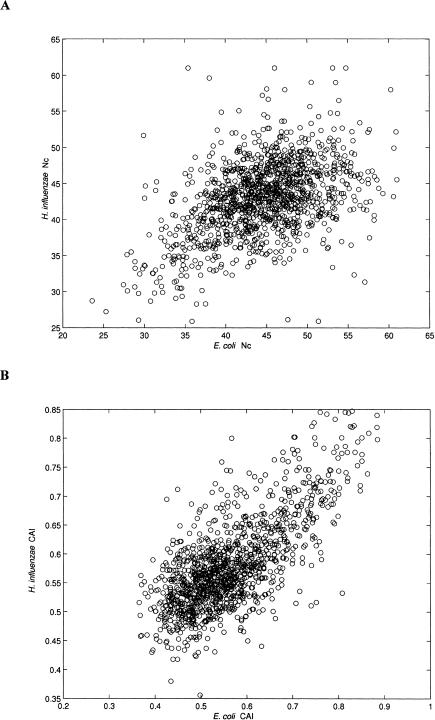
In the intergenomic analysis, the correlation coefficients between the codon bias measures of the 1271 orthologous proteins were found to be 0.64 (P < 0.001) for CAI and 0.42 (P < 0.001) for Nc (Fig. 5).
Figure 5.
Correlation between codon bias of orthologs. Codon bias is expressed either by Nc values (A) or by CAI values (B). Scatter plots of the corresponding values in 1271 E. coli and H. influenzae orthologs are shown.
Comparing the Influence of the mRNA SD Sequence and Codon Bias on Protein Expression
Both the intra- and intergenome analyses indicate that the biased codon usage (as measured by either CAI or Nc) has a greater influence on protein expression than the potential of the sequence upstream of the translational initiation site to base-pair with the rRNA. Below we provide a statistical assessment of this statement for Nc (summarized in Fig. 6). The same analysis holds for CAI.
Figure 6.
Determination of hierarchy of two different properties associated with translation. The results of the intra- and intergenome analyses for SD mRNA–16S rRNA base-pairing potential and codon bias are shown in steps that are described in the overview (Fig. 1).
The Nc value is separated to a greater extent between HE and NHE proteins than the SD free energy values. In addition, for both organisms, the correlation coefficient between Nc values and protein levels is greater than the correlation coefficient between the SD free energy values and protein levels. This result was statistically significant only for H. influenzae (as evident by the Wilcoxon signed rank test on absolute difference of ranks, P = 0.04). Thus, it seems that codon bias has a greater influence on protein expression. The correlation between Nc values of orthologs was also found to be significantly higher than the correlation between the SD free energy values of orthologs (P = 1.64e-4 for test on difference of correlation coefficients and P = 0.01 for the Wilcoxon signed rank test on absolute difference of ranks), and therefore we can conclude that biased use of codons in HE proteins is the more conserved property in the two organisms.
Stop Codon
The identity of the stop codon has been shown to affect the efficiency of translation termination (Poole et al. 1995). As done for the mRNA SD sequence and codon bias, we attempted to evaluate the influence of the stop codon identity on protein expression both by intra- and intergenome analyses. In the intragenome analysis, we searched for preferred stop codons in HE proteins. In the intergenome analysis, we examined the conservation of the stop codons among orthologs.
Because the base following the stop codon has been shown to be important for efficient termination, stop codons are often regarded as four bases (Poole et al. 1995). The stop codon UAAU has been shown to be the most efficient in E. coli (Poole et al. 1995). To see whether UAAU is indeed overrepresented in HE genes, we generated the corresponding contingency table (Table 1). For each genome, we tested by a χ2 test the null hypothesis that there was no difference between the frequencies of UAAU in HE and NHE genes. The null hypothesis was rejected for both genomes (P = 1.76e-18 for E. coli and P = 1.47e-6 for H. influenzae), indicating that there is indeed a significant preference for UAAU in HE genes. We can also represent the relationship between the UAAU stop signal and protein expression levels by the Φ coefficient, a measure of association that can be interpreted equivalently to the Pearson correlation coefficient. The Φ coefficient of UAAU for E. coli is 0.15, and for H. influenzae is 0.12. Note that the association measured by Φ does not regard the actual expression levels as does the correlation coefficient, but regards the association with the categories of HE or NHE proteins. Therefore, we will not compare the Φ values obtained here to the correlation coefficients obtained for the SD free energy values and codon bias measures.
Table 1.
UAAU Stop Signal: Comparison Between Highly and Not Highly Expressed Proteins
|
E. coli
|
H. influenzae
|
|||
|---|---|---|---|---|
| UAAUb | Othersc | UAAUb | Othersc | |
| Highly expresseda | 186 | 238 | 142 | 155 |
| Not highly expressed | 703 | 2250 | 409 | 834 |
Highly expressed proteins are those extracted from 2D-gel data
The number of UAAU stop codons
The number of non-UAAU stop codons
A similar analysis of the standard three-base stop codons (UAA, UAG, and UGA) in E. coli gave significant χ2 P-values (at a significance level of 0.05). For UAG and UGA, the Φ coefficients were slightly negative. We can conclude that UAA is slightly associated with higher expression levels, whereas UAG and UGA are associated with lower expression levels. For H. influenzae, the χ2 value was significant only for UAA and UGA. The Φ coefficient for these two stop codons are 0.11 and –0.11, respectively. Therefore, for H. influenzae, UAA is overrepresented in HE genes, whereas UGA is underrepresented.
The intergenome analysis indicated that the conservation of the stop codons is very low. We calculated the Φ coefficient for each stop codon (e.g., the contingency table for UAA is shown in Table 2, which demonstrates how often the stop codon is conserved and how often there is a different stop codon in the corresponding ortholog). We found that the only stop codon (including UAAU) that had a significant χ2 test was UAA (P = 0.008), with a Φ coefficient of 0.07. In this case, the Φ coefficient and the correlation coefficient comparing the orthologs regarding the other properties can be compared. We find that the correlation reflected by the Φ coefficient is lower than the correlation seen for the SD free energy (P = 2.59e-8). Therefore, we can conclude that in comparison to the codon bias and the SD free energy, the stop codon is the least conserved property associated with translation.
Table 2.
Conservation of UAA Stop Codon in Orthologs
UAA stop codon occurrence
Non-UAA stop codon occurrence
DISCUSSION
One of the major challenges in the postgenomic era is to extract new knowledge regarding gene expression from the accumulated sequence and functional data. Here we exploited different types of data to gain insight into sequence-dependent features that affect one stage in the pathway that leads from the gene to protein expression, namely, translation. These data included genomic data of coding sequences and measured levels of their protein products. From the genomic data, we were able to compute quantitative measures of various sequence-dependent features that influence translation efficiency. These quantitative measures were analyzed at two levels: First, we compared the influence of the different sequence-dependent properties on protein expression levels, and secondly, we compared the relative conservation of these properties between orthologs. Through these analyses we were able to compare the effect on translation of the mRNA SD sequence, the bias in codon usage, and the identity of the stop codon. We found that codon bias has the greatest influence on protein expression levels and is also the most conserved feature. The identity of the stop codon is the least conserved feature.
This hierarchy seems conceivable for several considerations. As translation elongation consumes a large amount of energy (Gold 1988), it seems vital that it be performed efficiently. The finding that the SD sequence has less of a direct influence on protein expression looks reasonable as well. The binding of the 16S ribosomal subunit has been found not to be essential for translation initiation (Calogero et al. 1988), and, in fact, there exist mRNAs without SD sequences (e.g., Boni et al. 2001), and even leaderless mRNAs (Moll et al. 2002). Furthermore, a favorable SD sequence combined with a weak termination signal has been suggested to lead to ribosome queuing, which leads to mRNA degradation (Jin et al. 2002). Therefore, another explanation for the weak correlation between protein levels and the mRNA–rRNA base-pairing potential is that favorable SD sequences may be involved in both HE and NHE genes. It should be noted that the effect of the SD sequence might be much more complicated than just its potential to base-pair with the rRNA. The free energy measure that was used by us actually expresses the potential of the SD mRNA to bind to the rRNA when the SD mRNA is in an open conformation. However, there may be differences in the folded states of the SD mRNA of different genes and these may affect its base-pairing potential with the rRNA as well (de Smit and van Duin 1994). Thus, until these different contributions can be taken into account, we should treat our conclusion of the lesser effect of the SD sequence with the appropriate reservations.
It is important to remark that the Shine-Dalgarno measure is much more sensitive to errors in start-site detection than codon bias. An incorrectly predicted translational start-site can lead to a completely incorrect Shine-Dalgarno parameter, while only slightly affecting the measure of codon bias. To avoid such errors, we attempted to use as accurate start-sites as possible (see Methods). Nevertheless, the possibility that some of the results may have been affected by incorrect start-site predictions cannot be completely excluded.
It has been shown that the identity of the stop codon influences the rate of translation termination. In prokaryotes, there are two codon-specific release factors that play a role in translational termination, Release Factor 1 (RF1) and Release Factor 2 (RF2), which lead to the release of the synthesized polypeptide chain. RF1 recognizes both the UAG and UAA stop codons, whereas RF2 recognizes the UGA and UAA stop codons (for review, see Nakamura et al. 1996). It has been shown experimentally that the identity of the nucleotide following the stop codon influences termination efficiency, and that UAAU leads to the most efficient termination in comparison to all other four-base stop signals (Poole et al. 1995). Indeed, we found a very significant overrepresentation of UAAU in highly expressed genes both in E. coli and in H. influenzae. However, its association with high expression as reflected by the Φ coefficient was found to be weak, and it was not conserved in orthologous genes. We believe that the weak conservation of the stop codon and its weak association with protein expression is caused by several aspects. First, fast release of the polypeptide chain should be a global phenomenon across all genes, as it leads to quicker recycling of the ribosomes, which is of crucial importance to fast-growing bacteria (for review, see Nakamura et al. 1996). Second, the choice of the stop codon has been proposed to be involved in the prevention of translational read-through (Sharp and Bulmer 1988). Third, many genes in E. coli overlap, and therefore the stop codon in one gene can be part of the SD sequence or start codon of the following gene (Eyre-Walker 1996). For example, more than half of the UGA stop codons were shown to overlap a coding sequence or the SD sequence (Eyre-Walker 1996). Because the identity of the stop codon is influenced by different factors, and efficient stop codons may be more of a global phenomenon in the cell allowing for ribosome recycling, it is logical that the identity of the stop codon plays a lesser role in determining protein levels than the two other examined properties.
We can examine our results for different functional categories of the proteins. Interestingly, there is one functional category where the properties determined by all three features were found to be more conserved than in other functional categories. These were the proteins classified in the COG database as involved in information storage and processing. This functional category includes proteins involved in replication, transcription, and translation. It therefore seems that proteins that are involved in these essential molecular mechanisms tend to pursue all strategies to achieve a higher level of protein expression.
One point that should be noted where one should be cautious, is that the protein expression levels were taken from specific growth conditions, and can be expected to vary in different conditions. In contrast, the sequence-dependent features are permanent features of the genes. Therefore, when looking for a connection between the sequence-dependent features and the levels of protein expression, we cannot extrapolate information on a gene-specific level. Nevertheless, as shown here, we could point out significant general trends.
The analysis presented here extends the concept of comparative genomics. Traditionally, studies in comparative genomics focus on sequence comparison and on aspects of sequence conservation. Here, we compare not just the sequences per se, but also quantitative properties that are sequence-dependent, that is, the base-pairing potential of the SD mRNA and 16S rRNA sequences and the codon bias. This enables us to assess the relative importance and the conservation of these properties, beyond the sequences involved. There are many other sequence-dependent features that affect gene expression. These include, for example, the DNA features involved in transcription and degradation signals that affect mRNA half-life. Whereas some sequence-dependent features have commonly used measures, others still need to be further studied, and their quantitative measures need to be developed. Upon achievement of these, a similar analysis that incorporates these features as well should provide additional insight into the relative contribution of the various features that affect gene expression.
METHODS
Data Sources
In this study two genomes were analyzed, the E. coli genome and the H. influenzae genome. The sequence data were taken from GenBank (http://www.ncbi.nlm.nih.gov). Protein levels were extracted from published 2D-gel data (VanBogelen et al. 1996; Link et al. 1997; Langen et al. 2000). Proteins in the 2D-gel data were considered to be HE, and proteins not in the 2D-gel data were considered to be NHE. Although it is possible that in the NHE set there exist HE proteins, it is likely to be dominated by true NHE proteins.
Pairs of orthologous proteins were taken from the COG database (Tatusov et al. 2001). Proteins not in COGs were removed from the NHE set because they may be falsely predicted open reading frames.
The mRNA Shine-Dalgarno (SD) Sequence
The mRNA SD sequence was evaluated by its ability to base-pair with the corresponding sequence in the rRNA (Schurr et al. 1993). Thus, the quantitative measure for an SD sequence was the free energy gain when such base-pairing occurs. These values were computed using the program of Osada et al. (1999). The last seven bases of the 16S rRNA and the 20 bases upstream of the start codon were taken for the calculation. These 20 bases were scanned to find the subsequence that best fits the 16S rRNA SD sequence, that is, gets the lowest free energy value when base-paired with the rRNA. This subsequence was determined as the mRNA SD sequence, and the computed free energy was the quantitative measure of that SD sequence.
For consistency, the translational start sites in both genomes were determined by the GeneMarkS gene predictions (Besemer et al. 2001; http://opal.biology.gatech.edu/GeneMark/GeneMarkS/index.html). To validate the GeneMarkS predictions, the start sites were compared with 860 proteins whose N termini were sequenced (EcoGene17, third release; Rudd 2000). Of these 852 were among the proteins in our database, and for 794 the N termini coincided with the predicted start sites. Because proteins can undergo processing, this is in fact likely to be an underestimate.
Codon Bias
Codon bias was evaluated by two measures:
The Codon Adaptation Index (CAI; Sharp and Li 1987), which relates the frequency of codons in a protein to their frequency in a reference set of highly expressed proteins. This is the most commonly used measure for codon bias. Conventionally, and also here, ribosomal proteins have been used as a reference set of highly expressed proteins. CAI values range between 0 and 1, and the higher the value the more biased is the codon usage of the particular amino acid sequence toward codons that frequently appear in the reference set.
The effective number of codons (Nc; Wright 1990), which measures how many different codons per amino acid are used in a protein. For proteins that contain all 20 amino acids, this measure ranges between 20 and 61, where a value of 20 means that only one codon is used per amino acid, and a value of 61 means that all possible codons are used. Thus, lower Nc values indicate a biased use of certain codons and should correspond to high values of CAI. Nc was chosen as an additional measure to be used for codon bias for two main reasons: Nc exhibits a lower coefficient of variation than CAI (Comeron and Aguade 1998). Also, because we were interested in studying the influence of codon bias on protein expression, defining a measure based on known highly expressed proteins will obviously lead to the conclusion that this group of proteins is highly expressed.
Statistical Tests
Distinction between HE and NHE proteins. The Wilcoxon rank sum test (equivalent to the Mann-Whitney test) was used to determine whether the values of a parameter differ in HE as opposed to NHE proteins.
Correlation between properties. Correlations were calculated both between a property and expression levels, and for the same property in orthologous genes. The Spearman rank correlation coefficient (rs) was used. However, because the data sets are large, this is equivalent to calculating the Pearson correlation coefficient. For data from a 2 × 2 contingency table, the Φ coefficient (Liebetrau 1983) was calculated to assess the association between two properties. The Φ coefficient ranges from -1 to 1 and can be interpreted similarly to the Pearson correlation coefficient, and they can therefore be compared.
Comparison of correlation coefficients. To compare which sequence feature is more conserved among orthologous genes, the correlation coefficients determined for the different features were statistically compared using the test described in Zar (1984). In addition, the Wilcoxon signed rank test on the absolute differences of ranks was used to compare correlations.
Acknowledgments
We thank Norman Grover for useful discussions and Ruti Hershberg, Yael Altuvia, and Itay Onn for their helpful comments. G.L. was supported by a doctoral fellowship from the Sudarsky Center. This study was supported by a strategic grant from the Israeli Ministry of Science and by the Israeli Science Foundation, administered by the Israeli Academy of Sciences and Humanities.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1485203.
References
- Besemer, J., Lomsadze, A., and Borodovsky, M. 2001. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29: 2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornsson, A., Mottagui-Tabar, S., and Isaksson, L.A. 1996. Structure of the C-terminal end of the nascent peptide influences translation termination. EMBO J. 15: 1696-1704. [PMC free article] [PubMed] [Google Scholar]
- Blattner, F.R., Plunkett III, G., Bloch, C.A., Perna, N.T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J.D., Rode, C.K., Mayhew, G.F., et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277: 1453-1474. [DOI] [PubMed] [Google Scholar]
- Boni, I.V., Artamonova, V.S., Tzareva, N.V., and Dreyfus, M. 2001. Non-canonical mechanism for translational control in bacteria: Synthesis of ribosomal protein S1. EMBO J. 20: 4222-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calogero, R.A., Pon, C.L., Canonaco, M.A., and Gualerzi, C.O. 1988. Selection of the mRNA translation initiation region by Escherichia coli ribosomes. Proc. Natl. Acad. Sci. 85: 6427-6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron, J.M. and Aguade, M. 1998. An evaluation of measures of synonymous codon usage bias. J. Mol. Evol. 47: 268-274. [DOI] [PubMed] [Google Scholar]
- de Smit, M.H. and van Duin, J. 1994. Control of translation by mRNA secondary structure in Escherichia coli. A quantitative analysis of literature data. J. Mol. Biol. 244: 144-150. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker, A. 1996. The close proximity of Escherichia coli genes: Consequences for stop codon and synonymous codon use. J. Mol. Evol. 42: 73-78. [DOI] [PubMed] [Google Scholar]
- Fleischmann, R.D., Adams, M.D., White, O., Clayton, R.A., Kirkness, E.F., Kerlavage, A.R., Bult, C.J., Tomb, J.F., Dougherty, B.A., Merrick, J.M., et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269: 496-512. [DOI] [PubMed] [Google Scholar]
- Futcher, B., Latter, G.I., Monardo, P., McLaughlin, C.S., and Garrels, J.I. 1999. A sampling of the yeast proteome. Mol. Cell. Biol. 19: 7357-7368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold, L. 1988. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu. Rev. Biochem. 57: 199-233. [DOI] [PubMed] [Google Scholar]
- Gygi, S.P., Rochon, Y., Franza, B.R., and Aebersold, R. 1999. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19: 1720-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideker, T., Thorsson, V., Ranish, J.A., Christmas, R., Buhler, J., Eng, J.K., Bumgarner, R., Goodlett, D.R., Aebersold, R., and Hood, L. 2001. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292: 929-934. [DOI] [PubMed] [Google Scholar]
- Ikemura, T. 1985. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 2: 13-34. [DOI] [PubMed] [Google Scholar]
- Jin, H., Bjornsson, A., and Isaksson, L.A. 2002. Cis control of gene expression in E. coli by ribosome queuing at an inefficient translational stop signal. EMBO J. 21: 4357-4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaya, S., Yamada, Y., Kudo, Y., and Ikemura, T. 1999. Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: Gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene 238: 143-155. [DOI] [PubMed] [Google Scholar]
- Karlin, S., Mrazek, J., Campbell, A., and Kaiser, D. 2001. Characterizations of highly expressed genes of four fast-growing bacteria. J. Bacteriol. 183: 5025-5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langen, H., Takacs, B., Evers, S., Berndt, P., Lahm, H.W., Wipf, B., Gray, C., and Fountoulakis, M. 2000. Two-dimensional map of the proteome of Haemophilus influenzae. Electrophoresis 21: 411-429. [DOI] [PubMed] [Google Scholar]
- Lee, T.I., Rinaldi, N.J., Robert, F., Odom, D.T., Bar-Joseph, Z., Gerber, G.K., Hannett, N.M., Harbison, C.T., Thompson, C.M., Simon, I., et al. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298: 799-804. [DOI] [PubMed] [Google Scholar]
- Liebetrau, A.M. 1983. Measures of association. In Quantitative applications in the social sciences, Sage University Paper series, 07-032 (eds. J.L. Sullivan and R.G. Niemi). Sage Publications, Beverly Hills and London.
- Link, A.J., Robison, K., and Church, G.M. 1997. Comparing the predicted and observed properties of proteins encoded in the genome of Escherichia coli K-12. Electrophoresis 18: 1259-1313. [DOI] [PubMed] [Google Scholar]
- Ma, J., Campbell, A., and Karlin, S. 2002. Correlations between Shine-Dalgarno sequences and gene features such as predicted expression levels and operon structures. J. Bacteriol. 184: 5733-5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll, I., Grill, S., Gualerzi, C.O., and Blasi, U. 2002. Leaderless mRNAs in bacteria: Surprises in ribosomal recruitment and translational control. Mol. Microbiol. 43: 239-246. [DOI] [PubMed] [Google Scholar]
- Mottagui-Tabar, S., Bjornsson, A., and Isaksson, L.A. 1994. The second to last amino acid in the nascent peptide as a codon context determinant. EMBO J. 13: 249-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, Y., Ito, K., and Isaksson, L.A. 1996. Emerging understanding of translation termination. Cell 87: 147-150. [DOI] [PubMed] [Google Scholar]
- Nakamura, Y., Gojobori, T., and Ikemura, T. 2000. Codon usage tabulated from international DNA sequence databases: Status for the year 2000. Nucleic Acids Res. 28: 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada, Y., Saito, R., and Tomita, M. 1999. Analysis of base-pairing potentials between 16S rRNA and 5′ UTR for translation initiation in various prokaryotes. Bioinformatics 15: 578-581. [DOI] [PubMed] [Google Scholar]
- Poole, E.S., Brown, C.M., and Tate, W.P. 1995. The identity of the base following the stop codon determines the efficiency of in vivo translational termination in Escherichia coli. EMBO J. 14: 151-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha, E.P., Danchin, A., and Viari, A. 1999. Translation in Bacillus subtilis: Roles and trends of initiation and termination, insights from a genome analysis. Nucleic Acids Res. 27: 3567-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd, K.E. 2000. EcoGene: A genome sequence database for Escherichia coli K-12. Nucleic Acids Res. 28: 60-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai, H., Imamura, C., Osada, Y., Saito, R., Washio, T., and Tomita, M. 2001. Correlation between Shine-Dalgarno sequence conservation and codon usage of bacterial genes. J. Mol. Evol. 52: 164-170. [DOI] [PubMed] [Google Scholar]
- Schurr, T., Nadir, E., and Margalit, H. 1993. Identification and characterization of E. coli ribosomal binding sites by free energy computation. Nucleic Acids Res. 21: 4019-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinger, D.W., Cheung, K.J., Mei, R., Johansson, E.M., Richmond, C.S., Blattner, F.R., Lockhart, D.J., and Church, G.M. 2000. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat. Biotechnol. 18: 1262-1268. [DOI] [PubMed] [Google Scholar]
- Sharp, P.M. and Bulmer, M. 1988. Selective differences among translation termination codons. Gene 63: 141-145. [DOI] [PubMed] [Google Scholar]
- Sharp, P.M. and Li, W.H. 1987. The codon Adaptation Index—A measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15: 1281-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine, J. and Dalgarno, L. 1974. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: Complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. 71: 1342-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman, P.T., Sherlock, G., Zhang, M.Q., Iyer, V.R., Anders, K., Eisen, M.B., Brown, P.O., Botstein, D., and Futcher, B. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9: 3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov, R.L., Mushegian, A.R., Bork, P., Brown, N.P., Hayes, W.S., Borodovsky, M., Rudd, K.E., and Koonin, E.V. 1996. Metabolism and evolution of Haemophilus influenzae deduced from a whole-genome comparison with Escherichia coli. Curr. Biol. 6: 279-291. [DOI] [PubMed] [Google Scholar]
- Tatusov, R.L., Natale, D.A., Garkavtsev, I.V., Tatusova, T.A., Shankavaram, U.T., Rao, B.S., Kiryutin, B., Galperin, M.Y., Fedorova, N.D., and Koonin, E.V. 2001. The COG database: New developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29: 22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjaden, B., Saxena, R.M., Stolyar, S., Haynor, D.R., Kolker, E., and Rosenow, C. 2002. Transcriptome analysis of Escherichia coli using high-density oligonucleotide probe arrays. Nucleic Acids Res. 30: 3732-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBogelen, R.A., Abshire, K.Z., Pertsemlidis, A., Clark, R.L., and Neidhardt, F.C. 1996. Gene–protein database of Escherichia coli K-12, edition 6. In Escherichia coli and Salmonella: Cellular and molecular biology (eds. F.C. Neidhardt et al.), pp. 2067-2117. ASM Press, Washington, DC.
- Washburn, M.P., Koller, A., Oshiro, G., Ulaszek, R.R., Plouffe, D., Deciu, C., Winzeler, E., and Yates III, J.R. 2003. Protein pathway and complex clustering of correlated mRNA and protein expression analyses in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 100: 3107-3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, F. 1990. The `effective number of codons' used in a gene. Gene 87: 23-29. [DOI] [PubMed] [Google Scholar]
- Zar, J.H. 1984. Comparing two correlation coefficients. In Biostatistical analysis, pp. 313-314. Prentice-Hall, Englewood Cliffs, NJ.
WEB SITE REFERENCES
- http://www.ncbi.nlm.nih.gov; NCBI home page.
- http://opal.biology.gatech.edu/GeneMark/GeneMarkS/index.html; GeneMarkS gene predictions.