Abstract
Introduction
Breast cancer is a genetically heterogenous disease with subtypes differing in prognosis and chemosensitivity. The basal-like breast cancer (BLBC) molecular subtype is associated with poorer outcomes, but is more responsive to taxane-based chemotherapy, which is used in multiple settings of breast cancer. Kinesins are intracellular transport proteins that interact with microtubules, which are also the mechanistic target for taxanes. We investigated the relationship between taxane resistance in BLBC and kinesins using both expression and functional studies.
Methods
Kinesin (KIF) expression was evaluated in three settings in relation to taxane resistance: (i) the NCI-60 cancer cell lines, (ii) pre-treatment samples from four BLBC patient cohorts receiving neoadjuvant chemotherapy regimens with and without taxanes, and (iii) post-treatment samples from residual breast cancer following neoadjuvant taxane-containing chemotherapy. We used a novel functional approach to gene modification, validation-based insertional mutagenesis, to select kinesin-overexpressing clones of BLBC cells for evaluation of related mechanisms of taxane resistance.
Results
In the NCI-60 cell line dataset, overexpression of the kinesin KIFC3 is significantly correlated with resistance to both docetaxel (p<0.001) and paclitaxel (p<0.001), but not to platinum-based chemotherapy, including carboplatin (p=0.49) and cisplatin (p=0.10). Overexpression of KIFC3 and KIF5A in pre-chemotherapy samples similarly predicted resistance to paclitaxel in the MDACC cohorts (p=0.01); no KIF predicted resistance to fluorouracil-epirubicin-cyclophosphamide or cisplatin in BLBC patient cohorts treated without taxanes. KIF12 is the most overexpressed KIF gene in post-chemotherapy taxane-resistant residual breast cancers (2.8 fold-change). Functional studies established that overexpression of KIFC3, KIF5A and KIF12 were specific in mediating resistance to docetaxel and not vincristine or doxorubicin. We demonstrated that mutation of the ATP-binding domain of a kinesin abolishes its ability to mediate docetaxel resistance.
Conclusions
We show that kinesin overexpression correlates with specific taxane resistance in BLBC cell lines and tissue. Our results suggest a novel approach to overcoming taxane resistance in breast cancer through concurrent or sequential use of kinesin inhibitors, highlighting the ATP-binding domain as a drug development target.
INTRODUCTION
The systemic therapy of breast cancer has seen many advances over the last few decades, with the introduction of taxanes, including paclitaxel and docetaxel, representing a major milestone. Taxanes are widely used in the adjuvant, neoadjuvant and metastatic settings of breast cancer. These agents interact with microtubules, which are heterodimers of α-tubulin and β-tubulin subunits, binding to β-tubulin to stabilise and prevent microtubule depolymerization. Thus, taxanes cause disruptions of mitotic spindle formation, inhibiting cell division and leading to cell death [1].
While taxanes are extremely successful in establishing cure or durable response in breast cancer, drug resistance, as manifested by relapse and tumor progression, remains a major challenge for breast oncologists. Several different mechanisms account for the taxane resistance observed in human tumors and tumor cell lines, including overexpression of the multidrug transporter P-glycoprotein, altered drug metabolism, decreased sensitivity to death-inducing stimuli, alterations in microtubule dynamics, and altered binding of taxanes to microtubule targets [2]. Various candidates have been investigated for prediction of response in breast cancer to taxanes, including proteins like βIII-tubulin [3], the microtubule-associated proteins MAP2 [4], MAP4 and TAU [5], and the microtubule-destabilizing phosphoprotein stathmin [6]. Genome-wide microarray studies have also been used to predict taxane response in breast cancer [7, 8]. While promising, none of these markers have been prospectively validated or integrated into the routine clinical practice of breast oncologists, and the functional role of most of these individual genes remains to be explored.
Recent research in breast cancer has been influenced by relatively new approaches to molecular subtyping of breast cancer, with emerging evidence that prognostic and predictive biomarkers may differ between subtypes [5, 8]. Little is known about how taxane resistance is mediated in these molecular subtypes of breast cancer, each of which has a distinct clinical and biological phenotype. These include luminal-A, luminal-B, HER2-overexpressing and basal-like breast cancers [9]. In particular, basal-like breast cancers (BLBC), expressing genes that are characteristic of basal myoepithelial cells in normal mammary glands, are endocrine-insensitive, and chemotherapy is the only systemic option for these cancers. Although BLBC are associated with aggressive clinical behavior, they also exhibit a higher response rate to chemotherapy, including taxanes [10]. Interestingly, while markers of taxane response have been identified in estrogen receptor-positive breast cancer, for example, the TAU protein [5], to date, no similar marker has been confirmed in BLBC.
Drug resistance is often achieved through the overexpression of a specific protein, while overexpression is often achieved through amplification of the corresponding gene or by epigenetic regulation. In our recent study, we utilized a novel validation-based insertional mutagenesis (VBIM) method [11] to better understand the regulatory mechanism underlying docetaxel resistance in breast cancer cells. Using this method, we discovered that overexpression of the kinesin KIFC3 confers docetaxel resistance in breast cancer cells [12], and subsequently, that overexpression of several kinesins individually, including both N- and C-kinesins, were associated with docetaxel resistance in breast cancer cells [12]. Kinesins are motor proteins that transport cargoes by walking unidirectionally along microtubule tracks, tracks, hydrolyzing one molecule of ATP at each step. In addition, kinesins are key participants in chromosomal and spindle movements during mitosis [13]. As such, kinesins represented very plausible targets for mediating taxane resistance. Indeed, they had already gained attention as possible mitotic drug targets. For example, in recent years, ispinesib (SB-715992), an allosteric small-molecule inhibitor of KSP (KIF11) ATPase activity, was the first small molecule kinesin inhibitor to enter clinical trials. However, despite this, the role of kinesins in drug resistance had not been explored until we showed that overexpression of four different kinesins (KIFC3, KIFC1, KIF1A and KIF5A) could mediate docetaxel resistance in BLBC cell lines and determined that three of these kinesins increase the fraction of soluble tubulin in cells, mechanistically demonstrating that they oppose the microtubule-stabilizing effect of docetaxel [12].
The above hypothesis-generating observations have led to this current study where we address these hypotheses from the in vitro insights, using microarray data derived from breast cancer patients exhibiting resistance to a taxane-based regimen, the NCI-60 cell lines, and pre-treatment samples from BLBC patients undergoing taxane-based neoadjuvant therapy. We also focus on the mechanistic insight into the relationship between kinesin overexpression and taxane resistance, emphasizing the role of the ATP-binding domain in mediating taxane resistance in BLBC.
MATERIALS AND METHODS
Subjects and samples
The Institutional Review Board for Human Subjects’ Protection of the Cleveland Clinic (CC) approved this study. Informed consent was obtained from all patients. Patients who did not achieve a pathologic complete response (pCR), i.e., those with evidence of residual invasive carcinoma following taxane-based chemotherapy (n=8), were identified from a review of the CC database (Table 1). We selected these residual cancer samples based on the clinical likelihood of high expression of genes mediating resistance being present. It should be noted that no post-chemotherapy cancer tissue is available from patients with complete response after neoadjuvant chemotherapy. The corresponding histologically benign breast tissue from 5 of the same 8 patients with post-therapy residual breast cancer was used as controls. These samples were frozen following resection and stored at −80°C. Total RNA was isolated from the samples using the RNEasy kit (Qiagen, Valencia, CA). Standard manufacturer’s processing techniques were used for downstream processing, and the resulting cDNA was hybridized on Illumina Human Ref-8 Version 3 microarrays targeting ~24,500 well annotated transcripts. The CC microarray dataset has been deposited in the Gene Expression Omnibus (GEO) under the accession GSE22796.
Table 1.
Clinical data of Cleveland Clinic patients who received neoadjuvant chemotherapy with incomplete pathologic response.
| Patient | Age | Tumor Type | Pre-treatment clinical T and N |
ER (% cells immunoreactive) |
PR (% cells immunoreactive) |
HER2 amplification (HER2/CEP 17 ratio) |
Pathologic grade |
Post-treatment pathologic T and N |
Residual Cancer Burden Class |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 41 | Ductal | T3 N1 | 0.9 | 0.7 | absent (1.3) | 2 | pT2 pN1 | RCB-III |
| 2 | 56 | Lobular | T3 N1 | 0.8 | 0.05 | absent (1.3) | 3 | pT2 pN1 | RCB-III |
| 3 | 61 | Ductal | T2 N1 | 0.9 | 0.9 | absent (1.4) | 3 | pT1 pN1 | RCB-III |
| 4 | 68 | Lobular | T2 N0 | 0.7 | 0.7 | absent (1.0) | 1 | pT2 pN1 | RCB-II |
| 5 | 65 | Ductal | T2 N1 | 0.9 | 0.7 | absent (1.4) | 2 | pT2 pN1 | RCB-III |
| 6 | 59 | Ductal | T3 N2 | − (<5%) | − (<5%) | absent (1.0) | 3 | pT3 pN3 | RCB-III |
| 7 | 44 | Ductal | T3 N1 | 0.7 | − (<5%) | absent (1.3) | 1 | pT1 pN1 | RCB-II |
| 8 | 35 | Ductal | T2 N0 | 0.8 | − (<5%) | absent (1.8) | 2 | pT2 pN1 | RCB-III |
Statistical analysis of microarrays
We acquired probe-level raw expression data from the NCI-60 cell line dataset (replicated Novartis experiments using the Affymetrix U95Av2 platform, available at the Developmental Therapeutics Program website of the National Cancer Institute, http://dtp.nci.nih.gov/mtargets/download.html) and four patient cohorts: two cohorts from the MD Anderson Cancer Center (MDACC) dataset (GSE20194) receiving taxane-based chemotherapy (T-FAC: paclitaxel followed by fluorouracil, doxorubicin and cyclophosphamide), a Dana-Farber Cancer Institute (DFCI) cohort with triple-negative breast cancer patients receiving neoadjuvant cisplatin (GSE18864) and a cohort from the European Organization for Research and Treatment in Cancer (EORTC) 10994 trial receiving neoadjuvant fluorouracil, epirubicin and cyclophosphamide (FEC) (GSE4779) from the Gene Expression Omnibus (GEO) public repository [14]. Analysis was done in R 2.10.0 and MatLab. For pre-processing, the robust multi-chip averaging (RMA) algorithm was used.
Cell line data set
Based on the NCI-60 cell line drug screen [15], cell lines were classified as either sensitive or resistant to taxanes (docetaxel, paclitaxel) and platinum-based drugs (cisplatin and carboplatin) [16]. Briefly, cell lines that were in the same extreme third in terms of drug sensitivity for both GI 50 (concentration required to reduce growth by 50%) and TGI (concentration required for total growth inhibition) for the same drug were classified as such, with ‘central third’ being defined robustly in terms of quantiles (Additional File 1). Subsequently, we evaluated on an a priori basis differences in expression profiles of tested KIFs, using individual univariate logistic regression with Bonferroni correction applied for statistical significance calculations (Additional File 2).
Chemotherapy-resistant and sensitive breast cancer data sets
For the MDACC data set, 230 pre-treatment samples from two clinical trial cohorts of breast cancer patients undergoing similar neoadjuvant taxane-based chemotherapy were evaluated (Affymetrix U133A). We selected the subset of samples that were BLBC through the use of a nearest centroids predictor for five intrinsic subtype mean expression profiles [17], eventually identifying 50 samples as BLBC. Essentially, this approach involves classifying each sample based on externally defined average expression of selected genes for each class. Response to preoperative taxane-based chemotherapy was categorized per definitions provided in the MDACC dataset, pathological complete response (pCR) representing sensitive samples and residual invasive cancer (RD) representing resistant samples. A similar approach was undertaken for the DFCI (Affymetrix U133Plus 2.0) and the EORTC (Affymetrix X3P) data sets, where 28 and 37 samples were classified as BLBC respectively. Penalized logistic regression is an effective tool in handling high-dimensional data with dichotomous outcomes (pCR/RD). We investigated α values ranging from 0 to 1 with granularity of 0.01. A set of regularization parameter (λ) values were evaluated by the coordinate descent algorithm and an optimal λ for each α was identified through 100-fold cross-validation. Values of the coefficients for these genes indicate the relative importance of the corresponding gene subset identified. For the PLR analysis, probe sets corresponding to the same gene were averaged.
Post-chemotherapy taxane-resistant samples
The CC dataset was exported from Illumina Beadstudio, pre-processed, normalized and filtered using a detection threshold of 0.05, resulting in selection of 15,573 of 18,401 genes. Quality control was performed on the raw expression data using visual examination of boxplots. For the CC dataset, to select a subset of candidate KIF genes highly expressed in residual tissue, and thus being good candidates for functional studies, prediction analysis of microarrays (PAM) was performed using five-fold cross validation, with default gene thresholds and offset percentage [18], with a threshold misclassification error of 0.15 [18].
Cells and reagents
MDA-MB-231 and MDA-MB-468 cells which are derived from BLBC [19], were kindly provided by Dr. John Pink (Case Western Reserve University, Cleveland, OH). Cells were grown in a humidified atmosphere containing 5% CO2 in RPMI 1640 medium, supplemented with 5% fetal bovine serum (HyClone Laboratories, Inc., Logan, UT). Docetaxel, doxorubicin and vincristine were obtained from Sigma (St Louis, MO). Anti-KIF12 was from Proteintech Group, Inc. (Chicago, IL) Anti-KIF5A was from Abcam Inc. (Cambridge, MA).
Plasmids
The KIFC3 and KIF5A plasmids were described in our previous report [12]. The full-length cDNA of KIF12 was made following the sequence published by Katoh [20]. The KIF12 sequence (accession # NM_138424.1) in the NCBI site is truncated. The protein sequence showed that the ATP-binding domain is missing from the N-terminal site. The missing part was synthesized from GenScript USA Inc. (Piscataway, NJ). The cDNA of the remaining part was purchased from Open Biosystems (clone ID 3865868) (Huntsville, AL). The full-length cDNA of KIF12 was made by 3-piece ligation cloning into the retroviral vector pLPCX.
PCR-mediated site-directed mutagenesis
To generate a point mutation resulting in K92R in the ATP-binding domain of wild-type (WT) KIF5A, we designed PCR primers with a phosphate group at the 5’ end. The primer sequences around the mutagenized sites were 5’-cgaacacataccatggaggga-3’ (forward) and 5’-ccctgaggatgtctgtccata-3’ (reverse). The KIF5A plasmid was taken as DNA template and PCR was performed according to the protocol in Quick Change 11 site-directed mutagenesis kit from Stratagene (La Jolla, CA). The amplified fragments were ligated and digested with Dpn1. Transformants were sequenced and aligned to WT KIF5A sequence revealing that the GKT of the KIF5A ATP-binding domain in WT KIF5A was mutated to GRT [21].
Retroviral transduction and cell infection
Retroviruses encoding cDNAs encoding KIF12, KIFC3, WT KIF5A and mutant KIF5A (K92R) or empty vector were produced by transient transfection of 293T cells [12]. Supernatant media containing virus were collected at 36 to 48 h, supplemented with 4 µg/mL of polybrene, filtered through a 0.22-µm filter, and added to MDA-MB-231 and MDA-MB-468 cells overnight. The cells expressing KIF12, and WT and K485R were selected with puromycin (1 µg/mL).
Cell survival assay
Cell survival assays were performed in MDA-MB-231 and MDA-MB-468 cells stably overexpressing KIF12, KIFC3, wild-type KIF5A or mutant KIF5A using techniques described previously [12].
Immunoblotting and mRNA expression
Western analyses were performed as described previously [22]. The membranes were probed with antibodies against KIF12, and KIF5A or anti–pan-actin. To determine mRNA expression levels, total RNA was extracted, and an RT reaction was performed using SuperScript III First-Strand Synthesis System. RT-PCR was done using KIF12-specific primers.
Statistical analysis
Values were expressed as means±s.d. P-values were based on the paired t-test and significance set at 0.05 (values that are significant are marked with an asterisk).
RESULTS
Specific KIFs are overexpressed in docetaxel and paclitaxel-resistant NCI-60 cell lines
We first sought to determine if the association of KIF overexpression with taxane resistance may be determined using well known in vitro models. We initially identified docetaxel-sensitive cell lines (COLO-205, HCC-2998, HL-60 (TB), HT-29, MDA-MB-435, NCI-H522, RPMI-8226, SF-539) and docetaxel-resistant cell lines (786-0, ACHN, CAKI-1, EKVX, IGROV1, OVCAR-4, SF-268). Paclitaxel-sensitive cell lines (COLO-205, HCC-2998, HS-578T, HT-29, NCIH23, RPMI-8226, SF-539, SNB-75) and paclitaxel-resistant cell lines (A498, EKVX, MALME-3M, NCI/ADR-RES, OVCAR-4, SK-MEL-28, T-47D, UACC-257, UO-31) were similarly identified. Consistent with our previous work reporting that overexpression of KIFC3 mediating docetaxel resistance [12], KIFC3 overexpression was found to be significantly correlated with resistance to both docetaxel (p<0.001) and paclitaxel (p<0.001), but not to carboplatin (p = 0.49) or cisplatin (p=0.10), with the latter two serving as our negative controls. Results of individual KIFs are reported in Additional File 2.
Specific kinesin overexpression in pre-chemotherapy tumors correlates with resistance to docetaxel, but not to non-taxane based regimens
We first divided the MDACC dataset of patients receiving paclitaxel-based chemotherapy into responders (n=22) and non-responders (n=28) so as to identify highly ranked genes that would classify one way or the other. For our penalized logistic regression analysis of BLBC tumors which select subsets of highly discriminating genes, 978 discriminating genes were identified at αof 0.02 and λ of 2.52 for convergence in the MDACC data set (Additional File 3), which included the kinesins KIFC3 and KIF5A. Individually, overexpression of each of these 2 genes was associated with resistance to paclitaxel on univariate logistic regression (O.R. 0.48 and 0.49, respectively, for response prediction, p=0.01 and 0.01, respectively) [Table 2]. For the DFCI dataset, comprising patients who received neoadjuvant cisplatin as single agent, at a similar range of α parameters, no KIF gene was found to be predictive of response, even at a similar α of 0.02, where 40 genes correlating with cisplatin response were identified (Additional File 4). For the EORTC dataset of patients receiving FEC chemotherapy without taxanes, there was no convergence at any α parameter, indicating that there were no selected group of transcripts from genes predictive of response. This implies that kinesin overexpression is specific for taxane resistance.
Table 2.
Correlation of KIF gene expression with taxane response in the MDACC data set
| Gene | Coefficient* | p-value |
|---|---|---|
| 'KIF11' | 0.07 | 0.76 |
| 'KIF13A' | −0.29 | 0.17 |
| 'KIF13B' | −0.21 | 0.54 |
| 'KIF14' | −0.29 | 0.31 |
| 'KIF15' | 0.12 | 0.72 |
| 'KIF16B' | 0.19 | 0.47 |
| 'KIF17' | −0.06 | 0.61 |
| 'KIF18A' | 0.35 | 0.15 |
| 'KIF18B' | −0.04 | 0.88 |
| 'KIF1A' | 0.00 | 0.98 |
| 'KIF1B' | 0.37 | 0.18 |
| 'KIF1C' | −0.40 | 0.14 |
| 'KIF20A' | 0.49 | 0.09 |
| 'KIF21B' | −0.40 | 0.14 |
| 'KIF22' | −0.24 | 0.60 |
| 'KIF23' | 0.11 | 0.50 |
| 'KIF24' | −0.12 | 0.40 |
| 'KIF25' | −0.26 | 0.21 |
| 'KIF26B' | −0.05 | 0.76 |
| 'KIF2A' | −0.89 | 0.05 |
| 'KIF2C' | −0.08 | 0.81 |
| 'KIF3A' | −0.04 | 0.94 |
| 'KIF3B' | −0.63 | 0.12 |
| 'KIF3C' | −0.26 | 0.40 |
| 'KIF4A' | 0.58 | 0.09 |
| 'KIF5A' | −0.72 | 0.01 |
| 'KIF5B' | 0.47 | 0.27 |
| 'KIF5C' | −0.32 | 0.15 |
| 'KIFAP3' | −0.03 | 0.88 |
| 'KIFC1' | 0.03 | 0.88 |
| 'KIFC3' | −0.73 | 0.01 |
| 'MPHOSPH1' | −0.18 | 0.35 |
A positive result indicates a positive correlation with taxane response and a negative result a positive correlation with taxane resistance. Genes selected from penalized logistic regression are highlighted in bold.
Specific KIFs are overexpressed in post-chemotherapy taxane-resistant residual breast cancer
The majority of KIF genes were overexpressed in tumor tissue relative to normal tissue (Figure 2). To identify candidate genes for functional analysis, we applied prediction analysis of microarrays, and identified a subset of 8 KIF genes that provided a cross-validated misclassification error of 0.15 (Table 3). KIF12 is the most overexpressed discriminating KIF gene in taxane-resistant breast cancer samples relative to normal matched breast tissue.
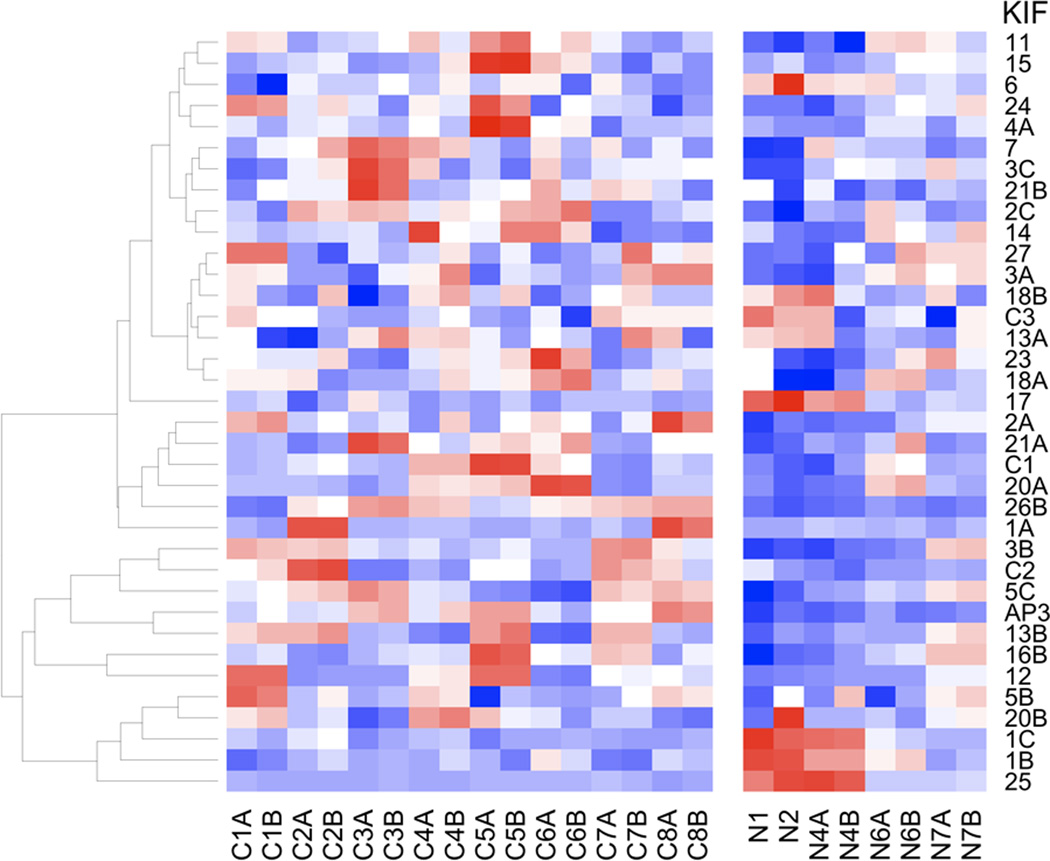
Figure 2. Heatmap of KIF genes from the CC data set.
Data are presented from patients with residual disease following neoadjuvant chemotherapy for locally advanced breast cancer, together with corresponding normal breast tissue, showing differential expression of KIFs between residual breast cancer samples of taxane resistant breast cancer (left block) and non-cancerous tissue (right block), including all technical replicates. Red represents relative over-expression and blue represents relative under-expression of genes.
Table 3.
Identification of differentially expressed KIF genes between taxane-resistant breast cancer and corresponding normal breast tissue
| Kinesin | Tumor Score* |
Normal Score* |
Fold change (tumor vs normal) |
|---|---|---|---|
| KIF12 | 0.0425 | −0.068 | 2.82 |
| KIF26B | 0.3101 | −0.4961 | 1.94 |
| KIF5C | 0.0251 | −0.0402 | 1.74 |
| KIF3B | 0.1372 | −0.2194 | 1.6 |
| KIF17 | −0.1186 | 0.1897 | 0.65 |
| KIF1B | −0.1344 | 0.2151 | 0.61 |
| KIF1C | −0.2438 | 0.3901 | 0.47 |
| KIF25 | −0.3468 | 0.5549 | 0.19 |
A score that discriminates between tumor and normal classes derived from prediction analysis of microarrays
Overexpression of kinesins increases resistance to docetaxel but not to anthracyclines or vincristine
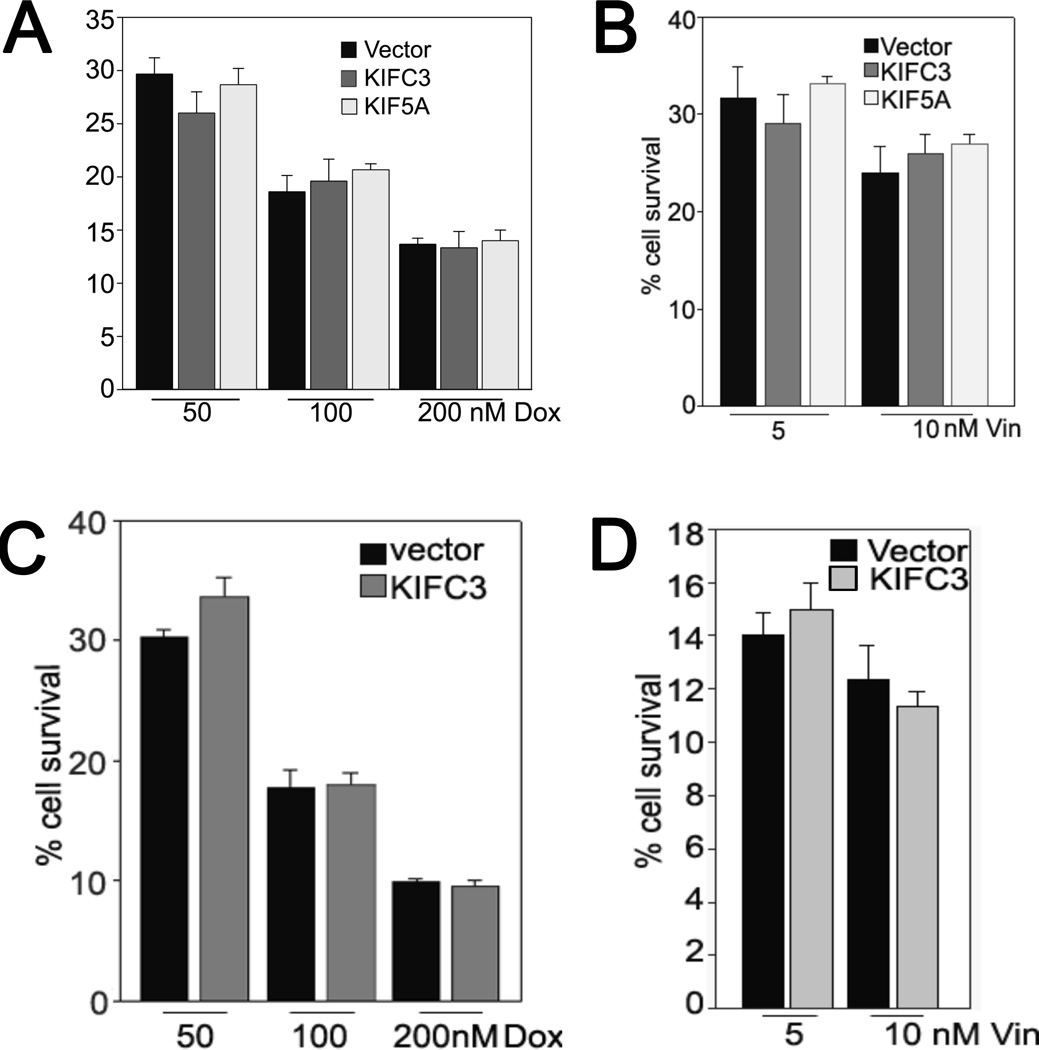
In our previous study [12], we showed that overexpression of KIFC3 and KIF5A mediates resistance to docetaxel in breast cancer cells. We evaluated here the specificity of the relationship between three specific kinesins (KIFC3, KIF5A and KIF12) and taxane resistance. Given that patients in the MDACC data set had received both taxane and anthracycline agents on a sequential basis, it was important to determine whether the overexpression of kinesins could also confer resistance to doxorubicin, an anthracycline. MDA-MB231 cells overexpressing KIFC3 or KIF5A and MDA MB468 cells overexpressing KIFC3 ([12], Figures 2–4) were treated with doxorubicin. There were no significant differences in the percentages of surviving control cells compared to cells overexpressing the kinesins upon exposure to doxorubicin at different concentrations (Figure 4). We also explored whether these two kinesins confer resistance to a microtubule targeting drug other than a taxane. Upon exposure to the microtubule depolymerizing drug vincristine, the percentage of surviving control cells was again similar to that of cells overexpressing the kinesins. Together, these results indicate that kinesins mediate taxane resistance specifically.
Figure 4. Over-expression of KIFC3 and KIF5A does not mediate resistance to doxorubicin or vincristine in breast cancer cells.
A & B, MDA MB231 cells. C & D, MDA MB468 cells. Cells infected with retroviral vectors encoding KIFC3 or KIF5A or with empty vector controls were treated with doxorubicin (Dox) or vincristine (Vin) for 24 hours. After 5–6 days, the cells were lysed and the A260 was measured. The percentages of surviving cells were calculated relative to untreated controls. The data represent means ± SD of two experiments in which each measurement was performed in triplicate. We chose the highest ranked KIFs from the MDACC data set of pre-treatment cancer for this in vitro experiment, given that the specific treatment regimen for the data set included taxanes and anthracyclines.
We determined experimentally with cell survival assays that overexpression of the kinesins KIFC3, KIF5A and KIF12 in MDA-MB231 and MDA-MB468 (KIFC3 and KIF5A) resulted in an increased percentage of surviving KIF-expressing cells, compared to control cells infected with vector alone (Figures 3 and 4). We selected the two highly ranked KIF genes whereby we have demonstrated that they confer resistance specifically to taxanes, and not to an anthracycline or to vinca alkaloids, the latter being a microtubule depolymerizing drugs (Figure 4B, D). These results indicate that even modest over-expression of a kinesin can specifically mediate significant taxane resistance in this in vitro system.
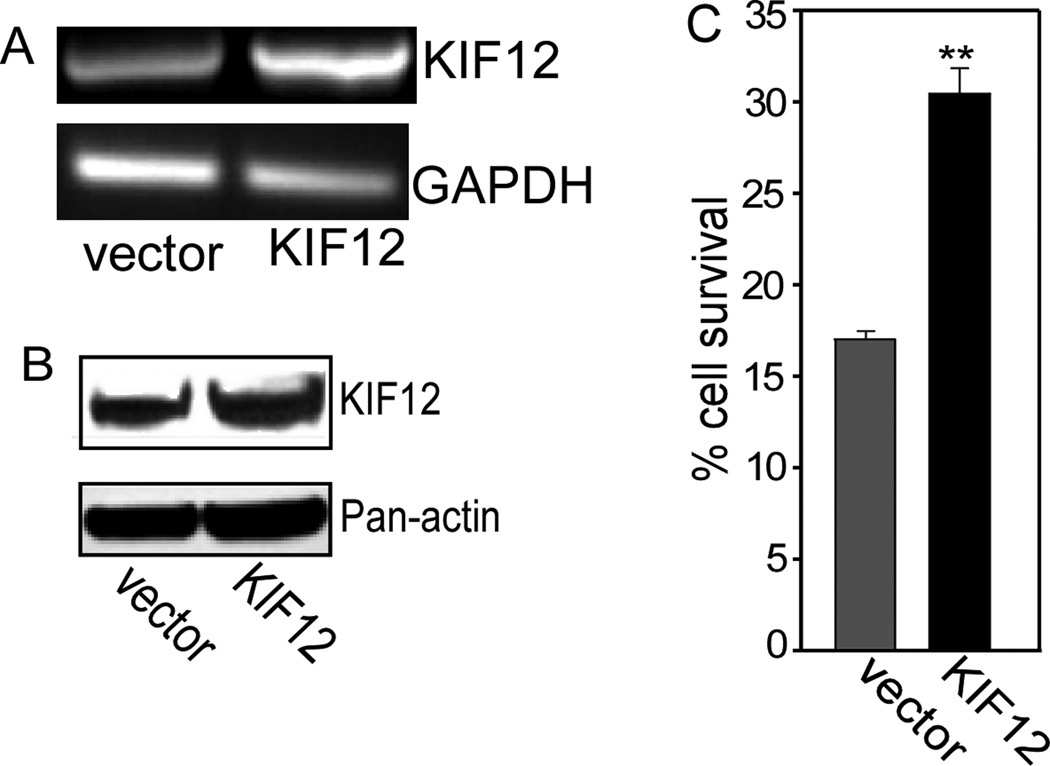
Figure 3. KIF12 over-expression mediates docetaxel resistance in MDA-MB-231 cells.
A. Cells were infected with empty vector, or a retroviral construct encoding KIF12. The mRNA expression levels were determined by RT-PCR, using KIF12-specific primers. GAPDH was used as a control. B. The protein level was determined by Western analysis, using total cell lysates. Actin was used as a control. C. Cell survival assays were performed in cells infected with empty vector or with a retrovirus encoding KIF12, followed by treatment with docetaxel. The fraction of surviving cells was determined by normalizing the data from docetaxel-treated cells to untreated controls. Data shown represent means ± SD of three experiments, in which each measurement was performed in triplicate. **, P < 0.005. Similar results were obtained for KIFC3 and KIF5A as previously reported [12].
The ATP-binding domain of kinesin is essential for resistance to docetaxel
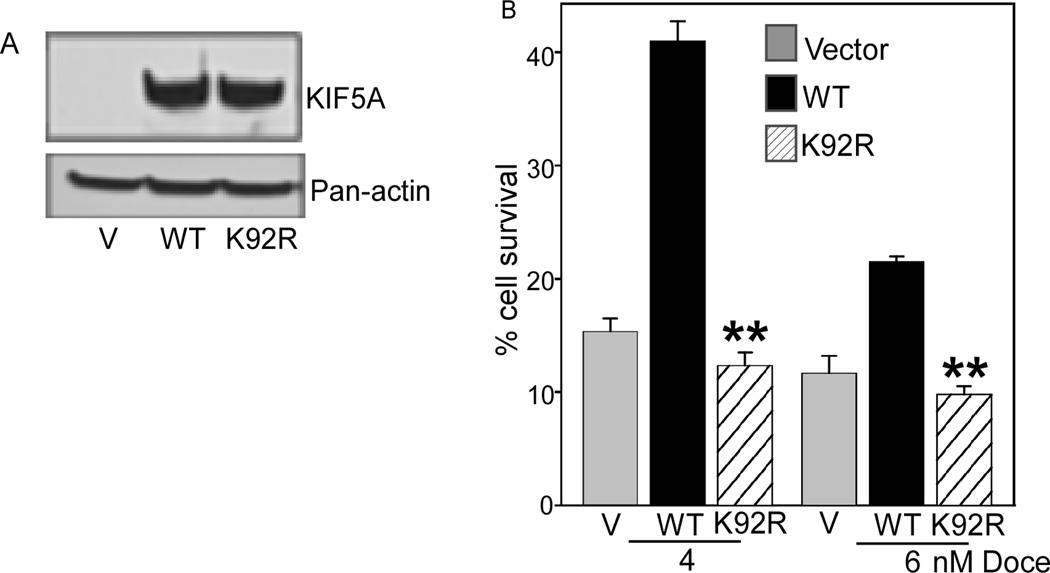
To determine whether a point mutation in the ATP-binding domain of a kinesin is required for it to mediate docetaxel resistance, we chose KIF5A for evaluation in the cell line MDA-MB-231. KIF5A was also identified in the MDACC dataset as a discriminating gene (at p=0.01). GKT, a highly conserved sequence in the ATP binding domains of many kinesins was mutated to GRT. This type of mutant kinesin binds to microtubules but does not hydrolyze ATP [23]. We generated a stable pool of MDA-MB-231 cell lines overexpressing mutant KIF5A. The expression level of mutant kinesin is equivalent to that of the wild-type protein (Figure 4A). Upon exposure to 4 or 6 nM docetaxel, increased survival was observed for cells expressing wild-type KIF5A compared with control cells infected with vector alone. In contrast, cell survival was decreased significantly (P<0.005) in cells expressing the mutant KIF5A, with approximately the same survival as control cells (Figure 4B). These results indicate that the ATP binding domain of a kinesin is essential for it to mediate resistance to docetaxel.
DISCUSSION
Our evaluation of these in vitro and in vivo datasets provides support for the previously unknown role of kinesins in mediating taxane resistance in breast cancer. The large sample size of the publicly available gene expression dataset from MDACC (n=230) permitted analysis of a relatively large number of patients with BLBC with available data on taxane response, highlighting specific kinesins in mediating drug resistance. It should be noted that the MDACC cohort received T-FAC, or paclitaxel followed by fluorouracil-doxorubicin (an anthracycline)-cyclophosphamide. Importantly, our analysis of the EORTC and DFCI cohorts, who received fluorouracil-epirubicin (an anthracycline)-cyclophosphamide and cisplatin respectively, did not demonstrate KIF expression as being predictive of response, suggesting that the effect of kinesins on drug resistance may be specific to taxanes. This in vivo observation is further supported by our functional work showing that kinesin overexpression mediates resistance to docetaxel in BLBC cell lines, but not to doxorubicin or vincristine.
These results have implications for personalization of medical therapy by breast oncologists. Our results, together with findings in estrogen-receptor positive breast cancer [5], provides support for the idea that pathways and corresponding markers of taxane resistance differ among biologically distinct subtypes of breast cancer. Indeed, we limited our analysis to BLBC to avoid a possible confounding of the relationship between gene expression and taxane resistance [24–26]. The clinical result that KIFC3 overexpression is highly predictive of resistance in BLBC matches our previous functional work showing that KIFC3 overexpression is associated with taxane resistance in two basal-like breast cancer cell lines, MDA-MB-231 and MDA-MB-468 [12].
Our results, highlighting KIFC3, KIF5A and KIF12 as mediators of taxane resistance in BLBC through gene expression studies of tissue with separate functional validation, provide translational support for the role of kinesins in taxane resistance, a novel and potentially clinically important area in oncology. With regard to appropriate risk profiling of individuals for therapy, the use of KIF profiles as a clinical predictor of taxane resistance in BLBC deserves further evaluation in the setting of a prospective trial. Most critically, other than the obvious implication of an additional therapeutic target, our results may have implications for the most appropriate timing for maximal clinical benefit for these agents, supporting concurrent use of these agents with taxanes. Synergistic kinesin inhibition with sequential delivery of taxanes is an interesting potential therapeutic approach. In a similar example, the current sequential delivery of taxanes and anti-metabolites was derived from in vitro observations of synergism [27].
Although our work is most directly related to breast cancer, our results suggest that a possibility of extension into other cancer types, given the results of our unbiased evaluation of a comprehensively characterized panel of cancer cell lines (NCI-60). We were able to determine that overexpression of KIFC3 was correlated with both docetaxel and paclitaxel resistance, but not cisplatin or carboplatin resistance, the latter two platinum-based drugs being negative controls. These cell lines are derived from diverse histologies, implying a possible general mechanistic relationship between specific kinesin overexpression and taxane resistance.
With regard to the potential mechanisms involved in this relationship between kinesins and taxane resistance in BLBC, our work in determining that a point mutation in the ATP-binding domain of a kinesin may restore taxane sensitivity suggests two interesting conclusions. Firstly, the ATP-binding domain is essential in mediating taxane resistance; and secondly, it may be possible to modulate taxane resistance with kinesin inhibition through one possible strategy of developing agents targeted to this specific domain. Such agents may be used either concurrently, or sequentially with taxanes. Additionally, our results are mechanistically consistent with recent reports that inhibition of KIF ATPase is a potential therapeutic approach in breast cancer cells [28] and in taxane-resistant models [29], pointing to a cytotoxic effect of KIF ATPase inhibition that is independent of its modulation of taxane resistance. Thus, our results provide insights into a mechanism of interest in breast cancer chemotherapy resistance, which may have therapeutic implications.
CONCLUSIONS
Taxane chemotherapy is a cornerstone of systemic therapy in breast cancer, and corresponding drug resistance is a challenge for breast oncologists. We have shown that overexpression of specific kinesins is associated with taxane resistance in breast cancer, and detailed a potential mechanism for inhibition. Our results highlight the potential opportunity for sequential or synergistic modulation of taxane resistance in BLBC using selective KIF inhibitors in the clinical setting, and provide mechanistic insight into the relationship between kinesin proteins and taxane resistance.
Supplementary Material
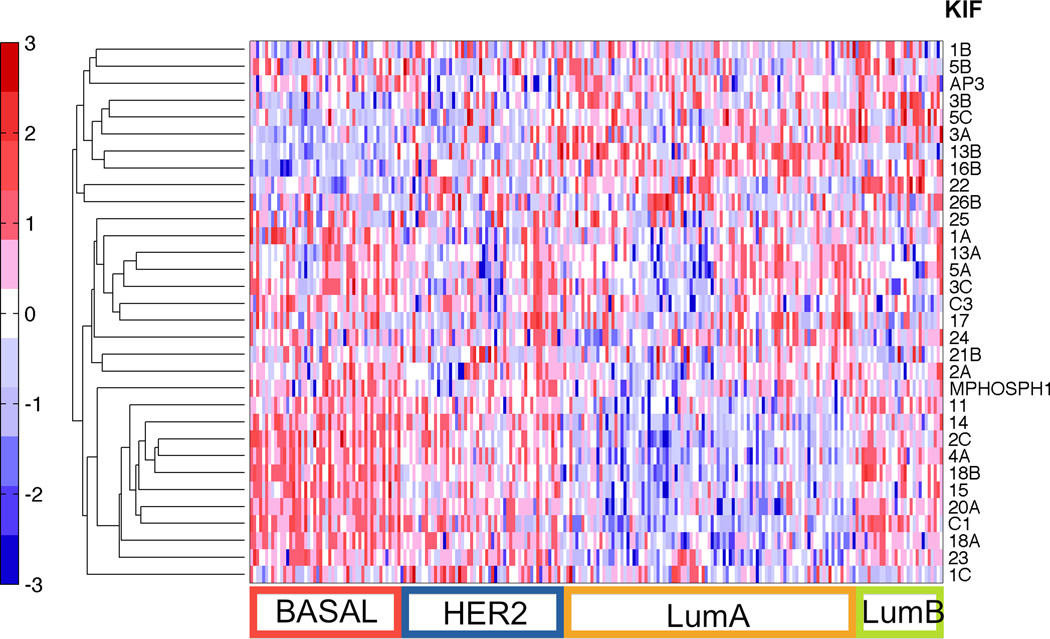
Figure 1. Heatmap of KIF genes from the MDACC data set (GSE20194).
Differential expression of the available KIF genes is shown in this data set of pre-chemotherapy tumor samples, with clusters of high and low expressing KIF genes evident and discriminating between the subtypes. Samples along the horizontal axis have been classified into the various molecular subtypes of breast cancer by centroids (refer to Methods for detailed description). This figure illustrates the variation of KIF expression across the different molecular subtypes of breast cancer, supporting our stratification of analysis to BLBC. Red represents relative over-expression and blue represents relative under-expression of genes.
Figure 5. A point mutation in the ATP-binding domain of KIF5A inhibits KIF5A-mediated docetaxel resistance.
A. Empty vector (V), or retroviral constructs with KIF5A (WT), or K92R, a point mutation into the ATP-binding domain of KIF5A, were each transfected in MDA-MB-231 cells. The level of expression was determined by Western analysis, using total cell lysates. Actin protein levels as manifested by probing with a pan-actin antibody was used as a control. B. Cell survival assays were performed in cells infected with empty vector or with retroviruses encoding KIF5A, or mutant form of KIF5A (K92R), after treatment with docetaxel. The fraction of surviving cells was determined by normalizing the data from docetaxel-treated cells to untreated controls. Columns, means of three experiments: each measurement was performed in triplicate; bars, SD, **, P < 0.005.
ACKNOWLEDGEMENTS
We thank the participating patients for their contributions. We also thank Charissa Peterson for her able technical assistance. The National Institutes of Health and the William Randolph Hearst Foundations funded this study.
No funding organization was involved in the design, conduct, analysis, writing or submission of this manuscript.
MHT is the Ambrose Monell Foundation Cancer Genomic Medicine Clinical Fellow and a Lee Foundation (Singapore) Fellow, and GB is an NCI R25T Fellow. GTB is the co-holder of the Zapis Chair of Breast Cancer Research; GRS is the Distinguished Scientist of the Cleveland Clinic; and CE is the holder of the Sondra J. and Stephen R. Hardis Chair of Cancer Genomic Medicine at the Cleveland Clinic and is an American Cancer Society Clinical Research Professor.
LIST OF ABBREVIATIONS
- ATP
Adenosine triphosphate
- BLBC
Basal-like breast cancer
- CC
Cleveland Clinic
- DFCI
Dana-Farber Cancer Institute
- EORTC
European Organization for Research and Treatment of Cancer
- GI50
Concentration required to reduce growth by 50%
- KIF
Kinesin superfamily
- MDACC
MD Anderson Cancer Center
- pCR
Pathological complete response
- RD
Residual disease (invasive cancer)
- RT-PCR
Reverse transcriptase-polymerase chain reaction
- TGI
Concentration required for total growth inhibition
- VBIM
Validation-based insertional mutagenesis
Footnotes
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
GTB, GRS and CE conceptualized, designed and directed the study. MHT, SD, GB, MSO and RW conducted the experiments, obtained and analyzed experimental and clinical data; EDK reviewed and prepared tissue specimens; MHT, SD, GB, GRS and CE interpreted the data; MHT and CE drafted the manuscript; and all authors critically reviewed the manuscript, provided revisions and approved the final manuscript. GRS and CE had access to all the data and are responsible for the conduct and content of the study.
REFERENCES
- 1.Shalli K, Brown I, Heys SD, Schofield AC. Alterations of beta-tubulin isotypes in breast cancer cells resistant to docetaxel. FASEB J. 2005;19(10):1299–1301. doi: 10.1096/fj.04-3178fje. [DOI] [PubMed] [Google Scholar]
- 2.Dumontet C, Sikic BI. Mechanisms of action of and resistance to antitubulin agents: microtubule dynamics, drug transport, and cell death. J Clin Oncol. 1999;17(3):1061–1070. doi: 10.1200/JCO.1999.17.3.1061. [DOI] [PubMed] [Google Scholar]
- 3.Kamath K, Wilson L, Cabral F, Jordan MA. BetaIII-tubulin induces paclitaxel resistance in association with reduced effects on microtubule dynamic instability. J Biol Chem. 2005;280(13):12902–12907. doi: 10.1074/jbc.M414477200. [DOI] [PubMed] [Google Scholar]
- 4.Bauer JA, Chakravarthy AB, Rosenbluth JM, Mi D, Seeley EH, De Matos Granja-Ingram N, Olivares MG, Kelley MC, Mayer IA, Meszoely IM, et al. Identification of markers of taxane sensitivity using proteomic and genomic analyses of breast tumors from patients receiving neoadjuvant paclitaxel and radiation. Clin Cancer Res. 2010;16(2):681–690. doi: 10.1158/1078-0432.CCR-09-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andre F, Hatzis C, Anderson K, Sotiriou C, Mazouni C, Mejia J, Wang B, Hortobagyi GN, Symmans WF, Pusztai L. Microtubule-associated protein-tau is a bifunctional predictor of endocrine sensitivity and chemotherapy resistance in estrogen receptor-positive breast cancer. Clin Cancer Res. 2007;13(7):2061–2067. doi: 10.1158/1078-0432.CCR-06-2078. [DOI] [PubMed] [Google Scholar]
- 6.McGrogan BT, Gilmartin B, Carney DN, McCann A. Taxanes, microtubules and chemoresistant breast cancer. Biochim Biophys Acta. 2008;1785(2):96–132. doi: 10.1016/j.bbcan.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Chang JC, Wooten EC, Tsimelzon A, Hilsenbeck SG, Gutierrez MC, Elledge R, Mohsin S, Osborne CK, Chamness GC, Allred DC, et al. Gene expression profiling for the prediction of therapeutic response to docetaxel in patients with breast cancer. Lancet. 2003;362(9381):362–369. doi: 10.1016/S0140-6736(03)14023-8. [DOI] [PubMed] [Google Scholar]
- 8.Juul N, Szallasi Z, Eklund AC, Li Q, Burrell RA, Gerlinger M, Valero V, Andreopoulou E, Esteva FJ, Symmans WF, et al. Assessment of an RNA interference screen-derived mitotic and ceramide pathway metagene as a predictor of response to neoadjuvant paclitaxel for primary triple-negative breast cancer: a retrospective analysis of five clinical trials. Lancet Oncol. 2010;11(4):358–365. doi: 10.1016/S1470-2045(10)70018-8. [DOI] [PubMed] [Google Scholar]
- 9.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 10.Martin M, Mackey J, Vogel C. Benefit from adjuvant taxanes and endocrine responsiveness in breast cancer. Breast. 2007;16(Suppl 2):S127–S131. doi: 10.1016/j.breast.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Lu T, Jackson MW, Singhi AD, Kandel ES, Yang M, Zhang Y, Gudkov AV, Stark GR. Validation-based insertional mutagenesis identifies lysine demethylase FBXL11 as a negative regulator of NFkappaB. Proc Natl Acad Sci U S A. 2009;106(38):16339–16344. doi: 10.1073/pnas.0908560106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De S, Cipriano R, Jackson MW, Stark GR. Overexpression of kinesins mediates docetaxel resistance in breast cancer cells. Cancer Res. 2009;69(20):8035–8042. doi: 10.1158/0008-5472.CAN-09-1224. [DOI] [PubMed] [Google Scholar]
- 13.Hirokawa N, Noda Y. Intracellular transport and kinesin superfamily proteins, KIFs: structure, function, and dynamics. Physiol Rev. 2008;88(3):1089–1118. doi: 10.1152/physrev.00023.2007. [DOI] [PubMed] [Google Scholar]
- 14.Lee JK, Coutant C, Kim YC, Qi Y, Theodorescu D, Symmans WF, Baggerly K, Rouzier R, Pusztai L. Prospective comparison of clinical and genomic multivariate predictors of response to neoadjuvant chemotherapy in breast cancer. Clin Cancer Res. 2010;16(2):711–718. doi: 10.1158/1078-0432.CCR-09-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6(10):813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 16.Coombes KR, Wang J, Baggerly KA. Microarrays: retracing steps. Nat Med. 2007;13(11):1276–1277. doi: 10.1038/nm1107-1276b. author reply 1277–1278. [DOI] [PubMed] [Google Scholar]
- 17.Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99(10):6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganapathy V, Ge R, Grazioli A, Xie W, Banach-Petrosky W, Kang Y, Lonning S, McPherson J, Yingling JM, Biswas S, et al. Targeting the Transforming Growth Factor-beta pathway inhibits human basal-like breast cancer metastasis. Mol Cancer. 2010;9(1):122. doi: 10.1186/1476-4598-9-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katoh M. Characterization of KIF12 gene in silico. Oncol Rep. 2005;13(2):367–370. [PubMed] [Google Scholar]
- 21.Nakata T, Hirokawa N. Point mutation of adenosine triphosphate-binding motif generated rigor kinesin that selectively blocks anterograde lysosome membrane transport. J Cell Biol. 1995;131(4):1039–1053. doi: 10.1083/jcb.131.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kan CE, Patton JT, Stark GR, Jackson MW. p53-mediated growth suppression in response to Nutlin-3 in cyclin D1 transformed cells occurs independently of p21. Cancer Res. 2007;67(20):9862–9868. doi: 10.1158/0008-5472.CAN-07-0259. [DOI] [PubMed] [Google Scholar]
- 23.Wordeman L, Wagenbach M, Maney T. Mutations in the ATP-binding domain affect the subcellular distribution of mitotic centromere-associated kinesin (MCAK) Cell Biol Int. 1999;23(4):275–286. doi: 10.1006/cbir.1999.0359. [DOI] [PubMed] [Google Scholar]
- 24.Farmer P, Bonnefoi H, Anderle P, Cameron D, Wirapati P, Becette V, Andre S, Piccart M, Campone M, Brain E, et al. A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat Med. 2009;15(1):68–74. doi: 10.1038/nm.1908. [DOI] [PubMed] [Google Scholar]
- 25.Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14(5):518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 26.Wennmalm K, Ostman A, Bergh J. Stromal signature identifies basal breast cancers. Nat Med. 2009;15(3):237–238. doi: 10.1038/nm0309-237. author reply 238. [DOI] [PubMed] [Google Scholar]
- 27.Smorenburg CH, Sparreboom A, Bontenbal M, Verweij J. Combination chemotherapy of the taxanes and antimetabolites: its use and limitations. Eur J Cancer. 2001;37(18):2310–2323. doi: 10.1016/s0959-8049(01)00309-4. [DOI] [PubMed] [Google Scholar]
- 28.Purcell JW, Davis J, Reddy M, Martin S, Samayoa K, Vo H, Thomsen K, Bean P, Kuo WL, Ziyad S, et al. Activity of the kinesin spindle protein inhibitor ispinesib (SB-715992) in models of breast cancer. Clin Cancer Res. 2010;16(2):566–576. doi: 10.1158/1078-0432.CCR-09-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woessner R, Tunquist B, Lemieux C, Chlipala E, Jackinsky S, Dewolf W, Jr, Voegtli W, Cox A, Rana S, Lee P, et al. ARRY-520, a novel KSP inhibitor with potent activity in hematological and taxane-resistant tumor models. Anticancer Res. 2009;29(11):4373–4380. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.