Abstract
One of the most powerful methods available to assign function to a gene is to inactivate or knockout the gene. Recently,we described the first target-selected knockout in zebrafish. Here,we report on the further improvements of this procedure,resulting in a highly efficient and easy method to do target-selected mutagenesis in zebrafish. A library of 4608 ENU-mutagenized F1 animals was generated and kept as a living stock. The DNA of these animals was screened for mutations in 16 genes by use of CEL-I-mediated heteroduplex cleavage (TILLING) and subsequent resequencing. In total,255 mutations were identified,of which 14 resulted in a premature stop codon,7 in a splice donor/acceptor site mutation,and 119 in an amino acid change. By this method,we potentially knocked out 13 different genes in a few months time. Furthermore,we show that TILLING can be used to detect the full spectrum of ENU-induced mutations in a vertebrate genome with the presence of many naturally occurring polymorphisms.
Over the years, the zebrafish has proven to be an excellent vertebrate model organism for studying many aspects of human biology and disease. The development of the embryo outside of the mother and the full transparency of the embryo, make it well suited for studying, for example, early developmental processes and organ formation. More importantly, however, due to its small size, large number of offspring, and relative short generation time, the zebrafish is well suited for genetic studies. At present, many dominant and recessive forward genetic screens have resulted in thousands of phenotypic mutants (Driever et al. 1996; Haffter et al. 1996), for which the genes involved are now being cloned.
Currently, the genome sequence of the zebrafish is being elucidated and a good draft version is expected soon. At that moment, we will know nearly the complete set of genes encoded by the zebrafish genome, leaving us with the question of what the function is for most of these genes. Although the forward genetic studies will undoubtedly result in extremely valuable data to this end, cloning of the responsible genes is still laborious. Therefore, reverse genetics techniques or knockout technology may become increasingly important tools for revealing gene functions.
A few years ago, RNA interference seemed to be a promising new approach for specifically inactivating genes in zebrafish (Wargelius et al. 1999; Li et al. 2000), but success rates in zebrafish were low and varying, with reports of major nonspecific effects on embryonic development reported as well (Oates et al. 2000; Zhao et al. 2001). The most widely used reverse genetics method in zebrafish is undoubtedly the use of morpholinos that are targeted to the translation initiation site of a specific transcript, thereby inhibiting its translation (Nasevicius and Ekker 2000). The disadvantage of this method is that it is a transient method, requiring repeated injection for each experiment, and only suited for early developmental stages. In addition, nonspecific side effects, like widespread cell death and neuronal degeneration, have been reported (for review, see Heasman 2002).
Attempts to implement a method for generating permanent knockouts similarly (as routinely done for the mouse) have not been successful yet for zebrafish. Such an approach requires homologous recombination in pluripotent embryonic stem (ES) cells and subsequent generation of chimeric embryos. Although ES-like cells for zebrafish have been described (Sun et al. 1995), and the generation of some chimeras from wild-type cells has been reported (Ma et al. 2001), no targeted knockouts have been obtained using this approach. An alternative to this might be to clone zebrafish by nuclear transfer of genetically modified cultured cells (Lee et al. 2002). However, this has also not resulted in a targeted knockout.
Recently, we have shown that reverse genetic analysis of gene function in zebrafish is possible by target-selected mutagenesis (Wienholds et al. 2002); random mutagenesis, followed by screening for mutations in target genes. This general approach, using different types of chemical mutagens and mutation-detection methods, has proven to be successful in Caenorhabditis elegans (Jansen et al. 1997), Drosophila (Bentley et al. 2000), plants (Arabidopsis and Lotus) (McCallum et al. 2000; Perry et al. 2003), mouse (Beier 2000; Coghill et al. 2002), and rat (Zan et al. 2003). For target-selected mutagenesis in zebrafish, male zebrafish are mutagenized using N-ethyl-N-nitrosourea (ENU), identical to that done for forward genetic screens. These males are used to generate a large population of F1 animals that consequently harbor many random heterozygous mutations in their genomes. Next, DNA from these animals is analyzed for the occurrence of mutations in a specific gene of interest. Proof of principle for the zebrafish was obtained by screening a library of DNA samples and matching frozen sperm samples derived from 2679 individual fish. By DNA resequencing, 15 mutations were identified in the rag1 gene. One of these introduced a premature stop codon in the essential core domain of the recombinase, resulting in a complete loss-of-function phenotype, as demonstrated by the lack of recombination of V(D)J segments at the immunoglobulin locus in animals that were homozygous for this mutation (Wienholds et al. 2002).
Here, we describe major modifications to the original method, now allowing efficient generation of knockouts in zebrafish. Firstly, a larger library of ENU-mutagenized animals was constructed and kept alive during the screening for mutations. Secondly, as resequencing is both laborious and expensive, we implemented an alternative method for detection of mutations—TILLING (targeting induced local lesions in genomes; McCallum et al. 2000). This method is based on enzymatic cleavage of heteroduplex DNA using the plant endonuclease CEL-I (Oleykowski et al. 1998). TILLING is used successfully for a similar objective in Arabidopsis (Colbert et al. 2001; Till et al. 2003). We report the identification of 255 mutations, including 21 that are most likely to result in a loss-of-function of 13 different genes. We thus generated 13 different potential knockout fish in a few months.
METHODS
Zebrafish Housing and Mutagenesis
Forty adult TL male zebrafish (4-months-old) were mutagenized by six consecutive treatments with 3.0 mM ENU as described (van Eeden et al. 1999). Of these, 28 surviving and fertile fish were outcrossed with heterozygous albino (alb/+) females, derived from a hybrid München/AB background for the single locus test, or with TL females to generate F1 progeny for the library. To construct a library of 4608 mutagenized fish, both healthy-looking F1 males and females were finclipped (tail) and grouped in 384 pools of 12 fish per tank at the age of from 6 to 8 mo. To recover the fish carrying a specific mutation, all 12 fish from a positive pool were finclipped again, housed separately, and genotyped. Subsequently, carriers were outcrossed against AB or München/AB hybrid fish.
Genomic DNA Isolation and DNA Library Construction
Genomic DNA for the library was isolated from finclips in deep 96-well plates (1 mL capacity per well) for the library and single tubes for the genotyping. Freshly cut finclips were transferred directly to plates/tubes that are kept on dry ice. Fins were lysed by overnight incubation at 55°C in 400 μL pre-warmed lysis buffer (100 mM Tris-HCl at pH 8-8.5, 200 mM NaCl, 0.2% SDS, 5 mM EDTA, and 100 μg/mL proteinase K). DNA was precipitated by adding 300 μL of isopropanol and centrifugation at >6000g, and washed using 70% ethanol. Finally, pellets were dissolved in 1 mL of water. For the DNA library, 5-μL aliquots were arrayed in 384-well PCR plates using a 96-channel pipettor (HYDRA-96, Robbins Scientific), covered with aluminum foil tape (3M) and stored at -20°C.
CEL-I-Mediated Library Screening
CEL-I enzyme was isolated from celery according to Oleykowski et al. (1998) and Yang et al. (2000), with minor modifications (see our Web site for a detailed protocol, http://cuppen.niob.knaw.nl). The enzyme activity for each batch of CEL-I was determined experimentally using a dilution series on control samples. Screening for ENU-induced mutations was done using CEL-I mediated heteroduplex cleavage, analogous as described for Arabidopsis (Colbert et al. 2001), but with several adaptations, as described below. All pipeting steps were done on a Genesis Workstation 200 (Tecan) and Microlab 2200 (Hamilton), or using multichannel pipets. Target genes were amplified by a nested PCR approach in 384-well plates. In the first PCR with gene-specific primers, a touchdown cycling program was used (94°C for 60 sec; 30 cycles of 94°C for 20 sec, 65°C for 30 sec with a decrement of 0.5°C per cycle, and 72°C for 60 sec; followed by 10 cycles of 94°C for 20 sec, 58°C for 30 sec, and 72°C for 60 sec, and an additional extension step of 72°C for 180 sec; GeneAmp9700, Applied Biosystems). PCR samples contained 5 μL of genomic DNA isolated from finclips, 0.2 μM forward (f1) and 0.2 μM reverse (r1) primer, 200 μM of each dNTP, 25 mM Tricine, 7.0% Glycerol (m/v), 1.6% DMSO (m/v), 2 mM MgCl2, 85 mM NH4Acetate (pH8.7), and 0.2 U Taq DNA polymerase in a total volume of 10 μL.
After the first PCR reactions, the samples were diluted with 20 μL of water, and 1 μL was used as template for the second, nested PCR reaction. This reaction contained a mixture of gene-specific forward (M13F-f2, 0.08 μM) and reverse (M13R-r2, 0.04 μM) primers that contain universal M13 adaptor sequences at their 5′ end, and the two corresponding universal M13F (5′-TGTAAAACGACGGCCAGT; 0.1 μM) and M13R (5 ′ AGGAAACAGCTATGACCAT; 0.1 μM) primers labeled with fluorescent dyes (IR Dye 700 and IR Dye 800, respectively) for detection. In addition, the PCR samples contained 200 μM of each dNTP, 20 mM Tris-Hcl (pH8.4), 50 mM KCl, 1.5 mM MgCl2, and 0.1 U Taq DNA polymerase in a total volume of 5 μL. Standard cycling conditions were used for the nested PCR reactions (30 cycles of 94°C for 20 sec, 58°C for 30 sec, and 72°C for 60 sec, followed by an additional extension step of 72°C for 180 sec).
Directly following the nested PCR, heteroduplex formation was done by incubating at 99°C for 10 min, and 70 cycles of 70°C for 20 sec with a decrement of 0.3°C per cycle. Next, 1.25 μL of aliquots of four individual PCR reactions were pooled (total volume of 5 μL) and incubated with 0.01 μL of CEL-I enzyme solution in a total volume of 15 μL (buffered in 10 mM Hepes at pH7.0, 10 mM MgSO4, 10 mM KCl, 0.002% Triton X-100, 0.2 μg/mL BSA) at 45°C for 15 min. CEL-I reactions were stopped by adding 5 μL of 75 mM EDTA. Fragments were purified using Sephadex G50 (medium coarse) minicolumns in 96-well filter plates (Multiscreen HV; Millipore) and eluted into plates prefilled with 5 μL of formamide loading buffer (37% [v/v] de-ionized formamide, 4 mM EDTA [pH8.0], 90 μg/mL bromophenol blue) per well. Samples were concentrated to about 1 μL by heating at 85°C for 45-60 min without cover. A total of 0.4 μL was applied to a 96-lane membrane comb (The Gel Company) and loaded on 25 cm of denaturing 6% polyacrylamide gels on LI-COR 4200 DNA analyzers. Raw TIFF-images produced by the analyzers were manipulated using Adobe Photoshop, and potential mutations were detected and scored manually.
Genotyping by Sequencing
For genotyping, PCR reactions were performed using the same conditions as for the CEL-I-mediated screening. In the nested PCR, only gene-specific primers (M13F-f2 and M13R-r2) were used at 0.2 μM. Nested PCR products were diluted with 30 μL of water, and 1 μL was used as template for the sequencing reactions. Sequencing reactions contained 0.5 μL of DYEnamic ET Terminator (Amersham Pharmacia Biotech), 3.5 μL of ET Terminator dilution buffer (Amersham Pharmacia Biotech), and 0.5 μM of M13F or M13R primer in a total volume of 10 μL. Cycling conditions were as recommended by the manufacturer. Sequencing products were purified using Sephadex G50 (superfine coarse) minicolumns, and analyzed on a 96-capillary 3700 DNA analyzer (Applied Biosystems).
RESULTS
Generation of a Living Library of Mutagenized F1 Fish
The success rate of both forward and reverse mutagenesis screens is mainly dependent on two factors, mutagenesis efficiency and the number of individuals screened. To find loss-of-function mutations in target genes, both factors should be as high as possible. In our first target-selected mutagenesis screen, we were successful in finding a knockout in the zebrafish rag1 gene (Wienholds et al. 2002). However, the library of mutagenized F1 fish we used only comprised 2679 individuals and had a molecular mutation frequency of about one mutation per 450,000 bp (Wienholds et al. 2002). These small numbers make the odds for finding at least one loss-of-function mutation in any average target gene relatively small. Therefore, we generated a novel and larger library. Founder animals were heavily mutagenized with ENU and 4608 healthy F1 male and female fish were raised and used to construct the library (Fig. 1). In contrast to the previous library, we did not generate a permanent resource by cryopreservation of sperm or testis samples. Instead, we maintained the library as a living resource that can be used for a limited time. There are several reasons for this. First of all, the cryopreservation of testis or sperm samples is labor intensive, taking several months to complete, whereas the construction of a living library only took 2 wk to complete. Secondly, the success-rate for the recovery of fish carrying mutations from frozen testis samples by in vitro fertilization (IVF) turned out to be highly variable, in some cases resulting in loss of alleles (E. Wienholds, unpubl.). To minimize the space requirements of a living library, the fish were grouped in 384 pools of 12. After identification of an interesting mutation, all the fish of the positive pool were finclipped and genotyped again to recover the carrier of this mutation.
Figure 1.
Efficient target-selected mutagenesis in zebrafish. Forty male zebrafish were mutagenized with ENU and outcrossed with wild-type females to generate a library of 4608 mutagenized F1 fish. Both males and females were finclipped and grouped in 384 pools of 12 fish per fish tank. DNA was isolated from the finclips and arrayed in twelve 384-well PCR plates. Amplicons of target genes were selected from local gene assemblies using GENOTRACE and primer picking software. Targets were amplified by PCR with gene-specific primers (1), followed by a nested PCR with internal gene-specific primers containing universal adaptor sequences (2), in combination with IR DYE-labeled universal M13 primers. Heteroduplexes were formed and samples were pooled fourfold. Pooled samples were incubated with CEL-I enzyme, and fragments were analyzed by denaturing polyacrylamide gel electrophoresis. Steps from PCR amplification to CEL-I incubation can be done in a completely unattended robotic setup. The four samples represented in a positive pool were reamplified from genomic DNA and subsequently resequenced. Fish carrying interesting mutations were recovered from pools of the F1 library by finclipping and resequencing each of the 12 fish of a pool.
Single locus tests were done to calculate the mutation frequency of the mutagenized founder fish. In total, we found 11 albinos in 3427 mutagenized genomes screened. This is comparable with previous efficient mutagenesis screens (Mullins et al. 1994). Preliminary data from a small-scale forward F3 screen with a subset of the animals also suggests that the mutagenesis efficiency for this new library is high (F. van Eeden, unpubl.). All fish of the library were screened for mutations in several target genes. The average per-base mutation frequency for these genes was 1 in 235,000 bp (Table 1). This is approximately twofold higher than for the previous library.
Table 1.
Summary of Amplicons Screened and Mutations Found
|
Gene
|
Amplicon
|
Screening
|
No. of mutations
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | Cds (bp) | Name | Length/cdsc(bp) | Success (%) | Screened (Mb) | Total | Recovered | Nonsense | Splice | Missense | Silent | Noncoding | Mutation frequency (bp/mutation) |
| gene1 | 690 | A | 569/264 | 88 | 2.3 | 16 | 6 | — | — | — | 1 | 5 | 384,553 |
| gene2 | 1434 | A | 777/381 | 74 | 2.6 | 28 | 15 | — | 1 | 6 | 2 | 6 | 176,634 |
| gene3 | 1821 | A | 639/327 | 80 | 2.4 | 22 | 18 | 1 | 2 | 7 | 3 | 5 | 130,867 |
| gene4 | 4860a | A | 911/879 | 81 | 3.4 | 21 | 14 | 1 | — | 8 | 5 | — | 242,878 |
| gene5 | 1239 | A | 812/327 | 76 | 2.8 | 24 | 18 | 2 | 1 | 2 | 3 | 10 | 157,983 |
| gene6 | 951 | A | 541/436 | 33d | 0.8 | 6 | 6 | 1 | — | 2 | 3 | — | 137,111 |
| gene7 | 2271b | A | 590/264 | 68 | 1.8 | 24 | 6 | — | 1 | 2 | 1 | 2 | 308,122 |
| gene8 | 3240b | A | 442/270 | 79 | 1.6 | 14 | 6 | — | 1 | 2 | 2 | 1 | 268,170 |
| gene9 | 1122 | A | 841/279 | 78 | 3.0 | 34 | 20 | — | — | 7 | 2 | 11 | 151,138 |
| B | 448/230 | 81 | 1.7 | 9 | 7 | — | 1 | 2 | 2 | 2 | 238,879 | ||
| gene10 | 1410 | B | 664/395 | 64 | 2.0 | 8 | 4 | 1 | — | 3 | — | — | 489,554 |
| gene11 | 4110 | A | 720/720 | 87 | 2.9 | 27 | 11 | — | — | 6 | 5 | — | 262,405 |
| gene12 | 480 | A | 496/469 | 87 | 2.0 | 13 | 6 | — | — | 3 | 3 | — | 331,407 |
| gene13 | 2151 | A | 816/443 | 75 | 2.8 | 17 | 9 | 1 | — | 4 | 1 | 3 | 313,344 |
| gene14 | 8532b | A | 953/953 | 83 | 3.6 | 28 | 16 | — | — | 14 | 2 | — | 227,805 |
| B | 873/873 | 80 | 3.2 | 22 | 13 | 1 | — | 7 | 5 | — | 247,556 | ||
| C | 813/813 | 91 | 3.4 | 20 | 10 | — | — | 5 | 5 | — | 340,914 | ||
| G | 921/881 | 87 | 3.7 | 22 | 11 | 1 | — | 6 | 4 | — | 335,659 | ||
| gene15 | 6912b | A | 955/955 | 89 | 4.4 | 25 | 18 | 2 | — | 12 | 4 | — | 244,480 |
| gene16 | 6524 | A | 798/775 | 90 | 3.3 | 26 | 17 | 2 | — | 9 | 6 | — | 194,674 |
| C | 752/418 | 93 | 3.2 | 32 | 16 | 1 | — | 8 | 2 | 5 | 201,416 | ||
| E | 736/526 | 81 | 2.7 | 20 | 8 | — | — | 4 | 2 | 2 | 343,388 | ||
| Average | 2984 | 730/540 | 79 | 2.7 | 20.8 | 11.6 | 0.6 | 0.3 | 5.4 | 2.9 | 2.4 | 234,625 | |
| Total | 47,747 | 16,067/11,878 | 59.8 | 458 | 255 | 14 | 7 | 119 | 63 | 52 | |||
Length of complete coding sequence/cds is based on Drosophila melanogaster homolog
Length of complete cds is based on Homo sapiens ortholog
Length of nested PCR amplicon, excluding primer sequences; length of cds in amplicon
Library not completely screened for this amplicon; false positives not counted
Amplicon Selection and Primer Design
As no fully assembled zebrafish genome is available yet, PCR amplicons for genes of interest were designed from cDNA or EST sequences. These sequences were used as input for GENOTRACE (Berezikov et al. 2002; http://genotrace.niob.knaw.nl) to determine the genomic organization by use of whole-genome sequencing trace archives and retrieve noncoding sequences flanking the exons. These noncoding sequences were used for primer design to amplify the coding sequences (Fig. 1). Preferably, amplicons were chosen such that the coding sequences from the 5′ end of the gene were amplified, maximizing the chance that a premature stop codon results in a full loss-of-function. Nested sets of primers were designed automatically using a specially designed PRIMER3-based (Rozen and Skaletsky 2000) Web application (http://primers.niob.knaw.nl). Using this setup, we have designed nested sets of primers for 29 amplicons, of which 22 turned out to be suitable for screening the library. Failure of the other seven amplicons is most likely due to the repetitive nature of the zebrafish genome and the large amounts of single nucleotide polymorphisms (SNPs; ∼1 in 200 bp; data not shown). The size of the amplicons that did work varied from 442 to 955 bp, with an average of 730 bp, including 450 bp of coding sequence (Table 1). For most target genes, we designed and screened only one amplicon, resulting in only 25% of the total coding sequences screened (11,878 bp of 47,747 total, Table 1).
High-Throughput Screening of the Mutant Library by TILLING
DNA from the 4608 F1 fish was isolated from finclips and arrayed in twelve 384-well PCR plates for automated robotic mutation discovery analysis. Although we initially used resequencing for mutation detection (Wienholds et al. 2002), here we switched to an enzyme-mediated heteroduplex cleavage approach (Oleykowski et al. 1998), because this is both faster and cheaper than sequencing. We adapted the protocol for CEL-I mediated heteroduplex cleavage from the originally described approach for mutation discovery in Arabidopsis (TILLING; Colbert et al. 2001) and introduced some minor modifications (Fig. 1). Firstly, we introduced a nested PCR to avoid the influence of the variation in quality and quantity of the genomic DNA isolated from finclips. Secondly, in the second PCR, we use a mixture of gene-specific primers with M13 adapters and fluorescently labeled universal M13 forward and reverse primers to reduce the costs for fluorescently labeled primers. Thirdly, after PCR amplification of individual samples, products are denatured and reannealed to form heteroduplexes, pooled only fourfold instead of eightfold, and subsequently treated with CEL-I. We found that due to the presence of many SNPs in the zebrafish strains that we used, further pooling might decrease the sensitivity and result in an increase of false negatives. The final CEL-I-digested and purified samples resulting from a single PCR amplicon for the whole library were analyzed on 12 denaturing polyacrylamide slab gels with 96 samples, representing 384 animals per gel (e.g., Fig. 2). The amplicons of the four samples of the positive pool were subsequently reamplified from genomic DNA, and the nature of the mutation was determined by resequencing (Fig. 1). We have set up a high-throughput mutation discovery pipeline that consists of a liquid handling robot with four 384-well PCR blocks incorporated, which runs the PCR amplification, pooling, and CEL-I digestion steps completely unattended within 20 h. Purified samples are subsequently analyzed on four LICOR analyzers that can be run at least three times a day. This setup can be operated by a single person, and allows the initial screening of the complete library for one amplicon of maximal 1000 bp within a single day.
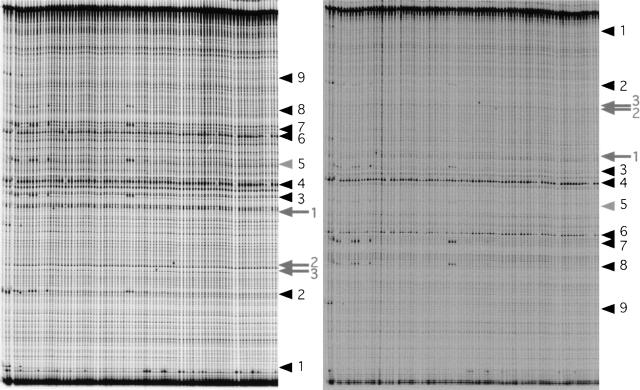
Figure 2.
Representative example of a gel used to screen 384 fish for mutations in the dicer gene. Each of the 96 lanes of the polyacrylamide gel contains fourfold-pooled CEL-1-digested PCR products of ∼830 bp. The IR Dye 700 and IR Dye 800 channels are shown at left and right, respectively. Arrowheads indicate nine SNPs present in the founder fish. SNP no. 5 is only visible in one channel, and therefore, indicated by a gray arrowhead. Arrows indicate the three potential mutations on this gel (all rank 1).
ENU-Induced Mutations Are Efficiently Identified by TILLING
The TILLING method for mutation discovery has been set up for ethylmethanesulfonate (EMS)-induced point mutations in the relative simple genome of Arabidopsis (Colbert et al. 2001). The organization of the zebrafish genome is much more complex than the Arabidopsis genome. There are many SNPs and repetitive DNA stretches that might negatively influence the discovery of ENU-induced mutations. In addition, the spectrum of ENU-induced mutations is much broader than for EMS (Fig. 3A). To determine the sensitivity of TILLING in identifying ENU-induced mutations in zebrafish, potential mutations were scored according to the criteria in Table 2 (Example is shown in Supplemental Figure 1, available online at www.genome.org). In total, 59.8 million bp was screened by TILLING (Table 1), in which we identified 435 unique potential mutations. These 429 suspected mutations could be distributed over the six different scoring classes (Table 2). Overall, the confirmation rate after resequencing is 59%, resulting in 255 real mutations. As expected, success rates for confirmation decreased from scoring rank 1 to 5. However, it remained sufficiently high in all classes (minimum of 23%), so that in future screens, classification according to these criteria might not be necessary. Aberrant-looking lanes and potential mutations were classified in rank 6. Confirmation of these mutations also turned out to be quite efficient (38%; Table 2), justifying the inclusion of this type of potential mutations in future screens. Thus, any difference observed in the pooled CEL-I-digested samples is worth further investigation by resequencing.
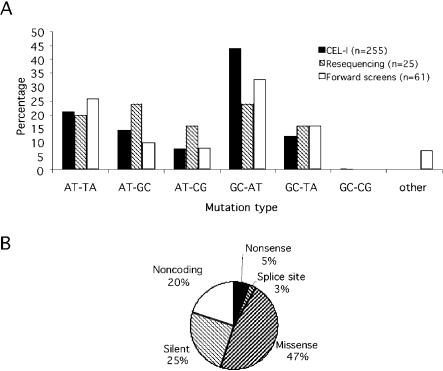
Figure 3.
Characterization of the ENU-induced mutations. (A) ENU mutation spectrum of this screen is similar to that of resequenced genes (Wienholds et al. 2002; E. Wienholds and R.H.A. Plasterk, unpubl.) and forward screens (Knapik 2000; http://zfin.org). Single nucleotide changes are indicated, and small deletions/insertions and chromosomal rearrangments are summarized under others. (B) Distribution of the mutations at the coding level.
Table 2.
Mutation Quality Classification
| Class | Scoring criteria | No. of mutations observed by TILLING | No. of mutations confirmed by sequencing |
|---|---|---|---|
| Rank 1 | Strong signal in channel 1, strong signal in channel 2 | 110 | 97 (88%) |
| Rank 2 | Strong signal in channel 1, weak signal in channel 2 | 74 | 50 (68%) |
| Rank 3 | Strong signal in channel 1, no signal channel 2 | 66 | 40 (61%) |
| Rank 4 | Weak signal in channel 1, weak signal in channel 2 | 40 | 16 (40%) |
| Rank 5 | Weak signal in channel 1, no signal channel 2 | 47 | 11 (23%) |
| Rank 6a | Aberrant | 92 | 35 (38%) |
| Totalb | 435 | 255 (59%) |
Gel files as shown in Fig. 2 were inspected manually and a rank was assigned to each of the potential mutations as observed by TILLING. Potential mutations were confirmed by resequencing.
Lanes in gel that look different than normal, contain multiple signals, have a signal at the same height as a background signal, etc
Including six unclassified mutations from gene6.
The confirmation rate varies from gene to gene. We do not find a correlation with the number of SNPs within an amplicon, but we cannot exclude an effect of SNPs in the primer regions, which may result in less-efficient PCR and increased Taq polymerase errors. When variation in product intensity per lane is seen, as for example for genes 7 and 1 (data not shown), indicative for an inefficient amplification of one or more of the samples in this pool, confirmation rates decrease.
The CEL-I enzyme has a slight preference for certain types of heteroduplexes (Oleykowski et al. 1998). TILLING has been proven to work nicely for the limited spectrum of ethylmethanesulfonate (EMS)-induced mutations (mainly GC to AT transitions, Colbert et al. 2001; Greene et al. 2003). We inspected whether it also efficiently recognized the full spectrum of ENU-induced mutations. We compared the spectrum found in this screen with the spectrum found by resequencing of the rag1 and two other genes (Wienholds et al. 2002; E. Wienholds and R.H.A. Plasterk, unpubl.; Fig. 3A). Overall, the spectra are quite similar. Differences do not reflect the CEL-I preferences and are likely statistical variations due to the small number of mutations (25) found in the resequencing screens. This indicates that we do not miss a certain class of mutations. The mutation spectrum is also very similar to the mutation spectrum of the ENU-induced mutations found in forward genetic screens (Knapik 2000; http://zfin.org; Fig. 3A). This indicates that we also do not miss a certain class of mutations that might give a phenotypic change. In conclusion, we think that TILLING is sensitive enough to identify mutations in all classes of the ENU mutation spectrum.
Potential Knockouts by Target-Selected Mutagenesis
We screened for mutations in 16 different target genes, and in most cases, only one amplicon per gene. Due to the genomic organization of most target genes, the amplicons also contained, on average, 26% noncoding sequences (4189 bp of 16,067 bp total; Table 1), like introns and untranslated regions. However, of the 255 confirmed mutations (Table 1; Fig. 3B), 52 (20%) were found in these noncoding sequences. ENU mutagenesis is expected to be random for coding and noncoding sequences. The discrepancy observed here (6%) is most likely caused by the fact that most of the noncoding sequences are located near the amplicons ends, in which mutations are more difficult to detect (Greene et al. 2003). Seven of these noncoding mutations (3%) affected splice donor or acceptor consensus sequences, and are likely to result in splice pattern alterations. Of the 196 mutations found in the coding regions, 63 (25%) are silent, 119 (47%) are missense, resulting in a variety of amino acid changes, and 14 (5%) are nonsense. These results are in full agreement with the changes that can be calculated from codon usage in zebrafish (Nakamura et al. 2000) combined with the molecular ENU spectrum found in our screen (data not shown). The mutations most likely to result in complete loss-of-function of the genes are the nonsense and splice donor/acceptor site mutations. In total, we found 21 of these types of mutations in 13 of the 16 target genes screened.
DISCUSSION
Recently, we have shown proof of principle that target-selected mutagenesis is possible in the zebrafish by screening ENU-mutagenized animals for point mutations (Wienholds et al. 2002). In this study, we implemented several changes to the original procedure to make it even more efficient, resulting in perhaps the method of choice for making knockouts in zebrafish.
First of all, we constructed a larger library of heavily mutagenized F1 fish. These fish were kept alive during the screening process. To minimize the space requirements, the fish were grouped in 384 pools of 12. As a consequence, the fish of individual pools had to be finclipped and genotyped again before we could recover the carrier of an interesting mutation. However, this can be done in a single day, which is diminishable in the timescale for creating a homozygous knockout. The carrier can be used immediately for large-scale outcrosses, gaining enough fish in the next generation to perform linkage analysis. Both males and females have been used to construct the library. This led to an approximately twofold reduction in the initial animals needed if compared with a library of frozen sperm or testis samples. In addition, interesting mutations found in opposite sexes can be crossed together straight away. For one of the target genes, we found two different, independent nonsense alleles, both disrupting most of the protein. Because these different mutations were found in a male and female, respectively, we were able to do phenotypic analysis of transheterozygotic embryos (without interference of homozygous background mutations) within 1 wk after recovery of the carriers.
A major holdback for creating a living library is that the library can only be screened for a limited time. This starts at the moment that fish can be finclipped (∼2-3 mo) up to their fertile lifetime (∼1.5 yr). This implies a scheduled mutagenesis and library construction each 1.5 yr. Because construction of the library only took us 2 wk to complete (mutagenesis not included), this is a fast alternative for constructing a permanent (cryopreserved) library, providing the availability of sufficient space. The latter can be addressed by the construction of several small libraries (e.g., 384 animals) at a regular interval. These libraries should then be screened for as many target genes as possible before a new library is ready for screening. Recovery of mutant animals from frozen sperm of the first library turned out to be very variable. In some cases, we were not able to recover a fish at all and lost the mutant allele (E. Wienholds, unpubl.). Keeping the library alive might circumvent this problem. However, diseases and other factors might cause fish to die during the screening process. We did see a substantial fraction of the library dying (∼25% over a half-year time period) in one part of the library. This was most likely caused by overcrowding and insufficient fresh water inflow, presumably resulting in toxification of the water. Improvement of housing conditions should abolish this problem in future libraries.
In the original method for target-selected mutagenesis in zebrafish, we identified mutations by resequencing (Wienholds et al. 2002). To both speed up and reduce the costs of target-selected mutagenesis in zebrafish, we incorporated the TILLING method (Colbert et al. 2001) for initial mutation detection. Subsequently, potential mutations were then confirmed by resequencing. We set up a high-throughput robotic pipeline that allows the screening of the complete library by TILLING for mutations in one amplicon within 1 d by a single person. To even further reduce the costs, nested PCR reactions with gene-specific primers coupled to universal adaptors and fluorescence-labeled universal primers were performed. Compared with sequencing, this reduced the costs ∼10-fold. Pooling samples more than fourfold (like eightfold in Arabidopsis; Colbert et al. 2001) might further speed up the screening process and reduce the costs. However, due to the presence of many SNPs and repetitive DNA stretches, it might be more difficult to detect the mutations, and therefore, the number of missed mutations might increase considerably. Although we used a high-throughput robotics setup for finding mutations in target genes by TILLING in a short time, this approach can also be adapted for use on a smaller scale by use of manual pipeting.
TILLING has originally been set up for the identification of EMS-induced mutations in Arabidopsis (Colbert et al. 2001). We found that ENU-induced mutations in zebrafish are also efficiently recognized. From the 435 potential mutations found by TILLING, 255 (59%) were confirmed by resequencing. There is a correlation with signal strength on the polyacrylamide gels and the ability to reconfirm the mutation. But the recovery is sufficiently high, even for the lowest quality class (23%), so that in future screens, any indication of a mutation being present should be followed up. The spectrum of ENU-induced mutations found by TILLING in zebrafish is very similar to the spectrum found by resequencing and forward screens. This indicates that TILLING can be used reliably to detect all classes of mutations introduced by ENU mutagenesis. Only the GC to AT class of transversions is considerably larger in our screen. This may be due to the low quality of DNA for some parts of the library (data not shown), resulting in cytosine deamination (Hofreiter et al. 2001). Although it is unlikely to identify such mutations twice in two completely independent assays (TILLING and resequencing), these types of mistakes end up in our final list of mutations, because this list is based upon the mutations that were mainly confirmed only once in the resequencing phase. To exclude this type of mistake from the list, samples have to be reamplified and sequenced once more. Other small differences with respect to the forward screens could reflect the bias for phenotypic alterations in forward screens. For example, nonsense codons (most likely to have a phenotypic effect) can never be found in the class of AT to GC transitions, but are found mainly in the AT to TA and GC to TA transversions and GC to TA transitions.
In the first target-selected mutagenesis screen in zebrafish, we screened extensively for mutations only in one target gene, the rag1 gene, and found one nonsense mutation. This turned out to be a loss-of-function allele (Wienholds et al. 2002). Here, we screened for mutations in 16 different target genes. The average molecular mutation frequency for these genes was almost twofold higher (1 in 235,00 bp) than for the rag1 gene in our previous screen (1 in 450,000 bp). Taking differences in mutagenesis efficiency into account, these results indicate that the success rate for finding mutations using TILLING is at least comparable or maybe even higher than for resequencing. Together with a larger library, this increased the chance of finding detrimental mutations considerably. In 13 of the 16 target genes we screened, we found 21 nonsense and/or splice acceptor/donor site mutations. These mutations are likely to result in loss-of-function or knockouts of the genes. On average, we screened only 25% of the coding regions of these target genes (Table 1). This implies that we could have found many additional nonsense and splice acceptor/donor site mutations if we had screened the complete coding sequences for a specific gene. Here, we focused on amplicons that were most suited for screening by TILLING and for which the PCR reactions worked under standard conditions (22 out of 29 amplicons). Despite the fact that most of these amplicons contained small exons, we were still able to find nonsense and splice acceptor/donor mutations in these exons. This indicates that it was worthwhile to screen these small exons for mutations.
Not only the nonsense and splice acceptor/donor site mutations are likely to be loss-of-function alleles, also several of the 199 missense mutations might be loss-of-function. Similar to forward genetic screens, in which about equal numbers of missense and nonsense mutations are responsible for the observed phenotypes (39% and 48%, respectively; Knapik 2000; http://zfin.org), ∼10-20 missense mutations in our set may be expected to result in a phenotypic difference. Furthermore, an allelic series of the remaining mutations may have more subtle, hypomorphic effects on the gene function, and help in the elucidation of specific protein (subdomain) function.
In summary, we have set up a fast, easy, and cheap high-throughput pipeline for making knockouts in zebrafish by target-selected mutagenesis. We show that this setup is well-suited for the detection of the full mutation spectrum induced by ENU in a vertebrate genome, and that the method is sensitive enough to detect novel mutations in the background of natural occurring SNPs, which is important when working with outbred animals such as zebrafish. We created 13 potential knockout animals in a few months. In addition, by use of this method, informative missense mutations may well be retrieved that can be very valuable when complete loss-of-function results in (embryonic) lethality. Furthermore, such mutations may be useful in protein-domain function studies. Taken together, we think that the target-selected mutagenesis approach described here is the method of choice for the generation of zebrafish knockouts.
Acknowledgments
We thank Steve Henikoff and Brad Till for the valuable help needed for setting up the TILLING method in our lab, Marco Koudijs, Adam Hurlstone, Anna-Pavlina Haramis, Marjo den Broeder, and Astrid Keijser for help with library construction and screening, and our collaborators for sharing unpublished sequences. This work was supported financially by a NWO genomics grant.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1725103. Article published online before print in November 2003.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Beier, D.R. 2000. Sequence-based analysis of mutagenized mice. Mamm. Genome 11: 594-597. [DOI] [PubMed] [Google Scholar]
- Bentley, A., MacLennan, B., Calvo, J., and Dearolf, C.R. 2000. Targeted recovery of mutations in Drosophila. Genetics 156: 1169-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov, E., Plasterk, R.H., and Cuppen, E. 2002. GENOTRACE: cDNA-based local GENOme assembly from TRACE archives. Bioinformatics 18: 1396-1397. [DOI] [PubMed] [Google Scholar]
- Coghill, E.L., Hugill, A., Parkinson, N., Davison, C., Glenister, P., Clements, S., Hunter, J., Cox, R.D., and Brown, S.D. 2002. A gene-driven approach to the identification of ENU mutants in the mouse. Nat. Genet. 30: 255-256. [DOI] [PubMed] [Google Scholar]
- Colbert, T., Till, B.J., Tompa, R., Reynolds, S., Steine, M.N., Yeung, A.T., McCallum, C.M., Comai, L., and Henikoff, S. 2001. High-throughput screening for induced point mutations. Plant Physiol. 126: 480-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever, W., Solnica-Krezel, L., Schier, A.F., Neuhauss, S.C., Malicki, J., Stemple, D.L., Stainier, D.Y., Zwartkruis, F., Abdelilah, S., Rangini, Z., et al. 1996. A genetic screen for mutations affecting embryogenesis in zebrafish. Development 123: 37-46. [DOI] [PubMed] [Google Scholar]
- Greene, E.A., Codomo, C.A., Taylor, N.E., Henikoff, J.G., Till, B.J., Reynolds, S.H., Enns, L.C., Burtner, C., Johnson, J.E., Odden, A.R., et al. 2003. Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics 164: 731-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffter, P., Granato, M., Brand, M., Mullins, M.C., Hammerschmidt, M., Kane, D.A., Odenthal, J., van Eeden, F.J., Jiang, Y.J., Heisenberg, C.P., et al. 1996. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 123: 1-36. [DOI] [PubMed] [Google Scholar]
- Heasman, J. 2002. Morpholino oligos: Making sense of antisense? Dev. Biol. 243: 209-214. [DOI] [PubMed] [Google Scholar]
- Hofreiter, M., Jaenicke, V., Serre, D., Haeseler Av, A., and Paabo, S. 2001. DNA sequences from multiple amplifications reveal artifacts induced by cytosine deamination in ancient DNA. Nucleic Acids Res. 29: 4793-4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, G., Hazendonk, E., Thijssen, K.L., and Plasterk, R.H. 1997. Reverse genetics by chemical mutagenesis in Caenorhabditis elegans. Nat. Genet. 17: 119-121. [DOI] [PubMed] [Google Scholar]
- Knapik, E.W. 2000. ENU mutagenesis in zebrafish-from genes to complex diseases. Mamm. Genome 11: 511-519. [DOI] [PubMed] [Google Scholar]
- Lee, K.Y., Huang, H., Ju, B., Yang, Z., and Lin, S. 2002. Cloned zebrafish by nuclear transfer from long-term-cultured cells. Nat. Biotechnol. 20: 795-799. [DOI] [PubMed] [Google Scholar]
- Li, Y.X., Farrell, M.J., Liu, R., Mohanty, N., and Kirby, M.L. 2000. Double-stranded RNA injection produces null phenotypes in zebrafish. Dev. Biol. 217: 394-405. [DOI] [PubMed] [Google Scholar]
- Ma, C., Fan, L., Ganassin, R., Bols, N., and Collodi, P. 2001. Production of zebrafish germ-line chimeras from embryo cell cultures. Proc. Natl. Acad. Sci. 98: 2461-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum, C.M., Comai, L., Greene, E.A., and Henikoff, S. 2000. Targeted screening for induced mutations. Nat. Biotechnol. 18: 455-457. [DOI] [PubMed] [Google Scholar]
- Mullins, M.C., Hammerschmidt, M., Haffter, P., and Nusslein-Volhard, C. 1994. Large-scale mutagenesis in the zebrafish: In search of genes controlling development in a vertebrate. Curr. Biol. 4: 189-202. [DOI] [PubMed] [Google Scholar]
- Nakamura, Y., Gojobori, T., and Ikemura, T. 2000. Codon usage tabulated from international DNA sequence databases: Status for the year 2000. Nucleic Acids Res. 28: 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasevicius, A. and Ekker, S.C. 2000. Effective targeted gene `knockdown' in zebrafish. Nat. Genet. 26: 216-220. [DOI] [PubMed] [Google Scholar]
- Oates, A.C., Bruce, A.E., and Ho, R.K. 2000. Too much interference: Injection of double-stranded RNA has nonspecific effects in the zebrafish embryo. Dev. Biol. 224: 20-28. [DOI] [PubMed] [Google Scholar]
- Oleykowski, C.A., Bronson Mullins, C.R., Godwin, A.K., and Yeung, A.T. 1998. Mutation detection using a novel plant endonuclease. Nucleic Acids Res. 26: 4597-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, J.A., Wang, T.L., Welham, T.J., Gardner, S., Pike, J.M., Yoshida, S., and Parniske, M. 2003. A TILLING reverse genetics tool and a Web-accessible collection of mutants of the legume Lotus japonicus. Plant Physiol. 131: 866-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen, S. and Skaletsky, H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132: 365-386. [DOI] [PubMed] [Google Scholar]
- Sun, L., Bradford, C.S., Ghosh, C., Collodi, P., and Barnes, D.W. 1995. ES-like cell cultures derived from early zebrafish embryos. Mol. Mar. Biol. Biotechnol. 4: 193-199. [PubMed] [Google Scholar]
- Till, B.J., Reynolds, S.H., Greene, E.A., Codomo, C.A., Enns, L.C., Johnson, J.E., Burtner, C., Odden, A.R., Young, K., Taylor, N.E., et al. 2003. Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res. 13: 524-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eeden, F.J., Granato, M., Odenthal, J., and Haffter, P. 1999. Developmental mutant screens in the zebrafish. Methods Cell Biol. 60: 21-41. [DOI] [PubMed] [Google Scholar]
- Wargelius, A., Ellingsen, S., and Fjose, A. 1999. Double-stranded RNA induces specific developmental defects in zebrafish embryos. Biochem. Biophys. Res. Commun. 263: 156-161. [DOI] [PubMed] [Google Scholar]
- Wienholds, E., Schulte-Merker, S., Walderich, B., and Plasterk, R.H. 2002. Target-selected inactivation of the zebrafish rag1 gene. Science 297: 99-102. [DOI] [PubMed] [Google Scholar]
- Yang, B., Wen, X., Kodali, N.S., Oleykowski, C.A., Miller, C.G., Kulinski, J., Besack, D., Yeung, J.A., Kowalski, D., and Yeung, A.T. 2000. Purification, cloning, and characterization of the CEL I nuclease. Biochemistry 39: 3533-3541. [DOI] [PubMed] [Google Scholar]
- Zan, Y., Haag, J.D., Chen, K.S., Shepel, L.A., Wigington, D., Wang, Y.R., Hu, R., Lopez-Guajardo, C.C., Brose, H.L., Porter, K.I., et al. 2003. Production of knockout rats using ENU mutagenesis and a yeast-based screening assay. Nat. Biotechnol. 21: 645-651. [DOI] [PubMed] [Google Scholar]
- Zhao, Z., Cao, Y., Li, M., and Meng, A. 2001. Double-stranded RNA injection produces nonspecific defects in zebrafish. Dev. Biol. 229: 215-223. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://cuppen.niob.knaw.nl; Detailed protocols for TILLING procedures and CEL-I isolation.
- http://genotrace.niob.knaw.nl; Interface to GENOTRACE for local genomic sequence assemblies from raw genome sequencing traces using EST or cDNA sequences as input.
- http://primers.niob.knaw.nl; Design of sets of nested (tailed) primers for TILLING.
- http://zfin.org; The zebrafish information network.