Abstract
Interleukin-6 (IL-6), a pro-inflammatory cytokine, is involved in prostate cancer progression, including androgen independence. Serum IL-6 levels also correlate with prostate tumor burden, prostate-specific antigen levels and metastasis. Since circulating cytokine levels vary considerably inter-individually, such variation could be linked to genetic factors, including genetic polymorphism. The -174G>C/rs1800795 polymorphism in the IL-6 promoter is functionally relevant in terms of transcriptional regulation and disease association. We investigated a possible association of the -174G/C polymorphism with prostate cancer. Since significant racial disparities exist in prostate cancer incidence, we also investigated this association between the -174G/C polymorphism and prostate cancer in Caucasians and African-Americans, separately. Direct sequencing of the PCR amplicon from genomic DNA was used for genotyping rs1800795 in all subjects [age-matched controls (N = 140) and prostate cancer patients (N = 164)]. Sample size and power was calculated using the PGA software. We found the GG genotype to be associated with increased risk of prostate cancer in Caucasian subjects, whereas the CC genotype was associated with increased risk in the African-American sample set. Such a dimorphic genotypic association with cancer and race is unique and suggests a complex gene-gene and gene-environment interaction.
Keywords: IL-6, Polymorphism, Prostate, Cancer, Disparities
INTRODUCTION
Interleukin-6 (IL-6), a pro-inflammatory cytokine is involved in the regulation of various cellular functions, i.e., proliferation, apoptosis, angiogenesis, differentiation, and regulation of immune response (Culig et al., 2005). IL-6 may also have a crucial role in the growth and differentiation of malignant tumors including prostate, melanoma, renal, lung, and myeloma (Lagmay et al., 2009). In prostate cancer, the role of IL-6 is particularly well established. Prostate tumor cells produce large amounts of IL-6 and its receptor, IL-6R (gp80), and gp130 (Knupfer and Preiss, 2008). IL-6 functions as a paracrine growth factor for the human LNCaP androgen-sensitive prostate cancer cells and an autocrine growth factor for the human DU145 and PC3 androgen-insensitive prostate cancer cells (Giri and Ittmann, 2001; Giri et al., 2001; Hobisch et al., 2001; Godoy-Tundidor et al., 2005). IL-6 can also transform benign prostate cells into the malignant phenotype and promote epithelial mesenchymal transition (Rojas et al., 2011).
IL-6 is also associated with androgen-independent prostate cancer. In the absence of androgens, IL-6 causes activation of the androgen receptor (AR), which is approximately 50% maximal activity induced by androgens. In low concentration conditions, androgen is potentiated by IL-6 leading to synergistic activation of AR (Culig, 2003). IL-6 levels also correlate well with prostate tumor burden, serum prostate-specific antigen (PSA) and clinically evident metastasis (Shariat et al., 2001).
Since circulating cytokine levels vary considerably inter-individually, such variation could be linked to genetic factors, most notably genetic polymorphism. Previous studies have shown that the IL-6 polymorphism (-174G>C) is associated with more aggressive disease in prostate cancer (Tan et al., 2005; Kesarwani et al., 2008). The -174G>C/rs1800795 polymorphism is functionally relevant both in terms of its location and its disease association. The rs1800795 polymorphism affects the transcription of the IL-6 gene, thus altering the serum IL-6 levels. More recent studies on the effect of rs1800795 on transcription factor binding have shown that -174G>C transversion gates the GATA1 access to IL-6 promoter, thereby linking this single nucleotide polymorphism (SNP) to differential risk of inflammation-related diseases such as prostate cancer (Cole et al., 2010). Several of the polymorphisms located in the IL-6 promoter region have also been studied in various disease conditions, i.e., coronary artery disease, asthma, rectal cancer, neuroblastoma, and juvenile arthritis (Corvol et al., 2009; Lagmay et al., 2009; Slattery et al., 2009; Ol et al., 2011). The effect of IL-6 promoter haplotype (-597G>A, -572G>C and -174G>C) has also been shown to have a significant effect on transcriptional regulation and disease association (Cussigh et al., 2011). On the basis of these studies, -174G>C has the highest impact on transcription regulation, making it a functional polymorphism (Falleti et al., 2010). Homozygotes for the G allele have been shown to have higher plasma levels of IL-6, higher IL-6 gene transcription activity and higher inducible IL-6 responses compared to subjects homozygous for the C allele (Pereira et al., 2011). In spite of the evidence supporting a positive association between -174G allele and higher IL-6 levels, studies have shown that it is the C allele that is associated with prostate cancer (Terry et al., 2000). Studies have also shown that the C allele is associated with increased risk of colorectal cancer, especially among alcohol users and those not taking anti-inflammatory drugs (Slattery et al., 2007). These results clearly suggest a complex interplay of additional regulatory mechanism or complex gene-environment interactions that modify IL-6 activity and expression in prostate and other cancers.
The -174G/C polymorphism is also associated with race. The non-Caucasian population has a higher frequency of the G allele as compared to the C allele, suggesting that a higher prevalence of the G allele may be associated with racial disparities in various diseases such as prostate cancer. The complex association of the IL-6 -174G/C polymorphism with prostate cancer and racial disparities between Caucasian and African-Americans was investigated in this study. The genotyping data were analyzed by log-linear modeling to reveal complex interactions between disease (prostate cancer), race (Caucasian and African-American) and age at diagnosis to establish the significance of the -174G/C polymorphism in prostate cancer disparities.
MATERIAL AND METHODS
Cell culture
The prostate cancer cell lines LNCaP, DU145 and PC3 were purchased and cultured according to the supplier instructions (American Type Culture Collection, Rockville, MD, USA).
Samples
The use of human samples and protocols for the study were approved by the institutional review board of Clark Atlanta University. Cancer samples and control samples constituted approximately 50% each of the total sample set (Table 1, N > 300). Retrospectively collected buffy coat samples were obtained from the Biospecimen Shared Resources at KU Cancer Center (University of Kansas Medical Center, Kansas City, KS, USA) and the Cooperative Human Tissue Network (Southern Division; National Cancer Institute, Bethesda, MD, USA). The purified genomic DNA samples were commercially obtained from BioServe Inc. (Beltsville, MD, USA). All samples were stored at -80°C until analysis. De-identified, comprehensive clinical information regarding age, ethnicity and stage was available for all samples. The ethnicity of individuals who provided the samples was as reported by the supplier, and no specific admixture analysis was performed. However, a family history of prostate cancer and PSA level were not available for all samples and were thus not included in the final statistical analysis.
Table 1.
Sample demographics [the age (means ± SE, or median and range) of all samples and samples stratified with race is shown].
| Total No. of samples analyzed | N | Age
|
||
|---|---|---|---|---|
| Mean + SE (years) | Median (years) | Range (years) | ||
| All | ||||
| Cancers | 164 | 63.707 ± 0.746 | 63.5 | 43–86 |
| Control | 140 | 60.214 ± 1.344 | 64 | 17–81 |
| Cancers | ||||
| Caucasian | 84 | 59.750 ± 0.888 | 59.5 | 37–83 |
| African-American | 80 | 67.862 ± 1.028 | 68 | 43–86 |
| Normal | ||||
| Caucasian | 78 | 57.244 ± 1.955 | 61 | 17–81 |
| African-American | 62 | 63.952 ± 1.6 | 67 | 20–74 |
The χ2 with corresponding P values shows the comparison between race and disease status control and cancer: χ2 = 0.61, P = 0.73.
DNA isolation
Genomic DNA was isolated from cultured cells or buffy coat using the AquaPure genomic DNA isolation kit (Bio-Rad, Hercules, CA, USA). On average, approximately 30 μg DNA was routinely isolated from 300 μL buffy coat.
Genomic PCR
Genomic PCR was performed in a 25-μL PCR mixture that consisted of 12.5 μL GoTaq Master Mix (Promega), 30 ng genomic DNA and 400 pmol of each of 5′- and 3′-primers (see below). PCR was carried out for 35 cycles with annealing temperature of 54.3°C. The following primers were used (NC_000007.13) for rs1800795 (forward: 5′-TTTCTCTTTGTAAAACTT CGTGC-3′ and reverse: 5′-GACCCTCAGACATCTCCAGTC-3′). The PCR product was subsequently sequenced with the nested primer: 5′-TAAAGGAAGAGTGGTTCTGC-3′.
SNP detection
The rs1800795 polymorphism was detected by direct sequencing of the PCR amplicon. An aliquot of the respective PCR was first analyzed on a 1.5% agarose gel to confirm the specificity and quality of the reaction in terms of band size and absence of any background PCR product. Once confirmed, the remaining PCR product was cleaned using ExoSAP-IT (USB Corporation, Cleveland, OH, USA) before sequencing on an AB sequencer (DNA Sequencing Laboratory, Morehouse School of Medicine, Atlanta, GA, USA). Sequencing was performed using the nested primer within the PCR product as described above. The sequences were scanned using an ABI sequence scanner (Applied Biosystems, Foster City, CA, USA), and the SNPs were detected manually. The amplicons that had low-quality reads were re-sequenced. The SNPs were also detected by using the SNP detector software (Zhang et al., 2005) to ensure that the SNPs were caused by heterozygous allelic variation and not sequencing artifacts. The sequences were also aligned with the NCBI reference sequence locus NG_011640.1 in CLUSTALW (Conway Institute, University College Dublin, Dublin, Ireland) to validate respective sequence reads.
Sample size calculation
The sample size required to achieve statistically significant associations were calculated using the power calculator for case-control genetic association studies (PGA) (Menashe et al., 2008). The parameters used in sample size calculation were as follows: a power of 0.95 (95%), an α level of 0.05 and prostate cancer disease prevalence of 250 in 100,000 men, with actual rates for all races, for whites, and for blacks of 156, 149.5, and 233.8 per 100,000 men, respectively, according to Surveillance, Epidemiology and End Result data. An rs1800795 G allele frequency of 50% was chosen as the disease allele frequency. On the basis of these variables, PGA estimated a sample size of <100 cases (the actual sample size would have been lower with exact prostate cancer disease prevalence numbers). On the basis of these calculations, we selected a sample size of approximately 1.5 times that estimated by PGA.
Statistical analysis
SNP rs1800795 was tested for its association with prostate cancer. Odds ratios (ORs) and 95% confidence intervals were calculated for the genotype association with prostate cancer. Correlation between genotype distribution and individuals stratified by race and age was performed using logistic regression models. The log-linear model (LLM) was used as a test for independence in a two-way contingency table for multifactor dimensionality reduction (MDR) and to infer true associative structure in a set of multiple categorical variables such as disease status, genotype, race, and age. The NCSS 2007 (version 07.1.19; NCSS Kaysville, UT, USA) and SigmaStat (version 3.5; SigmaStat, San Jose, CA, USA) software packages were used for statistical analyses. P < 0.05 was considered to be statistically significant.
RESULTS
IL-6 SNP rs1800795 is located -174 bp upstream of the IL-6 gene. In chromosomal context, the position of rs1800795 is at Chr7:22766221 (dbSNP build 135).
Prostate cancer cell line rs1800795 genotype
Genotyping was performed on prostate cancer cell line genomic DNA for process validation including genomic DNA preparation, PCR sequencing and SNP detection (manually and SNP detector). Following successful validation of the process, the rs1800795 genotype in three prostate cancer cell lines LNCAP, DU145, and PC3 was GG, GG, and GC, respectively. The cell line genotype data indicated that LNCaP and DU145 cells had the homozygous G allele, while PC3 cells had a heterozygous genotype.
Sample demographics
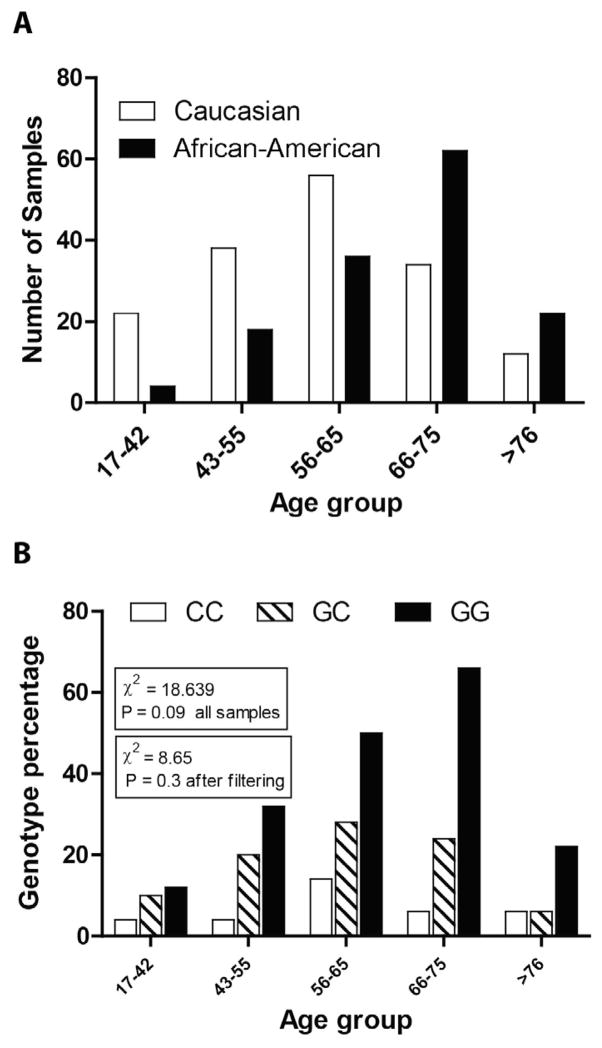
A retrospective case-control study was performed on 304 samples to analyze the rs1800795 polymorphism. The unrelated, clinically defined cancer samples (N = 164) comprised 53.9% of the total samples while the control samples (N = 140) comprised the remaining 46.1%, giving a statistically equal distribution in the two sample categories (P = 0.73). The mean age of the patients who provided cancer samples was 63.7 ± 0.746 years, and the mean age of individuals who were identified as disease-free (normal) was 60.2 ± 1.3 years. Both groups were considered age-matched (P = 0.273) (Table 1 and Figure 1A).
Figure 1.
A. Distribution of cases and controls across race in the study sample set. Chi-square is not significant indicating that the distribution is essentially similar. B. Distribution of the rs1800795 genotype frequency across age groups, which is similar (chi-square = 18.639, P = 0.09). The rs1800795 genotype distribution was also similar across race and age groups after excluding 17–42 age groups in which no cases were observed.
Comprehensive statistical analysis regarding the distribution of samples across age and race was also performed to determine the bias or possible difference in the incidence of prostate cancer between the racial groups and for different age groups. The initial chi-square analysis indicated that the distribution of cases and controls did not differ across the Caucasian (53.3%) and African-American sample sets (46.7%, P = 0.73) (Table 1 and Figure 1B).
Sample set frequency of the rs1800795 polymorphism
The frequency of rs1800795 genotype in the normal sample set was consistent with that reported for other control populations (NCBI dbSNP database). In this study, the rs1800795 allele frequency also conformed to Hardy-Weinberg equilibrium in the Caucasian population (chi-square = 3.715, d.f. = 2, P = 0.156). A marked deviation from Hardy-Weinberg equilibrium in the African-American sample set was observed due to the lack of the CC genotype. The frequency distribution of rs1800795 in our complete sample set (all samples) and samples stratified by race are listed in Table 2. The IL-6 genotype frequencies of 24 CC, 48 GC and 68 GG for the Caucasian subpopulation and 6 CC, 30 GC and 102 GG for the African-American subpopulation showed a statically significant difference in the distribution of genotypes in the two racial groups (chi-square = 18.68, P = 0.0001).
Table 2.
Association of the SNP rs1800795 genotype with prostate cancer.
| Genotype | Cancer (%) | Control (%) | Total (%) | OR | 95%CI | P |
|---|---|---|---|---|---|---|
| GG | 108 (65.8) | 74 (52.8) | 182 (59.8) | 1.72 | 1.083 to 2.733 | 0.0257 |
| GC | 44 (26.8) | 44 (31.4) | 88 (28.9) | 0.8 | 0.486 to 1.315 | 0.446 |
| CC | 12 (7.3) | 22 (15.7) | 34 (11.1) | 0.4 | 0.201 to 0.890 | 0.02 |
| Total | 164 | 140 | 304 | |||
| Caucasian cancer | Caucasian control | |||||
| GG | 50 (59.5) | 26 (33.3) | 76 (46.9) | 2.941 | 1.548 to 5.587 | 0.001 |
| GC | 28 (33.3) | 30 (35.7) | 58 (35.8) | 0.7 | 0.401 to 1.463 | 0.51 |
| CC | 6 (7.1) | 22 (28.2) | 28 (17.2) | 0.19 | 0.074 to 0.514 | 0.0007 |
| Total | 84 | 78 | 162 | |||
| African-American cancer | African-American control | |||||
| GG | 58 (72.5) | 48 (77.4) | 106 (74.6) | 0.7 | 0.355 to 1.664 | 0.562 |
| GC | 16 (20) | 14 (22.5) | 30 (21.1) | 0.8 | 0.381 to 1.925 | 0.0836 |
| CC | 6 (7.5) | 0 | 6 (4.2) | 10.91 | 0.602 to 197.6 | 0.0354 |
| Total | 80 | 62 | 142 |
Genotype distribution is shown in all, Caucasian and African-American samples. Percentages of all genotypes, odds ratio, 95% confidence interval and P values are indicated. (OR= odds ratio; 95%CI = 95% confidence interval; Af-Am = African American; Cauc = Caucasian).
rs1800795 distribution in the sample and its association with prostate cancer
Each one of the genotypes was tested for its association with prostate cancer for all samples and in samples stratified by race. The combined cancer and control groups revealed that GG was the major (dominant) genotype (59.8%), whereas CC was a minor genotype (11.1%; Table 2). The odds of having cancer with the GG genotype was found to be 72% higher than the odds with CC and GC genotypes (OR= 1.72, P = 0.0257, Table 2). On the other hand, the odds of the heterozygous GC genotype was found to be nonsignificant as compared to the odds of CC+GC (OR = 0.8, P = 0.446; Table 2). Alternatively, the GG genotype was overrepresented in the cancer case group (66%) as compared to the control group (53%), while the CC genotype was overrepresented in the control group (15.7%) compared with the cancer group (7.3%) (OR = 0.4, P = 0.02; Table 2).
Association of rs1800795 with prostate cancer and race
Sample stratification demonstrated that rs1800795 genotype distribution is associated with race. The distribution of the GG genotype in the cancer case and the control group in the Caucasian subpopulation was 59.5 to 33% (OR = 2.9, P = 0.001) whereas in the African- American subpopulation the difference was not significant (72.5 to 77%, OR = 0.7, P = 0.562). The OR of cancer incidence in the Caucasian group with the GG genotype was 2.9 (P = 0.001), implying that the odds for the cancer case is 190% higher in Caucasians with the GG genotype. The sample showed evidence that the GC genotype was essentially similar in distribution in both the Caucasian (OR = 1.2; P = 0.51) and African-American (OR = 0.8; P = 0.08) subpopulations as in the cancer and control groups (OR = 0.8; P = 0.44) and hence was not a risk factor for prostate cancer (Table 2). Unlike in the Caucasian subset, the CC genotype was absent in the African-American control group but was observed in the cancer group (0 vs 7.5%, OR = 10.91, P = 0.03). The genotype distribution suggested that C allele was significantly underrepresented in the African-American group as compared to the Caucasian group (25.3 vs 53%). The distribution of G allele was converse and it was overrepresented in African-American group as compared to the Caucasian group (95.7 vs 82.7%) but had no association with cancer incidence (Table 2). No significant association with stage, Gleason grade or IL-6 rs1800795 was observed (data not shown).
Interaction between the rs1800795 genotype and other variables (age and race)
The rs1800795 genotype frequency was not significantly different across age groups (chi-square = 18, P = 0.09; Figure 1B). The distribution of the data by age group and race (Figure 1B) showed that Caucasians comprised 67% of the data in the younger age group, 17 to 65, while the African-American subgroup comprised 65% of the data in the older age group, 66 and above. The 17–42 age group consisted primarily of Caucasians (85%) and no cases (including African-Americans). This age group (17–42) was then excluded in the later stages of the statistical analysis and age 41 to 50 was considered as baseline. After filtering, the genotype frequency remained statistically nonsignificant across age group (chi-square = 8.65, P = 0.3; Figure 1A and B). This filtering was performed to eliminate the possible confounding factor due to the inclusion of lower age group samples where there were no cases available. As stated above, this exclusion did not alter the genotype frequency distribution. After filtering, 278 samples (cases and controls) were available for analysis.
We used the LLM, an extension of the chi-square test for independence in a two-way contingency table for MDR and to infer true associative structure among a set of many categorical variables such as disease status, genotype, race, and age. Unlike logistic regression, no distinction is made between dependent and independent variables in the LLM model. In our first analysis, four factors were targeted: cancer (case and control), IL-6 (genotype: CC, GC and GG), race (African-Americans and Caucasians), and age group (stratified: 43–55, 56–65, 66–75, and 76 and above). The age variable was originally a continuous variable but was collapsed into five categories: 17–42, 43–55, 56–65, 66–75, and 76 and above. Since empty cells are not allowed in LLM, we have excluded the age group 17–43 (the youngest group in the data) as the cancer case cell for this age group had no value. The rest of the data satisfied the basic assumptions of LLM.
In selecting an appropriate model, the relative quality of each competing model was measured by its goodness of fit to the data as tested by either the Pearson chi-square statistic or the likelihood ratio-statistic - both of which are distributed as chi-square random variables with n-p degrees of freedom where n is the number of cells and p is the number of parameters in the model. The maximum possible number of terms for the four factors is a four-way term, but, our goal was to find a model with as few terms as possible MDR. The result of the multi-term test for the four factors (NCSS 2007 software) is shown in Table 3. On the basis of the significance level (probability), we concluded that the third-order terms would be the highest that would be needed in our model (P = 0.008), since there is lack of significance for a four-way table (P = 0.2186; Table 3). A three-way analysis includes three-way interaction, two-way interaction and single variable main effects also. We then performed a three-term LLM using a single-term test up to the third order (Table 3). The partial chi-square method was used to determine if a given term was necessary for achieving statistical significance. The partial chi-square statistic tests whether the term is significant after considering all other terms of the same order. The analysis in Table 4 shows that two three-way terms, ABC and ACD (A = cancer, B = IL-6, C = race, D = age group) were highly significant (Table 4), which was subsequently confirmed by the (NCSS 2007 software) step-down search algorithm (data not shown).
Table 3.
Log-linear model analysis for the assessment of main effects and all possible levels of interactions of the categorical variables - cancer status, race, genotype, and age group.
| Main effect and interactions | d.f. | Likelihood ratio, χ2 | P level |
|---|---|---|---|
| One-way and higher | 47 | 259.15 | 0 |
| Two-way and higher | 40 | 114.33 | 0 |
| Three-way and higher | 23 | 50.57 | 0.0008 |
| Four-way and higher | 6 | 8.28 | 0.2186 |
Table 4.
Main effects and up to three-way interaction detailed results for the four categorical variables (disease, genotype, race, and age).
| Main interactions | d.f. | Partial χ2 | P level |
|---|---|---|---|
| A (Cancer) | 1 | 8.19 | 0.0042 |
| B (IL-6) | 2 | 100.02 | 0 |
| C (Race) | 1 | 0.01 | 0.9091 |
| D (Age) | 3 | 36.61 | 0 |
| AB | 2 | 7.75 | 0.0207 |
| AC | 1 | 0.05 | 0.8247 |
| AD | 3 | 10.81 | 0.0128 |
| BC | 2 | 16.3 | 0.0003 |
| BD | 6 | 8.21 | 0.2234 |
| CD | 3 | 17.57 | 0.0005 |
| ABC | 2 | 10.61 | 0.005* |
| ABD | 6 | 9.82 | 0.1322 |
| ACD | 3 | 12.34 | 0.0063* |
| BCD | 6 | 7.42 | 0.2837 |
Values in bold mean significant three-factor interactions.
From Table 4, the two-way terms AC (distribution of case-control over race) and BD (IL- 6 and age group) were not significant, as expected. The other four two-factor interactions (AB, AD, BC, and CD) were significant but interpretations of LLM parameters refer to their highest order estimates where the main effects estimates (A, B, C, D) become irrelevant and the two-factor interactions represent only partial associations. On the basis of these results, we selected the best models and constructed tables for each of the significant three-factor interactions, ACD and ABC, and examined how the OR between any two variables varied across levels of the third variable.
The interactions between cancer incidence (A), genotype (B) and race (C) based on LLM as discussed above are shown in Table 5. From these results, we concluded that for African- Americans with genotype CC the odds of being diagnosed with cancer was 1700% (OR = 18) times more than Caucasians with the same CC genotype. On the other hand, the odds of prostate cancer incidence for an African-American with genotype GG was 52.5% (OR = 0.475) less than Caucasians with the GG genotype. Therefore, genotype CC is protective in the Caucasian population, whereas it is a risk factor in the African population. This role is interchanged for the GG genotype, where it is protective in the African-American population and a risk factor in the Caucasian population (Table 5). The genotype GC was found to be neutral for both races.
Table 5.
Odds ratio for IL-6 genotypes associated with race.
| IL-6 genotype | Race | Cancer (N) | Control (N) | OR | 95%CI lower | 95%CI upper |
|---|---|---|---|---|---|---|
| CC | African-American | 6 | 1 | 18.0 | 1.787 | 181.318 |
| Caucasian | 6 | 18 | ||||
| CG | African-American | 16 | 14 | 0.816 | 0.326 | 2.045 |
| Caucasian | 28 | 20 | ||||
| GG | African-American | 58 | 44 | 0.475 | 0.244 | 0.924 |
| Caucasian | 50 | 18 |
Reference: IL-6 genotype of Caucasians = 1.
DISCUSSION
In this study, we found an association between IL-6 promoter (-174 G/C) polymorphism and prostate cancer. The results are somewhat surprising when viewed in the context of samples stratified with race. Whereas the GG genotype was associated with increased risk of prostate cancer in Caucasian subjects, it was the CC genotype that was associated with increased risk in the African-American sample set. Such a dimorphic genotypic association with cancer and race is unique and suggests a complex gene-gene and gene-environment interaction.
In the African-America group, the CC genotype was absent in the control subjects and observed only in the cancer samples. This genotype distribution clearly demonstrates that the CC genotype is a risk factor for prostate cancer in African-Americans. The strong association of the CC genotype with prostate cancer is overall consistent with other population-based studies, which further supports the significance of this genotype as a risk factor for prostate cancer. The GG genotype is almost but not completely monomorphic in south Indians, Hubei Chinese and African-Americans. Thus, the lack of the CC genotype in the African-American control population is not surprising.
The results from our complete data set in general and Caucasian subjects in particular, demonstrating the association of the GG genotype with prostate cancer, differ from those of previous studies. In a study by Tan et al. (2005), a strong association between the C allele with various prostate cancer disease modalities was observed, although the ethnic identity of the subjects was not reported. A modest association between the C allele and prostate cancer in the Caucasian population was also observed in another study (Pierce et al., 2009). However, our data from the African-American sample set that showed a strong association of the CC genotype are consistent with these studies. The C allele was also shown to have a modest association with prostate cancer in African-American subjects (Zabaleta et al., 2008; Pierce et al., 2009). In the north Indian population, the “C” allele was also associated with a 2-fold risk of occurrence of metastasis in prostate cancer patients (Kesarwani et al., 2008). Although we did not specifically investigate statistical association between stage/grade of prostate cancer due to lack of this information for all subjects, all the African-American samples with the “C” allele were with Gleason grade 7 or higher, suggesting that this allele is associated with higher grade/metastatic prostate cancer at least in African-Americans. In African-Americans, prostate cancer is often more aggressive and highly metastatic. We speculate that the -174CC genotype in African-Americans could be a strong predictor of aggressive metastatic disease, whereas the GG genotype in this racial group could suggest a less aggressive disease, similar to ovarian and peritoneal carcinoma, in which the -174GG genotype has a favorable impact on survival (Garg et al., 2006).
No association between the -174G>C variant with prostate cancer risk, overall survival or transcriptional activity was observed in the Risk Factor for Prostate Cancer (RFPC) and Melbourne Collaborative Cohort Study (MCCS). Interestingly, this study reported a strong association between the -6331 “C” allele and prostate cancer in the RFPC study but not in the MCCS study, clearly suggesting the influence of demographics and/or gene-environment interaction on the outcome (Tindall et al., 2012).
IL-6, a pro-inflammatory cytokine, promotes chronic inflammation, which ultimately leads to proliferative inflammatory atrophy, the earliest known pre-neoplastic lesion in the prostate. Studies have also shown that IL-6 promotes androgen independence by promoting androgen synthesis in prostate cancer cells in an autocrine manner (Culig, 2003). Thus, the finding of increased IL-6 levels in prostate cancer supports the pro-tumorigenic role of IL-6. However, similar studies demonstrating increased serum IL-6 have been controversial, suggesting a more localized role of IL-6 in prostate cancer.
The -174G/C polymorphism in IL-6 promoter has been linked to its transcriptional activity also. Both C (Terry et al., 2000) and G (Fishman et al., 1998) alleles have been shown to promote IL-6 transcription in in vitro luciferase-based assays. The discrepancies between the effect of the -174G/C polymorphism on IL-6 transcription can be due to assembly or cooperativity with other regulatory complexes around this region in a cell type-specific manner that could influence IL-6 transcriptional activity (Fishman et al., 1998; Terry et al., 2000). It is therefore not surprising that both PC3 and DU145 cells secrete similar amounts of IL-6 in spite of different genotypes (GC and GG, respectively).
The strong association between the GG genotype and prostate cancer in Caucasian subjects is an unexpected finding as noted above. The rs1800795 (G) allele frequency is also higher in gastric cancer patients than in patients with chronic gastritis in a Brazilian population (Gatti et al., 2007). In a Caucasian study, the GG genotype is also a risk factor for type 2 diabetes (Vaxillaire et al., 2008), new onset of diabetes and higher C-reactive protein levels as compared to the CC genotype (Bamoulid et al., 2006). Metabolic disorders such as diabetes are also linked to increased risk of prostate cancer (Grossmann and Wittert, 2012). Higher C-reactive protein levels are also related to increased PSA and prostate cancer, further supporting the role of chronic inflammation in prostate cancer etiology (Eklund et al., 2009). The GG genotype has also been correlated with a higher level of IL-6 among patients with chronic obstructive pulmonary disease but not among control subjects. Patients carrying the IL-6 GG genotype have also shown higher serum levels of IL-6 and higher Pap compared to those carrying the CC or CG genotype (Eddahibi et al., 2006). Although serum IL-6 levels and their association with prostate cancer is debatable (Pierce et al., 2009), an association with IL-6 levels in prostate cancer tissue and genotype would most likely be required to strongly associate expression with genotype. We tried to analyze our data to find the association between cancer stage, Gleason score and the rs1800795 genotype. Although the majority of our patients with higher cancer stage and Gleason score had the GG genotype, this was not found to be statistically significant. The reason for the above finding could be due to non-homogeneity of our sample set, which renders the effects of such attributes variable across the data set, and possibly because of the differential effect we saw in terms of genotype association in the two ethnic groups studied.
In conclusion, our data support the role of the IL-6 -174 polymorphism in prostate cancer, but the association differs between the two ethnic populations studied. The prevalence of the -174G allele in the African-American population may contribute to the overall increased risk of prostate cancer, but the C allele could be associated with a more aggressive disease and poor survival. Although an individual’s risk of developing prostate cancer can be partly predicted on the basis of IL-6 genetic variants, complex gene-gene interactions, environmental factors, and epigenetic mechanisms are also major contributors to prostate cancer heterogeneity and incidence disparities. Our results clearly demonstrate that race, prostate cancer and age are confounders in relation to the IL-6 -174 polymorphism.
Acknowledgments
Research supported by NIH/NCMHD (#P20MD002285-01) and by NIH/NCRR/RCMI (#G12RR03062) in part for core facilities and additional resources. The authors wish to thank Ms. Qi Yang, DNA Sequencing Laboratory, Morehouse School of Medicine, Atlanta, GA, and the Research Cores at Morehouse School of Medicine supported through the Research Centers at Minority Institutions (RCMI) Program, NIH/NCRR/RCMI (Grant #G12-RR03034).
Footnotes
Conflicts of interest
The authors declare no conflict of interest.
References
- Bamoulid J, Courivaud C, Deschamps M, Mercier P, et al. IL-6 promoter polymorphism -174 is associated with new-onset diabetes after transplantation. J Am Soc Nephrol. 2006;17:2333–2340. doi: 10.1681/ASN.2006010066. [DOI] [PubMed] [Google Scholar]
- Cole SW, Arevalo JMG, Takahashi R, Sloan EK, et al. Computational identification of gene-social environment interaction at the human IL6 locus. Proc Natl Acad Sci U S A. 2010;107:5681–5686. doi: 10.1073/pnas.0911515107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corvol H, De Giacomo A, Eng C, Seibold M, et al. Genetic ancestry modifies pharmacogenetic gene-gene interaction for asthma. Pharmacogenet Genomics. 2009;19:489–496. doi: 10.1097/FPC.0b013e32832c440e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culig Z. Role of the androgen receptor axis in prostate cancer. Urology. 2003;62:21–26. doi: 10.1016/s0090-4295(03)00698-8. [DOI] [PubMed] [Google Scholar]
- Culig Z, Steiner H, Bartsch G, Hobisch A. Interleukin-6 regulation of prostate cancer cell growth. J Cell Biochem. 2005;95:497–505. doi: 10.1002/jcb.20477. [DOI] [PubMed] [Google Scholar]
- Cussigh A, Falleti E, Fabris C, Bitetto D, et al. Interleukin 6 promoter polymorphisms influence the outcome of chronic hepatitis C. Immunogenetics. 2011;63:33–41. doi: 10.1007/s00251-010-0491-7. [DOI] [PubMed] [Google Scholar]
- Eddahibi S, Chaouat A, Tu L, Chouaid C, et al. Interleukin-6 gene polymorphism confers susceptibility to pulmonary hypertension in chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2006;3:475–476. doi: 10.1513/pats.200603-038MS. [DOI] [PubMed] [Google Scholar]
- Eklund CM, Tammela TL, Schleutker J, Hurme M. C-reactive protein haplotype is associated with high PSA as a marker of metastatic prostate cancer but not with overall cancer risk. Br J Cancer. 2009;100:1846–1851. doi: 10.1038/sj.bjc.6605081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falleti E, Fabris C, Vandelli C, Colletta C, et al. Genetic polymorphisms of interleukin-6 modulate fibrosis progression in mild chronic hepatitis C. Hum Immunol. 2010;71:999–1004. doi: 10.1016/j.humimm.2010.06.006. [DOI] [PubMed] [Google Scholar]
- Fishman D, Faulds G, Jeffery R, Mohamed-Ali V, et al. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest. 1998;102:1369–1376. doi: 10.1172/JCI2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg R, Wollan M, Galic V, Garcia R, et al. Common polymorphism in interleukin 6 influences survival of women with ovarian and peritoneal carcinoma. Gynecol Oncol. 2006;103:793–796. doi: 10.1016/j.ygyno.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti LL, Burbano RR, Zambaldi-Tunes M, de-Labio RW, et al. Interleukin-6 polymorphisms, Helicobacter pylori infection in adult Brazilian patients with chronic gastritis and gastric adenocarcinoma. Arch Med Res. 2007;38:551–555. doi: 10.1016/j.arcmed.2006.12.011. [DOI] [PubMed] [Google Scholar]
- Giri D, Ittmann M. Interleukin-8 is a paracrine inducer of fibroblast growth factor 2, a stromal and epithelial growth factor in benign prostatic hyperplasia. Am J Pathol. 2001;159:139–147. doi: 10.1016/S0002-9440(10)61681-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri D, Ozen M, Ittmann M. Interleukin-6 is an autocrine growth factor in human prostate cancer. Am J Pathol. 2001;159:2159–2165. doi: 10.1016/S0002-9440(10)63067-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy-Tundidor S, Cavarretta IT, Fuchs D, Fiechtl M, et al. Interleukin-6 and oncostatin M stimulation of proliferation of prostate cancer 22Rv1 cells through the signaling pathways of p38 mitogen-activated protein kinase and phosphatidylinositol 3-kinase. Prostate. 2005;64:209–216. doi: 10.1002/pros.20235. [DOI] [PubMed] [Google Scholar]
- Grossmann M, Wittert G. Androgens, diabetes and prostate cancer. Endocr Relat Cancer. 2012;19:F47–F62. doi: 10.1530/ERC-12-0067. [DOI] [PubMed] [Google Scholar]
- Hobisch A, Ramoner R, Fuchs D, Godoy-Tundidor S, et al. Prostate cancer cells (LNCaP) generated after long-term interleukin 6 (IL-6) treatment express IL-6 and acquire an IL-6 partially resistant phenotype. Clin Cancer Res. 2001;7:2941–2948. [PubMed] [Google Scholar]
- Kesarwani P, Ahirwar DK, Mandhani A, Mittal RD. Association between -174 G/C promoter polymorphism of the interleukin-6 gene and progression of prostate cancer in North Indian population. DNA Cell Biol. 2008;27:505–510. doi: 10.1089/dna.2008.0742. [DOI] [PubMed] [Google Scholar]
- Knupfer H, Preiss R. sIL-6R: more than an agonist? Immunol Cell Biol. 2008;86:87–91. doi: 10.1038/sj.icb.7100113. [DOI] [PubMed] [Google Scholar]
- Lagmay JP, London WB, Gross TG, Termuhlen A, et al. Prognostic significance of interleukin-6 single nucleotide polymorphism genotypes in neuroblastoma: rs1800795 (promoter) and rs8192284 (receptor) Clin Cancer Res. 2009;15:5234–5239. doi: 10.1158/1078-0432.CCR-08-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menashe I, Rosenberg PS, Chen BE. PGA: power calculator for case-control genetic association analyses. BMC Genet. 2008;9:36. doi: 10.1186/1471-2156-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ol KK, Agachan B, Gormus U, Toptas B, et al. Cox-2 gene polymorphism and IL-6 levels in coronary artery disease. Genet Mol Res. 2011;10:810–816. doi: 10.4238/vol10-2gmr967. [DOI] [PubMed] [Google Scholar]
- Pereira DS, Garcia DM, Narciso FM, Santos ML, et al. Effects of 174 G/C polymorphism in the promoter region of the interleukin-6 gene on plasma IL-6 levels and muscle strength in elderly women. Braz J Med Biol Res. 2011;44:123–129. doi: 10.1590/s0100-879x2010007500152. [DOI] [PubMed] [Google Scholar]
- Pierce BL, Biggs ML, DeCambre M, Reiner AP, et al. C-reactive protein, interleukin-6, and prostate cancer risk in men aged 65 years and older. Cancer Causes Control. 2009;20:1193–1203. doi: 10.1007/s10552-009-9320-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas A, Liu G, Coleman I, Nelson PS, et al. IL-6 promotes prostate tumorigenesis and progression through autocrine cross-activation of IGF-IR. Oncogene. 2011;30:2345–2355. doi: 10.1038/onc.2010.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shariat SF, Andrews B, Kattan MW, Kim J, et al. Plasma levels of interleukin-6 and its soluble receptor are associated with prostate cancer progression and metastasis. Urology. 2001;58:1008–1015. doi: 10.1016/s0090-4295(01)01405-4. [DOI] [PubMed] [Google Scholar]
- Slattery ML, Wolff RK, Herrick JS, Caan BJ, et al. IL6 genotypes and colon and rectal cancer. Cancer Causes Control. 2007;18:1095–1105. doi: 10.1007/s10552-007-9049-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery ML, Wolff RK, Herrick J, Caan BJ, et al. Tumor markers and rectal cancer: support for an inflammation-related pathway. Int J Cancer. 2009;125:1698–1704. doi: 10.1002/ijc.24467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D, Wu X, Hou M, Lee SO, et al. Interleukin-6 polymorphism is associated with more aggressive prostate cancer. J Urol. 2005;174:753–756. doi: 10.1097/01.ju.0000168723.42824.40. [DOI] [PubMed] [Google Scholar]
- Terry CF, Loukaci V, Green FR. Cooperative influence of genetic polymorphisms on interleukin 6 transcriptional regulation. J Biol Chem. 2000;275:18138–18144. doi: 10.1074/jbc.M000379200. [DOI] [PubMed] [Google Scholar]
- Tindall EA, Severi G, Hoang HN, Southey MC, et al. Interleukin-6 promoter variants, prostate cancer risk, and survival. Prostate. 2012;72:1701–1707. doi: 10.1002/pros.22557. [DOI] [PubMed] [Google Scholar]
- Vaxillaire M, Veslot J, Dina C, Proenca C, et al. Impact of common type 2 diabetes risk polymorphisms in the DESIR prospective study. Diabetes. 2008;57:244–254. doi: 10.2337/db07-0615. [DOI] [PubMed] [Google Scholar]
- Zabaleta J, Lin HY, Sierra RA, Hall MC, et al. Interactions of cytokine gene polymorphisms in prostate cancer risk. Carcinogenesis. 2008;29:573–578. doi: 10.1093/carcin/bgm277. [DOI] [PubMed] [Google Scholar]
- Zhang J, Wheeler DA, Yakub I, Wei S, et al. SNPdetector: a software tool for sensitive and accurate SNP detection. PLoS Comput Biol. 2005;1:e53. doi: 10.1371/journal.pcbi.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]