Summary
Isolating spliceosomes at a specific assembly stage requires a means to stall or enrich for one of the intermediate splicing complexes. We describe strategies to arrest spliceosomes at different points of complex formation and provide a detailed protocol developed for isolating intact splicing complexes arrested between the first and second chemical steps of splicing. Briefly, spliceosomes are assembled on a radiolabeled in vitro-transcribed splicing substrate from components present in nuclear extract of HeLa cells. Spliceosome progression is arrested after the first step of splicing chemistry by mutating the pre-mRNA substrate at the 3′ splice site. The substrate also contains binding sites for the MS2 protein, which serve as an affinity tag. Purification of arrested spliceosomes is carried out in two steps: (1) size exclusion chromatography and (2) affinity selection via a fusion of MS2 and maltose binding protein (MBP). Complex assembly and purification are analyzed by denaturing poly-acrylamide gel electrophoresis.
Keywords: spliceosome, affinity purification, pre-mRNA splicing, MS2:MBP, nuclear extract, size exclusion
1. Introduction
The spliceosome is a large macromolecular machine responsible for removing introns in a process known as pre-mRNA splicing. It forms on each intron from over one hundred components including five nuclear ribonucleoproteins (U1, U2, U4, U5, and U6 snRNPs) and many non-snRNP proteins. Spliceosome assembly occurs in a stepwise manner through a series of intermediate splicing complexes that are characterized by their associated components and chemical state of the intron [1]. Briefly, U1 snRNP base pairs with the 5′ splice site in E complex and recruits U2 snRNP. In an ATP-dependent step U2 snRNP stably base pairs with the branchpoint sequence to form A complex. The addition of tri-snRNP (U5:U4/U6) and Prp19 complex leads to B complex. Several ATP-dependent rearrangements between RNA/RNA and RNA/protein interactions result in loss of U1 and U4 snRNPs and ready the spliceosome for catalysis as Bact complex forms. Additional rearrangements lead to B* complex and first step splicing chemistry in which the 2′ OH of the branchpoint adenosine in the intron attacks the phosphate bond at the 5′ splice site. This reaction leads to cleavage at the 5′ end of the intron and formation of lariat structure. Additional rearrangements and addition of proteins form C complex leads to second step chemistry where the 5′ OH of the upstream exon attacks at the 3′ splice site. This reaction cleaves the 3′ end of the intron and ligates the flanking exons. The resulting mRNA and lariat intron are then released from P complex.
Splicing can be recapitulated in vitro using a model pre-mRNA and cellular extract [2,3]. However, the dynamic nature of spliceosome assembly creates a challenge for capturing intermediate splicing complexes for further biochemical and structural studies. In S. cerevisiae, spliceosomes can be arrested and purified at specific stages by genetically manipulating proteins that are required for the next step in assembly [4,5]. In the human system, spliceosome assembly has been stalled by a variety of means including withholding ATP from the reaction, depleting or inactivating snRNPs with antisense oligonucleotides and manipulating the pre-mRNA substrate [6-14,2,15,3,16-18]. The latter provides the most efficient method to capture spliceosomes at distinct points of splicing catalysis. A pre-mRNA with a polypyrimidine tract less than 10 nt truncated before the 3′ splice site will accumulate Bact complex at a point before first step chemistry [18]. C complex can be stalled after first step chemistry on a pre-mRNA with a polypyrimidine tract more than 20 nt that is either truncated before the 3′ splice site [7] or that contains a 3′ splice site mutation [11,19]. Shortening the 3′ exon to less than 25 nt allows accumulation of P complex containing the unreleased splicing products (unpublished results, J.I, M.J.).
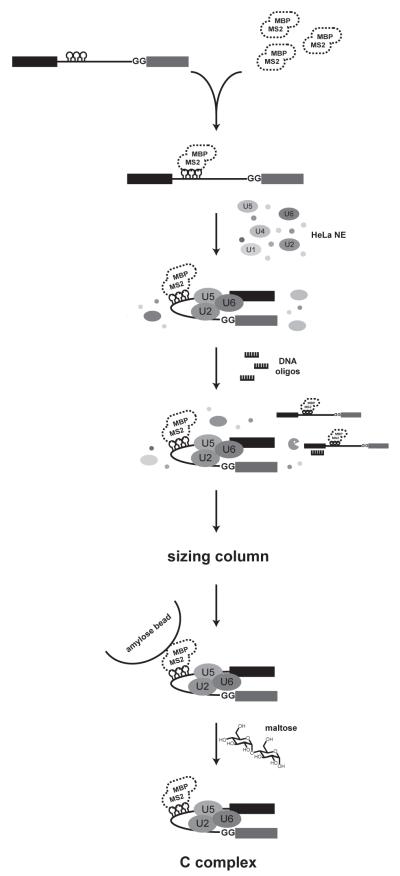
To purify stalled splicing complexes, the pre-mRNA can be further modified to incorporate an affinity tag. The most commonly used tag consists of three RNA hairpins containing the recognition sequence for the bacteriophage MS2 coat protein. These hairpins serve as a handle for amylose affinity selection by a fusion of MS2 to maltose binding protein (MBP), and this strategy has been extensively used to isolate and characterize splicing complexes [20,7,21-23,19,24-27,18]. In the protocol outlined below we detail conditions to assemble C complex spliceosomes in human nuclear extract and isolate the complexes by MS2:MBP affinity purification (Figure 1). The procedure may also be applied to spliceosome complexes stalled at other assembly intermediates.
Figure 1.
Schematic of MS2:MBP affinity purification of C complex spliceosomes with tagged pre-mRNA substrate.
2. Materials
To prevent contamination by RNases, all materials and equipment should be handled with gloves. All reagents should be prepared with RNase free water (see Note 1). Important: Radioactive materials should only be handled by authorized users with protective shielding, proper monitoring and appropriate attire in compliance with all state and federal regulations. Follow proper waste disposal procedures for all chemicals and radioisotopes.
2.1 In vitro transcription
1. DNA template: Linearized plasmid or PCR product containing a T7 promoter sequence followed by pre-mRNA sequence. (see Note 2).
2. Radioactive nucleotide: [α-32P] UTP at 3,000 Ci/mMol (see Note 3).
3. 5X transcription buffer (usually supplied with T7 polymerase): 200 mM Tris (pH 7.6-8.0), 30-40 mM MgCl2, 10 mM spermidine, 0-250 mM NaCl.
4. Nucleotide stocks: 10 mM ATP, 10 mM CTP, 10 mM GTP, 10 mM UTP.
5. CAP analog: 10 mM G(5′)ppp(5)′G (NEB).
6. 1 M diethiothreitol (DTT).
7. T7 RNA polymerase.
8. RNase Inhibitor (optional).
2.2 Denaturing polyacrylamide gel for pre-mRNA purification and analysis of splicing
1. 1X TBE: 0.09 M Tris-borate, 0.09 M Boric acid, 0.0025 M EDTA. Store at room temperature.
2. 15% denaturing polyacrylamide solution: 7 M Urea, 15% acrylamide (from AccuGel acrylamide solution 40% (w/v) 29:1 Acrylamide:Bis-acrylamide), 1X TBE. Store at 4°C.
3. 0.8 mm spacers and comb for thick gel and 0.4 mm spacers and comb for thin gel.
4. 20 × 27 cm glass plates, one should be notched to fit gel rig (Moliterno).
5. Large binder clips (2 inches).
6. Electrophoresis gel rig (Dan-Kar).
7. High voltage (>3000V) power supply.
8. RNA gel loading buffer: 95% formamide, 20 mM EDTA, 0.01% (w/v) bromophenol blue, 0.01% (w/v) xylene cyanol. Store at −20°C.
9. Gel extraction buffer: 0.3 M NaAc (pH 4.8), 1 mM EDTA, 10% phenol (pH 4.5). Store at 4°C.
2.3 Assembly of spliceosome complex
1. HeLa nuclear extract (see Note 4).
2. 1M glutamic acid monopotassium salt (KGlu, pH 7.5) (see Note 5).
3. 100 mM magnesium acetate (MgAc).
4. 100 mM ATP.
5. 250 mM creatine phosphate (CP).
6. 5 mg/mL yeast tRNA.
7. RNase Inhibitor.
8. Pre-mRNA transcript from in vitro transcription.
9. DNA oligonucleotides for RNase H digestion (see Note 6).
10. 10 mg/mL heparin.
11. 10-50 μM purified MS2-MBP protein (see Note 7).
2.4 Purification of spliceosomes
1. Sephacryl S-400 (GE Healthcare).
2. 1.0 × 10-cm glass column with stop-cock valve.
3. Amylose resin (NEB).
4. Mobicol spin column with small 35 μm filter.
5. Sizing column buffer (SCB-N): 150 mM KCl, 20 mM Tris-HCl (pH 7.9 at 4°C), 5 mM EDTA, 1 mM DTT, 0.5% NP-40. Make fresh for each spliceosome purification. Store at room temperature. (see Note 8)
6. Amylose column buffer (ACB): 150 mM KCl, 20 mM Tris-HCl (pH 7.9 at 4°C), 5 mM EDTA, 1 mM DTT. Make fresh for each spliceosome purification. Keep on ice.
7. Elution buffer: 150 mM KCl, Tris-HCl (pH 7.9 at 4°C), 5 mM EDTA, 1 mM DTT, 10 mM maltose.
8. Splicing dilution buffer: 100 mM Tris-HCl (pH 7.5), 10 mM EDTA, 1% SDS, 150 mM NaCl, 0.3 M NaAc (pH 4.8). Store at room temperature.
9. Phenol:chloroform:isomyl alcohol (25:24:1, pH 4.5).
3. Methods
3.1 In vitro transcription
A typical transcription reaction contains: 1X transcription buffer, 400 μM ATP, 400 μM CTP, 400 μM UTP, 200 μM GTP, 800 μM Cap analog, 40 ng linearized plasmid DNA template / μL of reaction (or 10-100 ng PCR product template), 1/10th volume [α-32P] UTP and 1/10th volume T7 RNA polymerase. (see Notes 2-3) A 50 μL reaction usually generates enough pre-mRNA transcript for two to three spliceosome preparations. Keep all reagents on ice unless specified.
1. To prepare a 50 μL transcription reaction, mix in order the following ingredients in a 1.5 mL microcentrifuge tube at room temperature: 17.5 μL water, 10 μL 5X transcription buffer, 2 μL 10 mM ATP, 2 μL 10 mM CTP, 2 μL 10 mM UTP, 1 μL 10 mM GTP, 1 μL 1M DTT, 4.5 μL CAP analog, 3 μL [α-32P] UTP, 2 μL 1 mg/mL linearized plasmid DNA template, 5 μL T7 RNA polymerase. Mix gently.
2. Dilute 1 μL of the reactions mixture into water and set aside. This will be used to calculate transcript concentration the next day in step 11.
3. Incubate transcription reaction at 37°C for 2-4 hrs.
4. During the incubation time pour a thick denaturing 5% polyacrylamide gel. Place 0.8 mm spacers between 20 × 27 cm glass plates and secure with large binder clips. Seal the bottom of the gel with tape or an additional spacer. In a 50 mL conical tube mix 15 mL 15% denaturing polyacrylamide solution and 30 mL 7 M urea in 1X TBE. Add 135 μL 20% ammonium persulfate and 45 μL TEMED just before pouring the gel. Insert a 0.8 mm comb with wells that can hold up to 60 μL sample. Let gel polymerize for at least 20-30 minutes on bench top.
5. If PCR product is used as a DNA template, following the reaction incubation add 1 μL RQ1 DNase to the reaction and incubate for an additional 20 minutes at 37°C. Otherwise skip to step 6.
6. Add 50 μL of RNA gel loading buffer to transcription reaction and set aside at room temperature.
7. Remove the bottom seal of the polymerized gel and clamp into an electrophoresis rig with an aluminum heat sink plate. Fill the top and bottom chambers with 1X TBE and be sure to remove any air bubbles at the bottom of the gel. Remove the comb and extensively rinse the wells with buffer using a syringe. Hook up the leads to a high voltage power supply and run the gel at constant wattage of 45 W for 20 minutes to pre-warm the gel. Meanwhile, heat samples at 95°C for 2 minutes and place on ice. Before loading the gel, rinse the wells again. Load 50 μL sample each into to neighboring lanes and run gel for 1 hour at constant wattage of 45 W.
8. Carefully take down the gel. Note that most of the unincorporated radioactive nucleotides will be in the bottom chamber buffer. Remove one of the glass plates and cover the gel supported by the other glass plate with plastic wrap. Place glow in the dark stickers on top of plastic wrap to orient the gel after exposure to film. Expose the gel for 1-2 minutes to X-ray film.
9. Using the X-ray film as a guide, cut out the transcript bands with a clean razor blade and transfer to 1.5 mL microcentrifuge tube. Add 400 μL gel extraction buffer and freeze tubes at −80°C for 10-20 minutes. Rotate tubes overnight at room temperature.
10. Next day, transfer gel extraction buffer with extracted transcript to 1.5 mL microcentrifuge tube and add 1 mL 100% ethanol. Discard the gel. Invert the tube a few times to mix and incubate at −80°C for 30 minutes. Centrifuge the tube at 14,000 rpm for 30 minutes at 4°C to pellet the transcript. Remove ethanol and wash the pellet with 100 μL 70% ethanol. Remove ethanol and let the pellet air dry. Resuspend pellet in 50 μL water and store at −20 °C.
11. To quantify the transcript, mix 1 μL with 3 mL of scintillation fluid in a scintillation tube. Repeat with 1 μL of the 1:100 reaction dilution from the previous day. Measure counts with a scintillation counter. Determine the transcript concentration with the following calculation: (cpm of transcript × nmoles of cold UTP in reaction × 104) / (# of U’s in transcript × reaction volume × 100 × cpm of reaction) = concentration of transcript in nM. Dilute the pre-mRNA transcript to 200 nM with water.
3.2 Spliceosome assembly
A typical splicing reaction contains: 1-10 nM pre-mRNA splicing substrate, 0-100 mM KGlu, 0-6 mM MgAc, 2 mM ATP, 5 mM CP, 0.1-0.5 mg/mL tRNA, and 40% HeLa nuclear extract. (see Notes 4-5) A 1 mL splicing reaction will generate 0.1-0.5 pmol spliceosomes depending on reaction efficiency and RNA degradation in the nuclear extract. Keep all reagents on ice unless specified.
1. For a 1 mL splicing reaction transfer 50 μL 200 nM pre-mRNA transcript into a microcentrifuge tube and heat at 95°C for 1 minute and then place on ice. Add 50 fold molar excess MS2:MBP fusion protein and incubate on ice for 5 minutes. For 50 μM MS2:MBP, this is 10 μL.
2. In a separate 1.5 mL microcentrifuge tube, mix in order 410 μL water, 60 μL 1M KGlu, 20 μL 100 mM MgAc, 20 μL 100 mM ATP, 20 μL 250 mM CP, and 10 μL 5 mg/mL tRNA (see Note 4). Mix this with the pre-mRNA and MS2:MBP and then add 400 μL HeLa nuclear extract. Splicing efficiency may be increased by splitting 100-200 μL of the reaction into separate 1.5 mL microcentrifuge tubes. Take a 10 μL aliquot into a new 1.5 mL microcentrifuge tube for a zero time point and save on ice.
3. Incubate splicing reaction at 30°C for 60 minutes. Take a 10 μL aliquot into a new 1.5 mL microcentrifuge tube for a 60′ time point and save on ice. If the splicing reaction was split into multiple tubes, combine them back into one tube at this point.
4. To digest excess unspliced pre-mRNA add 10 μL 100 μM DNA oligonucleotides for RNase H digestion to splicing reaction (see Note 6). Incubate at 30°C for an additional 20 minutes. Take a 10 μL aliquot into a new 1.5 mL microcentrifuge tube for an 80′ time point and save on ice.
5. Add 25 μL 10 mg/mL heparin to splicing reaction. Incubate at 30°C for 5 minutes and then transfer the splicing reaction to ice. (see Note 9)
3.3 Spliceosome purification
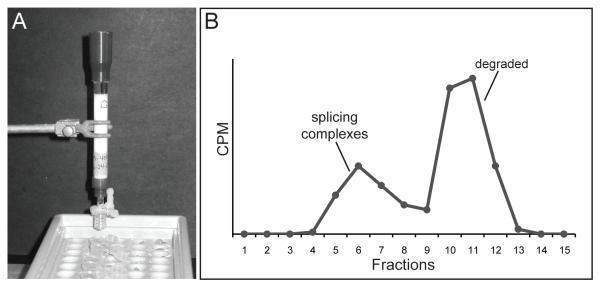
1. Prior to carrying out the purification pour a 5 mL sizing column of S-400 resin equilibrated in SCB-N into a 1.0 × 10 cm glass column (Figure 2A). Allow the resin to settle by gravity flow. This sizing column can be used multiple times by washing with 10 mL of SCB-N before each use.
Figure 2.
A. Image of size exclusion column. B. Representative analysis of sizing column fractions. Average cpm is plotted versus fraction number.
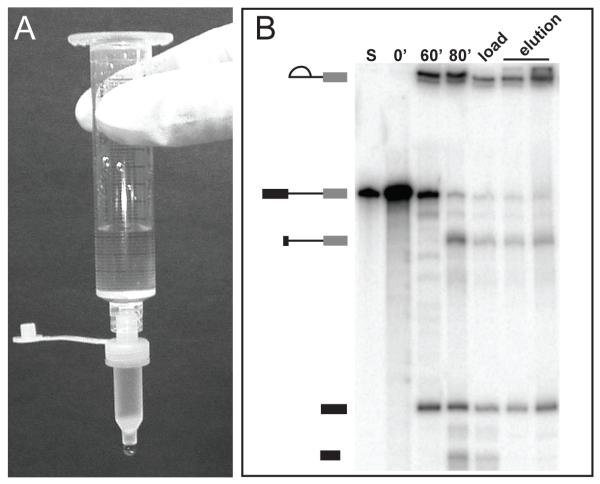
2. During the splicing reaction incubation prepare an amylose column. Fit a small 35 um filter into a Mobicol column. Add 100 μL of amylose resin equilibrated in ACB into the column (Figure 3A- see Note 10). Let resin settle by gravity. To get the column flowing a brief spin in a centrifuge at low speed may be necessary. Keep the column in a 1.5 mL microcentrifuge tube on ice.
Figure 3.
A. Image of amylose column attached to a syringe for washing. B. Denaturing PAGE analysis of RNA from in vitro splicing and affinity purification of C complex spliceosome. Lanes from left to right are the pre-mRNA standard used for quantification (S), time points taken during the splicing reaction (0 and 60 minutes) and after RNase H digestion (80 minutes), size exclusion peak fraction loaded onto amylose column (load), elution fractions from amylose column (elution). RNA species schematized on the left are, from top to bottom, lariat intermediate, pre-mRNA, 3′ RNase H digestion product, 5′ exon, and 5′ RNase H digestion product.
3. To start the purification, let buffer flow by gravity from the sizing column until the top of the resin bed is exposed. Carefully load the splicing reaction onto the sizing column being sure to not disturb the resin bed. Let the sample run into the column then load 500 μL SCB-N onto the resin bed and let it run into the column. Run an additional 10 mL of SCB-N through the column and collect 500 μL fractions on ice.
4. Use a Geiger counter to measure average cpm for each fraction. There should be two peaks of radioactivity (Figure 2B). The first peak is smaller and usually contained within the first 8 fractions and contains splicing complexes. The second peak is larger and contains degraded pre-mRNA transcript. Take a 10 μL aliquot from the first peak into a new 1.5 mL microcentrifuge tube and save on ice. Pool fractions from the first peak (see Note 11).
5. Load pooled fractions onto the amylose column by gravity flow and collect the flow-through in tubes on ice. We often reapply the column flow through two more times to maximize binding (see Note 12). To wash the column, attach a 10 mL syringe barrel to the top of the column using a luer adaptor cap and place in a 15 ml conical tube. Fill the syringe with 5 mL of cold ACB to wash the column by gravity flow at 4°C.
6. Elute complexes by applying 30 μL aliquots of elution buffer. Take the drip from the bottom of the column and place into a clean 1.5 mL microcentrifuge tube on ice (see Note 10). Repeat this 4-5 times. Check the average cpm with a Geiger counter to identify peak fractions containing the purified splicing complexes (see Note 10).
3.4 Denaturing gel analysis of spliceosome purification.
1. To prepare splicing time point and sizing column peak for denaturing gel analysis add 90 μL of splicing dilution buffer to each 10 μL sample. Then add 100 μL of phenol:chloroform:isomyl alcohol. Vortex well and spin for 10 minutes at 14,000 rpm at room temperature. Take 80 μL from the top aqueous layer, avoiding the interface, and put in a new 1.5 mL microcentrifuge tube. Add 300 μL 100% ethanol and invert a few times to mix. Incubate at −80°C for 30 minutes. Spin for 30 minutes in a microcentrifuge at 14,000 rpm at 4°C. Remove the ethanol and let the pellet air dry. Resuspend with 5 μL of RNA gel loading buffer. For amylose elution fractions, mix 1 μL of each elution fraction with 4 μL RNA gel loading buffer in a 1.5 mL microcentrifuge tube. For pre-mRNA standard dilute 1 μL of 200 nM pre-mRNA transcript in 39 μL RNA gel loading buffer.
2. Pour a thin denaturing 15% polyacrylamide gel. Place 0.4 mm spacers between 20 × 27 cm glass plates and secure with large binder clips. Seal the bottom of the gel with tape or an additional spacer. In a 50 mL conical tube take 25 mL 15% denaturing polyacrylamide solution. Add 75 μL 20% ammonium persulfate and 25 μL TEMED just before pouring the gel. Insert a 0.4 mm comb with wells that can hold up to 5 μL sample. Let gel polymerize for at least 20-30 minutes.
3. To run the gel, remove the bottom seal and clamp the gel into an electrophoresis rig with an aluminum heat sink plate. Fill the top and bottom chambers with 1x TBE and be sure to remove any air bubbles at the bottom of the gel. Remove the comb and extensively rinse the wells with buffer using a syringe. Hook up the leads to a high voltage power supply and run the gel at constant wattage of 30 W for 20 minutes to pre-warm the gel. Meanwhile, heat samples at 95°C for 1 minute and place on ice. Before loading the gel, rinse the wells again. Load 2.5 μL of each splicing time point and sizing column peak and 5 μL elution fraction samples into neighboring lanes. Also load 1 μL of pre-mRNA standard. Run gel for 2 hours at a constant wattage of 30 W.
4. Take down the gel. Remove one of the glass plates, lay down a used X-ray film on top of the gel and press down to adhere the gel to the film. Carefully peel the X-ray film with the gel from the glass plate, and then cover the gel supported by the film with plastic wrap. Place the gel in a phosphorimager cassette and expose overnight.
5. Using the appropriate software to analyze the phosphorimage of the gel, box out bands for pre-mRNA, one of the splicing intermediates and/or splicing products (Figure 3B). To correct for background, subtract the intensity of an equally sized box of a region in the lane above the band of interest from the band intensity. To normalize for the amount of label in each band, divide the corrected band intensity by the number of uridine residues in the corresponding RNA species. To quantify percentage of splicing efficiency for each lane separately, divide the intensity of the normalized bands for splicing intermediates or splicing products over the total intensity of bands for pre-mRNA plus splicing intermediates and splicing products. To quantify the concentration of spliceosomes in elution fractions, divide the intensity of a normalized splicing intermediate or splicing product band by the intensity of the normalized pre-mRNA standard band and multiply by 5 nM (or the concentration of pre-mRNA in the standard).
Footnotes
Contamination of reagents, tubes, equipment, etc. by RNases is always a concern when handling RNA. Gloves should be worn during the purification and reagents, pipette tips, tubes, etc. should be designated for RNA use only. We do not use DEPC treated water, but instead prefer glass distilled water stored in baked glassware.
Most commonly a derivative of the AdML gene product is employed as the pre-mRNA substrate for in vitro splicing in HeLa extract [2]. If the DNA template is contained in a plasmid, the plasmid must be linearized at the desired 3′ end by digestion with the appropriate restriction enzyme. Alternatively a PCR product may also be used as template.
We normally label RNAs with [a-32P] UTP, but other nucleotides can also be used if required. The specific activity of the RNA is controlled by modulating the concentrations of cold and hot UTP in the transcription reaction. We typically use 1/10th the volume of the transcription reaction for hot UTP and 400 μM cold UTP to obtain a “low” label that is sufficient to analyze splicing chemistry and detect complexes during purification.
HeLa nuclear extract is prepared as described in [28,29]. The splicing efficiency of the extract is dependent on the cell source and concentration of potassium and magnesium in the splicing reaction. We purchase HeLa cells that have been cultured for less than two weeks and shipped on wet ice from BioVest Intl. After preparing the extract, we freeze it in 200-400 μL aliquots at −80°C. The extract should be first tested with different concentrations of KGlu and MgAc to determine the best conditions for splicing [30]. We find that the range of optimal conditions lie between 0-100 mM KGlu and 0-6 mM MgAc. The nuclear extract should have at least 20% splicing efficiency to effectively purify splicing complexes.
Filter sterilize KGlu, MgAc, and heparin stocks. We divide these into 1 mL aliquots and store at −20°C
We use two 12 nt DNA oligonucleotides complimentary to the regions between 10-30 nt upstream of the 5′ splice site in the AdML pre-mRNA [19]. This region is accessible in unspliced pre-mRNA and the oligos form RNA/DNA hybrids, which allows endogenous RNase H to cleave the RNA. The region is protected from oligo binding in assembled spliceosomes.
MS2-MBP protein is expressed in Escherichia coli and purified first by amylose affinity followed by heparin chromatography as described in [31,18].
The buffer conditions for the purification were chosen with electron microscopy studies in mind. Often magnesium is thought to be important to stabilize ribonucleoprotein complexes. However, we found that splicing complexes tended to aggregate when 2 mM MgCl2 was present in the buffer, which was alleviated by addition of 5 mM EDTA. Although some proteins disassociate in the presence of EDTA (e.g. SR proteins), most core splicing components remain intact [19]. Different buffer conditions have been successfully used to purify splicing complexes and may be tested as desired.
Heparin is added to disrupt non-specific interactions between protein and nucleic acids and helps prevents splicing complexes from aggregating. However, it may also disrupt weaker specific interactions within the splicing complexes and may be omitted or used at a lower concentration if desired (e.g. [18,7]).
By using a small amount of affinity resin in a column geometry and minimizing the elution volume, spliceosomes elute at maximum concentration. We have not found any method to concentrate spliceosomes due to their “stickiness”. To elute splicing complexes in the smallest volume possible, use a pipette to suck out 30 μL elutions from the bottom “nib” of the Mobicol column. Usually the majority of purified spliceosomes peak in the second and third fraction at 5 to 15 nM concentration.
Depending on the downstream application for isolated splicing complexes we recommend taking only the first half of the splicing complex peak. The second half of the peak appears to contain additional proteins including excess MS2:MBP that we observe as additional background in EM images of the spliceosomes.
Reapplying flow-through maximizes binding of splicing complexes to the column. Nevertheless, we find that a significant percentage of the radioactivity does not bind the column.
Reference
- 1.Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3(7) doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Padgett RA, Hardy SF, Sharp PA. Splicing of adenovirus RNA in a cell-free transcription system. Proc Natl Acad Sci USA. 1983;80(17):5230–5234. doi: 10.1073/pnas.80.17.5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruskin B, Krainer AR, Maniatis T, Green MR. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984;38(1):317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- 4.Lardelli RM, Thompson JX, Yates JR, 3rd, Stevens SW. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA. 2010;16(3):516–528. doi: 10.1261/rna.2030510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warkocki Z, Odenwalder P, Schmitzova J, Platzmann F, Stark H, Urlaub H, Ficner R, Fabrizio P, Luhrmann R. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat Struct Mol Biol. 2009;16(12):1237–1243. doi: 10.1038/nsmb.1729. [DOI] [PubMed] [Google Scholar]
- 6.Barabino SML, Lamond AI. Antisense probes targeted to an internal domain in U2 snRNP specifically inhibit the second step of pre-mRNA splicing. Nucleic Acids Res. 1992;20(17):4457–4464. doi: 10.1093/nar/20.17.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bessonov S, Anokhina M, Will CL, Urlaub H, Luhrmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452(7189):846–850. doi: 10.1038/nature06842. [DOI] [PubMed] [Google Scholar]
- 8.Blencowe BJ, Sproat BS, Ryder U, Barabino S, Lamond AI. Antisense probing of the human U4/U6 snRNP with biotinylated 2′-OMe RNA oligonucleotides. Cell. 1989;59(3):531–539. doi: 10.1016/0092-8674(89)90036-6. [DOI] [PubMed] [Google Scholar]
- 9.Frilander M, Steitz J. Initial recognition of U12-dependent introns requires both U11/5′ splice-site and U12/branchpoint interactions. Genes Dev. 1999;13(7):851–863. doi: 10.1101/gad.13.7.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furman E, Glitz DG. Purification of the spliceosome A-complex and its visualization by electron microscopy. J Biol Chem. 1995;270:15515–15522. doi: 10.1074/jbc.270.26.15515. [DOI] [PubMed] [Google Scholar]
- 11.Gozani O, Patton JG, Reed R. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 1994;13:3356–3367. doi: 10.1002/j.1460-2075.1994.tb06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamond AI, Konarska MM, Sharp PA. A mutational analysis of spliceosome assembly: evidence for splice site collaboration during spliceosome formation. Genes Dev. 1987;1(6):532–543. doi: 10.1101/gad.1.6.532. [DOI] [PubMed] [Google Scholar]
- 13.Lamond AI, Sproat B, Ryder U, Hamm J. Probing the structure and function of U2 snRNP with antisense oligonucleotides made of 2′-OMe RNA. Cell. 1989;58(2):383–390. doi: 10.1016/0092-8674(89)90852-0. [DOI] [PubMed] [Google Scholar]
- 14.Nottrott S, Hartmuth K, Fabrizio P, Urlaub H, Vidovic I, Ficner R, Luhrmann R. Functional interaction of a novel 15.5kD [U4/U6.U5] tri-snRNP protein with the 5′ stem-loop of U4 snRNA. EMBO J. 1999;18(21):6119–6133. doi: 10.1093/emboj/18.21.6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reed R. The organization of 3′ splice-site sequences in mammalian introns. Genes Dev. 1989;3(12B):2113–2123. doi: 10.1101/gad.3.12b.2113. [DOI] [PubMed] [Google Scholar]
- 16.Ryder U, Sproat B, Lamond A. Sequence-specific affinity selection of mammalian splicing complexes. Nucleic Acids Res. 1990;18(24):7373–7379. doi: 10.1093/nar/18.24.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith CW, Nadal-Ginard B. Mutually exclusive splicing of alpha-tropomyosin exons enforced by an unusual lariat branch point location: implications for constitutive splicing. Cell. 1989;56(5):749–758. doi: 10.1016/0092-8674(89)90678-8. [DOI] [PubMed] [Google Scholar]
- 18.Bessonov S, Anokhina M, Krasauskas A, Golas MM, Sander B, Will CL, Urlaub H, Stark H, Luhrmann R. Characterization of purified human Bact spliceosomal complexes reveals compositional and morphological changes during spliceosome activation and first step catalysis. RNA. 2010;16(12):2384–2403. doi: 10.1261/rna.2456210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jurica M, Licklider L, Gygi S, Grigorieff N, Moore M. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA. 2002;8:426–439. doi: 10.1017/s1355838202021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Behzadnia N, Golas MM, Hartmuth K, Sander B, Kastner B, Deckert J, Dube P, Will CL, Urlaub H, Stark H, Luhrmann R. Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J. 2007;26(6):1737–1748. doi: 10.1038/sj.emboj.7601631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, Stark H, Urlaub H, Luhrmann R. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol Cell Biol. 2006;26(14):5528–5543. doi: 10.1128/MCB.00582-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herold N, Will CL, Wolf E, Kastner B, Urlaub H, Luhrmann R. Conservation of the protein composition and electron microscopy structure of Drosophila melanogaster and human spliceosomal complexes. Mol Cell Biol. 2009;29(1):281–301. doi: 10.1128/MCB.01415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ilagan J, Yuh P, Chalkley RJ, Burlingame AL, Jurica MS. The role of exon sequences in C complex spliceosome structure. J Mol Biol. 2009;394(2):363–375. doi: 10.1016/j.jmb.2009.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merz C, Urlaub H, Will CL, Luhrmann R. Protein composition of human mRNPs spliced in vitro and differential requirements for mRNP protein recruitment. RNA. 2007;13(1):116–128. doi: 10.1261/rna.336807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma S, Falick AM, Black DL. Polypyrimidine tract binding protein blocks the 5′ splice site-dependent assembly of U2AF and the prespliceosomal E complex. Mol Cell. 2005;19(4):485–496. doi: 10.1016/j.molcel.2005.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma S, Kohlstaedt LA, Damianov A, Rio DC, Black DL. Polypyrimidine tract binding protein controls the transition from exon definition to an intron defined spliceosome. Nat Struct Mol Biol. 2008;15(2):183–191. doi: 10.1038/nsmb.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419(6903):182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 28.Abmayr SM, Reed R, Maniatis T. Identification of a functional mammalian spliceosome containing unspliced pre-mRNA. Proc Natl Acad Sci USA. 1988;85:7216–7220. doi: 10.1073/pnas.85.19.7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dignam JD, Lebovitz RM, Roeder RD. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reichert V, Moore MJ. Better conditions for mammalian in vitro splicing provided by acetate and glutamate as potassium counterions. Nucleic Acids Res. 2000;28(2):416–423. doi: 10.1093/nar/28.2.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jurica MS, Moore MJ. Capturing splicing complexes to study structure and mechanism. Methods. 2002;28(3):336–345. doi: 10.1016/s1046-2023(02)00240-2. [DOI] [PubMed] [Google Scholar]