Abstract
To contribute to our understanding of the genome complexity of sugarcane, we undertook a large-scale expressed sequence tag (EST) program. More than 260,000 cDNA clones were partially sequenced from 26 standard cDNA libraries generated from different sugarcane tissues. After the processing of the sequences, 237,954 high-quality ESTs were identified. These ESTs were assembled into 43,141 putative transcripts. Of the assembled sequences, 35.6% presented no matches with existing sequences in public databases. A global analysis of the whole SUCEST data set indicated that 14,409 assembled sequences (33% of the total) contained at least one cDNA clone with a full-length insert. Annotation of the 43,141 assembled sequences associated almost 50% of the putative identified sugarcane genes with protein metabolism, cellular communication/signal transduction, bioenergetics, and stress responses. Inspection of the translated assembled sequences for conserved protein domains revealed 40,821 amino acid sequences with 1415 Pfam domains. Reassembling the consensus sequences of the 43,141 transcripts revealed a 22% redundancy in the first assembling. This indicated that possibly 33,620 unique genes had been identified and indicated that >90% of the sugarcane expressed genes were tagged.
Sugarcane is an important industrial crop of tropical and subtropical regions and is cultivated on close to 20 million hectares in more than 90 countries (FAO; http://apps.fao.org). Sugarcane belongs to the grass family (Poaceae), an economically important seed plant family that includes cereals such as maize, wheat, rice, and sorghum as well as many forage crops. The main product of sugarcane is sucrose, which accumulates in the stalk internodes. Sucrose, extracted and purified in specialized mill factories, is used as a raw material in human food industries or is fermented to produce ethanol, a low-pollution fuel. Ethanol is produced on a large scale by the Brazilian sugarcane industry.
The traditional cultivated sweet cane, Saccharum officinarum, has been improved by a century of breeding, the success of which has relied primarily on a series of interspecific crosses with a wild species, Saccharum spontaneum. S. officinarum and S. spontaneum are both polyploid. The former usually has 2n = 80, whereas the latter has two main cytotypes, 2n = 64 or 2n = 112 (Bremer 1961; Ewing et al. 1998). As a consequence, sugar cane varieties are at present highly polyploid and aneuploid, with a chromosome number of 100–130. Most of the chromosomes come from S. officinarum, with 10%–25% being inherited from S. spontaneum (D'Hont et al. 1996). The nonreplicated genome size of a somatic sugarcane cell (2C) is estimated to be 7440 Mb in S. officinarum. Because S. officinarum is octaploid, the size of a complete nonredundant chromosome set should be eightfold smaller, ∼930 Mb (D'Hont and Glaszmann 2001). This value is comparable to that of sorghum (∼760 Mb) and about twice that of rice (∼430 Mb).
The level of heterozygosity of all sugar cane varieties is high (Lu et al. 1994). Although the basic chromosome numbers are different for S. officinarum (x = 10) and S. spontaneum (x = 8; D'Hont et al. 1998), the two genomes are highly collinear. Thus, in a given cultivar, the basic gene set will be present in 10–14 copies, with one to four of these coming from S. spontaneum. This means that, for each gene, up to 14 distinct sequence haplotypes originating from the two species may be distinguished among transcripts.
Despite the genome complexity, the amount of information available for the sugarcane genome has been increasing. Genetic maps have been produced for S. spontaneum, S. officinarum, and modern cultivars (al Janabi et al. 1993; Grivet et al. 1996). Saturating the whole genome is an enormous task because of the high level of ploidy, and none of the maps is yet complete. Two-thirds of the genome is estimated to have been covered (Dufour et al. 1997). Synteny with other grasses has been described based on genetic map comparisons done with common marker sets. Such comparisons have shown that collinearity between sugarcane and sorghum chromosomes is high (Ming et al. 1998). A bacterial artificial chromosome (BAC) library has been constructed for cultivar R570 (Tomkins et al. 1999) and is presently used for the map-based cloning of a fungal resistance gene (D'Hont and Glaszmann 2001).
Gene discovery has been limited in sugarcane. Some genes, mostly those involved in sucrose metabolism, were identified in the 1990s (Kumar et al. 1992; Bugos and Thom 1993; Bucheli et al. 1996). More recently, an expressed sequence tag (EST) project provided a few hundred “single-pass” sequences of anonymous genes (Carson and Botha 2000). To date, a total of 786 DNA sequence entries of all types, derived from various species of the Saccharum genus, were listed in the GenBank release of February 2003.
EST projects provide a very useful means of quickly accessing gene sequence information. As robot throughput increases and cost-per-read drops, it is now reasonable to determine a sequence tag for a large proportion of genes using this random cDNA sequencing approach. For example, the availability of the complete genome sequence of Arabidopsis thaliana revealed that the 105,000 ESTs available at the end of the year 2000 were enough to tag 60% of the 25,500 genes (The Arabidopsis Genome Initiative 2000). When combined with breakthroughs in highly parallel designs for gene expression analysis, large-scale EST projects now offer new perspectives for understanding the molecular basis of important traits in plants of agricultural relevance (Duggan et al. 1999).
We report here the analysis of a collection of ∼240,000 ESTs generated by the Sugarcane EST Project (SUCEST; http://sucest.lad.ic.unicamp.br/public) from 26 cDNA libraries that were constructed from different sugarcane organs and tissues sampled at various developmental stages (Vettore et al. 2001). The ESTs were assembled into 43,141 distinct contigs using CAP3 (Huang and Madan 1999). This new collection of single-pass sequences targets a highly diverse set of sugarcane genes and constitutes an important new resource for the genomics of sugarcane and related species.
RESULTS
Sequencing, Assembling, and Analysis of Sugarcane ESTs
Standard, unidirectional cDNA libraries were generated from a set of sugarcane tissues sampled from different varieties at various developmental stages (Table 1; Vettore et al. 2001). Libraries were also generated from a mixture of calli tissues subjected to cold or heat treatment. Two libraries (AD1 and HR1) were constructed with plantlets inoculated with Gluconacetobacter diazotroficans or Herbaspirilum diazotroficans, both of which are endophytic nitrogen-fixing bacteria that colonize sugarcane tissues (Lee et al. 2000). From each cDNA library, between 3000 and 14,000 clones were sequenced. A total of 260,352 clones were processed, generating 259,325 5′-end sequences and 32,364 3′- end sequences equivalent to 291,689 ESTs (Table 2). After trimming low-quality and vector sequences and removing contaminant bacterial and ribosomal RNA sequences, the resulting data set contained 237,954 high-quality ESTs with a minimum of 140 bases, with a phred quality ≥20 and an average of 864 bp in length (Table 2). Mitochondrial and chloroplast sequences were not filtered, but they contributed with <0.4% of the entire data set (data not shown). Insert amplification of 400 random clones from each library revealed inserts ranging between 500 bp and 5000 bp, with an average size of 1250 bp (Table 2). The 237,954 high-quality ESTs were assembled using the CAP3 program. A total of 221,616 ESTs were assembled into 26,803 contigs, while 16,338 remained as singletons (Table 3). There are 22,378 EST pairs (44,756 reads) representing clones sequenced from both the 3′- and 5′-ends. From these, 13,257 (∼60%) are present in the same contig. The 3′- and 5′-read pairs that did not assemble in the same contig could be originated from long cDNA inserts in which the reads from opposite ends did not overlap. The combined set of contigs and singletons resulted in 43,141 sequences (hereafter referred to as SASs for Sugarcane Assembled Sequences) representing putatively different transcripts. The alignment of the SASs with known protein sequences in the Non-Redundant Protein (nr) GenBank database (Benson et al. 2002; BLASTX E ≤ 10-40) revealed that >33% of the SASs contain clones with inserts representing the full-length protein sequence (Table 3).
Table 1.
SUCEST cDNA Librariesa
| Library | Source | Sugarcane variety |
|---|---|---|
| ADI | Root-shoot transition zone, stalk and apical meristem of Plantlets maintained in vitro inoculated with Gluconacetobacter diazotroficans | SP70-1143b |
| AM1 | Apical meristem and surrounding tissues of mature plants | SP80-3280c |
| AM2 | Apical meristem and surrounding tissues of immature plants | SP80-3280c |
| CL6 | Pool of sugarcane calli submitted to low (4°C) and high (37°C) temperature stress | SP80-3280d |
| FL1 | Inflorescence at beginning of development (1 cm long) | SP80-87432d |
| FL3 | Base of developing inflorescence (5 cm long) | PB5211 × P57150-4d |
| FL4 | Developed inflorescence and rachis (20 cm long) | PB5211 × P57150-4d |
| FL5 | Developed inflorescence (20 cm long) without rachis | PB5211 × P57150-4d |
| FL8 | Developing inflorescence and rachis (10 cm long) | SP80-87432d |
| HR1 | Root-shoot transition zone, stalk and apical meristem of plantlets maintained in vitro inoculated with Herbaspirilum rubrisubalbicans | SP70-1143d |
| LB1 | Lateral buds from field-grown adult plants | SP80-3280c |
| LB2 | Lateral buds from adult plants grown in a greenhouse | SP80-3280d |
| LR1 | Leaf roll from field-grown adult plants (large insert library) | SP80-3280d |
| LR2 | Leaf roll from field-grown adult plants (small insert library) | SP80-3280d |
| LV1 | Etiolated leaves from plantlets grown in vitro | SP83-5077, SP80-185, SP87-396, SP80-3280, SP80-3280 × SP81-5441d |
| RT1 | Root tips (1—3 cm long) from adult plants (large insert library) | SP80-3280d |
| RT2 | Root tips (1—3 cm long) from adult plants (small insert library) | SP80-3280d |
| RT3 | Root apex from the stalk button of adult plants | SP80-3280c |
| RZ1 | Shoot—root transition zone from adult plants (large insert library) | SP80-3280c |
| RZ2 | Shoot—root transition zone from adult plants (small insert library) | SP80-3280c |
| RZ3 | Shoot—root transition zone from young plants | SP80-3280c |
| SB1 | Stalk bark from undeveloped, upper part of adult plants | SP80-3280c |
| SD1 | Developing seeds (large insert library) | CB47-89, RB855205, RB845298, RB805028e |
| SD2 | Developing seeds (small insert library) | CB47-89, RB855205, RB845298, RB805028e |
| ST1 | First apical stalk internode of adult plants | SP80-3280d |
| ST3 | Fourth apical stalk internode of adult plants | SP80-3280d |
Standard cDNA libraries were constructed from a range of tissues sampled from plants at different developmental stages and grown in vitro, in a greenhouse, or in the field. All sugarcane varieties used are derived from commercial hybrids (S. spontaneum × S. officinarum). The library code, the tissue source, and the variety used are indicated. Plants were grown at the locations indicated in footnotes b—e
Universidade Federal do Rio de Janeiro, Rio de Janeiro, RJ
Universidade Estadual de Campinas, Campinas, SP
Copersucar Experimental Station, Piracicaba, SP
Universidade Federal de Alagoas Experimental Station, Serra do Ouro, AL
Table 2.
SUCEST EST Summary
| Total sequences | 291,689 |
| cDNA clones sequenced (5′ or 3′) | 260,352 |
| 5′-end sequences | 259,325 |
| 3′-end sequences | 32,364 |
| Total high-quality sequences | 237,954 |
| Success index (%) | 81.6 |
| Average insert size (bp) | 1250 |
| Average sequence size (bp) | 864 |
| Bases with phred quality ≥20/read | 399 |
Numbers of sequenced cDNA clones and generated ESTs from 26 cDNA libraries. A total of 260,352 cDNA clones were sequenced (259,325 5′-end and 32,364 3′-end sequences). After trimming and the removal of low-quality sequences, rRNA, and bacterial sequences. 237,954 high-quality sequences with a minimum of 140 bases remained. The average insert size was evaluated by polymerase chain reaction (PCR) analysis of 400 clones from each cDNA library. The success index represents the percentage of high-quality sequences relative to the total number of sequences.
Table 3.
Sequence Assembly Results
| Total sequences analyzed | 237,954 |
| Number of contigs | 26,803 |
| Number of singletons | 16,338 |
| Number of sugarcane assembled sequences (SAS) | 43,141 |
| Number of assembled sequences matching known genes | 27,833 |
| Number of clones with full-length inserts | 14,409 |
High-quality ESTs were assembled using CAP3. A BLASTX match cutoff of ≤10—5 was used to assign similarity. Clones were considered as having a putative full-length insert when their sequences started within the first 15 amino acids of their best BLASTX match with an E = value ≤10—40.
To estimate the level of redundancy, the 43,141 SASs were compared with each other using parameters of the cross_match (http://www.phrap.org/) algorithm that allowed two sequences to be considered as originating from the same transcript when they have 98% nucleotide identity over a minimum of 100 bp. Assembly of the 43,141 SASs resulted in 28,361 singletons and 5259 contigs, indicating ∼22% redundancy. This value is similar to the redundancy level in the honeybee EST assembly (Whitfield et al. 2002) and indicates that the SUCEST project may have identified 33,620 expressed sugarcane genes. Sugarcane may have a gene number similar to rice, which is presently estimated to be ∼35,000 genes (Goff et al. 2002; Yu et al. 2002), a number also similar to that estimated for tomato (Van der et al. 2002). Thus, the genes identified by SUCEST may represent >90% of the sugarcane expressed genes.
Of the 43,141 SASs, 27,833 (65%) were similar to known protein sequences present in the GenBank nr database (TBLASTX, E ≤ 10-5; Table 3). Comparison of the 43,141 SASs with Arabidopsis and rice genomes (TBLASTX, E ≤ 10-5) revealed that 71% and 82% of SASs had a significant match with Arabidopsis and rice, respectively (M. Vincentz and C.F.M. Menck, in prep.).
Sequence Polymorphism
EST data generated from various chromosome haplotypes offer the opportunity to detect sequence polymorphism, as demonstrated for the human (Buetow et al. 1999; Picoult-Newberg et al. 1999) and maize (Rafalski 2002) genomes. In sugarcane, the main difficulty is to distinguish paralogous genes from alleles of the same gene when more than 10 distinct haplotypes inherited from two species can theoretically be found at each locus. We conducted a detailed analysis of the sugarcane ESTs of two small families of genes, the 6-phosphogluconate dehydrogenases (Grivet et al. 2001) and the alcohol dehydrogenases (Grivet et al. 2003). A mean number of 15.4 single-nucleotide polymorphisms (SNP) and 3.6 insertions/deletions (INDEL) were detected for each of the five genes analyzed. SNPs were observed throughout the gene sequence, whereas INDELs were restricted to the 5′ and 3′ noncoding regions. None of the assembled sequences corresponding to these genes ever contained ESTs from two distinct genes, but, as a rule, ESTs of the same gene were kept apart in different assemblies when they differed for a large INDEL or a series of SNPs.
Analysis of the gene families in Arabidopsis showed that <2% of nuclear genes had a paralogous counterpart with a sequence identity ≥95% over ≥90% of the coding region, that is, conditions susceptible to the introduction of mistakes in EST clustering. We assumed that an analysis of polymorphism within assembled sugarcane EST sequences would be conservative such that it would probably miss some real polymorphism but would reveal very few false ones. Using this premise, we conducted a global analysis of SNPs and INDELs in the SAS data set based on the parameters described by Picoult-Newberg et al. (1999). An SNP was declared present at a given position when an alternative base to the consensus sequence was present at least twice in an alignment window with a phred quality ≥20. No more than one SNP was admitted in every window of 5 bp. Only SASs with more than four reads were considered. In all, a total of 42,936 SNPs were detected for 14,445 SASs analyzed (mean of 2.97 SNP per SAS). This value is considerably higher than that reported for the human genome with similar data (Garg et al. 1999; Picoult-Newberg et al. 1999; Deutsch et al. 2001).
Based on sequence polymorphism data in modern sugarcane cultivars obtained using molecular markers (Grivet et al. 1996; Hoarau et al. 2001), it is possible that the high SNPs observed in the SUCEST data set may at least partially be due to contrasted S. officinarum versus S. spontaneum sequence differences in the interspecific hybrids used to prepare the cDNA libraries. Transitions (64%) were more frequent than transversions (36%). As expected, a significant positive correlation (P < 0.05) was observed between the number of SNPs detected in an SAS and the number of reads present in that SAS. INDELs were tentatively detected as series of adjacent positions along SASs, where the absence of any base was recorded at least twice within an alignment window with a phred quality ≥20. With this rule, 3631 INDELs of 1 to 24 bases were detected in the entire database.
A set of the predicted SNPs was tested experimentally. Polymorphism was verified through restriction analysis for 51 SNP sites on a panel of 55 genotypes, including representatives of S. officinarum, S. spontaneum, and modern cultivars (data not shown). Polymorphism among individuals was observed for 31 (61%) of these sites, indicating that a large majority of SNPs detected in our study are real.
Functional Annotation of Sugarcane ESTs
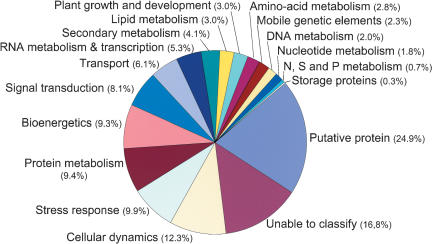
SASs were categorized with respect to functionally annotated genes in other organisms and grouped into 18 broad categories of biological roles (Fig. 1). Although several schemes of categorization may be equally valid, we have attempted to group together proteins that share similar functional characteristics or cellular roles, rather than follow a strict biochemical classification. Roles were assigned according to the known or putative involvement of a gene or a protein in a cellular process or pathway, as opposed to its participation in a specific binding or catalytic function. The sum of SASs per category did not add to 100%, because some SASs were classified in more than one category. Thus, for example, the enzyme phenylalanine ammonia-lyase was assigned to three categories (bioenergetics, stress response, and secondary metabolism).
Figure 1.
Sugarcane gene prediction classification. We classified 26,525 SASs with similarities to known protein sequences in the nonredundant protein (nr) database into 18 functional categories. The categories were generated either by automatic BLASTX of SASs against the categorized databases (cog.aa, http://www.ncbi.nlm.nih.gov/cgi-bin/COG/palog?fun=all; egad.aa, http://www.tigr.org/docs/tigr-scripts/egad_scripts/role_report.spl; kegg.aa, http://www.genome.ad.jp/kegg/kegg2.html; mips-at.aa, http://www.mips.biochem.mpg.-de/cgi-bin/proj/thal/filter_funcat.pl?all) or by manual inspection of BLASTX matches in the nr database by members of the SUCEST Consortium. In both cases, the BLASTX E-value cutoff was ≤10-5. The percentage of SASs found in each category is indicated next to the corresponding pie chart sector. The average percentages of SASs per category do not add up to 100% because some contigs or singletons appear in more than one category.
Examination of the primary BLAST matches revealed three major groups of SASs with varying potential to predict their cellular function. SASs in the first group, which matched sequences of known proteins with strong and nominal similarity, are likely to be transcripts of genes with similar functions (15,466 SASs; 36%). The function of the BLAST match was used to assign cellular roles to this group. The second class was formed by 15% of the total SASs (6614 or 24.9% of the categorized SAS in Fig. 1), and this group matched to “unknown protein,” “hypothetical protein,” or “putative protein,” with no indication of the function of the gene product. Most of the unknown proteins came from ESTs from other species that had been entered into the GenBank nonredundant database. The third group consisted of 15,308 SASs (35.6% of the total SASs) with no matches in the GenBank nr database. Almost 50% of all SASs annotated in the SUCEST database were associated with five broad categories: (1) cellular dynamics (biogenesis, organization and structure of the cell, cell division, cell growth, motility), (2) stress response (cell rescue activities, disease, virulence, and defense), (3) protein metabolism (folding and stabilization, modification, synthesis, targeting, sorting and translocation, proteolysis), (4) bioenergetics (C-compound and carbohydrate metabolism, photosynthesis), and (5) cellular communication/signal transduction (Fig. 1).
An “unable to classify” category (Fig. 1) was used for proteins and genes whose roles were unknown or could not be assigned with confidence based on searches of the literature. This category included 4445 SASs. This number corresponds to 17% of all categorized SASs or 10.3% of all SASs identified in the SUCEST database.
The SUCEST functional categories grouped according to the sugarcane tissues (organs) from which the mRNA was extracted are shown in Table 4. Although the proportion of unique genes involved in each cellular role was relatively uniform among the sugarcane organs, striking differences existed in the actual levels of gene expression. For example, developing seeds exhibited elevated expression of genes encoding storage proteins (fivefold above the average; Table 4) and relatively low levels of cellular communication transcripts compared with the other sugarcane organs. These may be because of the fact that storage proteins are typically expressed in high levels in cereal seeds (Leite et al. 1999). A higher proportion (more than twofold) of genes involved in nitrogen, sulfur, and phosphate metabolism was found in the root libraries, and reflected the involvement of this organ in nutrient uptake (Table 4). The number of transcripts involved in nucleotide metabolism, plant growth, and development was elevated in calli (Table 4). This may be associated with the high rate of cell proliferation in this tissue.
Table 4.
Percentage of SAS in 18 Broad Categories Across Eight Sugarcane Organs
| Functional Category | AD/HR | AM/LB | CL | FL | LV | RT | SD | ST | X ± SD (%) |
|---|---|---|---|---|---|---|---|---|---|
| Amino acid metabolism | 6.0 | 4.0 | 5.9 | 4.2 | 3.9 | 7.3 | 4.6 | 4.9 | 5.1 ± 1.2 |
| Bioenergetics | 17.3 | 12.6 | 17.5 | 12.6 | 12.0 | 21.9 | 16.9 | 15.8 | 15.8 ± 3.3 |
| Cellular communication/signal transduction | 10.7 | 11.1 | 9.8 | 11.0 | 10.1 | 10.8 | 7.9 | 11.8 | 10.4 ± 1.2 |
| Cellular dynamics | 18.5 | 19.7 | 15.0 | 19.1 | 18.6 | 18.3 | 15.1 | 20.1 | 18.1 ± 2.0 |
| DNA metabolism | 2.5 | 3.8 | 2.4 | 3.8 | 4.8 | 2.2 | 1.8 | 1.7 | 9 ± 1.1 |
| Lipid, fatty-acid, and isoprenoid metabolism | 4.0 | 3.5 | 3.2 | 3.8 | 3.7 | 4.4 | 4.6 | 3.5 | 3.8 ± 0.5 |
| Mobile genetic elements | 1.5 | 1.3 | 2.0 | 1.4 | 1.0 | 1.1 | 0.7 | 1.1 | 1.3 ± 0.4 |
| Nitrogen, sulfur, and phosphate metabolism | 0.9 | 0.7 | 0.8 | 0.6 | 0.7 | 1.7 | 0.7 | 0.7 | 0.9 ± 0.3 |
| Nucleotide metabolism | 2.5 | 2.5 | 3.1 | 2.5 | 2.2 | 2.4 | 2.2 | 2.6 | 2.5 ± 0.3 |
| Plant growth and development | 3.8 | 3.4 | 4.5 | 3.5 | 3.4 | 4.0 | 2.9 | 4.1 | 3.7 ± 0.5 |
| Protein metabolism | 16.9 | 18.7 | 17.2 | 18.4 | 21.7 | 16.5 | 20.5 | 16.5 | 18.3 ± 1.9 |
| RNA metabolism and transcription | 6.7 | 7.2 | 7.1 | 6.8 | 6.8 | 5.6 | 5.0 | 6.4 | 6.5 ± 0.8 |
| Secondary metabolism | 6.1 | 3.9 | 5.2 | 4.5 | 4.0 | 8.4 | 5.5 | 6.8 | 5.5 ± 1.5 |
| Storage proteins | 0.2 | 0.1 | 0.2 | 0.1 | 0.1 | 0.3 | 5.7 | 0.2 | 0.9 ± 1.9 |
| Stress response | 14.7 | 12.7 | 14.4 | 12.8 | 13.4 | 17.6 | 17.8 | 15.7 | 14.9 ± 2.0 |
| Transport | 8.5 | 6.7 | 6.3 | 7.1 | 6.7 | 8.0 | 9.2 | 7.8 | 7.5 ± 1.0 |
| Putative proteins | 15.2 | 16.2 | 15.8 | 16.8 | 15.1 | 14.0 | 12.7 | 15.8 | 15.2 ± 1.3 |
| Unable to classify | 6.3 | 6.1 | 6.4 | 6.7 | 6.3 | 5.8 | 5.3 | 6.3 | 6.1 ± 0.4 |
The 26 SUCEST cDNA libraries were grouped according to the similarity of the sugarcane organs used as source for mRNA extraction: AD/HR (AD1 and HH1 libraries); AM/LB (AM1, AM2, LB1, and LB2 libraries); CL (CL6 library); FL (FL1, FL3, FL4, FL8, and FL8 libraries); LV (LV1, LR1, and LR2 libraries); RT (RT1, RT2, and RT3 libraries); SD (SD1 and SD2 libraries); ST (ST1, ST3, SB1, RZ1, RZ2, and RZ3 libraries). The sum of the percentages per category (column X) is higher than 100% because some SAS appear in more than one category. SD, standard deviation.
Protein Domain Analysis
The distribution of conserved protein domains in the SUCEST database was analyzed by translating the SASs using the ESTScan algorithm (Iseli et al. 1999) and submitting for a domain search in the Pfam database (Bateman et al. 2000). For the 43,141 SASs, 40,756 proteins were assigned by ESTScan. Of these, 65 yielded two proteins, leading to 40,821 amino acid sequences ranging from 6 to 1782 amino acids (the average protein size being 211.96 ± 141.34). A high percentage (87%) of the 2385 SASs for which no protein could be assigned did not show significant similarity to sequences present at the public databases. This could indicate that they correspond to sequences of the 5′- or 3′-UTRs, which tend to be less conserved among organisms. Of the 40,821 protein sequences obtained, 12,921 presented at least one domain, totaling 1415 different domains.
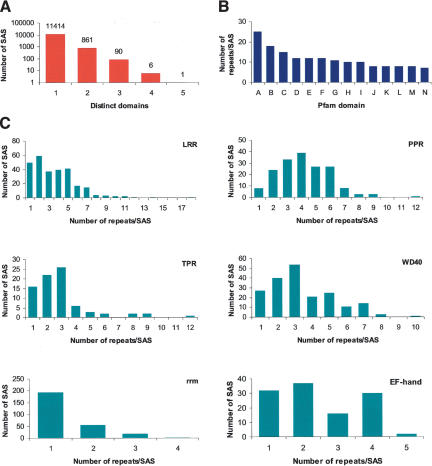
Most of the SASs identified in the SUCEST database coded for proteins containing one domain (11,414), but a significant number contained two or three domains (861 and 90, respectively; Fig. 2A). As ESTs are partial sequences in nature, the number of domains found in the SASs might not correspond to the real number of domains in the full-length protein sequences.
Figure 2.
Protein domain analysis. The 43,141 SASs were translated using the ESTScan algorithm, and the resulting 40,821 amino acid sequences were entered as queries in the Pfam database using the default settings of Pfam 7.0 [“global and local alignments merged” and “Pfam gathering threshold (GA)”]. A total of 12,921 SAS putative proteins produced significant matches with 1415 protein domain families of the Pfam database. (A) Number of distinct domains found for each SAS protein. The number of SAS proteins that contained one, two, three, four, or five distinct domains is shown. (B) Maximum number of repetitions for the top 14 repeated domains: nucleoporin FG (A), LRR (B), HEAT (C), M (D), PPR (E), TPR (F), XYPPX (G), WD40 (H), PC rep (I), ank (J), MORN (K), armadillo seq (L), PUF (M), and AT hook (N). The domains most often repeated in the same protein are shown along with the maximum number of repeats found for each domain. (C) Range of repetitions found for the LRR, PPR, TPR, WD40, rrm, and EF-hand domains. The domains with the most varied number of occurrences per SAS protein are indicated, along with the number of SAS proteins for each number of repeats.
The most repeated domain found in SUCEST database was the nucleoporin FG domain (Fig. 2B). The nucleoporin FG domain of the nuclear pore complex protein occurred 25 times in one SAS. The next most repeated domains were the leucine-rich repeat (LRR) and the HEAT repeat (related to armadillo/β-catenin-like repeats), both of which are involved in protein–protein interactions. Also common were the M repeats (found in multiple copies in bacterial M proteins) and the PPR repeat, of unknown function but frequently found in plants. The repeated domains represented by the largest number of SASs were also analyzed for their range and frequency of repetition. There were 274 occurrences of the LRR domain in the SUCEST database, and the number of repeats ranged from 1 to 18 (Fig. 2C). Also shown in Figure 2C are the range and frequency of repeats for 271 SASs with rrm domains (diagnostic of RNA-binding proteins), 196 SASs with WD40 repeats (characteristic of G-protein G-β subunits), 173 SASs with PPR repeats, 117 SASs with EF-hand domains, and 80 with TPR repeats. The number of repeats within the WD40-containing proteins of other systems varies from 5 to 8, but there is at least one example in sugarcane where 10 repeats are found.
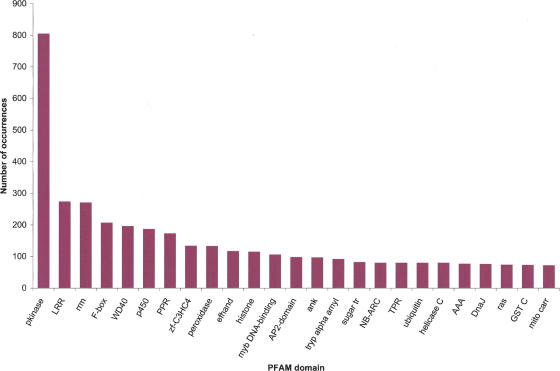
Figure 3 shows the top 25 domains found in sugarcane. The eukaryotic protein kinase and leucine-rich domains were the most frequent domains in sugarcane. These have also been shown to be enriched in A. thaliana. The top ranking domains in A. thaliana also include the RNA-recognition motif, the PPR repeats, the F-box domain, and the WD40 repeats, indicating a conservation of domain usage in plants (http://mips.gsf.de/proj/thal/db/tables/tables_func_frame.html).
Figure 3.
The number of occurrences for the 25 most common Pfam domains in SAS proteins. The 43,141 SASs were translated using the ESTScan algorithm, and the resulting 40,821 amino acid sequences were then entered as queries in the Pfam database. For protein domain analysis, the default settings of Pfam 7.0 [“global and local alignments merged” and “Pfam gathering threshold (GA)”] were used.
Tissue-Enriched Expression of Sugarcane Genes
An SAS was considered tissue-enriched if it contained at least three ESTs originating exclusively from a single sugarcane tissue. Of the 43,141 SASs, 1234 were tissue-enriched (Table 5; Supplemental material). These corresponded to 5716 ESTs (2.5% of the total ESTs). More than 80% of the tissue-enriched SASs (1020) contained three or four ESTs. The maximum number of ESTs in a tissue-enriched SAS was found for a prolamin that contained 360 ESTs. Developing seeds contained 1902 specific ESTs (33% of the total), with almost half of these (919) encoding prolamins, the major seed storage protein found in cereals. These ESTs included six putative new genes with a high level of expression in seeds (up to 32 ESTs/SAS).
Table 5.
Tissue-Enriched SAS
| Number of ESTs | Specific SAS | Best BLAST match | Library |
|---|---|---|---|
| 360 | 1 | (Y17556) α-kafirin [Sorghum bicolor] | SD |
| 103 | 1 | (A23207) zein zA1 [Zea mays] | SD |
| 76 | 1 | (Y17555) α-kafirin [Sorghum bicolor] | SD |
| 54 | 1 | (AF090447) zp22/D87 [Zea mays] | SD |
| 49 | 1 | (X57831) α-coixin 27 kDA [Coix lacryma-jobi] | SD |
| 43 | 1 | (Y17556) α-kafirin [Sorghum bicolor] | SD |
| 42 | 1 | (AF232008) β-glucosidase aggregating factor precursor [Zea mays] | RT |
| 38 | 1 | (Y17556) α-kafirin [Sorghum bicolor] | SD |
| 35 | 1 | (1107201B) zein A20 [Zea mays] | SD |
| 35 | 1 | (X16101) kafirin preprotein [Sorghum bicolor] | SD |
| 34 | 1 | (X53582) 19 kD zein [Zea mays] | SD |
| 32 | 1 | No match | SD |
| 31 | 1 | (S51798) γ-kafirin precursor [Sorghum bicolor] | SD |
| 28 | 1 | (X79885) β-coixin [Coix lacryma-jobi] | SD |
| 26 | 1 | No match | SD |
| 25 | 1 | (V01475) reading frame zein [Zea mays] | SD |
| 24 | 1 | (AC007789) putative low molecular early light-inducible protein [Oryza sativa] | SD |
| 24 | 1 | (1906392A) thaumatin-like protein [Zea mays] | SD |
| 22 | 1 | (M12144) 19kDa zein protein [Zea mays] and (AP002820) putative peroxidase [Oryza sativa] | SD |
| 22 | 1 | (AP002820) putative peroxidase [Oryza sativa] | RT |
| 21 | 1 | (X16101) kafirin preprotein [Sorghum bicolor] | SD |
| 21 | 1 | (X63113) α-coixin [Coix lacryma-jobi] | SD |
| 19 | 1 | (X56337) α-amylase [Oryza sativa] | CL |
| 19 | 1 | No match | SD |
| 18 | 1 | (AP000374) Cyclopropane fatty acid synthase [Arabidopsis thaliana] | FL |
| 17 | 1 | No match | SD |
| 16 | 1 | No match | SD |
| 16 | 1 | Unknown [Triticum aestivum] | RT |
| 14 | 2 | No match and (AB018375) OsENOD93a gene for early nodulin [Oryza sativa] | SD, RT |
| 13 | 4 | S.M. | SD, FL, RT |
| 12 | 6 | S.M. | SD, RT, HR, FL |
| 11 | 4 | S.M. | RT, CL, FL |
| 10 | 5 | S.M. | SD, FL, ST |
| 9 | 8 | S.M. | S.M. |
| 8 | 8 | S.M. | S.M. |
| 7 | 19 | S.M. | S.M. |
| 6 | 42 | S.M. | S.M. |
| 5 | 86 | S.M. | S.M. |
| 4 | 205 | S.M. | S.M. |
| 3 | 815 | S.M. | S.M. |
Tissue-specific SASs were ordered by the number of ESTs they contained. The best BLAST match for each tissue-enriched SAS is also shown, as well as their tissue of origin. Additional information on tissues-enriched SAS is available in the Supplemental Material (S.M.).
A search for the most common protein domains among the tissue-enriched SASs indicated that the protein kinase domain, followed by trypsin-α-amylase inhibitors, seed storage proteins, and lipid transfer protein, were the most frequent (Fig. 4). SASs with protein kinase domains include a MAP kinase, three wall-associated kinases, and five receptor protein kinases. The latter group is believed to transduce extracellular signals involved in pathogen perception, and one of the receptor-like kinases identified was specific to the library prepared from plantlets that were inoculated with the nitrogen-fixing bacteria Herbaspirillum. The F-box domain was also a top-ranking motif among tissue-enriched SASs. This motif serves as a link between a target protein and an ubiquitin-conjugating enzyme (Patton et al. 1998), and its specific expression may indicate tissue specific protein degradation. Cytochrome P450s are one of the most common domains specific to root SASs, and this is in accordance with their well-known role in the metabolism of environmental toxins.
Figure 4.
The most common domains in tissue-enriched SASs. The SASs were inspected for tissue specificity using the library of origin of their EST components. An SAS was considered tissue-enriched when it contained at least three ESTs found exclusively in a given library. Of the 43,141 SASs, 1234 were tissue-enriched and were inspected for the presence of conserved protein domains. The SASs were translated using the ESTScan algorithm. Domain occurrence was revealed by querying the Pfam database with the corresponding amino acid sequences using the default settings of Pfam 7.0 [“global and local alignments merged” and “Pfam gathering threshold (GA)”]. The most represented domains among the tissue-enriched SASs along with the number of SASs for each of then are shown.
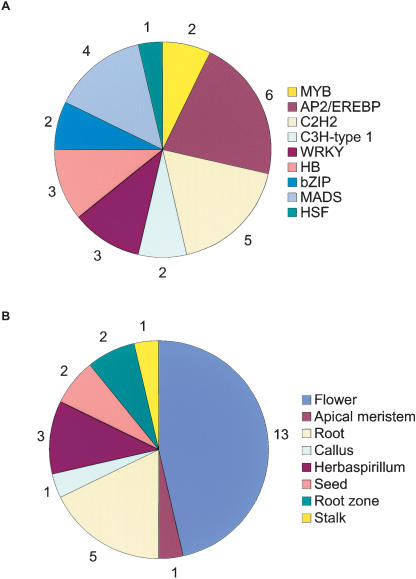
Cell type specification is commonly associated with the expression of specific transcription factors. Twenty-eight SASs with transcription factor domains were tissue-enriched (Fig. 5), among which the AP2-domain was the most represented. These elements are specific to plants and confer ethylene responsiveness (Ecker 1995). SASs containing the AP2-domain were specific for temperature-stressed calli, seeds, the root-to-shoot transition zone, flowers, stems, and roots. Within the core of the plant homeotic proteins is the MADS box found in the Agamous and Deficiens protein families involved in flower and fruit development (Ng and Yanofsky 2001). Four specific SASs with a MADS-box domain were found, three of which were flower-specific. Overall, 13 transcription factor families were found to be specific for flowers, five for roots, three for Herbaspirillum-inoculated plantlets, and two for developing seeds and other tissues (Fig. 5).
Figure 5.
The most common transcription factor domains in tissue-enriched SASs. The SASs were inspected for tissue specificity using the library of origin of their EST components. An SAS was considered tissue-enriched when it contained at least three ESTs found exclusively in a given library. The SASs were translated using the ESTScan algorithm. Domain occurrence was revealed by querying the Pfam database with the corresponding amino acid sequences using the default settings of Pfam 7.0 [“global and local alignments merged” and “Pfam gathering threshold (GA)”]. The 10 most frequent transcription factor domains were determined (see Fig. 6) and inspected for tissue specificity. (A) The most represented transcription factor domains among the tissue-enriched SASs along with the number of SASs in each of them. (B) The same data as in A showing the tissue origin and number of occurrences.
Regulation of Gene Expression
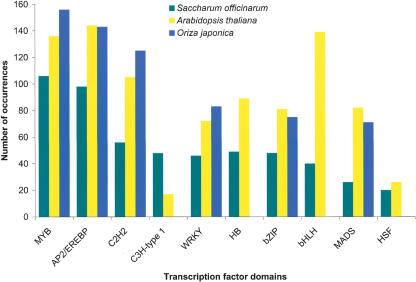
The unraveling of several eukaryotic genome sequences has allowed a comprehensive analysis of the transcription apparatus of cells and a comparison between plant and animal transcription factors (Riechmann et al. 2000). Around 5% of SASs (1416) encoded proteins with functions related to RNA metabolism and transcription (Fig. 1). This level was comparable to that of A. thaliana (Goff et al. 2002), in terms of number and most predominant transcription factor families present (Fig. 6). One interesting feature of the SUCEST database was the presence of a large group of SASs encoding proteins containing the C3H-type zinc-finger domains C-x8-C-x5-C-x3-H, which are not as numerous in Arabidopsis (Riechmann et al. 2000). In general, 60% of the transcription factors found in Arabidopsis or rice were also present in the SUCEST database.
Figure 6.
The 10 most common transcription factor Pfam domains in SAS proteins. The 43,141 SASs were translated using the ESTScan algorithm, and the resulting 40,821 amino acid sequences were then entered as queries in the Pfam database using the default settings of Pfam 7.0 [“global and local alignments merged” and “Pfam gathering threshold (GA)”]. The 10 most represented domains typical of transcription factors are indicated along with the number of occurrences for each one. The number of predicted transcription factors found in Arabidopsis thaliana (Riechmann et al. 2000) and Oryza japonica (Goff et al. 2002) for the corresponding domains are also indicated.
Signal Transduction
The increasing number of genomes being sequenced has shown that most of the signaling modules are conserved among animals and plants, although some are enriched in animals and completely absent in plants (McCarty and Chory 2000). A comprehensive analysis of all signal transduction components found in the SUCEST database revealed the same pattern observed in the Arabidopsis genome (The Arabidopsis Genome Initiative 2000). There was no evidence of seven-pass transmembrane receptors or an adenylyl cyclase in sugarcane. The main receptor class identified was the serine/threonine receptor kinase family containing the leucine-rich repeat (LRR), which is postulated to mediate the recognition of plant peptides and possibly pathogens. This domain was also found in disease-resistance genes (R-genes; Ellis et al. 2000). There were 274 LRR-domains-containing SASs in the SUCEST database (Fig. 3).
Signal transduction following receptor activation frequently involves heterotrimeric G-proteins. There were five SASs with the signature of G-α-proteins, 12 for β-subunits, and one for the γ-subunit. There were 74 SASs with Ras-like domains, as well as evidence for their modulators, ArfGAP (16 SASs), RhoGAP (5 SASs), and RasGEF (1 SAS). Adapter modules and proteins usually associated with signaling complexes such as the PH domain and 14–3–3 proteins also occurred in the SUCEST database with 13 and 16 SASs, respectively. There was little evidence of phosphotyrosine recognition by SH2 domains. Only one SAS with this signature was found and encoded a putative chromatin regulatory protein. SH3 domains, which may be associated with SH2 domains (Morton and Campbell 1994), occurred in three SASs. As for other plants, tyrosine kinases have not been found in the SUCEST database. However, protein-tyrosine phosphatases (three SASs) and dual specificity phosphatases (seven SASs) were present among the sugarcane ESTs. There were 805 SASs encoding protein kinases in the SUCEST database, which represented the largest domain family found. The importance of protein phosphorylation/dephosphorylation events was corroborated by the presence of 64 SASs encoding protein phosphatase 2C domains and 14 SASs for the regulatory B subunits of protein phosphatase 2A. Phosphorylation at histidine residues was also well represented in the SUCEST database, with nine SASs sharing similarity to the histidine kinase A (phosphoacceptor) domain, which includes transcripts similar to the ethylene and cytokinin receptors, and 26 SASs with the response regulator receiver domain.
Defense-Related Genes
As in other plants, sugarcane appears to have developed complex defense responses either to protect itself from attack by insects and microbial pathogens, or to profit from these interactions, as in the case of infection with diazothropic endophytes. In all, 80 SUCEST SASs encoded proteins with clear similarity to the NB-ARC domain, which is characteristic of one of the major classes of disease resistance genes (R genes). As stated above, >200 SASs encoded proteins with at least one LRR domain. The SUCEST database contained 46 SASs encoding WRKY transcription factor domains, which have been implicated in the regulation of genes associated with pathogen defense in plants. Other genes related to less specific defense responses (e.g., chitinases, β-1,3-glucanases, chalcone synthases, chalcone isomerases, isoflavone reductases, hydroxyproline-rich glycoproteins, proline-rich proteins, catalases, superoxide dismutases) have putative orthologs in sugarcane, which indicates a high conservation of defense strategies among plants.
DISCUSSION
Approximately two-thirds of the SASs were similar to other publicly available sequences (Table 3). The remaining sequences (∼36%) with no matches in the public databases at either the protein or nucleotide level may represent untranslated mRNAs, as well as new sugarcane genes. Shoemaker at al. (2002) demonstrated that 13% of the soybean ESTs returned no matches after BLASTX search on trimmed sequences against the GenBank nr database.
The two main groups of flowering plants, the monocots and dicots, diverged 200 million years ago (Wolfe et al. 1989), but despite this long period of independent evolution, plant genes display significant conservation. This is corroborated by the fact that most of the sugarcane SASs also occur in Arabidopsis (71%), maize (62%–68%), and rice (82%). The cereals have been evolving independently from a common ancestral species for 50–70 million years (Kellogg 1998). This shorter period, compared to the monocot/dicot divergence, could be the main reason for the synteny among grass genomes. Whereas 71% of the sugarcane and Arabidopsis proteins are similar, ∼82% of the sugarcane putative genes have a match in the rice genome. This implies that ∼11% of the sugarcane SASs may correspond to gene sequences that are either restricted to monocots (innovation or gene loss in dicots) or are fast-evolving sequences not retained by the fixed threshold. The remaining 18% of the SASs may represent proteins involved in the differentiation of sugarcane and rice, or may correspond to noncoding sequences that are under low selective pressure, which diverged rapidly (M. Vincentz and C.F.M. Menck, in prep.).
The abundance of SASs with predicted cellular roles in sugarcane was estimated by classifying the primary BLAST matches with similarity to known proteins into functional categories (Fig. 1). The proportion of transcripts involved in gene expression (the sum of the protein metabolism, RNA metabolism, and transcription categories in Table 4) was 24.9% (between 23% and 28.5% among sugarcane organs; Table 5). This compares favorably with an average of 21.9% in various human tissues (with most being between 20% and 27%; Adams et al. 1995).
In sugarcane, the putative genes related to cellular communication represent ∼12% of the total number of genes (signal transduction and secondary metabolism in Fig. 1), whereas ∼17% of the transcripts identified by the SUCEST may be involved in metabolism, including the SASs in five functional categories: amino acid metabolism, lipid, fatty-acid and isoprenoid metabolism, nitrogen, sulfur and phosphate metabolism, nucleotide metabolism, and bioenergetics (Fig. 1). According to Adams et al (1995), the proportion of genes related to cell signaling communication and metabolism in human tissues is ∼12.4% and ∼16.4%, respectively. The similarity in proportions in sugarcane and human cells implies that the cellular machinery involved in nonspecialized functions is highly conserved in eukaryotes.
The wealth of information generated in this work holds exciting prospects for plant scientists. Like most plants, sugarcane is exposed to biotic (viruses, bacteria, fungi, and a multitude of herbivores) and abiotic (drought, cold, and aluminum toxicity, e.g.) stress that contributes to decreased productivity. The SUCEST data set provides a fundamental basic resource for the understanding of the biology of this complex polyploid plant. In addition, this information may facilitate genomic analyses in other plants, particularly gene discovery, functional characterization, and annotation of cereal genomic sequences.
METHODS
Plant Material
Sugarcane tissues were obtained from the varieties SP70-1143, SP80-3280, SP80-87432, PB5211 × P57150-4, CB47-89, RB855205, RB845298, and RB805028 (Table 1). Plants were grown at the Copersucar experimental station (Piracicaba, SP, Brazil), at the Universidade Federal de Alagoas experimental station–Serra do Ouro (Murici, AL, Brazil), and at the Centro de Biologia Molecular e Engenharia Genética (Campinas, SP, Brazil). After harvesting, tissues were frozen in liquid nitrogen and stored at -80°C until RNA extraction.
cDNA Library
Total RNA was isolated from different sugarcane tissues with Trizol (Invitrogen) according to the manufacturer's instructions, or according to the method described by Manning (1991). Poly(A)+ was purified using Oligotex-dT (QIAGEN) according to the manufacturer's instructions. cDNA was synthesized using 1–5 μg of poly(A)+ RNA and directionally cloned as described in Vettore et al. (2001) using the SuperScript Plasmid System for cDNA Synthesis and Cloning kit (Invitrogen). Sepharose CL-2B (Pharmacia) columns (40 cm long, 1.5 mm ID) were used for the size fractionation of cDNAs. cDNA fractions containing fragments >500 bp were pooled and ligated into the vector pSPORT1 (Invitrogen) predigested with SalI and NotI. DH10B Escherichia coli cells (Invitrogen) were electroporated (25 μF; 200 Ω; 1.8 kV) with the resulting plasmids. The average titer of the libraries was ∼4.7 × 106 recombinants/μg cDNA. The transformants were spread on LB agar plates for direct picking without a library amplification step.
Template Preparation and DNA Sequencing
DNA templates were prepared in a 96-well format at all stages, from bacterial growth through to the purification after the sequencing reaction. The DNA was prepared using a 96-well alkaline lysis DNA method (http://sucest.lad.ic.unicamp.br/public). Sequencing reactions were done on plasmid templates using one-fourth of the standard volume of ABI Prism BigDye Terminator Sequencing Kits (Applied Biosystems). Reaction products were precipitated with 95% ethanol using 3 M sodium acetate and glycogen (1 g/L). Pellets were washed twice with 75% ethanol before drying under vacuum. The sequencing reaction products were analyzed on 377-96 ABI sequencers.
Sequence Analysis
The software phred (Ewing et al. 1998) was used to base call and to estimate error probability in the SUCEST chromatograms. The trimming process, which included the removal of ribosomal RNA, poly(A) tails, low-quality sequences, and vector and adapter regions, was conducted as described by Telles and da Silva (2001). The CAP3 assembler (Huang and Madan 1999), using read quality and default parameters, was used to cluster the trimmed reads. Similarities between SASs and sequences in the NCBI's nr, nt, and dbEST databases (Benson et al. 2002) were retrieved using BLAST (Altschul et al. 1997). The driver database for categorization consisted of 32,438 protein sequences from several sources divided into 30 categories (Telles et al. 2001). SASs that had BLAST matches with an E-value ≤e-10 and covering ≥70% of the hit protein present in the driver database (36% of all consensus sequences) were automatically assigned to the driver database protein categories. An annotation team manually categorized the remaining SASs using the BLASTX results. The SASs showing matches with the first 15 amino acids of a protein present in the GenBank database (BLASTX hit with E-values ≤e-40) were assumed to be full-length genes. For protein domain analysis, the default settings of Pfam 7.0 [“global and local alignments merged” and “Pfam gathering threshold (GA)”; Bateman et al. 2000] were used.
Acknowledgments
The authors thank the technicians and researchers who contributed to the sequencing effort and whose names are listed at the Web site http://sucest.lad.ic.unicamp.br/public. This project was jointly sponsored by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), and COPERSUCAR. We deeply regret the untimely death on July 8th, 2003 of Prof. Roberto V. Santelli, a very dedicated scientist and teacher.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1532103. Article published online before print in November 2003.
Footnotes
[Supplemental material is available online at http://sucest.lad.ic.unicamp.br/public. The sugarcane EST sequence data described in this paper have been submitted to GenBank under accession nos. CA064599–CA301538.]
References
- Adams, M.D., Kerlavage, A.R., Fleischmann, R.D., Fuldner, R.A., Bult, C.J., Lee, N.H., Kirkness, E.F., Weinstock, K.G., Gocayne, J.D., White, O., et al. 1995. Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence. Nature 377: 3-174. [PubMed] [Google Scholar]
- al Janabi, S.M., Honeycutt, R.J., McClelland, M., and Sobral, B.W. 1993. A genetic linkage map of Saccharum spontaneum L. `SES 208.' Genetics 134: 1249-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-815. [DOI] [PubMed] [Google Scholar]
- Bateman, A., Birney, E., Durbin, R., Eddy, S.R., Howe, K.L., and Sonnhammer, E.L. 2000. The Pfam protein families database. Nucleic Acids Res. 28: 263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson, D.A., Karsch-Mizrachi, I., Lipman, D.J., Ostell, J., Rapp, B.A., and Wheeler, D.L. 2002. GenBank. Nucleic Acids Res. 30: 17-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer, G. 1961. Problems in breeding and cytology of sugar cane. Euphytica 10: 59-78. [Google Scholar]
- Bucheli, C.S., Dry, I.B., and Robinson, S.P. 1996. Isolation of a full-length cDNA encoding polyphenol oxidase from sugarcane, a C4 grass. Plant Mol. Biol. 31: 1233-1238. [DOI] [PubMed] [Google Scholar]
- Buetow, K.H., Edmonson, M.N., and Cassidy, A.B. 1999. Reliable identification of large numbers of candidate SNPs from public EST data. Nat. Genet. 21: 323-325. [DOI] [PubMed] [Google Scholar]
- Bugos, R.C. and Thom, M. 1993. Glucose transporter cDNAs from sugarcane. Plant Physiol. 103: 1469-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson, D.L. and Botha, F.C. 2000. Preliminary analysis of expressed sequence tags for sugarcane. Crop Sci. 40: 1769-1779. [Google Scholar]
- Deutsch, S., Iseli, C., Bucher, P., Antonarakis, S.E., and Scott, H.S. 2001. A cSNP map and database for human Chromosome 21. Genome Res. 11: 300-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Hont, A. and Glaszmann, J.C. 2001. Sugarcane genome analysis with molecular markers, a first decade of research. Proc. Int. Soc. Sugarcane Technol. 24: 556-559. [Google Scholar]
- D'Hont, A., Grivet, L., Feldmann, P., Rao, S., Berding, N., and Glaszmann, J.C. 1996. Characterisation of the double genome structure of modern sugarcane cultivars (Saccharum spp.) by molecular cytogenetics. Mol. Gen. Genet. 250: 405-413. [DOI] [PubMed] [Google Scholar]
- D'Hont, A., Ison, D., Alix, K., Roux, C., and Glaszmann, J.C. 1998. Determination of basic chromosome numbers in the genus Saccharum by physical mapping of ribosomal RNA genes. Genome 41: 221-225. [Google Scholar]
- Dufour, P., Deu, M., Grivet, L., D'Hont, A., Paulet, F., Bouet, A., Lanaud, C., Glaszmann, J.C., and Hamon, P. 1997. Construction of a composite sorghum genome map and comparison with sugarcane, a related complex polyploidy. Theor. Appl. Genet. 94: 409-418. [Google Scholar]
- Duggan, D.J., Bittner, M., Chen, Y., Meltzer, P., and Trent, J.M. 1999. Expression profiling using cDNA microarrays. Nat. Genet. 21: 10-14. [DOI] [PubMed] [Google Scholar]
- Ecker, J.R. 1995. The ethylene signal transduction pathway in plants. Science 268: 667-675. [DOI] [PubMed] [Google Scholar]
- Ellis, J., Dodds, P., and Pryor, T. 2000. Structure, function and evolution of plant disease resistance genes. Curr. Opin. Plant Biol. 3: 278-284. [DOI] [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175-185. [DOI] [PubMed] [Google Scholar]
- Garg, K., Green, P., and Nickerson, D.A. 1999. Identification of candidate coding region single nucleotide polymorphisms in 165 human genes using assembled expressed sequence tags. Genome Res. 9: 1087-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff, S.A., Ricke, D., Lan, T.H., Presting, G., Wang, R., Dunn, M., Glazebrook, J., Sessions, A., Oeller, P., Varma, H., et al. 2002. A draft sequence of the rice genome (Oryza sativa L. spp. japonica). Science 296: 92-100. [DOI] [PubMed] [Google Scholar]
- Grivet, L., D'Hont, A., Roques, D., Feldmann, P., Lanaud, C., and Glaszmann, J.C. 1996. RFLP mapping in cultivated sugarcane (Saccharum spp.): Genome organization in a highly polyploid and aneuploid interspecific hybrid. Genetics 142: 987-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivet, L., Glaszmann, J.C., and Arruda, P. 2001. Sequence polymorphism from EST data in sugarcane: A fine analysis of 6-phosphogluconate dehydrogenase genes. Genet. Mol. Biol. 24: 161-167. [Google Scholar]
- Grivet, L., Glaszmann, J.C., Vincentz, M., da Silva, F.R., and Arruda, P. 2003. ESTs as a source for sequence polymorphism discovery in sugarcane: Example of the Adh genes. Theor. Appl. Genet. 106: 190-197. [DOI] [PubMed] [Google Scholar]
- Hoarau, J., Offmann, B., D'Hont, A., Risterucci, A.M., Roques, D., Glaszmann, J.C., and Grivet, L. 2001. Genetic dissection of a modern sugarcane cultivar (Saccharum spp). I. Genome mapping with AFLP markers. Theor. Appl. Genet. 103: 84-97. [DOI] [PubMed] [Google Scholar]
- Huang, X. and Madan, A. 1999. CAP3: A DNA sequence assembly program. Genome Res. 9: 868-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iseli, C., Jongeneel, C.V., and Bucher, P. 1999. ESTScan: A program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 138-148. [PubMed]
- Kellogg, E.A. 1998. Relationships of cereal crops and other grasses. Proc. Natl. Acad. Sci. 95: 2005-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, A.S., Moore, P.H., and Maretzki, A. 1992. Amplification and cloning of sugarcane sucrose synthase cDNA by anchored PCR. PCR Methods Appl. 2: 70-75. [DOI] [PubMed] [Google Scholar]
- Lee, S., Reth, A., Meletzus, D., Sevilla, M., and Kennedy, C. 2000. Characterization of a major cluster of nif, fix, and associated genes in a sugarcane endophyte, Acetobacter diazotrophicus. J. Bacteriol. 182: 7088-7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite, A., Cord Neto, G., Vettore, A.L., Yunes, J.A., and Arruda, P. 1999. Prolamins of sorghum, coix and millets. In Seed proteins (eds. R. Casey and P.R. Shewry), pp. 141-157. Kluwer Publishers, Dordrecht.
- Lu, Y., D'Hont, A., Paulet, F., Grivet, L., Arnaud, M., and Glaszmann, J.C. 1994. Molecular diversity and genome structure in modern sugarcane varieties. Euphytica 78: 217-226. [Google Scholar]
- Manning, K. 1991. Isolation of nucleic acids from plants by differential solvent precipitation. Anal. Biochem. 195: 45-50. [DOI] [PubMed] [Google Scholar]
- McCarty, D.R. and Chory, J. 2000. Conservation and innovation in plant signaling pathways. Cell 103: 201-209. [DOI] [PubMed] [Google Scholar]
- Ming, R., Liu, S.C., Lin, Y.R., da Silva, J., Wilson, W., Braga, D., van Deynze, A., Wenslaff, T.F., Wu, K.K., Moore, P.H., et al. 1998. Detailed alignment of Saccharum and Sorghum chromosomes: Comparative organization of closely related diploid and polyploid genomes. Genetics 150: 1663-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton, C.J. and Campbell, I. D. 1994. SH3 domains. Molecular `Velcro.' Curr. Biol. 4: 615-617. [DOI] [PubMed] [Google Scholar]
- Ng, M. and Yanofsky, M.F. 2001. Function and evolution of the plant MADS-box gene family. Nat. Rev. Genet. 2: 186-195. [DOI] [PubMed] [Google Scholar]
- Patton, E.E., Willems, A.R., and Tyers, M. 1998. Combinatorial control in ubiquitin-dependent proteolysis: Don't Skp the F-box hypothesis. Trends Genet. 14: 236-243. [DOI] [PubMed] [Google Scholar]
- Picoult-Newberg, L., Ideker, T.E., Pohl, M.G., Taylor, S.L., Donaldson, M.A., Nickerson, D.A., and Boyce-Jacino, M. 1999. Mining SNPs from EST databases. Genome Res. 9: 167-174. [PMC free article] [PubMed] [Google Scholar]
- Rafalski, A. 2002. Applications of single nucleotide polymorphisms in crop genetics. Curr. Opin. Plant Biol. 5: 94-100. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L., Heard, J., Martin, G., Reuber, L., Jiang, C., Keddie, J., Adam, L., Pineda, O., Ratcliffe, O.J., Samaha, R.R., et al. 2000. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290: 2105-2110. [DOI] [PubMed] [Google Scholar]
- Shoemaker, R., Keim, P., Vodkin, L., Retzel, E., Clifton, S.W., Waterston, R., Smoller, D., Coryell, V., Khanna, A., Erpelding, J., et al. 2002. A compilation of soybean ESTs: Generation and analysis. Genome 45: 329-338. [DOI] [PubMed] [Google Scholar]
- Telles, G.P. and da Silva, F.R. 2001. Trimming and clustering sugarcane ESTs. Genet. Mol. Biol. 24: 17-23. [Google Scholar]
- Telles, G.P., Braga, M.V.D., Dias, Z., Quitzau, J.A.A., da Silva, F.R., and Meidanis, J. 2001. Bioinformatics of the sugarcane EST project. Genet. Mol. Biol. 24: 8-15. [Google Scholar]
- Tomkins, J.P., Yu, Y., Miller-Smith, H., Frisch, D., Woo, S.S., and Wing, R.A. 1999. A bacterial artificial chromosome library for sugarcane. Theor. Appl. Genet. 99: 419-424. [DOI] [PubMed] [Google Scholar]
- Van der, H.R., Ronning, C., Giovannoni, J., Martin, G., and Tanksley, S. 2002. Deductions about the number, organization, and evolution of genes in the tomato genome based on analysis of a large expressed sequence tag collection and selective genomic sequencing. Plant Cell 14: 1441-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vettore, A.L., da Silva, F.R., Kemper, E.L., and Arruda, P. 2001. The libraries that made SUCEST. Genet. Mol. Biol. 24: 1-7. [Google Scholar]
- Whitfield, C.W., Band, M.R., Bonaldo, M.F., Kumar, C.G., Liu, L., Pardinas, J.R., Robertson, H.M., Soares, M.B., and Robinson, G.E. 2002. Annotated expressed sequence tags and cDNA microarrays for studies of brain and behavior in the honey bee. Genome Res. 12: 555-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, K.H., Gouy, M., Yang, Y.W., Sharp, P.M., and Li, W.H. 1989. Date of the monocot–dicot divergence estimated from chloroplast DNA sequence data. Proc. Natl. Acad. Sci. 86: 6201-6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., Hu, S., Wang, J., Wong, G.K., Li, S., Liu, B., Deng, Y., Dai, L., Zhou, Y., Zhang, X., et al. 2002. A draft sequence of the rice genome (Oryza sativa L. spp. indica). Science 296: 79-92. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://apps.fao.org; FAOSTAT Home Page.
- http://mips.gsf.de/proj/thal/db/tables/tables_func_frame.html; MATDB tables.
- http://sucest.lad.ic.unicamp.br/public; SUCEST Home Page.
- http://www.genome.ad.jp/kegg/kegg2.html; KEGG Encyclopedia.
- http://www.mips.biochem.mpg.de/cgi-bin/proj/thal/filter_funcat.pl?all; A. thaliana.Browse all contigs by functional catalog.
- http://www.ncbi.nlm.nih.gov/cgi-bin/COG/palog?fun=all; COGs Functional annotation.
- http://www.phrap.org/; The Phred/Phrap/Consed System Home Page.
- http://www.tigr.org/docs/tigr-scripts/egad_scripts/role_report.spl; EGAD Cellular Roles.