Summary
The fragile X syndrome, a common form of inherited intellectual disability, is caused by loss of the fragile X mental retardation protein FMRP. FMRP is present predominantly in the cytoplasm where it regulates translation of proteins important for synaptic function. We identify FMRP as a chromatin binding protein that functions in the DNA damage response (DDR). Specifically, we show that FMRP binds chromatin through its tandem Tudor (Agenet) domain in vitro, and associates with chromatin in vivo. We also demonstrate that FMRP participates in the DDR in a chromatin binding-dependent manner. The DDR machinery is known to play important roles in developmental processes such as gametogenesis. We show that FMRP occupies meiotic chromosomes and regulates the dynamics of DDR machinery during mouse spermatogenesis. These findings suggest that nuclear FMRP regulates genomic stability at the chromatin interface, and may impact gametogenesis and some developmental aspects of the fragile X syndrome.
Introduction
Chromatin is a complex biological entity comprised of DNA wrapped around histone octamers (Wolffe and Guschin, 2000). Posttranslational modifications of histone proteins serve as an interface for various chromatin “readers”, which are chromatin binding proteins that coordinate downstream processes, including the DNA damage response (DDR) and repair events (Costelloe et al., 2006; Downs et al., 2007; Stucki and Jackson, 2006). The mammalian DDR pathway is initiated by the activation of several conserved protein kinases, including ATM and ATR, which are members of the phosphatidylinositol 3-kinase-related kinase (PIKK) family. While ATM is activated by DNA double-strand breaks (DSB), ATR activity is triggered by stalled replication forks as well as single-strand DNA (Ciccia and Elledge, 2010). Upon activation, ATR phosphorylates histone H2A.X at serine 139 (termed γH2A.X) (Ciccia and Elledge, 2010; Liu et al., 2006; Ward and Chen, 2001) and the breast cancer associated tumor suppressor protein BRCA1 at serine 1423 (Gatei et al., 2001; Tibbetts et al., 2000). Both γH2A.X and BRCA1 are important regulators of genomic stability (Celeste et al., 2002; Nagaraju and Scully, 2007).
The fragile X mental retardation protein, FMRP, is an RNA-binding protein that mainly functions at the neuronal dendrites where it associates with specific mRNAs and modulates their translation, thus regulating a subset of proteins involved in synaptic function (Bassell and Warren, 2008; Brown et al., 2001). FMRP is critical for mGluR (metabotropic glutamate receptor)-dependent long-term depression, as well as other forms of synaptic plasticity. The lack of FMRP due to FMR1 gene silencing results in the fragile X syndrome, a common form of inherited intellectual disability and one of the leading causes of autism (Bear et al., 2004; Garber et al., 2008; Nelson, 1995; O’Donnell and Warren, 2002; Santoro et al., 2012; Warren and Nelson, 1994). Besides cognitive impairment, fragile X males also display macro-orchidism (Johannisson et al., 1987; O’Donnell and Warren, 2002) and female Fmr1 KO mice develop abnormal ovaries (Ascano et al., 2012) indicating an additional germ line or gonadal effect of disruption of Fmr1 expression.
Previous studies demonstrated a wide tissue distribution for FMRP, and established FMRP as largely a cytoplasmic protein with only about 4% FMRP in the nucleus (Feng et al., 1997), where its function remains unknown. However, several reports implicated a potential role for FMRP in the nucleus. Studies in Xenopus and zebrafish showed that at 2–3 hours post-fertilization, Fmrp is predominantly nuclear (Blonden et al., 2005; Kim et al., 2009; van’t Padje et al., 2005). In addition, Fmrp was found to decorate lampbrush chromosomes in Xenopus oocytes (Kim et al., 2009). Furthermore, nuclear FMRP interacting protein, NUFIP, associates with BRCA1 (Cabart et al., 2004), suggesting a potential functional relationship between FMRP and BRCA1 in the nucleus. FMRP has also been found in the PARP complexes, which heavily influence the DDR cascades (Helleday et al., 2005; Isabelle et al., 2010; Kedar et al., 2008; Poirier, 2010). Interestingly, mice lacking the DNA topoisomerase TOP3β, which is part of FMRP-containing mRNPs and is implicated in neuronal development, display progressive reduction in fecundity and aneuploidy (Kwan et al., 2003; Stoll et al., 2013). The fact that FMRP is present in DDR complexes and is predominantly nuclear in some gametes and early embryos led us to speculate that FMRP might have a novel nuclear function in the DDR during development.
In this study, we provide evidence that FMRP has an important role in the nucleus where it modulates the replication stress response at the chromatin interface. We show that FMRP regulates H2A.X phosphorylation, BRCA1 focus formation and accumulation of single strand DNA intermediates in a chromatin-binding dependent manner, and this nuclear role of FMRP is separable from its well-established role in translational regulation. We extend this nuclear function of FMRP to mammalian meiosis using mouse spermatocytes as a model. We show that FMRP decorates meiotic chromosomes and regulates γH2A.X induction, BRCA1 and ATR recruitment, and resolution of single-strand repair intermediates during meiosis. Taken together, our findings identify FMRP as a chromatin binding protein and demonstrate that it plays a previously unanticipated role in the DDR at the chromatin interface, which is independent from the canonical role of FMRP in translational regulation.
Results
Loss of FMRP compromises phosphorylation of H2A.X in response to replication stress
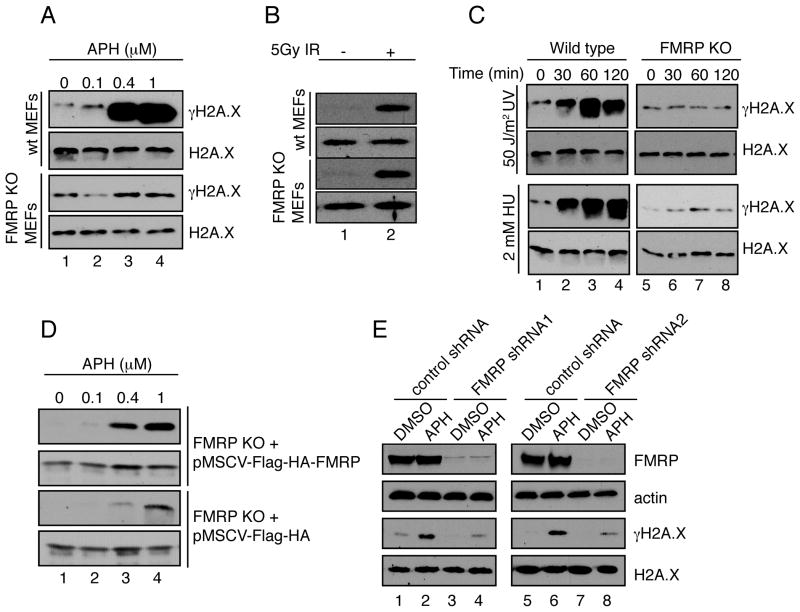
In order to determine whether FMRP has a role in the DDR, we analyzed γH2A.X induction in cells that lack FMRP. We first treated wild type and FMRP knockout (KO) MEFs with increasing concentrations of the replication stress inducer aphidicolin (APH), which largely triggers single-strand breaks, and ionizing radiation, which generates DSBs (Brown and Baltimore, 2003; Rogakou et al., 1998; Zhou and Elledge, 2000). In wild type but not FMRP KO MEFs, APH-induced replication stress elicited approximately 20-fold induction of γH2A.X (Fig. 1A, compare lanes 1–4 of the first and third panels), indicating a requirement for FMRP in the replication stress response. In addition, FMRP KO MEFs showed reduced formation of γH2A.X foci upon treatment with APH as compared to wild type MEFs (Fig. S1A–C). In contrast, FMRP KO cells showed comparable γH2A.X induction to that of the wild type MEFs in response to ionizing radiation, indicating an intact response to DSB (Fig. 1B, lane 2). In sum, FMRP KO MEFs showed distinct responses to different types of DNA damage, i.e., they responded to DSBs similarly to wild type MEFs but were defective in their response to replication stress.
Fig. 1. FMRP modulates histone H2A.X phosphorylation levels in response to replication stress.
(A) Wild type but not FMRP KO MEFs exhibited dose-dependent γH2A.X induction in response to APH (lanes 1–4). See also Fig. S1A–C. (B) Wild type MEFs and FMRP KO MEFs exhibited similar degrees of γH2A.X induction (5-fold) in response to 5Gy of IR (lanes 1 and 2). (C) Wild type but not FMRP KO MEFs exhibited time-dependent γH2A.X induction in response to 50 J/m2 of UV irradiation or 2mM of HU (10-fold induction at 60 min post-treatment) (compare lanes 1–4 to lanes 5–8). (D) FMRP KO MEFs reconstituted with wild type Flag-HA-FMRP (pMSCV-Flag-HA-FMRP) or vector alone (pMSCV-Flag-HA) were exposed to various concentrations of APH. See also Fig. S1D. pMSCV-Flag-HA-FMRP MEFs exhibited more pronounced γH2A.X induction compared to pMSCV-Flag-HA cells (12-fold in Flag-HA-FMRP cells and 4-fold in Flag-HA cells (lanes 1–4). (E) FMRP RNAi HeLa cells but not control cells showed diminished γH2A.X induction in response to APH (3.4-fold and 8-fold, respectively, compare lanes 1,2 to 3,4 and 5,6 to 7,8). See also Fig. S1E,F and Fig. S2.
To confirm that FMRP KO MEFs are defective in their response to replication stress, we subjected FMRP KO MEFs to additional sources of replication stress agents including hydroxyurea (HU) and UV irradiation. In both cases, FMRP KO MEFs failed to show a time-dependent increase of the γH2A.X level as compared to wild type MEFs (10-fold induction at 60 min post-treatment) (Fig. 1C, compare lanes 1–4 with 5–8 of the upper and lower panels). Importantly, FMRP KO MEFs reconstituted with a FLAG-HA epitope-tagged, wild type FMRP (Flag-HA-FMRP), conferred a more robust γH2A.X response to increasing concentrations of APH compared with the Flag-HA vector alone (Fig. 1D and Fig. S1D) (12-fold induction in Flag-HA-FMRP cells as compared to 4-fold induction in Flag-HA only cells). This was not a MEF-cell-specific effect since reduction of FMRP in HeLa cells by RNAi also resulted in a compromised induction of γH2AX in response to replication stress (Fig. 1E). In addition to H2A.X phosphorylation regulation, loss of FMRP also affected another ATR-dependent, replication response-specific phosphorylation event, phosphorylation of BRCA1 at Ser-1423 (Tibbetts et al., 2000) (Fig. S1E, F). Consistent with the potential role of FMRP in the replication stress response, FMRP RNAi knockdown HeLa cells reconstituted with Flag-HA vector alone, but not tagged wild type FMRP (Flag-HA-FMRP) were more sensitive to replication stress in the clonogenic survival assay (Fig. S2A, B), and FMRP KO MEFs were also more sensitive to replication stress compared to wild type MEFs (Fig. S2C). These findings are in line with previous reports describing a pro-survival role of FMRP (Jeon et al., 2012; Jeon et al., 2011; Liu et al., 2012). Taken together, the above findings link FMRP to replication stress-induced DNA damage response and indicate that FMRP may be part of the ATR-dependent signaling pathway.
FMRP is recruited to chromatin in response to replication stress
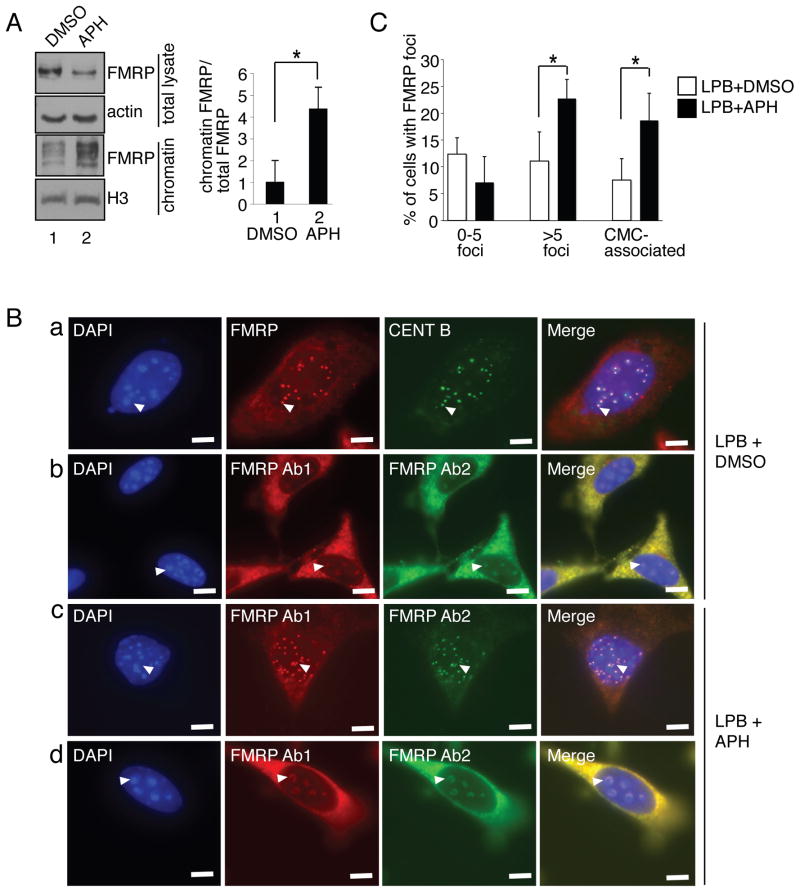
Many proteins that function in the DDR are recruited to chromatin in response to DNA damage, where they participate in the DDR events (Bostelman et al., 2007; Conde et al., 2009; Krum et al., 2010; Pei et al., 2011; Wakeman et al., 2012; Wysocki et al., 2005). We therefore investigated the possibility that the FMRP may function in the replication stress response through recruitment to chromatin. By chromatin fractionation, we detected association of FMRP with chromatin and this association was elevated by ~4-fold upon APH treatment (Fig. 2A compare lanes 1 and 2). Although biochemical fractionation allows detection of FMRP association with chromatin, direct visualization of FMRP in the nucleus is problematic due to the low level of nuclear FMRP (Fig. S3A). However, it is possible to raise nuclear FMRP levels by using leptomycin B (LPB), which inhibits nuclear protein export (Tamanini et al., 1999). As shown in Fig. 2B, in the presence of LPB, we detected FMRP foci in the vicinity of peri-centromeric domains (chromocenters (CMCs), which are easily recognizable in the mouse nuclei as large DAPI positive domains) (Figure 2B panels a and b). Consistently, FMRP staining overlapped with the centromeric protein B (CENT B) signal, which marks peri-centromeric heterochromatin (Fig. 2B panel a, arrowheads). In some cases FMRP formed larger structures wrapped around the CMCs (Fig. 2B panel b, arrowheads). The number of cells with FMRP foci as well as CMC-associated FMRP domains increased 2-fold after APH treatment (Fig. 2B panels c and d, Fig. 2C). In addition, we observed co-localization of FMRP and γH2A.X in MEFs treated with LPB (Fig. S3B, C). Although the significance of FMRP co-localization with CENT B, chromocenters, and γH2A.X foci requires further investigation, the above data nevertheless indicate that FMRP accumulates at specific chromatin domains and this accumulation can be increased upon replication stress, supporting our biochemical data (Fig. 2A).
Fig. 2. FMRP chromatin recruitment in response to replication stress.
(A) MEFs were treated with DMSO (lane 1) or APH (lane 2). Chromatin fractions were isolated and Western blotted for FMRP. Bar graph, relative ratio of chromatin-associated FMRP to total FMRP. Asterisk, p<0.05, Student t-test. Data are an average of 3 independent experiments with standard deviation. (B) Immunostaining of nuclear FMRP in APH treated or DMSO treated MEFs in the presence of leptomycin B (LPB). Panel a, FMRP co-localized with CENT B next to chromocenters (CMCs). Arrowheads, representative co-localized FMRP (red) and CENT B (green) foci docked near CMCs. Panel b, Representative FMRP signal (Ab-1: anti-FMRP (Abcam) antibody (red), Ab-2: anti-FMRP (Calbiotech) antibody (green)) enveloping CMCs in LPB treated MEFs. Panel c, Representative FMRP foci in LPB+APH treated cells. Panel d, representative FMRP signals enveloping CMCs in LPB+APH treated MEFs. Arrowheads, selected FMRP foci wrapped around CMCs. Scale bar, 10μm. (C) APH treatment resulted in doubling of the number of cells with 5 or more FMRP foci (>5) or FMRP CMCs. Asterisks, p<0.05, Student t-test. Data are an average of 3 independent experiments with standard deviation. See also Fig. S3.
FMRP binds chromatin via its N-terminal Agenet domain and this interaction is critical for FMRP function in the DDR
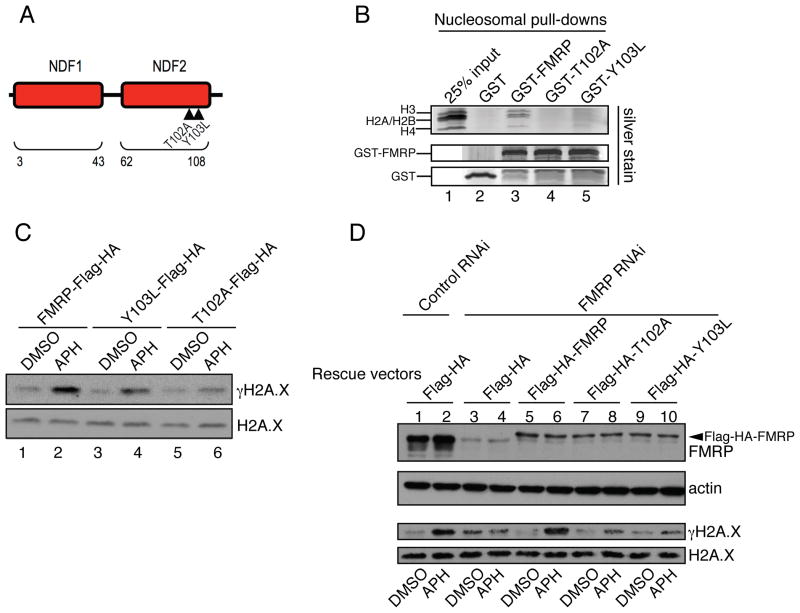
What is the molecular basis for the observed chromatin association of FMRP? FMRP contains an N-terminal Agenet domain (AgenetFMRP), which is a double-tudor domain that belongs to the Royal family of chromatin binding proteins (Maurer-Stroh et al., 2003; Ramos et al., 2006). Interestingly, the Agenet domain was recently shown to bind histone substrates methylated at various lysine residues (Adams-Cioaba et al., 2010; Sabra et al., 2013). This led us to hypothesize that FMRP might target chromatin through its Agenet domain. AgenetFMRP consists of two adjacent Tudor domains termed N-terminal domain of FMRP1 and N-terminal domain of FMRP2 (Ramos et al., 2006) (NDF1 and NDF2, respectively) (Fig. 3A). NMR studies identified residues T102 and Y103 on the surface of NDF2 as important for binding tri-methylated lysine (Ramos et al., 2006) (Fig. 3A). Mutating T102 and Y103 to A and L, respectively (T102A and Y103L), significantly compromised FMRP binding to native nucleosomes isolated from HeLa cells (Fig. 3B, compare lane 3 with 4 and 5) indicating that AgenetFMRP is required for FMRP association with nucleosomal substrates. We next explored the possibility of involvement of methyl-lysine recognition in FMRP binding to chromatin. We used a panel of recombinant Xenopus histones carrying methyl lysine analogs at various positions (Simon et al., 2007) in in vitro binding reactions with AgenetFMRP. AgenetFMRP did not show significant interaction with unmethylated histone H3 but bound histone H3 containing methyl-lysine analogs at several positions (Fig. S4A). Full length FMRP also bound methylated, but not unmethylated histone H3 (data not shown).
Fig. 3. FMRP docking to chromatin is essential for FMRP-dependent modulation of γH2A.X levels in response to replication stress.
(A) Diagram of AgenetFMRP. Mutations T102A and Y103L are demarcated by triangles. See also Fig. S4. (B) GST-FMRP or GST-FMRP carrying mutations in AgenetFMRP (GST-T102A and GST-Y103L) were incubated with isolated nucleosomes. Pull-down material was run on gradient gels followed by silver staining. A complete set of core nucleosomal histones including H3, H2A, H2B, and H4 were detected in wild type but not in mutant FMRP-mediated pull-downs (compare lanes 3–5). See also Fig. S5A. (C) Wild type FMRP (lanes 1 and 2) triggered more pronounced γH2A.X induction in FMRP KO MEFs in response to APH (12.8-fold) as compared to FMRP mutants (4-fold and 3-fold γH2A.X for Y103L and T102A mutants respectively) (lanes 3 and 4, 5 and 6). See also Fig. S1D. (D) FMRP RNAi in HeLa cells abolished γH2A.X induction in response to APH as compared to control RNAi (compare lanes 1,2 to lanes 3,4). Co-transfection with constructs expressing wild type but not mutant forms of FMRP restored the induction of γH2A.X in FMRP RNAi cells in response to APH (compare lanes 5,6 to lanes 7,8 and 9,10). The slower migrating band (in lanes 5–10) is Flag-HA-FMRP (indicated by an arrowhead).
We next carried out Microscale Thermophoresis (MST) (Jerabek-Willemsen et al., 2011; Wienken et al., 2010) in order to understand the binding dynamics of AgenetFMRP to various histone methylation marks. Consistent with the biochemical binding data, we found that AgenetFMRP exhibited higher affinity for histone H3 carrying lysine methylation mimics including H3Kc79me2 (Kd 135±28nM, Fig. S4B) and H3Kc27me1 (Kd 102±11nM, Fig. S4C) as compared to unmethylated H3 (Kd 1063±136nM, Fig. S4D). The biochemical and MST data both suggest that AgenetFMRP preferentially binds methylated histone H3 but does not display significant methyl site specificity in vitro. Importantly, AgenetFMRP mutations which abolish FMRP binding to native chromatin (Fig. 3A, B) also interfered with AgenetKHKHFMRP (Agenet and two adjacent nucleic acid binding domains) binding to the in vitro assembled methylated MLA nucleosomes (H3Kc79me2) (Fig. S5A, compare lanes 3, 4, and 5). Collectively, these data demonstrate that AgenetFMRP is necessary and sufficient for FMRP binding to chromatin, which might involve a sequence-independent methyl-lysine recognition function of AgenetFMRP.
FMRP binding to chromatin is required for FMRP-dependent modulation of γH2A.X levels in response to replication stress
We next carried out genetic complementation experiments to investigate potential functional roles of FMRP chromatin association in the DDR. FMRP KO MEFs were reconstituted with wild type or mutant forms of FMRP (T102A and Y103L), which are compromised in their ability to bind nucleosomes. Wild type FMRP (Fig. 3C, lanes 1 and 2) was more effective than the mutant forms of FMRP (Fig. 3C, lanes 3 and 4, 5 and 6 and Fig. S1D, which shows comparable expression of wild type and mutant FMRP proteins) in conferring the induction of H2A.X phosphorylation in the mouse FMRP KO MEF cells in response to APH treatment (12.8-fold γH2A.X increase with the wild type FMRP and 4- and 3-fold γH2A.X increase with the Y103L and T102A mutants, respectively). Similar results were obtained with HeLa cells in which the endogenous FMRP was inhibited by RNAi and which were then complemented with either the wild type FMRP or the FMRP Agenet domain mutants. As shown in Fig. 3D, wild type FMRP conferred a significantly higher level of γH2A.X response (9-fold induction, compare lanes 5 and 6, third panel from the top) than the Agenet point mutants (T102A and Y103L) (3-fold γH2A.X induction, compare lanes 7 versus 8 and 9 versus 10, third panel from the top). These findings suggest that the recruitment of FMRP to chromatin is critical for FMRP-dependent regulation of H2A.X phosphorylation.
FMRP mutants defective in supporting H2A.X phosphorylation are not compromised in their ability to modulate translation-dependent AMPAR trafficking
A well-documented role of FMRP is its ability to regulate activity-dependent synaptic translation of a specific subset of mRNAs, which is important for the maintenance of synaptic plasticity (Bassell and Warren, 2008; Bear et al., 2004; Brown et al., 2001; O’Donnell and Warren, 2002). Previous studies showed that a reduction of FMRP in dendrites leads to an excessive internalization of the alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor (AMPAR) subunit, GluR1 (Nakamoto et al., 2007), which is a critical process important for the maintenance of synaptic plasticity and the foundation for the mGluR theory of the fragile X syndrome (Bear et al., 2004). We asked whether the chromatin-binding defective FMRP point mutants were also compromised in their ability to dampen AMPAR internalization. As expected, immunofluorescence staining showed that FMRP KO neurons exhibited less AMPAR signal remaining on the surface and more internalized AMPAR signal relative to wild type neurons (Fig. S5B, compare panels 1 and 2). Quantitatively, the ratio of internalized to total AMPARs was increased in neurons isolated from Fmr1 KO mice as compared to wild type neurons (Fig. S5C, compare boxplots 1 and 2). Importantly, the FMRP chromatin-binding defective mutants were able to rescue this AMPAR trafficking defect similar to the wild type FMRP (Fig. S5B, panels 3–5 and Fig. S5C compare boxplots 1 versus 2 and 3–5). These findings indicate that the newly identified role of FMRP in the DDR is mechanistically distinct from its canonical function in modulating synaptic strength.
FMRP patient mutant R138Q is defective in mediating DDR events but retains normal translation-dependent AMPAR internalization
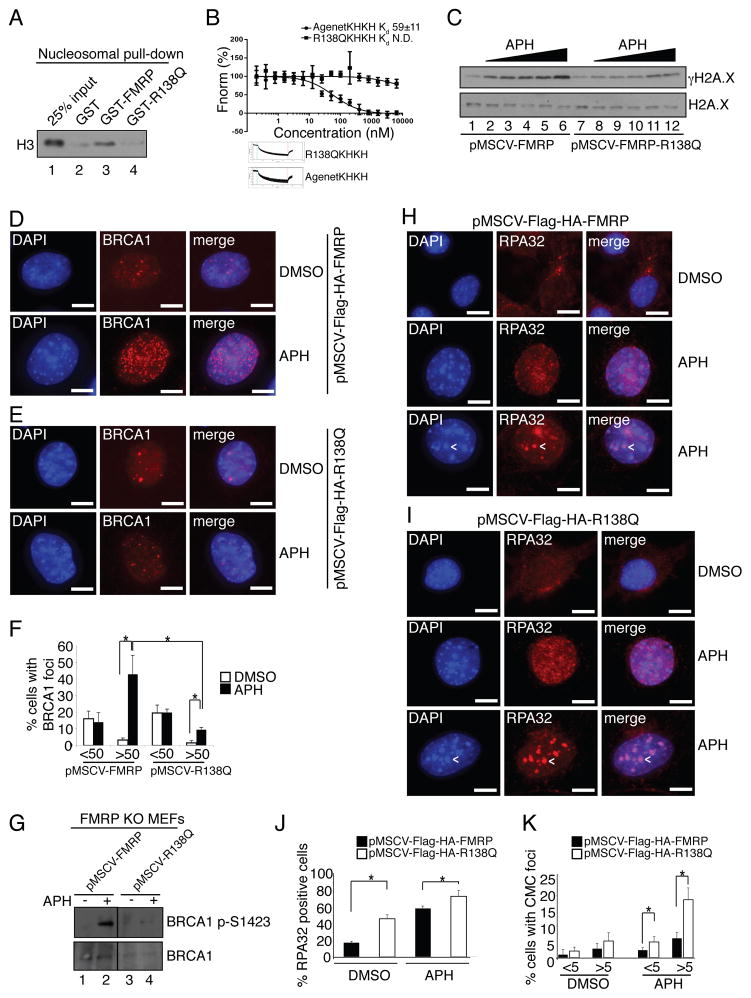
Recently, a novel FMRP sequence variant, R138Q, was found in a developmentally delayed male without the typical CGG-repeat expansion in the 5′ UTR of the FMR1 gene (Collins et al., 2010). Because the R138Q mutation lies near the extreme C-terminus of AgenetFMRP (Fig. S6A), we investigated whether this patient mutation affects FMRP nucleosomal binding. As shown in Fig. 4A, FMRP R138Q mutant failed to bind native nucleosomes (compare lanes 3 and 4) as well as recombinant H3Kc79me2 nucleosome (Fig. 4B, S5A lane 6, and S6B, which shows comparable levels of wild type and R138Q recombinant proteins used for the binding assays). Importantly, the R138Q mutant also failed to confer γH2A.X induction in the FMRP KO MEFs in response to replication stress (Fig. 4C compare lanes 1–6 with lanes 7–12 and Fig. S6C, which shows comparable levels of expression of wild type and R138Q reconstituted in the FMRP KO MEF cells). In addition to the γH2A.X defect, the R138Q FMRP mutant did not effectively support formation of BRCA1 foci and phosphorylation of BRCA1 at Ser-1423 in FMRP KO MEFs in response to APH treatment as compared to wild type FMRP (Fig. 4D–G and Fig. S6D). In addition, we observed an increased incidence of single strand DNA intermediates as indicated by RPA32 staining in FMRP KO MEFs rescued with the R138Q mutant as compared to wild type FMRP, suggesting a repair defect (Fig. 4, compare panels H and I, quantification J and K). Importantly, RPA32 staining associated with CMCs was also increased in FMRP KO MEFs complemented with R138Q, suggesting a possible functional significance of FMRP targeting to CMCs in the context of the DDR (Fig. 4H and I, bottom panels, arrows). FMRP KO MEFs reconstituted with the R138Q mutant were also more sensitive to the increasing concentrations of hydroxyurea (HU) as compared to wild type FMRP reconstituted cells in the clonogenic survival assay (Fig. S6E). In contrast, the FMRP R138Q mutant functioned similarly to wild type FMRP in suppressing excessive AMPAR internalization in FMRP KO neurons (Fig. S6F, compare panels 3 and 4, and Fig. S6G, compare boxplots 3 and 4). Taken together, these results suggest the tantalizing possibility that this newly identified nuclear function of FMRP in the DDR, when abrogated, may lead to a DDR-dependent clinical phenotype.
Fig. 4. Patient mutant R138Q is defective in γH2A.X induction and BRCA1 foci formation, and promotes excessive RPA retention on chromatin.
(A) Unlike wild type FMRP, the R138Q FMRP mutant failed to bind nucleosomes in vitro (compare lanes 3 and 4). (B) Equilibrium binding analysis using recombinant MLA nucleosomes di-methylated at H3K79 and wild type AgenetKHKH (Kd=59 nM) or R138QKHKH (binding not detected). See also Fig. S5A and S6B. (C) FMRP KO MEFs rescued with wild type FMRP but not the R138Q FMRP patient mutant exhibited a dose-dependent γH2A.X response triggered by APH (0.05μM, 0.1μM, 0.3μM, 0.5μM, 1μM). See also Fig. S6C. (D,E) BRCA1 foci formation in FMRP KO MEFs rescued with wild type FMRP (D) in response to APH was more pronounced as compared to FMRP KO MEFs rescued with the R138Q FMRP patient mutant (E). See also Fig. S6D. (F) 40% of FMRP KO MEFs rescued with wild type FMRP exhibited >50 BRCA1 foci per cell upon APH treatment, compared to 10% in MEFs rescued with the R138Q patient mutant. (G) BRCA1 S1423 phosphorylation in FMRP KO MEFs rescued with wild type FMRP in response to APH was more pronounced as compared to rescue with the R138Q FMRP patient mutant (compare lanes 2 and 4). (H, I) RPA32 foci formation in FMRP KO MEFs rescued with wild type FMRP in response to APH was less pronounced as compared to FMRP KO MEFs rescued with the R138Q patient mutant (compare middle panels in H and I). Note the accumulation of a subset of RPA32 foci at CMCs (arrowheads, lower panels). (J, K) Quantification of total (J) and CMC-associated (K) RPA32 foci in FMRP KO MEFs rescued with wild type FMRP and R138Q patient mutant in response to APH. Percentage of cells positive for RPA32 increased from 10% to 50% upon APH treatment after rescue with wild type FMRP and from 40% to 70% after rescue with the R138Q mutant. Note increased numbers of RPA32 positive cells in the case of R138Q mutant rescue MEFs even in the absence of APH treatment. (K) 17 % of R138Q mutant rescue MEFs and 6% of wild type FMRP rescue MEFs had >5 CMC-associated RPA32 foci upon APH treatment. Asterisks, p<0.05, Student t-test. Data are an average of 3 independent experiments with standard deviation. Scale bars, 10 μm. See also Fig. S6D.
FMRP is loaded onto chromosomes during male meiosis and regulates placement of γH2AX
The above findings provide strong support for a role of FMRP in the DDR via its association with chromatin. However, the biological significance of this finding was unclear. In this regard, mammalian meiosis represents perhaps the most relevant biological process where extensive DNA damage and recombinogenic events normally occur. In wild type meiotic cells, DSBs are generated during prophase by the topoisomerase-like enzyme SPO11 and form sites for homologous recombination and crossing over. DSBs accumulate γH2A.X and recruit many components of the somatic DDR machinery, including ATR and BRCA1. Repair then occurs in a highly regulated fashion, accompanied by pairing of homologous chromosomes (synapsis) and recombination between homologs (Blanco-Rodriguez, 2012; Garcia-Cruz et al., 2009; Turner et al., 2004; Turner et al., 2005). Importantly, in addition to defects in synaptic signaling in neurons, male fragile X patients exhibit macroorchidism, and Fmr1 KO mouse ovaries display premature follicular over-development (Ascano et al., 2012; Turner et al., 1980; Turner et al., 1975). Meiotic germ cells are therefore a relevant biological context in which to analyze the association of FMRP with chromatin in the DDR in vivo.
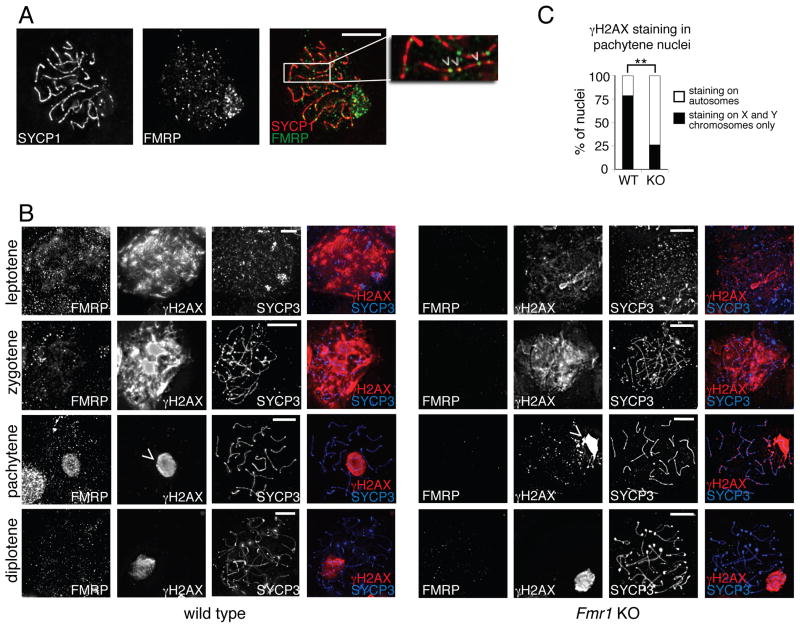
We used a mouse Fmr1 KO model to investigate whether FMRP is associated with chromatin and the DDR during mammalian meiosis. We first asked whether FMRP is present in the germ cell nucleus during meiosis. We performed immunostaining on chromosome spreads of adult male spermatocytes in meiotic prophase. Strikingly, we identified distinct FMRP puncta on condensed pachytene-stage chromosomes (Fig. 5A). These puncta were aligned along the chromosomes, as visualized by co-staining for the synaptonemal complex component SYCP1. FMRP puncta were not found on the chromosomes in Fmr1 KO cells, confirming the specificity of the antibody staining (Fig. S7A). We conclude that FMRP is present in the nucleus during meiotic pachytene and is localized on or near the chromatin at this stage.
Fig. 5. FMRP is present on meiotic chromosomes and regulates placement of γH2A.
Immunofluorescence staining was performed on spread chromosomes from adult male primary spermatocytes, and cells were imaged by deconvolution microscopy. (A) Pachytene stage nucleus showing FMRP puncta along the chromosomes. SYCP1 marks the full length of the autosomes during the pachytene stage. Inset shows FMRP puncta (green) aligned along SYCP1-stained chromosomes (red). See also Fig. S7A. (B) γH2A.X and FMRP staining in wild type (left) and Fmr1 KO (right) primary spermatocyte nuclei at leptotene, zygotene, pachytene, and diplotene stages of meiotic prophase. SYCP3 accumulates on chromosomes beginning in leptotene and is present along their full length during pachytene. In Fmr1 KO cells, accumulation of γH2A.X is delayed in the leptotene stage. At the pachytene stage, γH2A.X is restricted to the sex chromosomes (arrowheads) in wild type cells, but remains at some autosomal locations in Fmr1 KO cells. Scale bars, 10 μm. (C) Percentage of cells retaining γH2A.X outside of the sex chromosomes in WT and KO pachytene spermatocytes. **P<0.01, Fisher’s exact test. See also Fig. S7B.
In wild type meiotic cells, γH2A.X accumulates throughout the nucleus during the leptotene and zygotene stages of prophase, concomitant with DSB formation, but is removed from the chromosomes as repair proceeds and is absent from the autosomes by the pachytene stage. In males, the X and Y chromosomes retain γH2A.X during pachytene because these two chromosomes lack homologs and cannot fully synapse, and repair is delayed (Handel and Schimenti, 2010). Analogously, in mutants with defective repair and synapsis machinery, γH2A.X and other components of the DDR pathway are retained at unrepaired regions on the autosomes (Turner et al., 2005). We asked whether deposition of γH2A.X during meiotic prophase was impaired in Fmr1 KO cells. Fmr1 KO spermatocytes exhibited two distinct defects in γH2A.X accumulation: (1) reduced deposition of γH2A.X during the leptotene stage, and (2) inappropriate retention of γH2A.X on autosomes during pachytene (Fig. 5B). This phenotype was not the result of delayed or impaired formation of double-strand breaks, since there was no difference in SPO11 staining between wild type and KO cells (Fig. S7B). These defects were evident in only a subset of cells (Fig. 5C), perhaps explaining the preserved fertility of the Fmr1 KO males.
Fmr1 mutant mice exhibit defective chromosome synapsis and defective resolution of single-strand intermediates during meiotic prophase
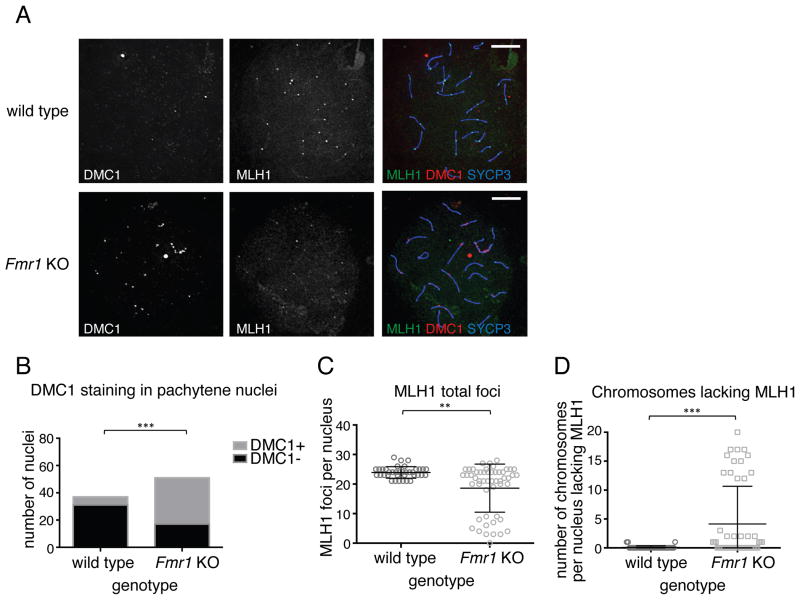
In wild-type meiotic cells, the RAD51 homolog DMC1 associates with the single-strand intermediates produced during DSB repair and facilitates invasion of the homologous chromosome, allowing recombination (Pittman et al., 1998; Schwacha and Kleckner, 1997; Yoshida et al., 1998). This process occurs during the zygotene stage and is largely complete by pachytene, by which time most DMC1 has dissociated from the chromosomes. Successful strand invasion catalyzed by DMC1 is required to proceed with repair and crossing over, including recruitment of the MLH1/MLH3 heterodimer during mid-to-late pachytene (Moens et al., 2002; Pittman et al., 1998; Yoshida et al., 1998). To determine whether single-strand intermediates were resolved in meiotic cells in the absence of FMRP, we co-stained pachytene nuclei with DMC1 and MLH1. We found that Fmr1 KO mid-pachytene spermatocytes inappropriately retained high levels of DMC1 on the chromosomes (Fig. 6A, B), associated with reduced recruitment of MLH1 (Fig. 6A, C, D). These findings suggest that resolution of single-strand DNA repair intermediates is delayed in meiotic germ cells in the absence of FMRP, resulting in impaired crossover formation.
Fig. 6. Fmr1 KO spermatocytes exhibit DNA repair defects and delayed resolution of single-strand intermediates at the pachytene stage.
Staining of chromosome spreads was performed as in Fig. 5. (A) Co-staining of DMC1, MLH1, and the synaptonemal complex component SYCP3, showing retention of DMC1 and reduction of MLH1 in Fmr1 KO cells at mid-pachytene. (B) Numbers of WT and KO cells positive for DMC1 staining at mid-pachytene. ***P<0.0001, Fisher’s exact test. (C) MLH1 foci per mid-pachytene nucleus in WT and KO. **P<0.01, Mann-Whitney U test. (D) Number of chromosomes per mid-pachytene nucleus lacking MLH1 foci. In WT cells, there is at least one MLH1 focus per chromosome. ***P<0.0001, Mann-Whitney U test. Scale bars, 10 μm.
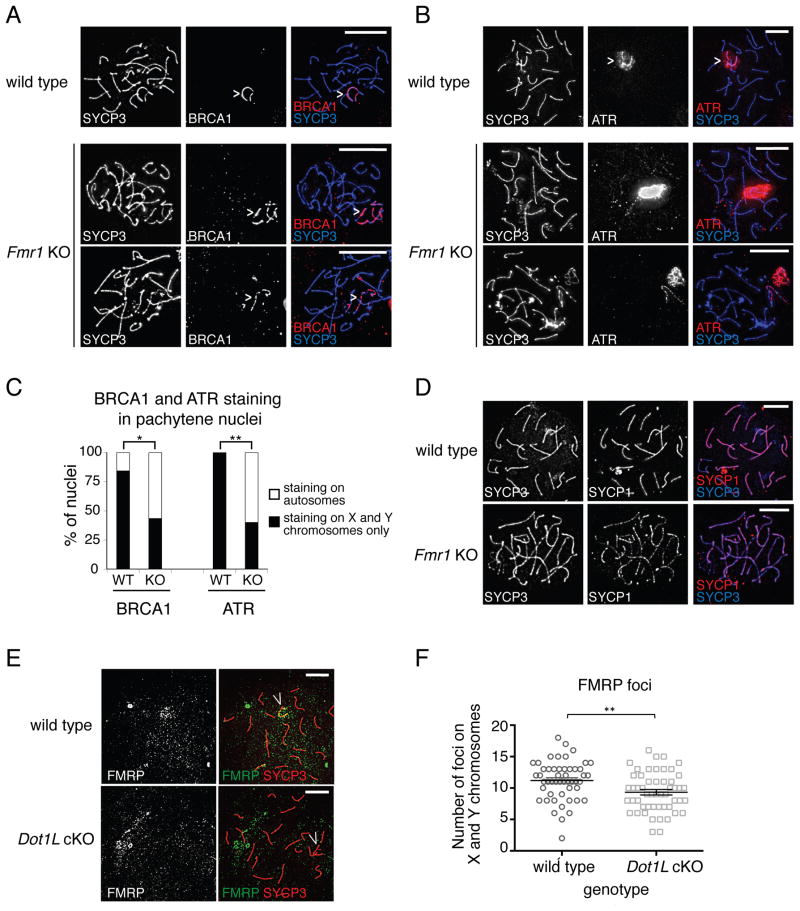
Consistent with a failure to repair DNA breaks, we found that BRCA1 and ATR were also inappropriately retained on the chromosomes in pachytene spermatocytes. BRCA1 and ATR were restricted to the unpaired X and Y chromosomes in wild-type spermatocytes, but were present on regions of the autosomes in Fmr1 KO spermatocytes (Fig. 7A–C). BRCA1 and ATR staining on the sex chromosomes was also discontinuous in Fmr1 KO spermatocytes, but continuous in wild type cells. Similar to the defects in γH2A.X deposition, DMC1 retention and MLH1 recruitment, these BRCA1 and ATR localization phenotypes varied between cells: some KO cells exhibited autosomal BRCA1 and ATR staining while others resembled wild type cells (Fig. 7C).
Fig. 7. Abnormal BRCA1 and ATR loading and synapsis defects in Fmr1 KO spermatocytes.
(A) Sample images of BRCA1 staining in pachytene spermatocytes in WT and KO animals. In wild type, BRCA1 staining is continuous and restricted to the sex chromosomes (arrowhead); in KO, it is discontinuous and frequently present on the autosomes. SYCP3 marks the chromosomes. (B) Sample images of ATR staining in pachytene spermatocytes in WT and KO animals. In wild type, ATR staining forms a cloud around the sex chromosomes (arrowhead) and is absent from the autosomes. In KO, ATR staining is retained in puncta on the autosomes and sometimes coats a complete autosome (bottom panels). SYCP3 marks the chromosomes. (C) Percentage of cells retaining BRCA1 or ATR outside of the sex chromosomes in WT and KO spermatocytes. *P<0.05; **P<0.01, Fisher’s exact test. (D) Co-staining of lateral (SYCP3) and central (SYCP1) elements of the synaptonemal complex shows discontinuous SYCP1 staining in Fmr1 KO cells, indicating defective synaptonemal complex formation. (E,F) Methylated H3K79 helps to recruit FMRP to chromatin. (E) Staining of FMRP in pachytene spermatocyte spreads from WT and Dot1L cKO mutants. Chromosome-associated FMRP signal is reduced in cKO cells, especially near the X and Y chromosomes. SYCP3 marks the chromosomes. (F) Quantitation of X- and Y-chromosome-associated FMRP foci. **P<0.01, unpaired t-test. Scale bars, 10 μm. See also Fig. S1C and S7C–E.
Because failure to resolve double strand breaks and to form interhomolog crossovers is also associated with defective synapsis, we next asked if Fmr1 KO spermatocytes also displayed synapsis defects. SYCP3, a lateral element of the synaptonemal complex (SC), assembles on unpaired chromosomes during early prophase, while SYCP1, a central element of the SC, assembles only on synapsed chromosomes (Fraune et al., 2012). We found that, whereas wild type pachytene nuclei had continuous SYCP1 staining along the chromosomes, many Fmr1 KO nuclei had discontinuous SYCP1 staining, indicating that SC formation is not complete (Fig. 7D) (Bishop et al., 1992; Pittman et al., 1998; Yoshida et al., 1998). Taken together, these findings suggest that resolution of single-strand repair intermediates, crossing over, and subsequent pairing of homologous chromosomes during meiotic prophase is incomplete in a subset of spermatocytes lacking FMRP.
Histone H3K79 methylation plays a role in the recruitment of FMRP to chromatin in vivo
As described above, both the AgenetFMRP and a full-length FMRP bind histone substrates in a methyl lysine-dependent manner (Fig. S4 and data not shown). However, it remains unclear whether FMRP binds methyl histones with some specificity in vivo and which methyltransferases are necessary for FMRP chromatin recruitment. Dot1, the H3K79 methyltransferase, has been shown to play a role in yeast meiosis (Ontoso et al., 2013a; San-Segundo and Roeder, 2000). In addition, recent reports demonstrated an increase in H3K79me2 and H3K79me3 levels in mouse spermatocytes in pachytene with H3K79me3 specifically enriched at the sex chromosomes and centromeres (Ontoso et al., 2013b). As a first step towards understanding the role of histone methylation in FMRP recruitment, we generated mice conditionally lacking Dot1L (Dot1L cKO, Fig. S7C–E) (Bernt et al., 2011), the only known mammalian H3K79 methyltransferase, in the germ cells, and stained meiotic spreads for FMRP. We found a small but significant reduction in the number of chromatin-associated FMRP puncta in the Dot1L cKO. This effect was especially evident on the X and Y chromosomes, where FMRP is particularly abundant during pachytene (Fig. 7E, F). Importantly, similar to FMRP KO MEFs, Dot1L mutant MEFs exhibited reduced γH2A.X foci formation in response to APH (Fig. S1C, right panel) as well as an increased sensitivity to increasing concentrations of APH compared to wild type MEFs (Fig. S2C). We conclude that methylated H3K79 might function in the same DDR pathway as FMRP, and help to recruit or retain FMRP at chromatin associated with DNA damage repair intermediates.
Discussion
We have identified FMRP as a chromatin binding protein and uncovered a novel and unanticipated function for FMRP in the nucleus where it regulates the DNA damage response. In addition, we uncovered a biological role for the DDR function of FMRP during mammalian spermatogenesis. We provided strong evidence that the Agenet domain binds histone H3 in a methylation dependent manner, without displaying overt preference towards a specific methyl lysine site. However, the binding specificity can conceivably be enhanced in vivo. Consistently, our preliminary data showed that the histone H3K79 methyltransferase DOT1L is important for FMRP chromatin association during meiosis, suggesting that H3K79 methylation may play a role in FMRP chromatin targeting in vivo. Our current data do not exclude the possibility that FMRP may also be capable of binding other methylated targets, such as nucleic acids.
This newly identified function of FMRP in the replication stress response appears independent of the classical role of FMRP in maintaining synaptic plasticity via translational regulation. Instead, nuclear FMRP may function in the DNA repair pathways through chromatin association. Our finding is consistent with published observations, including the report that FMRP interacts with poly(ADP-ribose) glycohydrolase (PARG) and poly (ADP-ribose) polymerase (PARP), which are major modulators of genomic stability (Gagne et al., 2005; Isabelle et al., 2010, Ciccia and Elledge, 2010; Haince et al., 2007). The fact that the DNA topoisomerase TOP3β is present in the FMRP-containing mRNPs, is involved in neuronal development and genomic stability, and also contributes to germ cell development, represents yet another intriguing connection between FMRP and the DDR. It is interesting to note that similar to FMRP, TOP3β is also associated with XY bivalents during pachytene (Kwan et al., 2003). We speculate that FMRP performs a docking function to regulate chromatin accessibility of DDR proteins. Although the detailed molecular mechanisms of FMRP-dependent DDR await further clarification, our data on the connection of FMRP with chromatin in the DDR represents an important advance in our understanding of FMRP function.
Importantly, DDR events such as γH2A.X induction and ATR/BRCA1 signaling heavily influence meiosis, specifically crossover formation and synapsis (Turner et al., 2004; Turner et al., 2005). Defects in synapsis can lead to chromosome nondisjunction, resulting in impaired gamete development or the generation of aneuploid gametes and developmental defects in the resulting embryo (Handel and Schimenti, 2010). Our findings suggest that the rate of germline chromosomal instability among Fmr1 knockout mice or fragile X patients at sites outside the fragile X locus may be elevated. This hypothesis is supported by recent findings describing increased rates of DNA damage and apoptosis in spermatocytes of Fmr1 KO mice (Tian et al., 2013). In addition, low FMRP levels were correlated with spermatogenesis defects in maturation arrest (MA) patients (Tian et al., 2013). Thus, our findings provide a potential molecular mechanism for the DNA damage, apoptosis and spermatogenesis defects observed in mice and patients lacking FMRP.
Interestingly, in yeast Dot1 mutants, meiotic cells exhibit increased levels of unrepaired DNA damage, and proceed through sporulation to produce mature spores with poor viability (San-Segundo and Roeder, 2000). In mouse spermatocytes, DOT1L chromatin loading and H3K79 methylation are dynamically regulated during meiosis. In particular, H3K79me3 and DOT1L protein accumulate at the sex chromosomes, and H3K79me3 accumulates at centromeres during the pachytene stage (Ontoso et al., 2013b). Our finding that FMRP is depleted at the sex chromosomes in Dot1L conditional mutants supports an interaction between FMRP and methylated histones during meiosis and raises that possibility that H3K79 methylation may be important for FMRP chromatin association in vivo. Interestingly, the Tudor domain of Survival Motor Neuron protein (SMN), which carries a methyl-lysine interacting surface similar to that of the FMRP Agenet domain (Ramos et al., 2006), was recently shown to interact with H3K79me1/2 in a DOT1L-dependent manner (Sabra et al., 2013)
Macroorchidism is a hallmark of the fragile X syndrome, but little is known with respect to its etiology. Malformed spermatids have been observed in both human fragile X patients and Fmr1 KO mice, suggesting a defect in sperm development (Slegtenhorst-Eegdeman et al., 1998; Johannisson et al., 1987). Adult male patients carrying the full fragile X repeat expansion produce sperm that carry a contracted pre-mutation but never the full expansion (Reyniers et al., 1993), implying that sperms carrying a full mutation are selected against at a pre-meiotic stage, allowing only those with a contracted FMR1 repeat to reach maturity (Bachner et al., 1993; Malter et al., 1997). Our finding that spermatocytes lacking FMRP exhibit defects in chromosome synapsis during meiotic prophase lends support to this model, and suggests a mechanism for this effect.
The idea of a functional involvement of FMRP in the DDR is especially appealing given recent evidence pointing to FMRP as a pro-survival protein. The absence of FMRP promotes apoptosis (Jeon et al., 2011) and telomere erosion in the fragile X patients, which is a major hallmark of genomic instability (Jenkins et al., 2008). In addition, fragile X patients have been reported to display a lower incidence of cancer (Schultz-Pedersen et al., 2001), whereas an increase in FMRP levels promotes tumor metastasis (Luca et al., 2013). Lastly, given that the loss of FMRP function leads to a common form of intellectual disability and autism, it is tempting to speculate that the role of FMRP in the DDR might represent a novel, previously unappreciated contributing factor to the development of the fragile X syndrome. Interestingly, a forward genetic screen in Drosophila identified 26 mis-sense mutations in the N terminus of dFMRP that affect axonal development (Reeve et al., 2005). Some of these mutations are localized to the dFMRP Agenet domain and predicted to impact the ability of dFMRP to bind chromatin. It has been suggested that the Agenet domain may also play a role in the translation-independent function of FMRP in synaptic signaling (Deng et al., 2013). Therefore, it remains to be determined if, in addition to its role in germ cell meiosis reported here, this nuclear function of FMRP also affects neuronal development and if the loss of FMRP has any DDR-related consequences in patients with the fragile X syndrome.
Experimental Procedures
Native nucleosome binding reactions
Reactions were performed in the presence of binding buffer (50mM Tris pH 7.5, 150mM NaCl, 2mM MgCl2, 0.1% Triton-X100) using 100 ng of GST fusion proteins and 5 μg of native nucleosomes isolated from HeLa cells at +4C°, and rotated for 2 hours before addition of glutathione agarose beads (GE Healthcare). Beads were washed 4 times with binding buffer. Three independent experiments were performed.
MLA nucleosome binding reactions
Mononucleosomes were prepared as described (Lu et al., 2008). The reactions were performed similarly to native nucleosome binding reactions, but using 2μg of MLA nucleosomes. Three independent experiments were performed.
γH2A.X induction rescue experiments
HeLa cells were transfected with FMRP shRNA or control scramble shRNA. Scramble shRNA was co-transfected with empty backbone vector (POZ-Flag-HA). FMRP shRNA was co-transfected with Flag-HA vector alone or with rescue vectors expressing either wild type or mutant forms of FMRP (Flag-HA-FMRP, Flag-HA-T102Y, or Flag-HA-Y103L). 3 days post transfection cells were treated with DMSO or APH (0.5μM) for 24 hours, then lysed in SDS sample buffer, and samples were subjected to Western blotting. FMRP KO MEF rescue experiments were performed identically to HeLa rescue experiments, except that rescue constructs were introduced into cells using the pMSCV-Flag-HA viral system. Three independent experiments were performed.
FMRP chromatin recruitment experiments
Chromatin fractionation experiments were adopted from (Méndez and Stillman, 2000). Briefly, after 1μM APH treatment of MEFs chromatin was isolated by resuspending cells in buffer A (10mM HEPES (pH 7.9), 10mM KCl, 1.5mM MgCl2, 1mM DTT, 3mM EDTA, 0.5% Triton-X100, protease inhibitor cocktail (Roche)) and nuclei were collected by low-speed centrifugation (4 min, 1,300 × g), washed once in buffer A and lysed in buffer B (3mM EDTA, 0.2mM EGTA, 1mM DTT and protease inhibitor cocktail (Roche)). Insoluble chromatin was collected by centrifugation (4 min 1,700 × g), washed again in buffer B and centrifuged again. The final chromatin pellet was resuspended in Laemmli buffer, sonicated and boiled for 15 min. Total protein lysate for determination of total protein levels was aliquoted from cells still resuspended in buffer A. All procedures were performed at +4°C. Three independent experiments were performed.
Immunofluorescence experiments
MEFs were treated with 10 ng/ml of leptomycin B (LPB) for 24 hours in the presence or absence of 0.5 μM APH. Cells were then fixed with ice cold methanol, stained with the antibodies of interest, and mounted using DAPI mounting medium (Vectashield). MEFs were counted according to the number of nuclear FMRP foci or large CMC-associated FMRP domains (CMCs) after LPB+DMSO or LPB+APH treatment (24 hrs). When Bethyl anti-BRCA1 and anti-RPA32 rabbit antibodies were used for staining cells were extracted with CSK buffer (10mM HEPES pH 7.4, 300 mM sucrose, 100mM NaCl, 3mM MgCl2, 0.5% Triton X-100) for 30 minutes at RT and then fixed in 4% paraformaldehyde (PFA) for 10 minutes followed by washes in PBS and immunostaining. 100 cells were counted in three independent experiments.
Preparation of meiotic chromosome spreads
Male Fmr1KO/Y and +/Y or Dot1LΔ/Δ and Dot1Lfl/+ littermates were sacrificed at 7 weeks of age. At least two individuals of each genotype were used for each experiment. The spread preparation protocol was modified from (Peters et al., 1997). Testes were immersed in DMEM and the tunicae were removed. The separated tubule suspension was spun for 8 minutes at 1000xg, and cells were resuspended in 1 ml hypo-buffer (30 mM Tris-HCl pH 8.2, 50 mM sucrose pH 8.2, 17 mM sodium citrate) and incubated for 7 minutes at room temperature. The cell suspension was split into 5 tubes and spun for 8 minutes at 1000 rpm, then resuspended in 170 μl 0.1M sucrose and dropped onto the slides, and allowed to spread for 2–3h. Slides were prepared with 1% paraformaldehyde with 0.1% TritonX-100, pH 9.2. For staining, slides were blocked in 3% BSA for 1 hour, incubated with primary antibody in 1% BSA overnight at 4°C, and then incubated with secondary antibody in 1% BSA for 1h at room temperature. Imaging was performed on a DeltaVision Elite deconvolution imaging system (Applied Precision) at 60x or 100x magnification. Stacks were compressed and analyzed using ImageJ software. Morphology of SYCP3-stained chromosomes was used to determine the stage of prophase.
Supplementary Material
Highlights.
Fragile X mental retardation protein FMRP binds chromatin via its Agenet domain
FMRP participates in the DNA damage response in a chromatin-dependent manner
FMRP occupies chromosomes and regulates DNA damage machinery in male mouse meiosis
Lack of FMRP results in meiotic defects, such as incomplete chromosome pairing
Acknowledgments
We thank S. Elledge and B. Yankner for advice and helpful suggestions; A. Ciccia for help with DNA damage experiments; D. Reinberg and P. Voigt for help with nucleosomal preparations; J. Griesbach, A. Lazic, and S. Duhr from NanoTemper Technologies and K. Yamagata, Y. Zheng and X. Shang for help with microscale thermophoresis and kinetic data analysis; E. Greer, A. Alekseyenko and G. Shanower for logistical support. We also thank J. Mowrey, S. Ceman, S. Keeney, and U. Fischer for reagents; M. Bear for Fmr1 KO mice; S. Armstrong for Dot1Lfl/fl mice; K. Luger, G. Narlikar, S. Ceman and U. Fischer for helpful comments and M. Goodheart, D. Cooper, E. Derby, L. Elow, K. Igarashi, L. Kolinski, L. Pomponi, and M. Schuck for technical assistance. We also thank our collaborators who contributed to this project but whose work was not included in the manuscript: B. Ren, U. Wagner and the Ren laboratory for FMRP genome-wide analysis and K. Zhao and his group for H3K79me2 genome-wide analysis; A. Vaquero for Suv39h double null MEFs; and G. Schotta and T. Jenuwein for Suv4-20h double null MEFs. This work was supported by NCI118487 and MH080129 to Y.S., by National Research Training Grant AG00222-7 to R.A, NRSA HD075591 to B.J.L., by HHMI funding to D.C.P. and in part by HD020521 and HD024064 to S.T.W. Y.S. is an American Cancer Society Research Professor.
Footnotes
Author Contributions
R.A. and Y.S. designed the study and co-wrote the manuscript. R.A. analyzed the data, performed histone pull-downs, cloning, mutagenesis, in vitro binding experiments, microscale thermophoresis, FMRP RNAi experiments, DNA damage experiments, MEF reconstitution experiments, survival assays, chromatin recruitment experiments, and immunofluorescence experiments. B.J.L. performed experiments using Fmr1 KO and Dot1L cKO mice, performed meiotic chromosome spreads and immunostaining, analyzed data, and co-wrote the manuscript. M.N.K. performed AMPAR internalization experiments and data analysis, and contributed to the writing of the manuscript. A.B. performed BRCA1 rescue experiments, and S.C., M.A., and C.X. performed in vitro biding assays with MLA histones and histone peptides and FMRP and Agenet. J.M. developed FMRP KO rescue cell lines. S.P. analyzed MST data. M.D.S., K.J.A, and A.S. designed and assembled MLA nucleosomes. C.R.V. developed Dot1L mutant MEFs and commented on the manuscript. T.G.K. and J. Min supervised histone peptide binding assays and W.F. and R.E.K. supervised nucleosome binding and commented on the manuscript. D.C.P. and S.T.W. supervised experiments and contributed to the writing of the manuscript. Y.S. supervised and directed the studies.
Conflict of Interest Statement
Y.S. is a co-founder of Constellation Pharmaceuticals and a member of its advisory board.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams-Cioaba MA, Guo Y, Bian C, Amaya MF, Lam R, Wasney GA, Vedadi M, Xu C, Min J. Structural studies of the tandem Tudor domains of fragile X mental retardation related proteins FXR1 and FXR2. PloS One. 2010;5:e13559. doi: 10.1371/journal.pone.0013559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascano M, Jr, Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492:382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, Feng Z, Punt N, Daigle A, Bullinger L, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20:66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blonden L, van’t Padje S, Severijnen LA, Destree O, Oostra BA, Willemsen R. Two members of the Fxr gene family, Fmr1 and Fxr1, are differentially expressed in Xenopus tropicalis. Int J Dev Biol. 2005;49:437–441. doi: 10.1387/ijdb.051974lb. [DOI] [PubMed] [Google Scholar]
- Brown EJ, Baltimore D. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev. 2003;17:615–628. doi: 10.1101/gad.1067403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabart P, Chew HK, Murphy S. BRCA1 cooperates with NUFIP and P-TEFb to activate transcription by RNA polymerase II. Oncogene. 2004;23:5316–5329. doi: 10.1038/sj.onc.1207684. [DOI] [PubMed] [Google Scholar]
- Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Molecular Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SC, Bray SM, Suhl JA, Cutler DJ, Coffee B, Zwick ME, Warren ST. Identification of novel FMR1 variants by massively parallel sequencing in developmentally delayed males. American Journal of Medical Genetics Part A. 2010;152A:2512–2520. doi: 10.1002/ajmg.a.33626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng PY, Rotman Z, Blundon JA, Cho Y, Cui J, Cavalli V, Zakharenko SS, Klyachko VA. FMRP regulates neurotransmitter release and synaptic information transmission by modulating action potential duration via BK channels. Neuron. 2013;77:696–711. doi: 10.1016/j.neuron.2012.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs JA, Nussenzweig MC, Nussenzweig A. Chromatin dynamics and the preservation of genetic information. Nature. 2007;447:951–958. doi: 10.1038/nature05980. [DOI] [PubMed] [Google Scholar]
- Feng Y, Gutekunst CA, Eberhart DE, Yi H, Warren ST, Hersch SM. Fragile X mental retardation protein: nucleocytoplasmic shuttling and association with somatodendritic ribosomes. J Neurosci. 1997;17:1539–1547. doi: 10.1523/JNEUROSCI.17-05-01539.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraune J, Schramm S, Alsheimer M, Benavente R. The mammalian synaptonemal complex: protein components, assembly and role in meiotic recombination. Experimental Cell Research. 2012;318:1340–1346. doi: 10.1016/j.yexcr.2012.02.018. [DOI] [PubMed] [Google Scholar]
- Garcia-Cruz R, Roig I, Robles P, Scherthan H, Garcia Caldes M. ATR, BRCA1 and gammaH2AX localize to unsynapsed chromosomes at the pachytene stage in human oocytes. Reprod Biomed Online. 2009;18:37–44. doi: 10.1016/s1472-6483(10)60422-1. [DOI] [PubMed] [Google Scholar]
- Isabelle M, Moreel X, Gagne JP, Rouleau M, Ethier C, Gagne P, Hendzel MJ, Poirier GG. Investigation of PARP-1, PARP-2, and PARG interactomes by affinity-purification mass spectrometry. Proteome Science. 2010;8:22. doi: 10.1186/1477-5956-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins EC, Tassone F, Ye L, Gu H, Xi M, Velinov M, Brown WT, Hagerman RJ, Hagerman PJ. Reduced telomere length in older men with premutation alleles of the fragile X mental retardation 1 gene. American Journal of Medical Genetics Part A. 2008;146A:1543–1546. doi: 10.1002/ajmg.a.32342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon SJ, Han SH, Yang SI, Choi JW, Kwon KJ, Park SH, Kim HY, Cheong JH, Ryu JH, Ko KH, et al. Positive feedback regulation of Akt-FMRP pathway protects neurons from cell death. Journal of Neurochemistry. 2012;123:226–238. doi: 10.1111/j.1471-4159.2012.07886.x. [DOI] [PubMed] [Google Scholar]
- Jeon SJ, Seo JE, Yang SI, Choi JW, Wells D, Shin CY, Ko KH. Cellular stress-induced up-regulation of FMRP promotes cell survival by modulating PI3K-Akt phosphorylation cascades. Journal of Biomedical Science. 2011;18:17. doi: 10.1186/1423-0127-18-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannisson R, Rehder H, Wendt V, Schwinger E. Spermatogenesis in two patients with the fragile X syndrome. I. Histology: light and electron microscopy. Hum Genet. 1987;76:141–147. doi: 10.1007/BF00284911. [DOI] [PubMed] [Google Scholar]
- Kim M, Bellini M, Ceman S. Fragile X mental retardation protein FMRP binds mRNAs in the nucleus. Mol Cell Biol. 2009;29:214–228. doi: 10.1128/MCB.01377-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KY, Moens PB, Wang JC. Infertility and aneuploidy in mice lacking a type IA DNA topoisomerase III beta. Proc Natl Acad Sci U S A. 2003;100:2526–2531. doi: 10.1073/pnas.0437998100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Jiang F, Bi X, Zhang YQ. Drosophila FMRP participates in the DNA damage response by regulating G2/M cell cycle checkpoint and apoptosis. Human Molecular Genetics. 2012;21:4655–4668. doi: 10.1093/hmg/dds307. [DOI] [PubMed] [Google Scholar]
- Luca R, Averna M, Zalfa F, Vecchi M, Bianchi F, Fata GL, Del Nonno F, Nardacci R, Bianchi M, Nuciforo P, et al. The Fragile X Protein binds mRNAs involved in cancer progression and modulates metastasis formation. EMBO Molecular Medicine. 2013;5:1523–1536. doi: 10.1002/emmm.201302847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malter HE, Iber JC, Willemsen R, de Graaff E, Tarleton JC, Leisti J, Warren ST, Oostra BA. Characterization of the full fragile X syndrome mutation in fetal gametes. Nature Genetics. 1997;15:165–169. doi: 10.1038/ng0297-165. [DOI] [PubMed] [Google Scholar]
- Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- Nagaraju G, Scully R. Minding the gap: the underground functions of BRCA1 and BRCA2 at stalled replication forks. DNA Repair. 2007;6:1018–1031. doi: 10.1016/j.dnarep.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DL. The fragile X syndromes. Semin Cell Biol. 1995;6:5–11. doi: 10.1016/1043-4682(95)90009-8. [DOI] [PubMed] [Google Scholar]
- O’Donnell WT, Warren ST. A decade of molecular studies of fragile X syndrome. Annu Rev Neurosci. 2002;25:315–338. doi: 10.1146/annurev.neuro.25.112701.142909. [DOI] [PubMed] [Google Scholar]
- Ontoso D, Kauppi L, Keeney S, San-Segundo PA. Dynamics of DOT1L localization and H3K79 methylation during meiotic prophase I in mouse spermatocytes. Chromosoma. 2013b doi: 10.1007/s00412-013-0438-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A, Hollingworth D, Adinolfi S, Castets M, Kelly G, Frenkiel TA, Bardoni B, Pastore A. The structure of the N-terminal domain of the fragile X mental retardation protein: a platform for protein-protein interaction. Structure. 2006;14:21–31. doi: 10.1016/j.str.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Reeve SP, Bassetto L, Genova GK, Kleyner Y, Leyssen M, Jackson FR, Hassan BA. The Drosophila fragile X mental retardation protein controls actin dynamics by directly regulating profilin in the brain. Curr Biol. 2005;15:1156–1163. doi: 10.1016/j.cub.2005.05.050. [DOI] [PubMed] [Google Scholar]
- Reyniers E, Vits L, De Boulle K, Van Roy B, Van Velzen D, de Graaff E, Verkerk AJ, Jorens HZ, Darby JK, Oostra B, et al. The full mutation in the FMR-1 gene of male fragile X patients is absent in their sperm. Nat Genet. 1993;4:143–146. doi: 10.1038/ng0693-143. [DOI] [PubMed] [Google Scholar]
- Sabra M, Texier P, El Maalouf J, Lomonte P. The Tudor protein survival motor neuron (SMN) is a chromatin-binding protein that interacts with methylated lysine 79 of histone H3. Journal of Cell Science. 2013;126:3664–3677. doi: 10.1242/jcs.126003. [DOI] [PubMed] [Google Scholar]
- Santoro MR, Bray SM, Warren ST. Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Annual Review of Pathology. 2012;7:219–245. doi: 10.1146/annurev-pathol-011811-132457. [DOI] [PubMed] [Google Scholar]
- Schultz-Pedersen S, Hasle H, Olsen JH, Friedrich U. Evidence of decreased risk of cancer in individuals with fragile X. American Journal of Medical Genetics. 2001;103:226–230. [PubMed] [Google Scholar]
- Tamanini F, Bontekoe C, Bakker CE, van Unen L, Anar B, Willemsen R, Yoshida M, Galjaard H, Oostra BA, Hoogeveen AT. Different targets for the fragile X-related proteins revealed by their distinct nuclear localizations. Hum Mol Genet. 1999;8:863–869. doi: 10.1093/hmg/8.5.863. [DOI] [PubMed] [Google Scholar]
- Tian H, Cao YX, Zhang XS, Liao WP, Yi YH, Lian J, Liu L, Huang HL, Liu WJ, Yin MM, et al. The targeting and functions of miRNA-383 are mediated by FMRP during spermatogenesis. Cell Death & Disease. 2013;4:e617. doi: 10.1038/cddis.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner G, Daniel A, Frost M. X-linked mental retardation, macro-orchidism, and the Xq27 fragile site. J Pediatr. 1980;96:837–841. doi: 10.1016/s0022-3476(80)80552-x. [DOI] [PubMed] [Google Scholar]
- Wolffe AP, Guschin D. Review: chromatin structural features and targets that regulate transcription. J Struct Biol. 2000;129:102–122. doi: 10.1006/jsbi.2000.4217. [DOI] [PubMed] [Google Scholar]
- Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.