Abstract
Chlamydomonas reinhardtii is the organism in which intraflagellar transport (IFT) was first visualized and in which the composition of IFT particles was originally elucidated. As the universality of IFT among ciliated/flagellated cells was uncovered, the diversity of organisms used to study IFT has grown. Still, because of the ease of isolation of flagella from Chlamydomonas and the battery of temperature-sensitive mutants affecting IFT proteins and motors, this unicellular alga remains the principal model for biochemical studies of IFT motors and cargo; furthermore, the long, exposed flagella of this cell are ideally suited for observing IFT in real time with GFP-tagged components of IFT.
I. Introduction
IFT was first observed by video-enhanced DIC light microscopy in flagella of Chlamydomonas as the movement of varicosities along the length of the flagella [Kozminski et al. (1993); see Kozminski (1995) for methods]. It was in Caenorhabditis elegans, however, that IFT was first visualized using GFP-tagged proteins (Orozco et al., 1999). Before that time, expression of foreign genes, including genes encoding GFP-tagged protein, in Chlamydomonas had been problematic, but in that same year a GFP gene was synthesized using the Chlamydomonas codon bias and incorporating mutations known to accelerate processing, increase fluorescence, and produce a single excitation peak at 489 nm in the encoded GFP (Fuhrmann et al., 1999). This engineered GFP was successfully expressed in Chlamydomonas and has since been used to tag several flagellar proteins including a subunit of the anterograde IFT motor (Mueller et al., 2005), an IFT protein (Qin et al., 2007), and a potential membrane cargo protein (Huang et al., 2007).
The steps required to image the movement of IFT particles in the flagella of Chlamydomonas are similar to those required for imaging of any GFP-tagged protein: genetically tagging and expressing the protein of interest in the cell, exciting the GFP, and imaging the fluorescent signal. Some of these steps will only be dealt with in a general way here, with more emphasis on some challenges specific to imaging GFP in Chlamydomonas flagella.
Whereas the movement of IFT components tagged with GFP has been visualized, the movement of cargo has been more elusive. CrPKD2, the Chlamydomonas orthologue of polycystin-2, a component of ciliary and flagellar membranes that plays a role in the development of polycystic kidney disease in humans, has been tagged with GFP and seen to move in an IFT-dependent manner (Huang et al., 2007), but whether it is transported directly by IFT is not certain. Components of the axoneme—dynein arms, radial spokes, proteins of the central pair complex and tubulin itself—make up a major class of putative cargo carried by IFT; yet to date no one has directly visualized any of these cargos moving by IFT. Instead, less direct biochemical methods have been used to study transport of these proteins into and out of flagella.
One axonemal component whose behavior within the flagellar matrix has been studied biochemically is the radial spoke. The intact radial spoke attached to the flagellar axoneme is a complex of 23 proteins that sediments at 20 S (Yang et al., 2001, 2006). In the cell body, however, the more prominent form of the spoke is a smaller complex composed of a subset of RSPs that sediment at 12 S (Qin et al., 2004). Both complexes are present in the soluble matrix fraction of the flagella: the 12 S complex on its way to the tip to assemble with other RSPs onto the axoneme; and the 20 S complex as a product of axonemal turnover, returning to the cell body. Analysis of flagellar matrix proteins in sucrose gradients for the presence of these two complexes can be used to differentiate anterograde from retrograde IFT cargo. Such analysis combined with various temperature-sensitive IFT mutants can be used to examine how deficiencies in the IFT machinery affect retrograde and anterograde movement of axonemal precursors. Methods will be described to analyze flagellar extracts on sucrose gradients to test for the presence of these complexes.
II. Methods for GFP Imaging of IFT Proteins
A. General Considerations in Tagging IFT Proteins
In the best of all worlds one could track the movement of an intracellular protein in its native state. Preferably a protein tag would have no effect on the activity or distribution of the protein and the tagged protein would be expressed at the same level as the endogenous protein that it replaces; but even if these standards are not completely met, a lot can be learned through GFP tagging. Frequently tags are placed at either the N- or C-terminus of the protein by use of a suitable vector. Our laboratory has designed such a vector (Huang et al., 2009), which contains two tandem sequences encoding GFP separated by two flexible linkers. As diagrammed in Fig. 1, judicious placement of unique restriction sites allows the 5′ or 3′ GFP sequence to be replaced by the coding sequence of choice, resulting in the encoded protein being fused at its C- or N-terminus to GFP via a flexible linker. Using this vector, transcription is driven by the PsaD promoter, which can transcribe cDNAs as well as genomic constructs in Chlamydomonas. A potential drawback of this vector is that to ensure the fidelity of expression of the tagged gene, use of its native promoter may be preferable.
Fig. 1.
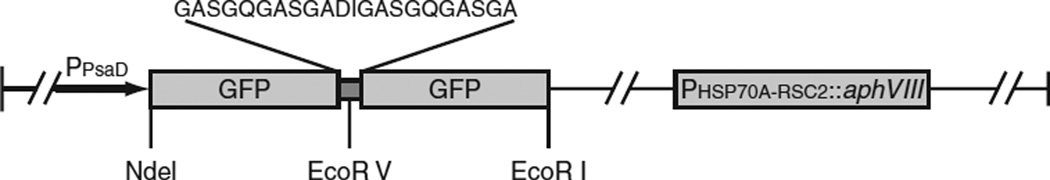
The vector designed for tagging Chlamydomonas genes with GFP, pHK85, has two copies of the GFP gene separated by two tandem linker sequences. The first or second GFP can be replaced by a cDNA or genomic sequence encoding a protein to be tagged with GFP at either its C- or N-terminus, respectively. Transcription is driven by the PsaD promoter and is terminated by the PsaD 3′ genomic sequence. The plasmid also encodes aphVIII as a selectable marker in Chlamydomonas.
To maintain the proper level of expression, ideally one would rescue a null mutant with a genomic clone encoding the tagged protein. In this way expression is driven by the native promoter and the tagged protein does not have to compete with the endogenous protein. This approach also has the advantage that rescue of the mutant phenotype ensures that the tagged protein is fully functional. Though ideal, null mutants are not necessary for satisfactory tagging of an IFT protein and observing its movement in flagella: the KAP subunit of the FLA10 heterotrimeric kinesin, which drives anterograde IFT, FLA3, has been successfully tagged with GFP in the temperature-sensitive mutant fla3-1 (Mueller et al., 2005); and the small GTPase IFT27 has been tagged with GFP and expressed in a wild-type background (Qin et al., 2007). In both cases the tagged protein was imaged moving in flagella at rates characteristic of IFT.
B. Preparing Cells for Observing Movement of GFP-Tagged Proteins
Motile Chlamydomonas often adhere to a coverslip by their flagella and remain quiescent, but they are unlikely to remain stationary in the light required for GFP imaging; therefore, in practice, paralyzed flagella are much more cooperative for making videos of GFP in flagella. Kozminski (1995) has detailed the advantages of using various paralyzed mutants and using mechanical or chemical means to paralyze flagella for observing IFT by video-enhanced DIC microscopy. For this purpose paralyzed flagella that remain flaccid are essential, so they can attach to a coverslip. Such flagella are found in motility mutants including pf1 (Kozminski et al., 1995) and pf15 (Iomini et al., 2001). The optical requirements for observing IFT by immunofluorescence are not as stringent as for DIC and stiff, paralyzed flagella, such as are found in pf18, are also suitable for observing the movement of intraflagellar GFP-tagged proteins if the cells are immobilized in 0.75–1% low melt agarose. One consideration when beginning a project of GFP tagging an IFT protein is, therefore, whether the recipient cells should be a paralyzed strain to facilitate the subsequent analysis of movement. When not complementing an IFT mutant with the tagged construct, using a paralyzed mutant as the recipient strain obviates the need for crossing the transformed strain. If transforming a flagellaless IFT mutant crossing the IFT mutant would be easier after transformation, when expression of the tagged protein has restored flagella to the transformant.
Although paralyzed mutants make the imaging of IFT more convenient, imaging can also be done in motile strains using a variety of methods to inhibit flagellar motility. We induce flagellar paralysis using 20 mM LiCl or 20 mM sodium pyrophosphate, which do not affect the rate or frequency of IFT (Dentler, 2005). Unlike sodium pyrophosphate, which causes flagellar shortening (Lefebvre et al., 1978), the flagella actually elongate in LiCl (Nakamura et al., 1987). We can obtain good videos of IFT27::GFP in cells with motile flagella with the following protocol. Pipette 3 µl of cells (~2 × 106 cells/ml) onto an 18 × 18-mm coverslip and add 1.2 µl of 200 mM LiCl. Pipette 8 µl of 1.5% low melt agarose maintained at 37°C onto the cells and gently mix it into the cell solution with the pipette tip. Carefully invert the coverslip onto a slide and gently tap it down so the solution covers the entire surface of the cover slip. Cells stabilized in the agar with straight, immotile flagella parallel to the coverslip can be located and used to visualize IFT. If the flagella begin to curl or swell at the tip as may occur after about 30 min, make a new slide.
C. Illumination
While mercury arc lamps can be used for exciting GFP and observing IFT in the flagella of living Chlamydomonas, an argon 488-nm laser excites GFP more effectively. Observing a GFP signal in the flagella is complicated by the overwhelming autofluorescence generated from the cell body, which can severely restrict the portion of the flagellum that can be clearly seen. To circumvent this problem we use a Mosaic digital illumination system (Photonics Instruments, St. Charles, IL) to control the area of the specimen that is illuminated. With this system we can illuminate only the flagella, thereby limiting the signal from the cell body. Another method of reducing the excessive fluorescence from the cell body is the use of confocal microscopy to limit detection of emitted light to a thin focal plane, as was done for KAP::GFP (Mueller et al., 2005), or TIRF microscopy in which only those GFP molecules very near the coverslip surface are excited (see Chapter 9 by Engel and Marshall, this volume). For the latter technique the flagella must be attached to the coverslip.
D. Optics and Image Capture
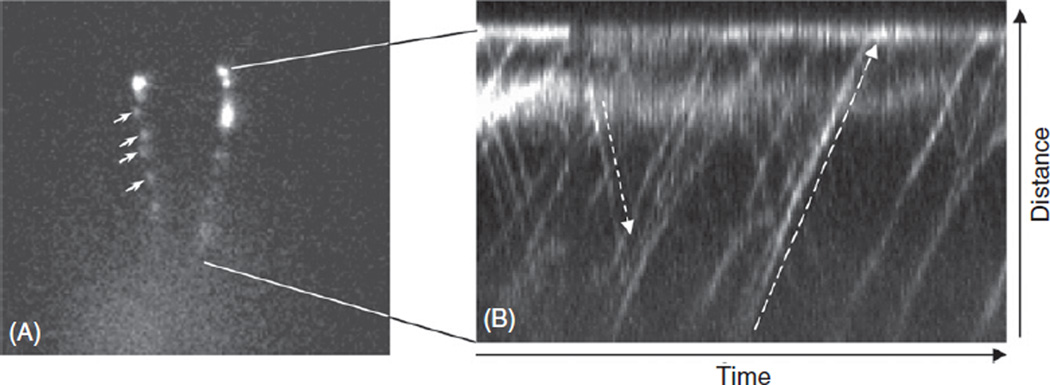
For imaging we use a Nikon Eclipse TE2000 inverted microscope equipped with a 100×, 1.4 NA Plan Apo objective lens. Samples are illuminated with a 300 mW 488 nm wavelength argon laser (Argon Ion Laser; National Laser Co., Salt Lake City, Ut) operating at 38 mW. The light is further attenuated by a prism that transmits only 70% of the incident light to the specimen. Using a tungsten lamp, preparations are scanned for a cell with stationary, preferably straight flagella parallel to the focal plane. Once one is found the Mosaic (or MetaMorph, (Molecular Devices, Sunnyvale, Ca)) software is used to draw the area to be illuminated over one or both flagella, avoiding getting too close to the cell body. Illumination is switched to the laser and images are collected with a Cascade 512B CCD camera (Photometrics, Tucson, AZ) with a back-illuminated 512 × 512 pixel array. The camera is controlled by MetaMorph software (version 7.1), and typically movement of flagellar IFT27::GFP can be imaged with the camera operating at 5 MHz with 4× gain, and an exposure time of 50 ms. For a full frame image this results in a frame rate of 118 ms. Faster frame rates can be obtained by decreasing the exposure time; operating the camera at 10 MHz, which also utilizes on-chip electron multiplier (EM) gain; and/or by decreasing the vertical size of the frame to a minimum. An example of a video of IFT27::GFP in the flagella of Chlamydomonas is shown in the supplementary material (http://www.elsevierdirect.com/companions/9780123749734, supplementary video) and one frame from the video is shown in Fig. 2.
Fig. 2.
Visualization of IFT27::GFP in the flagella of Chlamydomonas. (A) IFT27::GFP is visible in IFT trains (arrows) in the two flagella of the cell. A video of the IFT movement of this cell is presented in the supplementary material. (B) Traces of IFT trains are visible as lines on this kymograph of the video shown in the supplement. The downward pointing dotted line shows retrograde IFT and the upward pointed dotted line shows anterograde IFT.
To analyze movement, kymographs are generated using the kymograph function of MetaMorph by tracing a line over the flagellum wide enough to cover all of the fluorescent IFT trains and creating the kymograph based on maximum intensity (Fig. 2). Movement rates are computed from the slope of individual traces once the measurements of height and width are converted from number of pixels to distance in microns and time in seconds.
III. IFT Cargo
IFT cargo might be defined as molecules that are not part of the IFT machinery itself that are moved into, throughout, or out of the flagella by IFT. Evidence that axonemal proteins are cargo of IFT has been obtained through dikaryon rescue experiments (Piperno et al., 1996), sedimentation gradients (Piperno and Mead, 1997; Qin et al., 2004), immunoprecipitation (Qin et al., 2004), and genetic methods (Hou et al., 2007). Yet, to date, the membrane protein CrPKD2 is the only potential cargo whose movement in the flagella of Chlamydomonas actually has been measured to be the rate of IFT.
A. Analysis of IFT Cargo by Sucrose Density Gradients
Although we have not visualized radial spokes moving by IFT in flagella we can analyze factors that influence the accumulation of these putative cargos in the soluble fraction of the flagella. For example, the ratio of 12 S radial spoke complex entering flagella to 20 S complex exiting the flagella, increases during flagellar regeneration (Qin et al., 2004). This ratio is reversed (with relatively more 20 S exiting the flagella) when the flagella is resorbing due to inactivation of the anterograde motor in fla10 cells (Qin et al., 2004), which harbor a temperature sensitive mutation in the anterograde motor, FLA10.
B. Materials
HMDEK: 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes), pH 7.4, 5 mM MgSO4, 0.5 mM ethylene glycol bis(b-aminoethyl ether) N,N′-tetraacetic acid EGTA, 1 mM dithiothreitol, 25 mM KCl. We include the following protease inhibitors in HMDEK: 2 µg/ml aprotinin, 20 µg/ml benzamidine; 1 µg/ml leupeptin, 1 µg/ml pepstatin, and 50 µg/ml trypsin inhibitor. HMDEK + 10 or 30% sucrose.
C. Procedure
Harvest flagella from 32 to 40 l of Chlamydomonas according to published protocols (King, 1995; Witman, 1986; Witman et al., 1972). Regardless of which protocol is used, do not add sucrose to the cells before deflagellation (add it just after deflagellation instead) because 4% sucrose can affect the ratio of 12 S and 20 S spoke complexes and increases the amount of IFT proteins in the flagella. We generally use the pH shock method of deflagellation and obtain 0.25–0.50 ml of packed flagella when they are ultimately collected in a microfuge tube. All steps following deflagellation are done on ice or at 4°C.
Resuspend the flagellar pellet in an equal volume of HMDEK, add NP-40 to 0.5%, and incubate on ice for 10 min. Alternatively, the flagella can be frozen and thawed, releasing the matrix proteins but leaving much of the membrane in the axonemal pellet.
Sediment the insoluble material in a microfuge for 10 min.
Further clarify the supernatant at 100,000 × g for 10 min.
Load the clarified supernatant (less than 0.5 ml) onto a 12-ml 10–30% sucrose gradient in HMDEK. We pour gradients using a piston gradient former (Jule Inc., Milford, CT, USA) though satisfactory results can be obtained with an open air former.
Centrifuge the gradients for 16 h at 178,000g (38,000 rpm) in an SW41Ti rotor (Beckman Coulter).
Gradients are typically fractionated into 24–26 0.5-ml aliquots by pumping them from the bottom. A 25-µl capillary tube (Sigma) attached to 0.76-mm inner diameter pump tubing (Rainin) is inserted into the centrifuge tube. The tubing passes through a peristaltic pump and leads to a fraction collector. There may be a pellet in the bottom of the tube and if the capillary tube reaches the bottom of the tube fragments of the pellet may contaminate fractions. To avoid this problem put a piece of tape on the capillary tube to keep it suspended above the pellet.
Separate 15 µl aliquots of each fraction in an SDS-PAGE gel, transfer to blotting paper, and probe for a radial spoke protein.
The gradient profile should have two peaks of the radial spoke complexes at 12 S and 20 S. There may be some smaller complex or monomer as well. Standards should be used to calculate S-values: BSA (4.4 S), aldolase (7.35 S), catalase (11.3 S), and thyroglobulin (19.4 S) on parallel, or in the same, gradient. The 12-ml gradients fractionated into 24 fractions offers good resolution, but if this is not required, larger fractions can be collected or smaller gradients can be run. For example, 2-ml gradients can be run in 4 h to analyze 0.1 ml of soluble flagellar protein (Pan and Snell, 2003).
Biochemistry cannot directly show movement of molecules within the flagella so GFP imaging of axonemal precursors moving in the flagella at rates comparable to IFT would be a welcome advance. We have expressed RSP3::GFP in pf14 cells, but initial efforts to see its movement in the flagellum were hampered by the signal from the abundant copies of RSP3::GFP already attached to the axoneme. This problem may be solved by watching tagged spokes enter spokeless flagella following mating of the tagged strain with spokeless pf14 cells. Alternatively, photobleaching the tagged protein present on the axoneme or photoactivating a tagged protein inside the cell may allow precursors in transit to the flagellar tip to be seen.
Acknowledgments
I thank Dr. Kaiyao Huang for help with the microscopic techniques described here and for suggestions on the manuscript and Dr. Joel Rosenbaum for comments on the text and support of this work (National Institutes of Health grant GM014642).
References
- Dentler W. Intraflagellar transport (IFT) during assembly and disassembly of Chlamydomonas flagella. J. Cell Biol. 2005;170:649–659. doi: 10.1083/jcb.200412021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann M, Oertel W, Hegemann P. A synthetic gene coding for the green fluorescent protein (GFP) is a versatile reporter in Chlamydomonas reinhardtii. Plant J. 1999;19:353–361. doi: 10.1046/j.1365-313x.1999.00526.x. [DOI] [PubMed] [Google Scholar]
- Hou Y, Qin H, Follit J, Pazour GJ, Rosenbaum JL, Witman GB. Functional analysis of an individual IFT protein: IFT46 is required for transport of outer dynein arms into flagella. J. Cell Biol. 2007;176:653–665. doi: 10.1083/jcb.200608041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Diener D, Mitchell A, Pazour GJ, Witman GB, Rosenbaum JL. Function and dynamics of PKD2 in Chlamydomonas reinhardtii flagella. J. Cell Biol. 2007;179:501–514. doi: 10.1083/jcb.200704069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Diener DR, Rosenbaum JL. The ubiquitin conjugation system is involved in the disassembly of cilia and flagella. J. Cell Biol. 2009;186:601–613. doi: 10.1083/jcb.200903066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iomini C, Bebaev-Khaimov V, Sassaroli M, Piperno G. Protein particles in Chlamydomonas flagella undergo a transport cycle consisting of four phases. J. Cell Biol. 2001;153:480–491. doi: 10.1083/jcb.153.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SM. Large-scale isolation of Chlamydomonas flagella. In: Dentler WL, Witman GB, editors. Methods in Cell Biology. Vol. 47. San Diego: Academic Press; 1995. pp. 9–12. [DOI] [PubMed] [Google Scholar]
- Kozminski KG. High-resolution imaging of flagella. In: Dentler WL, Witman GB, editors. Methods in Cell Biology. Vol. 47. San Diego: Academic Press; 1995. pp. 263–271. [DOI] [PubMed] [Google Scholar]
- Kozminski KG, Beech PL, Rosenbaum JL. The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J. Cell Biol. 1995;131:1517–1527. doi: 10.1083/jcb.131.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. USA. 1993;90:5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre PA, Nordstrom SA, Moulder JE, Rosenbaum JL. Flagellar elongation and shortening in Chlamydomonas IV. Effects of flagellar detachment, regeneration, and resorption on the induction of flagellar protein synthesis. J. Cell Biol. 1978;78:8–27. doi: 10.1083/jcb.78.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller J, Perrone CA, Bower R, Cole DG, Porter ME. The FLA3 KAP subunit is required for localization of kinesin-2 to the site of flagellar assembly and processive anterograde intraflagellar transport. Mol. Biol. Cell. 2005;16:1341–1354. doi: 10.1091/mbc.E04-10-0931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Takino H, Kojima M. Effect of lithium on flagellar length in Chlamydomonas reinhardtii. Cell Struct. Funct. 1987;12:369–374. [Google Scholar]
- Orozco JT, Wedaman KP, Signor D, Brown H, Rose L, Scholey JM. Movement of motor and cargo along cilia. Nature. 1999;398:674. doi: 10.1038/19448. [DOI] [PubMed] [Google Scholar]
- Pan J, Snell W. Kinesin II and regulated intraflagellar transport of Chlamydomonas aurora protein kinase. J. Cell Sci. 2003;116:2179–2186. doi: 10.1242/jcs.00438. [DOI] [PubMed] [Google Scholar]
- Piperno G, Mead K. Transport of a novel complex in the cytoplasmic matrix of Chlamydomonas flagella. Proc. Natl. Acad. Sci. USA. 1997;94:4457–4462. doi: 10.1073/pnas.94.9.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Mead K, Henderson S. Inner dynein arms but not outer dynein arms require the activity of kinesin homologue protein KHP1FLA10 to reach the distal part of flagella in Chlamydomonas. J. Cell Biol. 1996;133:371–379. doi: 10.1083/jcb.133.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Diener DR, Geimer S, Cole DG, Rosenbaum JL. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell Biol. 2004;164:255–266. doi: 10.1083/jcb.200308132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Wang Z, Diener D, Rosenbaum J. Intraflagellar transport protein 27 is a small G protein involved in cell-cycle control. Curr. Biol. 2007;17:193–202. doi: 10.1016/j.cub.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman GB. Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 1986;134:280–290. doi: 10.1016/0076-6879(86)34096-5. [DOI] [PubMed] [Google Scholar]
- Witman GB, Carlson K, Berliner J, Rosenbaum JL. Chlamydomonas flagella I. Isolation and electrophoretic analysis of microtubules, matrix, membranes, and mastigonemes. J. Cell Biol. 1972;54:507–539. doi: 10.1083/jcb.54.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Diener DR, Rosenbaum JL, Sale WS. Localization of calmodulin and dynein light chain LC8 in flagellar radial spokes. J. Cell Biol. 2001;153:1315–1326. doi: 10.1083/jcb.153.6.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Diener DR, Yang C, Kohno T, Pazour GJ, Dienes JM, Agrin NS, King SM, Sale WS, Kamiya R, Rosenbaum JL, Witman GB. Radial spoke proteins of Chlamydomonas flagella. J. Cell Sci. 2006;119:1165–1174. doi: 10.1242/jcs.02811. [DOI] [PMC free article] [PubMed] [Google Scholar]