Abstract
Prostate-specific Ag (PSA) is a serine protease that is expressed exclusively by normal and malignant prostate epithelial cells. The continued high-level expression of PSA by the majority of men with both high- and low-grade prostate cancer throughout the course of disease progression, even in the androgen-ablated state, suggests that PSA has a role in the pathogenesis of disease. Current experimental and clinical evidence suggests that chronic inflammation, regardless of the cause, may predispose men to prostate cancer. The responsibility of the immune system in immune surveillance and eventually tumor progression is well appreciated but not completely understood. In this study, we used a mass spectrometry–based evaluation of prostatic fluid obtained from diseased prostates after removal by radical prostatectomy to identify potential immunoregulatory proteins. This analysis revealed the presence of Igs and the complement system proteins C3, factor B, and clusterin. Verification of these findings by Western blot confirmed the high-level expression of C3 in the prostatic fluid and the presence of a previously uncharacterized C-terminal C3 cleavage product. Biochemical analysis of this C3 cleavage fragment revealed a putative PSA cleavage site after tyrosine-1348. Purified PSA was able to cleave iC3b and the related complement protein C5. These results suggest a previously uncharacterized function of PSA as an immunoregulatory protease that could help to create an environment hospitable to malignancy through proteolysis of the complement system.
Prostate-specific Ag (PSA) is a serine protease that is a unique differentiation product of prostate tissue. PSA is one of the most abundant proteins in the seminal plasma, where it is present at milligram-per-milliliter concentrations. Although the exact physiologic role of PSA remains unknown, its major substrates in the seminal plasma are the gel-forming proteins semenogelin I and II (1–3). PSA is able to maintain the seminal plasma in a semiliquid state through cleavage of these gel-forming proteins. PSA is also produced in high amounts by prostate cancer cells. A role for PSA in the pathobiology of prostate cancer has been proposed based on its effect on prostate cancer growth (4) and its ability to cleave several important growth regulatory proteins (5). However, the exact role for PSA in prostate cancer has yet to be defined clearly. PSA is not expressed by any other tissue in the adult human man and leaks from prostate cancer sites with disrupted tissue architecture. On this basis, PSA has utility as a biomarker for prostate cancer. The overwhelming majority of men with prostate cancer, even those with poorly differentiated, high-grade disease, continue to express PSA at high levels throughout the course of disease progression.
The word prostate is derived from Greek and literally means “one who stands before” or “protector” (6). Although the exact role of the prostate gland is not clear, it is the guardian of the genitourinary tract and prevents foreign materials from entering the reproductive apparatus of the male. In light of this role, the prostate of the aging man exhibits significant chronic inflammation that can lead to the development of prostate cancer (7). The prostate tissue may be proinflammatory, but the prostatic fluid is not, as evidenced by the fact that men with prostatitis commonly have no or minimal inflammatory cells in the prostatic secretions. Immunoregulation within the prostatic fluid must also be finely balanced. The fluid must have the capability to eliminate foreign bacteria and viruses entering the genitourinary tract through the urethra. It must also shield the sperm from immune destruction within the vaginal tract while not eliminating cells within the reproductive tract of the female. In this regard, seminal plasma is devoid of complement activity and actually has a strong anti-complement activity (8–10).
In this study, we used a mass spectrometry–based evaluation of prostatic fluid obtained from cancer-containing prostates after removal by radical prostatectomy to identify potential immunoregulatory proteins. This analysis revealed the presence of Igs, as well as complement system proteins C3, factor B, and clusterin. Verification of these findings by Western blot analysis confirmed the high-level expression of C3 and a previously uncharacterized C-terminal C3 cleavage product. Biochemical analysis of this C-terminal cleavage fragment revealed a putative PSA cleavage site that was confirmed using purified PSA and C3. Additional studies revealed PSA to preferentially cleave iC3b, itself a cleavage product resulting from complement activation. We then tested whether this activity had functional consequences on CR3 activation, but could not detect any. Finally, we determined that the evolution-related complement protein C5, but not C4, is a substrate of PSA as well. PSA-mediated proteolysis of C5 inhibits complement pathway activity. These results suggest a previously unknown function of PSA as an immunoregulatory protease that could help to create an environment that is hospitable to malignancy through inactivation of the complement system. Finally, these findings suggest that PSA also has immunoregulatory activity in the seminal plasma to aid in normal fertility that could have been co-opted by prostate cancer cells as a means to avoid immune destruction.
Materials and Methods
Patient samples and cell lines
Prostatic fluid samples were collected from radical prostatectomy specimens as described previously according to a protocol approved by an institutional review board (11). Seminal plasma was obtained from discarded clinical samples. The RAW 264.7 macrophage cell line was obtained from American Type Culture Collection (Manassas, VA).
Mass spectrometric sample preparation and analysis
Individual prostatic fluid samples were loaded into the wells of a 4–12% Bis-Tris NuPage gel. Following electrophoretic separation, the gel was stained with SimplyBlue SafeStain (Invitrogen). Individual gel lanes were excised into 12 similarly sized pieces, and each piece was placed into a separate microcentrifuge tube. The stain of the gel slices was destained with water before being immersed into 500 μl of 100 mM ammonium bicarbonate. In-gel tryptic digestion was performed on all gel slices (1:20 ratio trypsin enzyme:substrate) for 18 h at 37°C. Mass spectrometric analysis and subsequent protein identifications were performed as described previously (4).
Western blot
Prostatic fluid samples stored at −80°C were thawed and centrifuged, and protein concentrations in the supernatant were determined using the bicinchoninic acid method. Proteins (5 μg) were separated by SDS-PAGE and then transferred to polyvinylidene difluoride (PVDF) membrane (Bio-Rad). Membranes were blocked with 4% nonfat milk in TBS-Tween 0.1%. Primary and secondary Abs were prepared in the same diluent. The membrane was probed with monoclonal anti-human–C3b-α (1:10,000; clone H206) from Millipore and ECL-anti-mouse IgG (1:8000) from GE Healthcare. The membrane was incubated with SuperSignal West Pico Substrate (Pierce) then exposed to x-ray film.
Immunoaffinity purification
Polyclonal anti-C3 (Complement Technology) was covalently linked to AminoLink Coupling Resin (Pierce) by following the manufacturer’s instructions. Briefly, 16.5 mg Ab was diluted into 2 ml coupling buffer before adding 40 μl sodium cyanoborohydride. This mixture was added to 2 ml resin and incubated for 5 h under gentle agitation. The column was washed, and remaining active sites were blocked before additional washing. Four prostatic fluid samples were pooled then diluted to 1.5 ml in TBS. Samples were added to the prepared column, and binding occurred for 1 h. The column was washed, and then elution buffer was added and 1-ml fractions were collected. Fractions containing relevant protein were concentrated using an Amicon Ultra-4 Centrifugal Filter Unit with an Ultracel-10 membrane (Millipore).
Edman degradation
Concentrated immunopurified prostatic fluid was separated on a 4–15% gel and transferred to a PVDF membrane. The membrane was cut in half where a small amount of immunopurified sample was probed with the anti-human–C3b-α Ab as described above. The remaining membrane was incubated with Coomassie stain before a brief destaining. The x-ray film was overlaid onto the Coomassie-stained membrane to identify the correct band that was then excised and sent to the Johns Hopkins Synthesis and Sequencing Facility for Edman degradation. The first seven N-terminal amino acids were determined with a Perkin-Elmer/Applied Biosystems Procise Protein Sequencing System.
Coincubation of C3/C3b/iC3b and PSA
Purified human C3, C3b, and iC3b (Complement Technology) were incubated with enzymatically active PSA (AbD Serotec) in the presence of 10 μM aprotinin (Sigma). PSA inhibitor (1 μM) was added to control reactions. Reactions took place in PSA buffer (50 mM Tris, 100 mM NaCl, pH = 7.5) overnight at 37°C. Reactions were stopped by the addition of sample loading buffer. Proteins were separated by SDS-PAGE and transferred to PVDF membrane as described above. Membranes were stained with Coomassie blue, briefly destained, and digitally imaged. The band at ~37 kDa was excised and sent for Edman degradation as described above.
Determination of cofactor activity
Purified human C3b was incubated with enzymatically active PSA and an increasing amount of factor H (Quidel). Reactions took place in PSA buffer overnight at 37°C. Reactions were stopped by the addition of sample loading buffer. Proteins were separated by SDS-PAGE and transferred to PVDF membrane as described above.
C3b/iC3b deposition assay
Sheep erythrocytes (ES) were opsonized with C3b as described (12). iC3-bopsonized ES were prepared by incubating Ab-sensitized sheep erythrocytes (EA) with C5-depleted serum. Approximately 2 million EA (Complement Technology) were mixed with 10 μL normal human serum stripped of C5 by immunoaffinity chromatography (C5 [−] NHS) in triplicate. After 20 min at 37°C, erythrocytes were washed twice with PBS. ES and EA were resuspended in PSA or BSA (125 μg/ml) in the presence of aprotinin (10 μM) and then incubated at 37°C for 2 h on a rotisserie mixer. Cells were washed once with PBS and resuspended in a 10-μg/ml solution of anti-human-C3b-α (clone H206) and incubated for 1 h on ice. Cells were washed once with PBS and resuspended in a 10-μg/ml solution of anti-mouse IgG Alexa Fluor 488 and incubated for 30 min on ice in the dark. Cells were washed with PBS and fixed with formalin. Levels of C3b-α were measured with a BD FACSCalibur at the Sidney Kimmel Comprehensive Cancer Center Flow Cytometry Core Facility.
CR3-mediated phagocytosis of EA-iC3b
Assessment of complement-mediated phagocytosis was performed as described (13, 14). EA-iC3b were prepared as described above. EA-iC3b were incubated with enzymatically active PSA or BSA in PSA buffer overnight at 37°C. RAW 264.7 cells were propagated in DMEM supplemented with 10% FBS in a humidified atmosphere of 5% CO2 at 37°C. The cells were seeded on polylysine-coated 96-well plates such that they were 90% confluent on the day of experimentation. RAW 264.7 cells were stimulated with 125 ng/μl PMA (Promega) for 10 min at 37°C. EA-iC3b pretreated with PSA or BSA were added to the stimulated RAW 264.7 cells, and phagocytosis proceeded for 75 min at 37°C. Phagocytosis was quantified colorimetrically by the conversion of 2,7-diaminofluorene by hemoglobin into a product that absorbs at 620 nm. The RAW 264.7 cells were washed twice in PBS. Erythrocytes that had bound but not been internalized were lysed by a brief incubation in 0.2% NaCl. The RAW 264.7 cells were again washed twice in PBS before being lysed with 50 μl of 6 M urea in 0.2 M Tris-HCl (pH 7.4). The cell lysates were mixed with 75 μl of working solution (10 volumes 0.2-M Tris-HCl pH 7.4, 1 volume 2,7-diaminofluorene stock, and 0.02 volumes 30% hydrogen peroxide). Absorbance at 620 nm was monitored with a plate reader.
Coincubation of C4/C5 and PSA
Purified human C4 and C5 (Complement Technology) were incubated with enzymatically active PSA in the presence of 10 μM aprotinin in PSA buffer. After overnight incubation at 37°C the reaction products were separated by SDS-PAGE and stained with SimplyBlue SafeStain.
C5 supplementation of C5 (−) NHS
Purified human C5 was mixed with enzymatically active PSA or BSA and incubated overnight at 37°C. The next day, 50 μl EAwas supplemented with 2 μl C5 (−) NHS. The C5 pretreated with PSA or BSA was added to the erythrocytes and incubated at 37°C for 20 min. Reactions were centrifuged at 1000 × g, and the supernatants were collected. The absorbance of the supernatant at 415 nm was recorded.
Comparison of C5 levels in serum, prostatic fluid, and seminal plasma
A Western blot analysis was performed as described above. The membrane was probed with polyclonal anti-human-C5 (1:2,000) from Complement Technology and donkey anti-goat IgG-HRP (1:20000) from Santa Cruz Biotechnology.
Addition of C5 to fresh seminal plasma
Purified human C5 in PSA buffer was incubated with fresh seminal plasma for 2 h at 37°C. PSA inhibitor (10 μM) was added to control reactions. Reactions were stopped by the addition of sample loading buffer. A Western blot was performed as described above. The membrane was probed with polyclonal anti-human-C5 (1:2,000) and donkey anti-goat IgG-HRP (1:20,000).
Results
Mass spectrometric based identification of 95 proteins in prostatic fluid
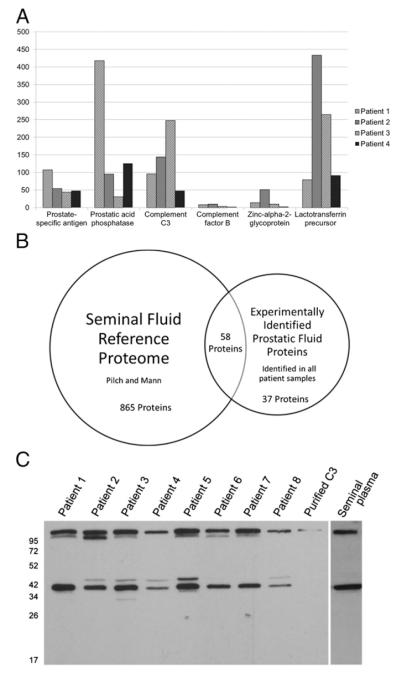
All protein species identified from the prostatic fluid of each of the four patients were introduced into the proteomic platform Protein Center (Thermo Fisher Scientific) as individual patient proteome files using the methodology outlined by Williams et al. (4). A comparative analysis was performed to determine which proteins had been identified in all analyzed patient samples (Fig. 1A and Supplemental Table I). The 95 proteins common to all four patients were introduced as an independent data set. Using the seminal plasma proteome published by Pilch and Mann (15) as a reference database, the subset of common experimentally identified proteins was compared with the reference database. Of the 95 proteins included in the experimental data set, 58 had been identified previously in seminal plasma (Fig. 1B). Both our dataset and the dataset of Pilch and Mann included proteins known to be expressed by the prostate at high levels such as PSA and prostatic acid phosphatase; inclusion of these proteins served as internal validation. Complement system proteins C3, factor B, and clusterin were detected in all four patient samples. These three proteins were also present in the reference database. Additional complement proteins present in the reference database but not in our dataset included C1, C2, C4, C9, and complement factor I.
FIGURE 1.
Proteomic analysis of prostatic fluid samples from radical prostatectomy specimens of men with prostate cancer. (A) Examples of the 95 proteins identified in each of four prostatic fluid samples. Y-axis indicates number of peptide “hits” for each protein from mass spectrometric analysis. (B) The prostatic fluid experimental dataset has considerable overlap with the previously described seminal plasma reference database. A Venn diagram was constructed showing overlap between protein species identified in our screen of prostatic fluid samples and the seminal plasma reference proteome of Pilch and Mann (12). Protein species found in our screen must have been identified in all four prostatic fluid samples to be included in the comparison. (C) Patient prostatic fluid and normal seminal plasma contains native C3 and a C3 fragment at ~37 kDa. Western blot of eight random prostatic fluid samples, purified human C3, and the seminal plasma of a healthy donor were probed for the presence of C3 with a monoclonal anti-human–C3b-α (clone H206) Ab.
C3 and a previously uncharacterized C3 fragment are present in diseased prostatic fluid and normal seminal plasma
To confirm the results from our proteomic study, we analyzed eight additional prostatic fluid samples from men with prostate cancer by Western blot to confirm the presence of C3. The Ab for this analysis, monoclonal anti-human-C3b-α (clone H206), is directed toward an epitope present on the α-chain of the C3 protein. The exact epitope recognized by this Ab is not known, but it is able to detect both C3b and C3c, which is consistent with detection of an epitope toward the C terminus of the C3 α-chain (16). C3 was detected in all eight prostatic fluid samples and in the seminal plasma of a healthy man (Fig. 1C). While equal amounts of protein were loaded for each sample, varying levels of C3 were detected by Western blot. Six of the eight prostatic fluid samples also tested positive for a C3 fragment of ~105 kDa that most likely represented C3b, which is evidentiary of complement activation. All eight prostatic fluid samples and the seminal plasma from a healthy donor also tested positive for a 37-kDa fragment using the anti-C3b-α Ab. This 37-kDa fragment was not detected in the serum of healthy individuals or in the serum of patients with prostate cancer (data not shown). Of the previously described C3 cleavage fragments, this 37-kDa fragment appears to be closest in size to C3c α-chain fragment 2.
Characterization of the novel 37 kDa C3 fragment
C3 is a well-characterized protein whose activation and degradation are tightly regulated. After conversion to C3b by the C3 convertase complex, C3b is subsequently inactivated by the proteolytic activity of factor I in the presence of cofactor molecules factor H, CR1, or CD46/membrane cofactor protein (MCP). Factor I cleavage generates multiple previously characterized cleavage fragments that include C3c, C3dg, and C3f (17) (Fig. 2). To better characterize this putative C3 fragment, immunoaffinity purification was used to purify the 37-kDa fragment from prostatic fluid for further characterization. Purification was achieved using a polyclonal C3 Ab. Because prostatic fluid sample volume was limited, four samples were pooled before purification. Seven cycles of Edman degradation on the purified pooled prostatic fluid samples revealed the amino acid sequence of the N-terminus of the 37-kDa fragment to be “HAKAKDQ.” Comparison to the C3 reference sequence indicates the that 37-kDa fragment is indeed a previously undescribed C3 fragment that maps to the 36.5-kDa C-terminal portion of the C3 α-chain (Fig. 2). This 37-kDa fragment is detectable under reducing conditions by Western blot because of its release from the N-terminal portion of the C3 α-chain following reduction of the disulfide bond. Further inspection of the sequence flanking of the cleavage site revealed that the new N-terminus was created by a chymotrypsin-like protease with cleavage after tyrosine-1348 in the C3 protein. In contrast, all other previously described C3 cleavage fragments are produced following cleavage by trypsin-like proteases. Furthermore, cleavage at tyrosine-1348 to generate the 37-kDa fragment, like every other previously described C3 fragment, is the result of cleavage within the C3 α-chain. C3 β-chain cleavage fragments have not been described.
FIGURE 2.
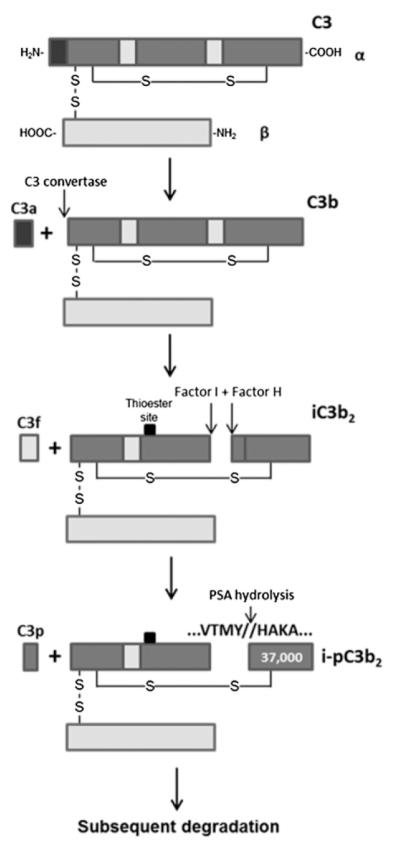
Schematic of complement C3 activation and degradation. After activation by the convertase, C3 is subject to normal degradation involving sequential factor I cleavage with factor H cofactor activity. iC3b is then subject to PSA cleavage after tyrosine-1348 and potentially other uncharacterized sites. PSA cleavage results in the production of a new 37-kDa fragment. The black square indicates the thioester site within C3.
PSA can cleave iC3b and generate the 37 kDa fragment in vitro
PSA is the major chymotrypsin-like serine protease in the seminal plasma and prostatic fluid. Therefore, we hypothesized that PSA was cleaving C3 based on sequence similarity between known PSA substrates and the cleavage sequence N-terminal to the tyrosine-1348 within C3, “TLSVVTMY/HAKAKDQ” (Fig. 2). To test this hypothesis, we incubated purified human C3 with purified enzymatically active PSA. The addition of a potent and specific PSA inhibitor (18) served as a negative control. Reducing gel electrophoresis revealed no significant cleavage of the C3 α-chain (Fig. 3A). This finding led us to hypothesize that tyrosine-1348 was part of a cryptic site exposed after proteolytic activation of C3 into C3b or iC3b. To test this hypothesis, we incubated purified human C3b and iC3b with purified enzymatically active PSA, again using a PSA inhibitor as a negative control. Reducing gel electrophoresis revealed degradation of both fragments of the iC3b α-chain; however, no effect was observed with C3b (Fig. 3A). Cleavage of iC3b resulted in a fragment at an m.w. similar to that observed after similar analysis of the prostatic fluid samples. To confirm that this cleavage product was the same proteolytic fragment detected in the prostatic fluid, we excised and sequenced the 37-kDa band by Edman degradation. The N-terminus of the PSA generated fragment was confirmed to be “HAKAKDQ,” consistent with cleavage after tyrosine-1348.
FIGURE 3.
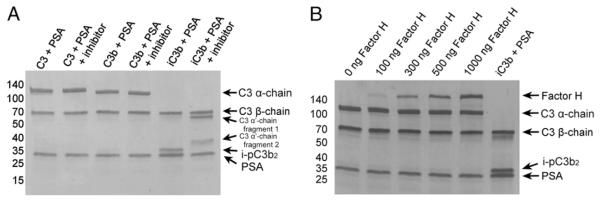
(A) PSA preferentially cleaves iC3b. Purified human C3, C3b, and iC3b were incubated with enzymatically active PSA in the presence of 10 μM aprotinin. PSA inhibitor (1 μM) was added to control reactions. Coomassie staining of a gel run under reducing conditions revealed a cleavage product at ~37 kDa that was generated in the absence of PSA inhibitor. (B) Factor H does not have cofactor activity for PSA-mediated cleavage of C3b. Purified human C3b was incubated with enzymatically active PSA and an increasing amount of factor H. Proteins were separated by SDS-PAGE and transferred to PVDF membrane before staining with Coomassie blue.
Factor H does not have cofactor activity to facilitate PSA-mediated cleavage of C3b
Factor I is unable to cleave C3b in the absence of the cofactor factor H. Therefore, we hypothesized that factor H might also have cofactor activity for PSA, enabling it to cleave C3b. To test this hypothesis, we repeated our C3b proteolysis assay with PSA in the presence of complement factor H (Fig. 3B). Results show that factor H does not impart any cofactor activity on PSA to mediate cleavage of C3b.
PSA cleaves iC3b, but not C3b, deposited on the surface of sheep erythrocytes
The next experiments were performed to determine whether PSA could cleave C3b or iC3b in a more relevant cellular context. ES were opsonized with C3b using purified C3 and alternative pathway enzymes factor B and factor D in the absence of factor I and H to prevent cleavage of C3b to iC3b. EA were opsonized with iC3b by brief incubation with C5-depleted normal human serum. The addition of C5-depleted serum ensures that the complement activation pathway only proceeds through deposition of C3b on the cell membrane and prevents the formation of the membrane attack complex and subsequent cell lysis. Factors I and H present in the C5-depleted serum convert C3b into iC3b. ES-C3b and EA-iC3b were incubated with enzymatically active PSA at 37°C. The erythrocytes were collected and labeled with monoclonal anti-human–C3b-α (clone H206) and analyzed by flow cytometry. Analysis revealed a decrease in C3b-α Ab signal when EA-iC3b were treated with 125 μg/ml PSA (approximately a 10-fold lower level of PSA than that observed in the prostatic fluid) (19) compared with the signal observed when cells were treated with the same concentration of BSA (Fig. 4A). Treatment of ES-C3b with PSA did not result in a decrease of the C3b-α Ab signal (Fig. 4B). To determine whether PSA was releasing the 37-kDa iC3b fragment into the supernatant, we collected and tested it for the presence of C3 fragments using Western blot. Two C3 fragments, one at 37 kDa and another at 39 kDa, were detected, which is consistent with a combination of factor I and PSA cleavage (Fig. 4C).
FIGURE 4.
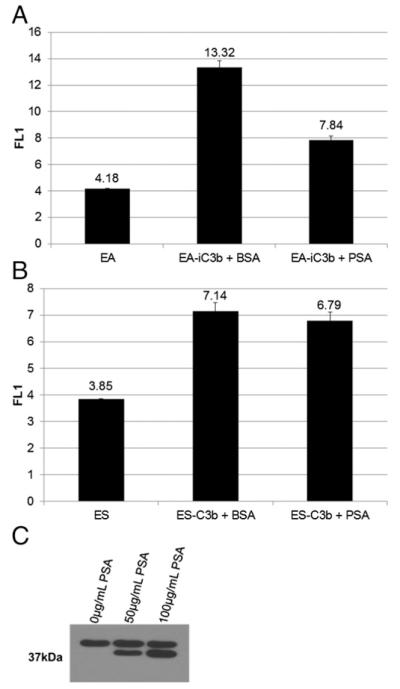
PSA is able to remove the 37-kDa C3 fragment from the surface of sheep erythrocytes opsonized with iC3b, but not C3b. (A) EA were coated with iC3b using C5-depeleted serum before treatment with equal amounts of PSA or BSA. (B) ES were coated with C3b using purified alternative pathway proteins and then treated with equal amounts of PSA or BSA. Flow cytometric analysis was performed to assess the amount of iC3b or C3b on the surface using an anti-C3 Ab. Because the cell population was homogenous, all cells were included in the gate. (C) The supernatant was isolated from PSA-treated iC3b-opsonized erythrocytes and probed for the presence of the 37-kDa fragment and other C3 fragments under reducing conditions by Western blot with the H206 Ab.
PSA-mediated cleavage of EA-iC3b does not alter complement-dependent phagocytosis
After conversion of C3b to its inactivated form, iC3b can no longer bind factor B and act as a C3 convertase. However, iC3b and its degradation product C3dg are active molecules that trigger specialized immune responses by interacting with complement receptors on leukocytes (20). Complement-dependent phagocytosis is an important mechanism of the host defense system and is primarily mediated by complement receptor CR3, and to a lesser extent CR1 and CR4. CR3 is expressed on many immune cells, including macrophages, monocytes, and neutrophils. C3b does not interact with CR3, and iC3b is predicted to interact with CR3 through binding sites that become exposed upon unfolding of the CUB domain after cleavage of the C3b α-chain (21). We hypothesized that the ability of PSA to cleave iC3b between the CUB and MG8 domain on the α-chain might interfere with CR3 binding. To test this hypothesis, we used an established protocol to measure complement-dependent phagocytosis (14). In this assay, the CR3+ RAW 264.7 macrophage cell line internalizes iC3b opsonized sheep erythrocytes. PSA-treated EA-iC3b were prepared as usual and were added to prestimulated RAW cells at a 20:1 ratio. Phagocytosis was stopped, and bound cells that had not internalized were lysed by addition of a hypotonic solution. A sensitive colorimetric assay that relies on the pseudoperoxidase activity of hemoglobin was used to evaluate the phagocytic efficiency (13). Cells were lysed, and hemoglobin was released from internalized EA-iC3b. The relative internalization can be measured by the pseudoperoxidase activity of hemoglobin, which converts 2,7-diaminofluorene into fluorene blue and can be measured spectrophotometrically. This sensitive method of detection did not demonstrate any difference in the degree of phagocytosis between PSA-treated and control EA-iC3b (Fig. 5).
FIGURE 5.
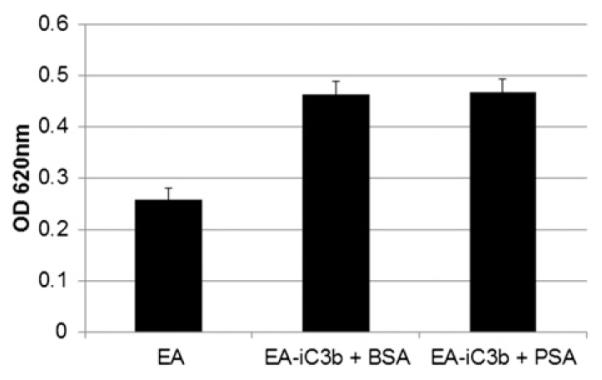
PSA-mediated cleavage of iC3b does not affect CR3-dependent phagocytosis. Sheep erythrocytes opsonized with iC3b were treated with PSA (100 μg/ml) or an equal amount of BSA overnight. RAW 264.7 cells were stimulated with 125 nM PMA for 10 min, after which erythrocytes were added (~20:1). The erythrocytes were phagocytised for 75 min. Erythrocytes bound but not internalized were lysed, and the number of erythrocytes phagocytosed were quantified by the colorimetric conversion of 2,7-diaminofluorene to fluorene blue (OD620) by the pseudoperoxidase activity of hemoglobin.
PSA also cleaves the homologous C5 protein
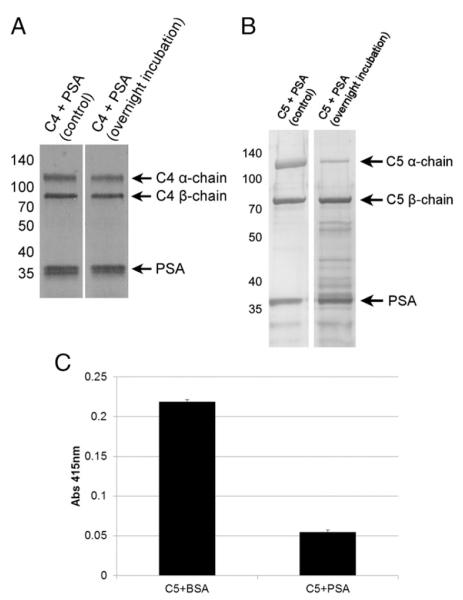
The complement system is a collection of >30 different proteins. Three key components (C3, C4, and C5) are thought to have evolved from a common ancestor, and they all share a similar m.w. and chain structure (22). Because of the similarities among the three proteins, we were curious whether C3 was uniquely cleaved by PSA or whether all were substrates of PSA. We treated C4 and C5 with enzymatically active PSA and looked for cleavage products by electrophoresis. We could not detect any significant proteolysis of the C4 α- or β-chains (Fig. 6A). The α-chain of C5 exhibited significant proteolytic degradation, whereas the β-chain was left intact (Fig. 6B), similar to what we observed with C3.
FIGURE 6.
(A) PSA does not cleave C4. (B) PSA cleaves the C5 α-chain, leaving the β-chain intact. Purified human C4 and C5 was incubated with enzymatically active PSA in the presence of 10 μM aprotinin. Coomassie staining of a gel run under reducing conditions revealed proteolysis of the C5 α-chain. (C) PSA-mediated cleavage of C5 is inhibitory. C5-depleted normal human serum was supplemented with C5 that had been incubated with PSA or BSA overnight. This serum was added to EA. Complement activity was quantified by absorbance of the supernatant at 415 nm after hemolysis.
PSA-mediated cleavage of C5 has functional consequences
We were curious whether PSA-mediated cleavage of C5 had functional consequences on the integrity of the complement cascade. To test this possibility, we used EA to assay total complement hemolytic activity. C5 was incubated with PSA overnight. The following day, we supplemented C5-depleted normal human serum with PSA-treated C5 or control C5 and added it to EA. We observed significantly less complement activity in the sample supplemented with PSA-treated C5 compared with control C5, indicating that PSA-mediated proteolysis of C5 negatively regulates the complement pathway (Fig. 6C).
Proteolysis of C5 in the seminal plasma can be abrogated by a PSA inhibitor
Seminal plasma is a rich source of proteins, including proteins of the complement system (15). However, unlike serum, this fluid is not a source of fully functional complement, likely because of the presence of complement inhibitory proteins and the absence of certain complement factors. Notably missing in the seminal plasma is C5 (Fig. 7A). We were curious whether the lack of C5 in seminal plasma might be due in part to PSA proteolytic activity. To answer this question, we supplemented seminal fluid with purified human C5 in the presence or absence of a PSA inhibitor. We then determined C5 levels by Western blot with a polyclonal Ab. In the absence of a PSA inhibitor, seminal plasma was able to degrade the α-chain of C5, leaving the β-chain intact (Fig. 7B).
FIGURE 7.
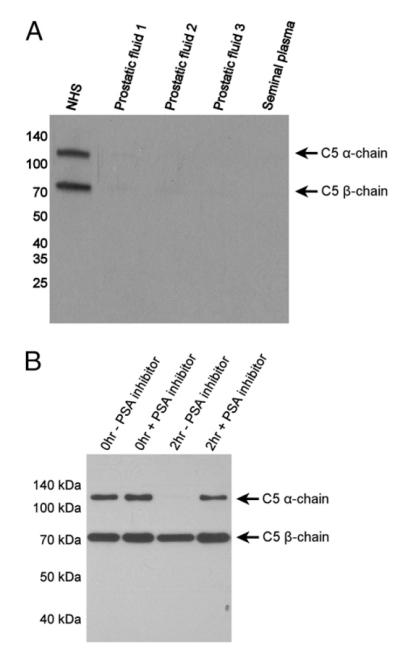
(A) C5 is not present in diseased prostatic fluid or healthy seminal plasma. Proteins (5 μg) were separated by SDS-PAGE and then transferred to PVDF membrane. The membrane was probed with polyclonal anti-human-C5. (B) Proteolysis of C5 in the seminal plasma can be abrogated by a PSA inhibitor. Seminal plasma was supplemented with purified human C5 in the presence or absence of a PSA inhibitor. After a 2-h incubation, C5 levels were determined by Western blot with a polyclonal C5 Ab.
Discussion
Complement is regarded as one of the first lines of immunologic defense, defending the host from foreign invaders by one of three pathways of activation known as the classical pathway, alternative pathway, and lectin pathway (23). Complement factor C3 has a central role in the complement cascade and supports the activation of all three pathways. Human C3 is the most abundant complement protein in the serum and, based on our proteomic studies of the prostatic fluid, is also one of the most abundant proteins in the seminal plasma. C3 is highly regulated before and after activation by C3 convertases. Cleavage by C3 convertases releases the anaphylatoxin C3a and generates C3b. Once formed, C3b rapidly attaches via covalent bond formation to various acceptors on the surface of bacteria and host cells. Because C3b does not have the ability to discriminate between self and non-self, it has the potential to damage host cells. Therefore, membrane-bound C3b activity must be regulated by other complement proteins. In this regard, C3b expresses multiple binding sites for other complement components that either amplify its convertase activity (factor B and properdin in the presence of factor D) or inactivate its activity (proteolysis by factor I in the presence of factor H, CR1, or CD46). C3b’s factor I mediated degradation product, iC3b, has an equally interesting biology. iC3b interacts with CR2, CR3, and CR4, the first of which plays a role in enhancing B cell immunity. iC3b’s other receptor binding partners, CR3 and CR4, have roles in clearance of pathogens by phagocytosis. In this study, we provide initial evidence that human PSA, via its chymotrypsin-like serine protease activity, can modulate the complement system through degradation of iC3b to produce new C3 degradation fragments and through degradation of the complement protein C5, thereby inactivating the complement cascade.
In this study, PSA was shown to cleave iC3b and was unable to cleave C3 or C3b. C3 is known to undergo a significant conformational change upon activation into C3b and then again following deactivation into iC3b by sequential proteolysis (21, 24–27). High-resolution crystal structures exist for both C3 and C3b documenting these conformational changes. These crystal structures detect a conformational change of up to 95Å and the exposure of cryptic binding sites. Examination of the crystal structures of C3 and C3b (PDB ID 2A73 and 2I07, respectively) reveal that the PSA cleavage site at tyrosine-1348 is part of a β-strand facing the interior of the protein, making it an inaccessible substrate of PSA. Unfortunately, we have a limited understanding of the structure of iC3b. The conversion from C3b to iC3b likely results in significant shifts and the generation of cryptic binding sites much like the earlier conversion from C3 to C3b. iC3b, but not C3b, interacts with CR2, CR3, and CR4, so these sites must be hidden in C3b but made accessible upon conversion to iC3b. Low resolution 3D-electron microscopy analysis of iC3b confirms a significant conformational change upon conversion from C3b to iC3b, but cannot provide atomic resolution (26). Our results indicate only iC3b to be a substrate of PSA, suggesting conversion into iC3b makes tyrosine-1348 accessible to the solvent and thus PSA-mediated proteolysis.
To confirm that this proteolytic activity could be duplicated in a more relevant cellular context, we repeated the assay with C3b and iC3b covalently attached to the surface of sheep erythrocytes (ES-iC3b and EA-iC3b, respectively) and analyzed it using flow cytometry. Following C3 activation, C3b becomes attached to cell membranes because of the formation of a covalent bond between the C3b protein and the cell surface. This bond is formed when exposed hydroxyl and amine groups on cell surface proteins and carbohydrates interact with the reactive thioester bond within the C3b protein. C3b is subject to factor I proteolysis resulting in iC3b, itself an important protein that interacts uniquely with CR2, CR3, and CR4. We treated both EA-iC3b and ES-C3b with PSA, but observed a decrease in Ab signal only with EA-iC3b, consistent with removal of part or all of iC3b from the erythrocyte surface. After PSA treatment, the 37-kDa iC3b fragment could be detected in the supernatant. PSA-mediated cleavage of iC3b after tyrosine-1348 alone would not liberate the 37-kDa fragment from the surface of the cell because of disulfide bonds linking the iC3b α-chain fragments 1 and 2. Electrophoretic analysis indicates additional PSA-mediated cleavage of iC3b (Fig. 3A), including cleavage of the α-chain fragment 1, which would release the 37-kDa fragment from the surface of the erythrocyte. Unfortunately cleavage of the α-chain fragment 1 appears to be nearly complete, making characterization of these cleavage fragments technically challenging. In the absence of PSA, the α-chain fragment 2 (39.5 kDa) can also be detected in the supernatant (Fig. 4C), indicating that additional proteolysis is occurring, perhaps by factor I and the appropriate cofactor.
Complement-dependent phagocytosis is an important mechanism of the host defense system and is primarily mediated by complement receptor CR3, and to a lesser extent CR1 and CR4. Many leukocytes express CR3, including professional phagocytes such as macrophages, monocytes, and neutrophils. C3b does not interact with CR3, and iC3b is predicted to interact with CR3 through binding sites, which become exposed upon unfolding of the CUB domain after cleavage of the C3b α-chain. We hypothesized that the significance of PSA’s ability to cleave iC3b between the CUB and MG8 domain on the α-chain might be that it results in interference with CR3 binding and subsequent phagocytosis. However, we could not detect any difference in phagocytosis between PSA-treated and control EA-iC3b. Thus, PSA-mediated proteolysis of purified iC3b could have other effects. In particular, we are currently exploring whether the 37-kDa C3 fragment generated by the combination of factor I and PSA cleavage has unique effects within the immune system.
C3, C4, and C5 are key components of the complement system and are similar in size (~ 200 kDa) and subunit structure. These homologous complement factors belong to the same gene family as the serum proteinase inhibitor α-2-macroglobulin and are the result of gene duplication (22). Interestingly, α-2-macroglobulin is the primary inhibitor of PSA in the serum. Owing to the similarities among this group of proteins, we were curious whether C3 was uniquely cleaved by PSA or all family members were substrates of PSA. C5, but not C4, was degraded by PSA in a manner similar to that of C3, although the cleavage products were too numerous for further analysis. PSA’s lack of proteolytic activity toward C4 confirms that PSA is not a promiscuous protease, and the activity toward C3 and C5 is likely specific. Similar to C3, cleavage of C5 was limited to the α-chain, leaving the β-chain entirely intact. In this study, C5 was readily degraded and inactivated upon addition to seminal plasma. This degradation could be blocked through the addition of a specific PSA inhibitor. These results suggest that PSA present in the seminal and prostatic fluid has the ability to degrade C5 present in the male reproductive tract. In addition, it is possible that PSA may has an additional immunosuppressive role by cleaving and inactivating C5 that is known to be present in the female reproductive tract, thus protecting spermatozoa from complement-mediated injury. The continued high-level expression of PSA by localized and meta-static prostate cancer cells, even after progression into disease castration resistant state, suggests that PSA might have a role in the initiation or progression of prostate cancer (5). Previous studies demonstrated that PSA could modulate a variety of cytokines and growth factors. PSA was first shown to cleave insulin-like growth factor binding proteins, resulting in the release of reactive insulin growth factor 1 (IGF-1) (28) and to release TGF-β from the small latent complex (29). PSA can also cleave parathyroid hormone-related protein to produce a fragment that, through recruitment of other factors, could promote an osteoblastic phenotype (30). However, in each of these studies, PSA cleavage was demonstrated only in biochemical assays using purified proteins. It is not known whether any of these proteins are relevant PSA substrates in vivo. In contrast, in this study we have been able to demonstrate the ability of PSA to proteolyze complement proteins in patient prostatic fluid samples in vivo. In this context, further study is necessary to understand the significance of PSA’s ability to cleave iC3b and C5 as it relates to the avoidance or inhibition of native immune suppression of prostate cancer growth and progression. These findings could also have bearing on the potential development of Ab and cell-based therapeutics for prostate cancer. Finally, it is necessary to determine whether these new PSA-induced C3 fragments can be detected in serum and have potential utility as biomarkers for prostate cancer.
Supplementary Material
Acknowledgments
We thank the Middle Atlantic Mass Spectrometry Laboratory at The Johns Hopkins University, Lizamma Antony for excellent technical support, and Nenoo Rawal and Michael Pangburn at Complement Technology for discussions.
This work was supported by Department of Defense (DOD) Idea Award W81XWH-10-PCRP-IDA (to S.R.D.), DOD Predoctoral Fellowship W81XWH-09-1-0219 (to M.L.M.), a DOD Postdoctoral Fellowship (to M.B.K.), and Prostate Specialized Project of Research Excellence Grant 2P50CA58236 (to S.R.D.).
Abbreviations used in this article
- EA
Ab-sensitized sheep erythrocyte
- ES
sheep erythrocyte
- PSA
prostate-specific Ag
- PVDF
polyvinylidene difluoride
Footnotes
Disclosures The authors have no financial conflicts of interest.
The online version of this article contains supplemental material.
References
- 1.Watt KW, Lee PJ, M’Timkulu T, Chan WP, Loor R. Human prostate-specific antigen: structural and functional similarity with serine proteases. Proc. Natl. Acad. Sci. USA. 1986;83:3166–3170. doi: 10.1073/pnas.83.10.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lilja H. A kallikrein-like serine protease in prostatic fluid cleaves the predominant seminal vesicle protein. J. Clin. Invest. 1985;76:1899–1903. doi: 10.1172/JCI112185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lilja H, Abrahamsson PA, Lundwall A. Semenogelin, the predominant protein in human semen. Primary structure and identification of closely related proteins in the male accessory sex glands and on the spermatozoa. J. Biol. Chem. 1989;264:1894–1900. [PubMed] [Google Scholar]
- 4.Williams SA, Jelinek CA, Litvinov I, Cotter RJ, Isaacs JT, Denmeade SR. Enzymatically active prostate-specific antigen promotes growth of human prostate cancers. Prostate. 2011;71:1595–1607. doi: 10.1002/pros.21375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams SA, Singh P, Isaacs JT, Denmeade SR. Does PSA play a role as a promoting agent during the initiation and/or progression of prostate cancer? Prostate. 2007;67:312–329. doi: 10.1002/pros.20531. [DOI] [PubMed] [Google Scholar]
- 6.Josef Marx F, Karenberg A. History of the term prostate. Prostate. 2009;69:208–213. doi: 10.1002/pros.20871. [DOI] [PubMed] [Google Scholar]
- 7.De Marzo AM, Platz EA, Sutcliffe S, Xu J, Grönberg H, Drake CG, Nakai Y, Isaacs WB, Nelson WG. Inflammation in prostate carcinogenesis. Nat. Rev. Cancer. 2007;7:256–269. doi: 10.1038/nrc2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brooks GF, Lammel CJ, Petersen BH, Stites DP. Human seminal plasma inhibition of antibody complement-mediated killing and opsonization of Neisseria gonorrhoeae and other gram-negative organisms. J. Clin. Invest. 1981;67:1523–1531. doi: 10.1172/JCI110183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petersen BH, Lammel CJ, Stites DP, Brooks GF. Human seminal plasma inhibition of complement. J. Lab. Clin. Med. 1980;96:582–591. [PubMed] [Google Scholar]
- 10.Tarter TH, Alexander NJ. Complement-inhibiting activity of seminal plasma. Am. J. Reprod. Immunol. 1984;6:28–32. doi: 10.1111/j.1600-0897.1984.tb00105.x. [DOI] [PubMed] [Google Scholar]
- 11.Fujita K, Ewing CM, Sokoll LJ, Elliott DJ, Cunningham M, De Marzo AM, Isaacs WB, Pavlovich CP. Cytokine profiling of prostatic fluid from cancerous prostate glands identifies cytokines associated with extent of tumor and inflammation. Prostate. 2008;68:872–882. doi: 10.1002/pros.20755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rawal N, Pangburn M. Formation of high-affinity C5 convertases of the alternative pathway of complement. J. Immunol. 2001;166:2635–2642. doi: 10.4049/jimmunol.166.4.2635. [DOI] [PubMed] [Google Scholar]
- 13.Montaño RF, Morrison SL. A colorimetric-enzymatic microassay for the quantitation of antibody-dependent complement activation. J. Immunol. Methods. 1999;222:73–82. doi: 10.1016/s0022-1759(98)00181-1. [DOI] [PubMed] [Google Scholar]
- 14.Chow C-W, Downey GP, Grinstein S. Measurements of phagocytosis and phagosomal maturation. Curr Protoc Cell Biol Chapter. 2004;15 doi: 10.1002/0471143030.cb1507s22. Unit 15.7. [DOI] [PubMed] [Google Scholar]
- 15.Pilch B, Mann M. Large-scale and high-confidence proteomic analysis of human seminal plasma. Genome Biol. 2006;7:R40. doi: 10.1186/gb-2006-7-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burger R, Bader A, Kirschfink M, Rother U, Schrod L, Wörner I, Zilow G. Functional analysis and quantification of the complement C3 derived anaphylatoxin C3a with a monoclonal antibody. Clin. Exp. Immunol. 1987;68:703–711. [PMC free article] [PubMed] [Google Scholar]
- 17.Sahu A, Lambris JD. Structure and biology of complement protein C3, a connecting link between innate and acquired immunity. Immunol. Rev. 2001;180:35–48. doi: 10.1034/j.1600-065x.2001.1800103.x. [DOI] [PubMed] [Google Scholar]
- 18.LeBeau AM, Singh P, Isaacs JT, Denmeade SR. Potent and selective peptidyl boronic acid inhibitors of the serine protease prostate-specific antigen. Chem. Biol. 2008;15:665–674. doi: 10.1016/j.chembiol.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Denmeade SR, Sokoll LJ, Chan DW, Khan SR, Isaacs JT. Concentration of enzymatically active prostate-specific antigen (PSA) in the extracellular fluid of primary human prostate cancers and human prostate cancer xenograft models. Prostate. 2001;48:1–6. doi: 10.1002/pros.1075. [DOI] [PubMed] [Google Scholar]
- 20.van Lookeren Campagne M, Wiesmann C, Brown EJ. Macrophage complement receptors and pathogen clearance. Cell. Microbiol. 2007;9:2095–2102. doi: 10.1111/j.1462-5822.2007.00981.x. [DOI] [PubMed] [Google Scholar]
- 21.Nishida N, Walz T, Springer TA. Structural transitions of complement component C3 and its activation products. Proc. Natl. Acad. Sci. USA. 2006;103:19737–19742. doi: 10.1073/pnas.0609791104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hughes AL. Phylogeny of the C3/C4/C5 complement-component gene family indicates that C5 diverged first. Mol. Biol. Evol. 1994;11:417–425. doi: 10.1093/oxfordjournals.molbev.a040123. [DOI] [PubMed] [Google Scholar]
- 23.Walport MJ. Complement. First of two parts. N. Engl. J. Med. 2001;344:1058–1066. doi: 10.1056/NEJM200104053441406. [DOI] [PubMed] [Google Scholar]
- 24.Janssen BJC, Christodoulidou A, McCarthy A, Lambris JD, Gros P. Structure of C3b reveals conformational changes that underlie complement activity. Nature. 2006;444:213–216. doi: 10.1038/nature05172. [DOI] [PubMed] [Google Scholar]
- 25.Wiesmann C, Katschke KJ, Yin J, Helmy KY, Steffek M, Fairbrother WJ, McCallum SA, Embuscado L, DeForge L, Hass PE, van Lookeren Campagne M. Structure of C3b in complex with CRIg gives insights into regulation of complement activation. Nature. 2006;444:217–220. doi: 10.1038/nature05263. [DOI] [PubMed] [Google Scholar]
- 26.Alcorlo M, Martínez-Barricarte R, Fernández FJ, Rodríguez-Gallego C, Round A, Vega MC, Harris CL, de Cordoba SR, Llorca O. Unique structure of iC3b resolved at a resolution of 24Å by 3D-electron microscopy. Proc. Natl. Acad. Sci. USA. 2011;108:13236–13240. doi: 10.1073/pnas.1106746108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janssen BJC, Huizinga EG, Raaijmakers HCA, Roos A, Daha MR, Nilsson-Ekdahl K, Nilsson B, Gros P. Structures of complement component C3 provide insights into the function and evolution of immunity. Nature. 2005;437:505–511. doi: 10.1038/nature04005. [DOI] [PubMed] [Google Scholar]
- 28.Cohen P, Graves HC, Peehl DM, Kamarei M, Giudice LC, Rosenfeld RG. Prostate-specific antigen (PSA) is an insulin-like growth factor binding protein-3 protease found in seminal plasma. J. Clin. Endocrinol. Metab. 1992;75:1046–1053. doi: 10.1210/jcem.75.4.1383255. [DOI] [PubMed] [Google Scholar]
- 29.Dallas SL, Zhao S, Cramer SD, Chen Z, Peehl DM, Bonewald LF. Preferential production of latent transforming growth factor β-2 by primary prostatic epithelial cells and its activation by prostate-specific antigen. J. Cell. Physiol. 2005;202:361–370. doi: 10.1002/jcp.20147. [DOI] [PubMed] [Google Scholar]
- 30.Cramer SD, Chen Z, Peehl DM. Prostate specific antigen cleaves parathyroid hormone-related protein in the PTH-like domain: inactivation of PTHrP-stimulated cAMP accumulation in mouse osteoblasts. J. Urol. 1996;156:526–531. doi: 10.1097/00005392-199608000-00076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.