Abstract
The aryl hydrocarbon receptor (AhR) mediates responses elicited by 2,3,7,8-tetrachlorodibenzo-p-dioxin by binding to dioxin response elements (DRE) containing the core consensus sequence 5′-GCGTG-3′. The human, mouse and rat genomes were computationally searched for all DRE cores. Each core was then extended by 7bp upstream and downstream, and matrix similarity (MS) scores for the resulting 19bp DRE sequences were calculated using a revised position weight matrix constructed from bona fide functional DREs. In total, 72,318 human, 70,720 mouse and 88,651 rat high-scoring (MS ≥ 0.8437) putative DREs were identified. Gene encoding intragenic DNA regions had ~1.6-times more putative DREs than the non-coding intergenic DNA regions. Furthermore, the promoter region spanning ±1.5kb of a TSS had the highest density of putative DREs within the genome. Chromosomal analysis found that the putative DRE densities of chromosomes X and Y were significantly lower than the mean chromosomal density. Interestingly, the 10kb upstream promoter region on chromosome X of the genomes were significantly less dense than the chromosomal mean, while the same region in chromosome Y was the most dense. In addition to providing a detailed genomic map of all DRE cores in the human, mouse and rat genomes, these data will further aid the elucidation of AhR-mediated signal transduction.
Introduction
Cis-regulatory elements located in the promoter region of genes are transcription factor binding sites that regulate gene expression. Most transcription factors have a preferred response element sequence to which they bind. The identification and location of these elements is important in elucidating transcription factor binding, signal transduction, and ultimately, their gene expression networks. Binding to elements in the proximal promoter region stabilizes the general transcriptional machinery at the transcriptional start site (TSS) to regulate gene expression. However, global location analyses of transcription factor binding using ChIP-chip and ChIP-seq technologies have demonstrated transcription factor binding at sites distant from the TSS (1-4). A comprehensive map of transcription factor binding element locations and distribution within a genome provides important complementary information for elucidating and modeling the gene expression network of a transcription factor.
The AhR is a ligand activated transcription factor belonging to the basic-helix-loop-helix-PAS (bHLH-PAS) family of proteins that serve as environmental sensors to different stimuli (5). 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) is the prototypical ligand, a widespread environmental contaminant that elicits diverse species-specific effects, including tumor promotion, teratogenesis, hepatotoxicity, modulation of endocrine systems, immunotoxicity and enzyme induction (6, 7). These effects are a result of changes in gene expression mediated by the AhR (8). The binding of TCDD and related compounds to the cytosolic AhR triggers a conformational change and translocation of the activated receptor to the nucleus where it heterodimerizes with the aryl hydrocarbon nuclear translocator (ARNT), another bHLH-PAS family member. The heterodimer then binds to dioxin response elements (DREs) containing the 5′-GCGTG-3′ core, to regulate gene expression (8, 9). Evidence indicates that nucleotides adjacent to the core consensus sequence modulate DNA-binding affinity and enhancer function (10-12).
Position weight matrices (PWMs) provide a similarity assessment of a motif or putative response element (13). When compared to a consensus sequence they have been used to rank and prioritize potential transcription factor binding site preferences. However, PWMs suffer from high false positive prediction rates since the probability of any nucleotide at any position within the binding site is assumed to be independent of all other positions. Fortunately, the DRE PWM is based on the 5′-GCGTG-3′ core, thus reducing false positive frequency (14).
We have previously identified the location and distribution of DREs relative to the TSS for a limited number of genes (14) based on prior builds of the human, mouse and rat genome assemblies (14). Improvements and innovations in sequencing technologies have since provided higher quality data with significantly fewer sequence gaps (15-17) in the most recent genome builds resulting in more accurate annotation. In addition, the latest mouse and rat genome builds were used to construct a revised PWM based on updated sequence information for 13 bona fide functional DREs. Consequently, we have expanded the scope of our initial DRE analysis to include the entire human, mouse and rat genomes using an improved PWM. This includes analyses of the intragenic (10 kb upstream to end of 3′ UTR) and intergenic DNA regions, chromosome and gene regions (10 kb upstream of a TSS, 5′ and 3′ untranslated regions [UTRs], and coding sequence [CDS]). Collectively, these results provide a detailed genomic map for all putative DREs in the human, mouse and rat genomes that will serve as an important resource for the further elucidation of AhR gene expression networks.
Experimental Procedures
Position Weight Matrix
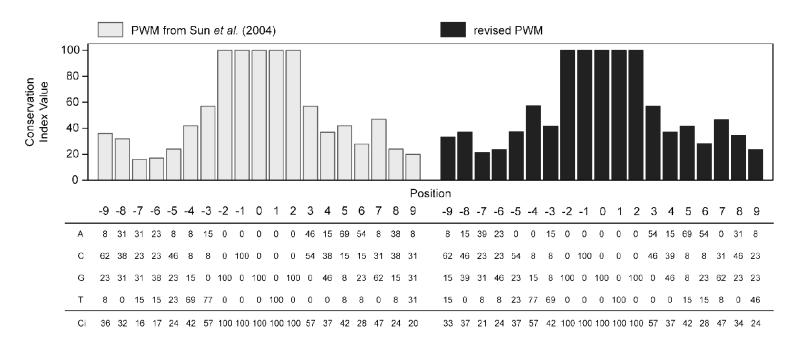
We have previously constructed a PWM using 13 bona fide functional DRE sequences from previous assembly builds of the mouse (mm3) and rat (rn2) genomes (11, 14, 18-23). These sequences were updated using the sequence information from the current genome assemblies for the mouse (mm9) and rat (rn4) (Table 1, updated sequences are underlined). Additionally, the previously identified sequence for the bona fide rat Aldh3a1 DRE could no longer be found in the rn4 genome build and was replaced with a functional DRE located 6,787 bp upstream of the TSS (24). Also note that the gene names for GstYa and Ugt1a1 have changed to Gsta2 and Ugt1a6, respectively, in the latest rat assembly. Updated sequences were used to develop a revised PWM using the bona fide 19 bp DRE-centered sequences (Figure 1). The replaced rat Aldh3a1 DRE sequence had the lowest matrix similarity (MS) score (0.8473), which was subsequently used as a threshold value to define 19 bp DRE sequences as putative DREs that were functional.
Table 1.
Bona fide DRE sequences used to construct the reviseda position weight matrix.
| Species | Gene Symbol |
RefSeq Identifier |
Position Relative to TSS |
Bona Fide DRE Sequenceb | Matrix Similarity Score |
Reference |
|---|---|---|---|---|---|---|
| Cyp1a1 | NM_001136059 | −491 | caagctcGCGTGagaagcg | 0.9466 | (11) | |
| Cyp1a1 | NM_001136059 | −871 | cctgtgtGCGTGccaagca | 0.9128 | (11) | |
| Mouse | Cyp1a1 | NM_001136059 | −984 | cggagttGCGTGagaagag | 0.9598 | (11) |
| Cyp1a1 | NM_001136059 | −1,059 | ccagctaGCGTGacagcac | 0.9260 | (11) | |
| Cyp1a1 | NM_001136059 | −1,206 | cgggtttGCGTGcgatgct | 0.9610 | (11) | |
| Cyp1b1 | NM_009994 | −872 | cccccttGCGTGcggagct | 0.9514 | (23) | |
|
| ||||||
| Cyp1a1 | NM_012540 | −1,045 | cggagttGCGTGagaagag | 0.9598 | (20) | |
| Cyp1a1 | NM_012540 | −1,120 | ccagctaGCGTGacagcac | 0.9260 | (20) | |
| Aldh3a1 | NM_031972 | −6,787 | tgccctgGCGTGactttgtc | 0.8473d | (24) | |
| Rat | Nqo1 | NM_017000 | −400 | tccccttGCGTGcaaaggc | 0.9332 | (19) |
| Sod1 | NM_017050 | −274 | gaggcctGCGTGcgcgcct | 0.8481 | (22) | |
| Gsta2e | NM_017013 | −910 | gcatgttGCGTGcatccct | 0.8728 | (21) | |
| Ugt1a6e | NM_057105 | −3,856 | agaatgtGCGTGacaaggt | 0.8950 | (18) | |
bona fide DRE sequences were updated using builds mm9 and rn4 genome builds
sequences used in Sun et. al., 2004 were updated with the mm9 and rn4 genome builds; revised sequences are underlined
replaces previous DRE sequence for rat Aldh3a1
denotes the MS score used as the threshold score
Gsta2 and Ugt1a6 were previously named GstYa and Ugt1a1 respectively, and were renamed within the rn4 genome build
Figure 1.
Comparison of the previously published position weight matrix (PWM) and conservation index (Ci) for dioxin response elements (DREs) with the revised PWM. The matrix and plot of the Ci on the left (light grey bars) was previously published by Sun et al. (2004). The matrix and plot (black) on the right is the revised PWM and Ci using the current mouse (mm9) and rat (rn4) genome assemblies from the UCSC Genome Browser. The matrix (bottom) shows the percentage of occurrence for a specific nucleotide at that given position. For example, positions −2 to 2 define the 5′-GCGTG-3′ DRE core, each nucleotide within the core has a Ci value of 100. The histogram (top) is a graphical representation of the Ci values, which are listed below the PWM. The Ci provides a measure of conservation at each base pair position. If a PWM is 100% conserved at a position, the Ci value is 100, whereas if the position is truly random (A=25%, C=25%, G=25%, T=25%) then the Ci value is 0.
Whole-Genome Identification of DREs
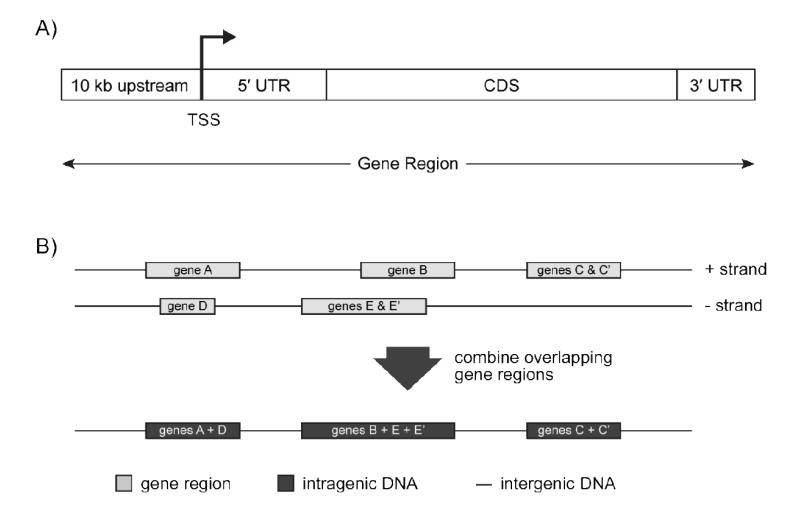
Sequences for human (hg19), mouse (mm9) and rat (rn4) genome assemblies and associated annotation within the refGene and refLink databases were downloaded from the UCSC Genome Browser (25). Individual segments of a gene region (i.e. the 10 kb sequence upstream of a TSS, the 5′ and 3′ UTRs and the CDS) for each mature gene encoding reference sequence (RefSeqs with NM prefixed identifiers), were determined using the genomic coordinates within the refGene databases (Figure 2A). Intragenic DNA regions within the genomes were computationally identified by merging overlapping gene regions (defined in Figure 2A) from both strands of the genome, and the DNA between adjacent intragenic regions are defined as the non-transcribed intergenic DNA regions (Figure 2B). The lengths for each of these defined regions and the number of RefSeqs on each chromosome are provided for the human, mouse and rat genomes in Supplementary Table 1. In total, 28,906 human, 24,327 mouse and 15,737 rat mature RefSeqs were searched. Gene annotation associated with each RefSeq sequence was derived from the refLink database in the UCSC Genome Browser.
Figure 2.
Defining the various genomic regions used for DRE location analysis. A) Genomic locations from the UCSC Genome Browser refGene database were used to obtain sequences for 10 kb region upstream of the TSS, the 5′ and 3′ UTRs, and the CDS of every known human, mouse and rat RefSeq sequence. A gene region is defined as the sequence spanning the region 10 kb upstream of a TSS through to the end of the 3′ UTR. B) Intragenic DNA regions in a genome were determined by combining the non-overlapping gene regions. For example, gene regions of tissue specific isoforms of a gene that have different TSS positions were merged to determine the longest spanning range (genes C & C’ and genes E & E’). Additionally, overlapping genes on both strands of the genome were also merged (genes B + E + E’). Non-transcribed DNA segments that span the regions between adjacent intragenic regions are defined as the intergenic DNA regions.
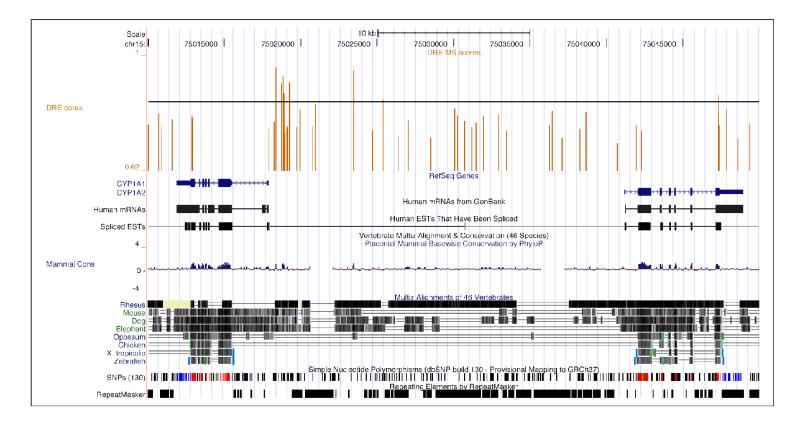
The sequence of each individual chromosome was computationally searched for the 5′-GCGTG-3′ core sequence using a previously described search algorithm (14). Each core was then extended by 7 bp upstream and downstream of the core. MS scores for the 19 bp DRE sequences were calculated using the revised PWM. For genomic location analysis, the position of a DRE core is defined as the center base (5′-GCGTG-3′) of the 5 bp core sequence (underlined). Putative DRE densities were calculated based on the number of putative DREs occurring in an interrogated region (e.g. intergenic DNA region or 5′ UTR) divided by the total sum of the region length. Results from the computational genome-wide DRE search can be downloaded as bedGraph track format (Supplementary file 5-7) and uploaded to the UCSC Genome Browser for visualization (Figure 3).
Figure 3.
Visualization of DRE sequence locations in the UCSC Genome Browser for human CYP1A1 and CYP1A2 gene regions and adjacent intergenic regions. The genomic location and MS score for each identified 19 bp DRE sequence has been loaded into the UCSC Genome Browser as a bedGraph track (see DRE cores track at top). The vertical bars represent the 5 bp DRE core and the height of the bar provides an indication of the MS score for the 19 bp DRE core containing sequence. The horizontal black line within the DRE cores track indicates the threshold MS score (0.8473) to assist with the identification of putative functional DREs.
Putative DRE densities from the different defined genomic regions (i.e. intergenic, intragenic, 10 kb upstream, UTRs and CDS) were identified as non-Gaussian using Q-Q plots (car package; qq.plot). The Wilcoxon Rank Sum Test (non-parametric t-test) was used to compare intergenic and intragenic putative DRE densities within species. The Kruskal-Wallis test (non-parametric one-way ANOVA), followed by the Nemenyi-Demico-Wolfe-Dunn Test (non-parametric Tukey’s test; nemenyi.test.R) was used to compare the putative DRE densities in the 10 kb upstream, 5′ UTR, CDS, and 3′ UTR DRE densities within species. All analyses were performed in R (version 2.12.0).
Random Sequence Comparison
To investigate the random frequency of DRE cores within each genome, 25,000 random sequences of 15 kb in length were computationally generated by randomly selecting A, C, G or T’s. These sequences were then searched for DRE cores, and the 19 bp DRE sequence MS score was calculated using the described algorithm (14) with the revised PWM.
Microarray Analysis
Whole-genome microarray analysis of hepatic tissue from mice orally gavaged with 30 μg/kg TCDD was performed using 4×44k whole genome oligonucleotide arrays from Agilent Technologies (Santa Clara, CA). The same RNA from a previous study was used for the gene expression profiling (26). Changes in gene expression due to TCDD treatment were conducted according to the manufacturer’s Two-Color Microarray-Based Gene Expression Analysis protocol Version 5.0.1. Microarray data were normalized using a semiparametric method (27), and statistically analyzed using an empirical Bayes method (28). Differentially expressed genes were determined by both a fold change and a statistical cutoff (|fold change| ≥ 1.5 and P1(t) ≥ 0.999).
Results
Position Weight Matrix (PWM)
Our previous PWM used bona fide DRE sequence information from earlier drafts of the mouse (mm3) and rat (rn2) genomes (Figure 1). These sequences have since been updated with the most current information available from the mouse (mm9) and rat (rn4) genome assemblies (Table 1). As a result, the sequence of two bona fide DREs in the promoter region of the mouse and rat Cyp1a1 gene have changed (Table 1, see footnote b). Additionally, the previously used DRE for rat Aldh3a1 could no longer be found in the latest rat genomic sequence, and was replaced with a recently characterized DRE located 6.8 kb upstream of the TSS (24) (Table 1). These updates altered the PWM and the conservation index (Ci) vector, which represents the degree of conservation of the individual nucleotide position, primarily in the 7 bp flanking 5′ arm of the consensus sequence (Figure 1). Recalculation of MS scores for the bona fide DREs identified the rat Aldh3a1 motif as having the lowest score, 0.8473, which was subsequently used to characterize computationally identified sequences as putative DREs.
Genome-Wide Distribution of DREs
Our previous computational search for the 5′-GCGTG-3′ DRE core was limited to sequences 5 kb upstream and 2 kb downstream of a TSS for known RefSeqs in previous genome builds (14). This current study extended the search to the entire human, mouse and rat genomes, including the non-transcribed intergenic DNA regions (Figure 2B). Computational searches identified 1.65, 1.04 and 1.07 million DRE cores in the human, mouse and rat genomes, respectively (Table 2). After extending these cores by the 7 bp upstream and downstream flanking sequences, MS scores were calculated using the revised PWM. A total of 72,318 human, 70,720 mouse and 88,651 rat 19 bp DRE sequences had a MS score greater than or equal to 0.8473, and were classified as putatively functional DREs (Table 2). The density of putative DREs with respect to the total length of the genomes were 23.4, 26.6, and 32.6 DREs per million base pairs (Mbp) in the human, mouse and rat, respectively. These values were determined from searching 3.10 billion human, 2.66 billion mouse and 2.72 billion rat base pairs (Table 2).
Table 2.
Distribution of DRE cores, putative DREs and putative DRE densities across the human, mouse and rat genomes.
| Species | Genome | Intergenic DNAa |
Intragenic DNAa |
Gene Regionb |
||||
|---|---|---|---|---|---|---|---|---|
| 10kb upstreamc | 5′ UTRc | CDSc | 3′ UTRc | |||||
| Region length (Mbpd) | Human | 3,096 | 1,836 | 1,260 | 295 | 236 | 1,599 | 44 |
| Mouse | 2,655 | 1,588 | 1,067 | 247 | 149 | 1,040 | 31 | |
| Rat | 2,719 | 1,973 | 746 | 159 | 74 | 569 | 18 | |
| Total number of DRE cores |
Human | 1,648,651 | 759,030 | 889,621 | 303,016 | 172,235 | 1,054,383 | 26,925 |
| Mouse | 1,036,996 | 492,703 | 544,293 | 154,921 | 82,306 | 510,060 | 16,546 | |
| Rat | 1,070,366 | 676,193 | 394,173 | 94,514 | 41,455 | 292,526 | 9,711 | |
| Total number of putative DREse |
Human | 72,318 | 34,322 | 37,996 | 13,272 | 7,606 | 46,004 | 1,123 |
| Mouse | 70,720 | 33,018 | 37,702 | 10,176 | 6,024 | 35,819 | 1,220 | |
| Rat | 88,651 | 54,888 | 33,763 | 7,962 | 3,515 | 24,954 | 870 | |
| Putative DRE density (per Mbpd) |
Human | 23.4 | 18.7 | 30.2 | 45.0 | 32.2 | 28.8 | 25.6 |
| Mouse | 26.6 | 20.8 | 35.3 | 41.3 | 40.5 | 34.5 | 39.5 | |
| Rat | 32.6 | 27.8 | 45.3 | 50.2 | 47.6 | 43.9 | 49.4 | |
intergenic and intragenic DNA region are defined in Figure 2B
gene region is defined as the transcribed gene plus 10 kb usptream of the TSS as depicted in Figure 2A
regions are defined using the genomic locations in the refGene database from the UCSC Genome Browser
Mbp = million basepairs
putative DREs defined as the 19 bp DRE centered core containing sequence with a MS score ≥ 0.8473
Approximately 40% of the human, 40% of the mouse and 27% of the rat genomes are comprised of intragenic DNA (Figure 2B), 53% of all putative DREs in the human and mouse genomes were identified in these regions while only 38% of all putative DREs mapped to the intragenic DNA in the rat (Table 2). This difference is likely a result of the relatively fewer number of rat RefSeqs (15,737) compared to the human (28,906) and the mouse (24,327). Relative putative DRE densities (i.e., intragenic/intergenic DNA putative density ratio) suggest that intragenic regions have ~1.6-times greater putative DRE density compared to intergenic DNA regions in each genome. For example, the human genome had putative DRE densities per Mbp of 30.2 and 18.7 in the intragenic and intergenic DNA regions, respectively (30.2/18.7 = 1.6). This suggests that there is a greater likelihood of putative DREs in the intragenic regions of the genome as opposed to the non-transcribed intergenic DNA regions. However, the density of putative DREs was generally higher in the rat genome (Table 2), likely due to the relative immaturity of gene annotation associated with the rat genome. The location and MS score for each identified 19 bp DRE sequence has been loaded into the UCSC Genome browser and can be visualized as a bedGraph track (Figure 3). The non-parametric Wilcoxon rank sum test of the mean chromosomal intragenic and intergenic putative DRE densities for each species (Table 3) identified significant intragenic enrichment with respect to the intergenic DNA regions. Further examination of DRE distribution within defined gene region segments (i.e., 10 kb upstream, 5′ and 3′ UTRs and CDS; Figure 2A) found that segment-specific putative DREs densities were comparable in human and mouse regions. Kruskal-Wallis non-parametric tests of the mean chromosomal putative DRE densities (Table 4) confirmed significantly higher density of putative DREs in the 10 kb upstream and 5′ UTR relative to the CDS in the human and mouse genomes. Although these same regions in the rat genome possessed a higher density of putative DREs relative to the CDS, statistical analyses was not able to detect any significant differences in the densities.
Table 3.
Chromosomal density of putative DREsa (per Mbpb) within the intergenic and intragenic DNA regionsc of the human, mouse and rat genomes.
| Chromosome | Human |
Mouse |
Rat |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | Intergenic DNA |
Intragenic DMA |
Total | Intergenic DNA |
Intragenic DNA |
Total | Intergenic DNA |
Intragenic DNA |
|
| 1 | 22.7 | 17.5 | 28.9 | 24.0 | 20.1 | 30.1 d | 32.9 | 28.4 | 42.3 |
| 2 | 22.9 | 20.0 | 27.0 d | 28.2 | 23.3 | 33.9 | 26.5 d | 23.0 d | 38.8 d |
| 3 | 19.8 d | 16.1 d | 24.2 d | 23.7 | 18.7 | 33.9 | 35.2 | 31.1 | 43.9 |
| 4 | 19.3 d | 17.2 | 23.1 d | 28.8 | 21.5 | 39.5 e | 30.1 | 26.1 | 39.1 d |
| 5 | 21.0 | 19.0 | 24.4 d | 32.3 e | 24.3 e | 42.4 e | 31.6 | 26.9 | 45.4 |
| 6 | 20.9 | 13.8 | 23.9 d | 26.2 | 21.2 | 32.8 | 32.8 | 28.2 | 46.2 |
| 7 | 25.3 | 20.7 | 30.6 | 26.7 | 19.7 | 36.7 | 34.8 | 29.2 | 49.2 |
| 8 | 24.3 | 21.6 | 28.6 | 29.6 | 23.2 | 39.9 e | 36.1 | 30.9 | 47.5 |
| 9 | 22.0 | 16.7 d | 31.3 | 30.1 | 23.9 e | 36.9 | 33.5 | 30.1 | 42.8 |
| 10 | 26.4 | 22.9 e | 30.5 | 28.6 | 22.1 | 37.6 | 43.8 e | 37.4 e | 53.9 e |
| 11 | 24.7 | 18.9 | 31.2 | 33.9 e | 26.7 e | 40.9 e | 29.6 | 24.4 d | 44.0 |
| 12 | 24.0 | 20.0 | 28.5 | 27.0 | 22.3 | 35.7 | 62.7 e | 52.0 e | 81.3 e |
| 13 | 16.7 d | 13.4 d | 25.3 d | 26.6 | 23.7 e | 31.6 | 31.3 | 26.4 | 44.0 |
| 14 | 19.9 d | 15.1 d | 28.9 | 24.8 | 21.0 | 31.1 | 32.4 | 29.0 | 43.1 |
| 15 | 23.2 | 18.6 | 30.1 | 26.7 | 19.3 | 37.8 | 29.8 | 26.1 | 41.2 |
| 16 | 35.0 e | 25.6 e | 48.0 e | 24.6 | 19.4 | 32.9 | 33.3 | 27.6 | 48.3 |
| 17 | 34.8 e | 26.6 e | 40.7 e | 31.0 e | 22.7 | 40.9 e | 36.1 | 33.6 | 43.1 |
| 18 | 23.3 | 19.6 | 29.8 | 26.7 | 20.2 | 37.7 | 30.7 | 26.7 | 43.3 |
| 19 | 43.6 e | 29.7 e | 53.3 | 30.8 e | 23.2 | 38.4 | 43.2 e | 38.6 e | 53.5 |
| 20 | 32.5 e | 27.0 e | 38.7 e | 45.0 e | 34.4 e | 68.5 e | |||
| 21 | 23.0 | 15.6 d | 44.2 e | ||||||
| 22 | 33.3 e | 21.5 | 51.5 e | ||||||
| X | 19.4 d | 16.9 | 24.7 d | 12.6 d | 11.1 d | 16.4 d | 16.0 d | 14.5 d | 25.5 d |
| Yf | 9.5 d | 8.1 d | 27.9 | 4.5 d | 3.7 d | 16.4 d | |||
|
| |||||||||
| Mean density | 24.5 | 19.5 | 32.3 | 26.1 | 20.5 | 34.5 | 34.6 | 29.8 | 46.9 |
| Std. Dev. | 7.1 | 4.7 | 8.9 | 6.5 | 4.9 | 6.9 | 8.9 | 7.2 | 11.1 |
putative DREs defined as the 19 bp DRE centered core containing sequence with a MS score ≥ 0.8473
Mbp = million basepairs
intergenic and intragenic DNA region are defined in Figure 2B
putative DRE density is less than the lower limit of the 99% confidence interval of the mean
putative DRE density is greater than the upper limit of the 99% confidence interval of the mean
no sequence data for chromosome Y is available in rn4 build of the rat genome
Table 4.
Chromosomal density of putative DREsa (per Mbpb) within the 10kb upstream, 5′ and 3′ UTRs, and CDS regionsc of RefSeq sequences in the human, mouse and rat genomes.
| Chromosome | Human |
Mouse |
Rat |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10kb upstream |
5′ UTR | CDS | 3′ UTR | 10kb upstream |
5′ UTR | CDS | 3′ UTR | 10kb upstream |
5′ UTR | CDS | 3′ UTR | |
| 1 | 35.8 d | 27.5 | 27.3 | 22.1 d | 39.8 | 30.6 d | 29.6 | 32.8 | 43.3 d | 52.6 | 41.7 | 34.6 d |
| 2 | 62.2 e | 40.2 | 27.1 | 26.2 | 38.8 | 41.2 | 34.0 | 36.1 | 49.7 | 34.9 d | 37.1 d | 36.2 d |
| 3 | 51.4 | 18.1 d | 22.9 d | 24.8 | 41.9 | 42.6 | 31.2 | 40.3 | 48.4 | 33.6 d | 44.0 | 61.9 |
| 4 | 57.4 e | 29.2 | 28.6 | 27.5 | 49.4 e | 46.1 | 37.2 | 43.1 | 42.6 d | 36.1 d | 38.2 | 42.9 d |
| 5 | 54.0 | 43.8 | 28.9 | 30.4 | 49.1 e | 58.7 e | 38.8 e | 34.0 | 53.7 | 53.4 | 43.5 | 50.8 |
| 6 | 41.4 | 28.4 | 26.8 | 23.8 | 35.0 d | 37.7 | 32.4 | 38.5 | 49.2 | 54.6 | 44.7 | 39.1 d |
| 7 | 49.9 | 32.5 | 27.2 | 30.4 | 36.5 | 41.6 | 39.1 e | 44.9 | 60.2 e | 49.9 | 49.0 | 51.8 |
| 8 | 52.2 | 27.6 | 27.0 d | 29.7 | 48.7 e | 33.6 d | 39.6 e | 50.4 e | 50.5 | 49.1 | 45.2 | 50.4 |
| 9 | 46.1 | 28.9 | 30.3 | 35.1 e | 40.2 | 42.9 | 36.4 | 44.8 | 47.4 | 51.5 | 41.2 | 66.4 e |
| 10 | 50.8 | 29.4 | 23.2 | 24.5 | 40.3 | 33.6 d | 34.2 | 40.1 | 54.3 | 66.4 e | 51.6 | 48.7 |
| 11 | 34.9 d | 37.4 | 27.7 | 24.4 | 48.2 e | 52.0 e | 39.5 e | 47.2 e | 48.8 | 33.9 d | 44.8 | 77.6 e |
| 12 | 42.4 | 27.4 | 26.0 d | 16.4 d | 42.3 | 38.5 | 36.4 | 50.9 e | 77.7 | 69.1 e | 84.5 e | 61.1 |
| 13 | 84.8 e | 47.6 e | 31.8 | 48.1 e | 34.0 d | 36.2 | 30.3 | 37.6 | 42.5 d | 54.6 | 42.6 | 55.2 |
| 14 | 56.7 e | 32.7 | 26.2 d | 30.2 | 40.6 | 40.7 | 31.7 | 40.6 | 42.0 d | 55.3 | 42.3 | 67.1 e |
| 15 | 47.0 | 27.0 d | 34.1 | 27.3 | 48.4 e | 41.9 | 39.1 e | 41.4 | 46.5 | 42.7 d | 39.3 | 73.3 e |
| 16 | 51.6 | 41.3 | 49.4 e | 47.9 e | 39.8 | 40.9 | 29.9 | 41.3 | 65.1 | 63.3 e | 43.9 | 65.9 e |
| 17 | 33.8 d | 60.5 e | 31.5 | 18.2 d | 46.4 e | 46.4 | 41.9 e | 39.8 | 46.3 | 53.8 | 41.5 | 37.4 d |
| 18 | 76.0 e | 30.1 | 35.7 | 35.1 e | 40.4 | 38.3 | 37.8 | 37.0 | 53.8 | 48.8 | 39.3 | 25.2 d |
| 19 | 25.0 d | 50.9 e | 52.7 e | 27.6 | 43.0 | 51.3 e | 37.3 | 38.4 | 55.4 | 48.9 | 52.4 | 58.1 |
| 20 | 49.9 | 33.7 | 37.0 e | 20.7 d | 64.0 e | 58.9 e | 73.7 e | 78.3 e | ||||
| 21 | 41.6 | 19.2 d | 32.5 | 21.7 d | ||||||||
| 22 | 42.9 | 51.4 e | 41.3 e | 28.3 | ||||||||
| X | 26.8 d | 25.0 d | 21.4 d | 12.9 d | 20.5 d | 19.8 d | 14.8 d | 14.5 d | 31.0 d | 30.3 d | 23.2 d | 34.4 d |
| Yf | 45.2 | 98.9 e | 42.6 e | 16.7 d | 26.7 d | 84.1 e | 13.3 d | 0.0 d | ||||
|
| ||||||||||||
| Mean density | 48.3 | 37.0 | 31.6 | 27.1 | 40.5 | 42.8 | 33.5 | 37.8 | 51.1 | 49.6 | 45.9 | 53.2 |
| Std. Dev | 13.6 | 16.8 | 8.0 | 8.5 | 7.3 | 12.5 | 7.4 | 11.4 | 9.9 | 10.9 | 12.6 | 15.3 |
putative DREs defined as the 19 bp DRE centered core containing sequence with a MS score ≥ 0.8473
Mbp = million basepairs
regions are defined using the genomic locations in the refGene database from the UCSC Geriome Browser
putative DRE density is less than the lower limit of the 99% confidence interval of the mean
putative DRE density is greater than the upper limit of the 99% confidence interval of the mean
no sequence data for chromosome Y is available in rn4 build of the rat genome
Chromosome Level Analysis of Putative DREs
In order to further investigate putative DRE distribution across the genomes, chromosomal level analysis was performed (Tables 3 and 4). Examination of individual chromosomes identified examples where the putative DRE density was significantly different than the mean chromosomal value (outside the 99% confidence interval of the mean; Table 3, see footnotes d and e). For example, putative DRE densities for rat chromosome 2 and human chromosome 13 were 26.5 and 16.7 per Mbp, respectively, which were significantly less than the mean value for each genome (34.6 and 24.5 per Mbp, for the rat and human, respectively). Furthermore, human chromosomes 16 and 17 had significantly greater putative DREs density than the mean chromosomal density. There are also instances where the putative DRE densities in the intergenic DNA (Table 3), or in a specific gene region segment (i.e. 10 kb upstream region, CDS and UTRs; Table 4), were significantly different than the chromosomal mean for that region. These data suggest that there are chromosome- and segment-specific biases in putative DRE densities across the genome that may have biological relevance in AhR-mediated responses.
Interestingly, putative DRE densities in chromosome X and Y of the human and mouse were significantly lower than the chromosomal average (Tables 3 and 4; there currently is no sequence data available for chromosome Y in the rat). For example, mouse chromosome Y has an intragenic putative DRE density of 16.4 per Mbp, almost half the density of any other mouse chromosome for the same region. In contrast, the putative density in the 5′ UTR for chromosome Y was 84.1 per Mbp, nearly double the chromosomal average in the mouse genome. Human chromosome Y was similar with a lower putative DRE density in the intragenic region, but the 5′ UTR density was more than 2.6-times greater than the mean chromosomal value. Similar to chromosome Y, the putative densities in the intragenic regions of chromosome X were significantly lower than the mean in each genome. However, unlike chromosome Y, the density in the 5′ UTR was also lower than the mean chromosome value. Such region differences in chromosomes X and Y may contribute to sex-specific AhR-mediated responses. It is important to recognize that the lower total putative DRE densities in the sex chromosomes are likely due to the lower chromosomal contribution of intragenic DNA. For example, intragenic DNA accounts for only 6% of the total DNA on human chromosome Y compared to 36% on human chromosome 9. Supplementary Tables 2 and 3 provide a complete chromosomal summary of the total number of putative DRE in the intergenic and intragenic DNA regions, the UTRs and the CDS for the human, mouse and rat genomes.
Random Sequence Analysis
To examine the chance occurrence of putative DREs, 25,000 random sequences of 15 kb were generated and searched for DREs. The computational search found 731,636 core sequences and extending these sequences by 7 bp on both ends, identified 108,210 chance occurrences of putative DREs (MS score ≥ 0.8473). In total, 375 Mbp were searched resulting in 288.6 putative DREs per Mbp. This chance occurrence of putative DRE density is significantly greater than the calculated densities in each genome both at the global and chromosomal level, suggesting that regions with a high density of putative DREs have a greater likelihood of being biologically significant.
Putative DRE Density Proximal to the TSS
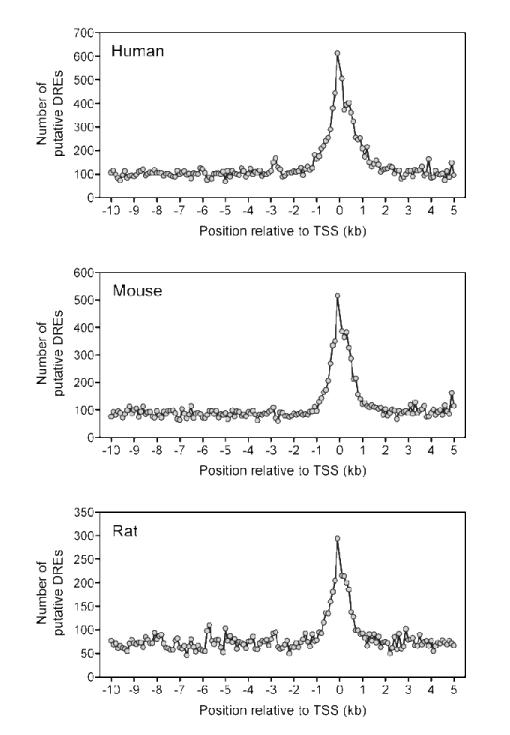
Putative DRE densities across genomes and chromosomes provide a gross estimate of occurrence. Finer analysis of different gene region segments generally found greater putative DRE density in the 10 kb upstream and 5′ UTR regions. To further investigate these segments, the number of putative DREs in non-overlapping 100 bp windows spanning the region 10 kb upstream and 5 kb downstream of a TSS were plotted (Figure 4). Putative DREs were not equally distributed within this 15 kb region, with the highest density occurring within ±1.5 kb of a TSS. In each species, the density was the greatest at approximately 100 bp directly upstream of the TSS. A sharp 3′ drop from the maximum was observed followed by a secondary peak 200-400 bp downstream of the TSS before putative DRE occurrence returned to basal levels.
Figure 4.
Distribution of putative DREs in the regions 10 kb upstream to 5 kb downstream of a TSS for all RefSeq sequences. The −10 kb to 5 kb region of a TSS were divided into non-overlapping 100 bp windows. The total number of putative DREs (MS score ≥ 0.8473) were determined for each 100 bp window and graphed. The density of putative DREs was greatest in the 3 kb region centered around the TSS.
Gene Level Analysis of DREs
Unique Entrez Gene identifiers for mature gene-encoding RefSeqs (NM prefixed RefSeq identifiers) were obtained from the UCSC Genome Browser refLink database and used to determine the distribution of putative DREs associated with 18,893 human, 20,018 mouse and 15,342 rat annotated genes (Table 5). The majority of all known genes had at least one DRE core present within 10 kb upstream of the TSS and the transcribed gene. However, 55 human, 343 mouse, and 327 rat genes did not have a DRE core within this same region. It is surprising to identify so many genes without a DRE core since the average gene region length (10 kb upstream of a TSS plus the transcribed gene) in the different genomes is 61 kb and the 5′-GCGTG-3′ sequence is expected to occur once every 512 bp. The lack of a DRE core in these genes may suggest that they are not targets of AhR regulation. However, 7 of the 343 mouse genes without a DRE core were differentially regulated in the temporal microarray data set. These responses may be regulated by AhR-independent mechanisms or via distally located DREs. Subsequent statistical analysis using a Chi-squared test resulted in a p-value < 0.001 (α = 0.05) illustrating a significant difference in the number of genes with and without a DRE core. Although there are a significant number of genes not containing a DRE core within the region 10 kb upstream of a TSS plus the transcribed gene, distal DREs in the intragenic DNA regions may also have functional importance, consistent with reported DRE-independent AhR mediated gene expression (29).
Table 5.
noAnalysis of DRE core and putative DRE containing RefSeq sequences and genes in the human, mouse and rat genomes.
| Human |
Mouse |
Rat |
||||
|---|---|---|---|---|---|---|
| RefSeqs | Genes | RefSeqs | Genes | RefSeqs | Genes | |
| Genomea | 28,906 | 18,893 | 24,327 | 20,018 | 15,737 | 15,342 |
| With a DRE core | 28,871 | 18,858 | 23,982 | 19,675 | 15,400 | 15,015 |
| With a putative DREb | 20,502 | 13,050 | 15,885 | 12,623 | 10,105 | 9,809 |
based RefSeqs and Entnez Gene IDs stored in the refGene and refLink databases from the UCSC Genome Browser
putative DREs defined as the 19 bp DRE centered core containing sequence with a MS score ≥ 0.8473
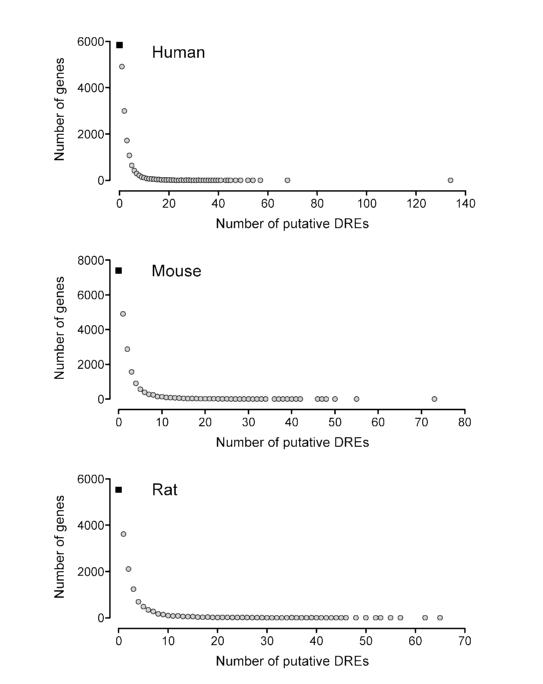
Further restricting this analysis to the 19 bp DRE sequences with a MS score ≥ 0.8473 (i.e., putative DREs) identified 69%, 63% and 64% of all human, mouse and rat genes, respectively, had at least one putative DRE (Table 5). Moreover, approximately 60% of all human, mouse and rat genes have 1 to 10 putative DREs (Figure 5). Interestingly, the maximum number of putative DREs was found in human PTPRN2 with 134, mouse Wwox with 73, and rat Odz2 with 65. Orthologs of these genes also had a high number of putative DREs. For example, there were 24 and 25 putative DREs in the mouse and rat PTPRN2, respectively. However, neither gene has been explicitly investigated for their responsiveness to TCDD nor are these genes responsive in our or any other TCDD microarray datasets (30-34). Unfortunately, the global gene expression effects of TCDD have been investigated in a limited number of models (e.g., in vitro and in vivo human, mouse and rat hepatic tissue, human breast cancer cells, mouse uterus). Gene expression is species-, sex-, age-, tissue- and cell-specific, and therefore the effects of TCDD on PTPRN2, Wwox and Odz2 gene expression warrant further investigation in other models to determine their potential AhR regulation.
Figure 5.
Frequency of putative DREs within known human, mouse and rat genes. For each species, the gene region (10 kb upstream of a TSS through to the end of the 3′ UTR) was searched for putative DREs. Approximately 35% of all known genes did not contain a putative DRE (black box) while nearly 60% of all genes had between 1 and 10 putative DREs. Approximately 5% of all genes have more than 10 putative DREs.
Global hepatic temporal gene expression analysis at 2, 4, 8, 12, 18, 24, 72, and 168 h identified 1,896 genes that were differentially expressed (|fold change| ≥ 1.5 and P1(t) ≥ 0.999) at one or more time points following a single oral dose of 30 μg/kg TCDD in immature, ovariectomized C57BL/6 mice. Of these, 1,247 had putative DREs within the 10 kb upstream or transcribed regions (includes 5′ and 3′ UTR and CDS). Genes that exhibited significant differential expression in the mouse liver included Fabp12 with 8 putative DREs (23.5-fold induction), and Cyp1a1 with 7 putative DREs (205-fold induction). The remaining 649 differentially expressed genes, which included unannotated and hypothetical genes, did not have a putative DRE. Examining only well-annotated genes found 593 TCDD responsive genes without a putative DRE within the region 10 kb upstream or transcribed region. The responses of some these genes include the up-regulation of Chad (+6.88-fold) and Olfr114 (+9.97-fold), and the repression of Serpina7 (−7.98-fold). The complete microarray data set is available in Supplementary Table 4. The responses of Olfr114 and Serpina7 have previously been reported to be AhR-dependent (35-37), however it is unclear if the responses of these genes are directly mediated by the activated AhR complex, or secondary responses.
Differentially regulated genes indentified through microarray analysis of TCDD-treated immature, ovariectomized Sprague Dawley rats (31, 32) were also searched for putative DREs. From those studies, 604 genes were responsive (|fold change| ≥ 1.5 and P1(t) ≥ 0.99) at 2 or more time points and 528 had at least one putative DRE within the 10 kb upstream or transcribed regions. This current mouse microarray study and the previous rat studies covered 5,451 orthologous genes, and only 52 of those were responsive in both models and possessed at least one putative DRE. These results are consistent with our previous orthologous promoter analysis that demonstrated that few human, mouse and rat orthologs had positionally conserved DRE upstream of a TSS (14).
Discussion
Genome-wide identification of potential cis-acting regulatory elements provides important information for elucidating signaling networks. Many computational and traditional in vitro approaches have generally focused on relatively few genes and a small segment of a target gene promoter, while neglecting more distal elements, which may also have important regulatory roles (14, 38-43). In order to fully elucidate the signaling transduction of transcription factors, both proximally and distally located response elements need to be identified and characterized.
The structure and function of the AhR as well as its mode of action are highly conserved, with homologs found in nearly all vertebrates. AhR activation by TCDD results in target gene expression via the DRE core sequence, 5′-GCGTG-3′. Our previous DRE computational analysis was limited to the proximal promoter regions (5 kb upstream to 2 kb downstream of a TSS) of known genes in earlier drafts of the human, mouse and rat genomes (14). This current study leverages the availability of higher quality finished human and mouse assemblies (15, 44), as well as the most current build of the rat genome to establish a revised PWM and calculate MS scores for all DRE core containing sequences located throughout the human, mouse and rat genomes, including the non-transcribed intragenic DNA regions.
Approximately 60% of the human and mouse genomes consist of stretches of non-transcribed intergenic DNA, while we define the remaining 40% as intragenic regions that include the 10 kb upstream promoter region, the 5′ and 3′ UTRs and the CDS (Table 2). Despite these differences in length, the total number of DRE core sequences and putative DREs were comparable in the intergenic DNA and intragenic regions. The draft assembly of the rat genome consists predominantly of intergenic DNA (73%), reflecting the immaturity of its annotation. Consequently, the intergenic DNA bias in the rat resulted a greater number of identified DRE cores and putative DREs in the intergenic DNA regions compared to intragenic DNA. Even within intragenic regions, putative DREs were found in CDS and 3′ UTR regions, and not limited to proximal-promoters (Table 4).
It has been suggested that the relative location of a bound transcription factor may have different roles in regulating gene expression. For example, the estrogen receptor (ER), p53 and forkhead box protein A1 (1-4), interact with proximal and distal response elements located throughout the genome, including the intergenic DNA. Transcription factor binding at the core promoter is presumed to stabilize the basal transcriptional machinery, while more distal motifs exert regulation through a looping mechanism or by altering chromatin structure (45-47). Consequently, a comprehensive map of potential binding sites throughout the genome provides important information for elucidating the AhR gene expression network.
Computational searches identified putative DREs in all genome regions. However, once the size of each region was taken into consideration, the density of putative DREs was found to be highest in the intragenic DNA regions of all three species. Moreover, putative DRE densities varied dramatically across chromosomes with some chromosomes having significantly higher densities (e.g., human chromosome 19, mouse chromosome 5, and rat chromosome 12) compared to the mean chromosomal density, while others (e.g., human chromosome 13, and chromosome rat 2) were significantly less dense. Interestingly, the sex chromosomes, and especially chromosome Y, the rat genome withstanding, were the least dense in terms of the total putative DREs amongst all the other chromosomes. Putative DRE densities within the 10 kb upstream region, the UTRs and the CDS were also substantially different from the mean chromosomal value for those regions. TCDD elicits sex-specific physiological and gene expression responses in rodents (7, 48, 49). These differences in sensitivity and physiological responses may be influenced by DREs differentially regulating gene expression on the sex chromosomes. Note that no sequence information for chromosome Y is currently available in the rat draft assembly. This will likely be resolved in the next phase of the rat genome sequencing effort (16, 50).
Within human and mouse chromosomes putative DRE densities were highest in the 5′ UTR and the region 10 kb upstream of the TSS. In contrast, DRE densities in rat genes were slightly higher in the 3′ UTR compared to either the 10kb upstream region or the 5′ UTR. However, as previously mentioned, limited annotation of the rat genome may have biased the identification of DREs to the 3′ UTR. A more finite analysis of the density around the proximal promoter found the greatest putative DRE density within ±1.5 kb of the TSS of known RefSeq sequences for all three species, with the maximum density occurring 100 bp upstream of a TSS. This coincides with 70% of all RNA polymerase II (Pol II) binding (2, 3), suggesting that proximal AhR binding recruits and stabilizes Pol II binding at the TSS. Additionally, due to the GC rich nature of the DRE core sequence, the putative DRE density profile mirrors the CpG island frequency in the proximal promoter region (51). Consequently, methylation status of putative DRE cores within CpG islands may affect gene expression. However, in a recent study inhibition of DNA methylation by AzaC in human MCF-7 cells did not affect TCDD-induced CYP1A1 expression (52).
Searching the region 10 kb upstream of a TSS and the transcribed region for all known genes in the genomes found that approximately 65% of all genes contained at least one putative DRE. However, gene expression is species-, sex-, age-, tissue-, cell and promoter context-dependent. Moreover, many responses may be secondary, thereby not involving direct interaction with the AhR. Consequently, the presence of a putative DRE within the gene region is not sufficient to elicit a transcriptional response. Although our use of a MS score ≥ 0.8473 to define a 19 bp DRE sequence as putative is based on experimental data indicating it is the lowest scoring bona fide functional DRE (i.e., rat Aldh3a1 DRE), recent protein-binding microarray studies indicate that more degenerative sites also bind transcription factors and have important functional roles in regulating gene expression (53, 54).
Transcription factors can also indirectly regulate gene expression by tethering to other proximally bound transcription factors. For example, progesterone receptor tethers to Sp1, Stat5 and AP1 to regulate genes independent of a progesterone response element (55-57). Moreover, the AhR is recruited to estrogen-responsive regions in a gene-specific (58) and DRE-independent manner (59). Furthermore, AhR:ARNT heterodimers regulate target gene expression by interacting with an alternate response element sequence, independent of the DRE core consensus sequence (60, 61). All of these factors must be taken into context in order to fully comprehend AhR-mediated gene regulation.
Computationally searching the human, mouse and rat genome assemblies has revealed that putative DREs are not randomly distributed. Our detailed genomic map has identified putative DREs in intergenic and intragenic DNA regions. Furthermore, putative DRE distributions vary across specific genome regions. This suggests that AhR binding to putative DREs in different genomic locations may have differing roles in regulating gene expression. Complementary studies are in progress to investigate AhR complex binding to DREs located in intergenic and intragenic regions.
Supplementary Material
Acknowledgments
Funding Support
The authors would like to thank Dr. Jason Matthews for critically reviewing this manuscript. This work was supported by the National Institute of Environmental Health Sciences Superfund Basic Research Program [P42ES04911].
References
- (1).Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- (2).Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M. Genome-wide analysis of estrogen receptor binding sites. Nature Genetics. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- (3).Lin C-Y, Vega VB, Thomsen JS, Zhang T, Kong SL, Xie M, Chiu KP, Lipovich L, Barnett DH, Stossi F, Yeo A, George J, Kuznetsov VA, Lee YK, Charn TH, Palanisamy N, Miller LD, Cheung E, Katzenellenbogen BS, Ruan Y, Bourque G, Wei C-L, Liu ET. Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet. 2007;3:e87. doi: 10.1371/journal.pgen.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Wederell ED, Bilenky M, Cullum R, Thiessen N, Dagpinar M, Delaney A, Varhol R, Zhao Y, Zeng T, Bernier B, Ingham M, Hirst M, Robertson G, Marra MA, Jones S, Hoodless PA. Global analysis of in vivo Foxa2-binding sites in mouse adult liver using massively parallel sequencing. Nucleic Acids Res. 2008;36:4549–4564. doi: 10.1093/nar/gkn382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Gu Y, Hogenesch J, Bradfield C. The PAS superfamily: sensors of environmental and developmental signals. Annu Rev Pharmacol Toxicol. 2000;40:519–561. doi: 10.1146/annurev.pharmtox.40.1.519. [DOI] [PubMed] [Google Scholar]
- (6).Denison MS, Heath-Pagliuso S. The Ah receptor: a regulator of the biochemical and toxicological actions of structurally diverse chemicals. Bulletin of environmental contamination and toxicology. 1998;61:557–568. doi: 10.1007/pl00002973. [DOI] [PubMed] [Google Scholar]
- (7).Poland A, Knutson JC. 2,3,7,8-tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: examination of the mechanism of toxicity. Annu Rev Pharmacol Toxicol. 1982;22:517–554. doi: 10.1146/annurev.pa.22.040182.002505. [DOI] [PubMed] [Google Scholar]
- (8).Hankinson O. The aryl hydrocarbon receptor complex. Annu Rev Pharmacol Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- (9).Swanson H, Chan W, Bradfield C. DNA binding specificities and pairing rules of the Ah receptor, ARNT, and SIM proteins. J Biol Chem. 1995;270:26292–26302. doi: 10.1074/jbc.270.44.26292. [DOI] [PubMed] [Google Scholar]
- (10).Gillesby BE, Stanostefano M, Porter W, Safe S, Wu ZF, Zacharewski TR. Identification of a motif within the 5′ regulatory region of pS2 which is responsible for AP-1 binding and TCDD-mediated suppression. Biochemistry. 1997;36:6080–6089. doi: 10.1021/bi962131b. [DOI] [PubMed] [Google Scholar]
- (11).Lusska A, Shen E, Whitlock JP. Protein-DNA interactions at a dioxin-responsive enhancer. Analysis of six bona fide DNA-binding sites for the liganded Ah receptor. J Biol Chem. 1993;268:6575–6580. [PubMed] [Google Scholar]
- (12).Shen ES, Whitlock JP. Protein-DNA interactions at a dioxin-responsive enhancer. Mutational analysis of the DNA-binding site for the liganded Ah receptor. J Biol Chem. 1992;267:6815–6819. [PubMed] [Google Scholar]
- (13).Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Sun YV, Boverhof DR, Burgoon LD, Fielden MR, Zacharewski TR. Comparative analysis of dioxin response elements in human, mouse and rat genomic sequences. Nucleic Acids Res. 2004;32:4512–4523. doi: 10.1093/nar/gkh782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Church DM, Goodstadt L, Hillier LW, Zody MC, Goldstein S, She X, Bult CJ, Agarwala R, Cherry JL, DiCuccio M, Hlavina W, Kapustin Y, Meric P, Maglott D, Birtle Z, Marques AC, Graves T, Zhou S, Teague B, Potamousis K, Churas C, Place M, Herschleb J, Runnheim R, Forrest D, Amos-Landgraf J, Schwartz DC, Cheng Z, Lindblad-Toh K, Eichler EE, Ponting CP, Consortium MGS. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol. 2009;7:e1000112. doi: 10.1371/journal.pbio.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Worley KC, Weinstock GM, Gibbs RA. Rats in the genomic era. Physiol Genomics. 2008;32:273–282. doi: 10.1152/physiolgenomics.00208.2007. [DOI] [PubMed] [Google Scholar]
- (17).Zody MC, Jiang Z, Fung H-C, Antonacci F, Hillier LW, Cardone MF, Graves TA, Kidd JM, Cheng Z, Abouelleil A, Chen L, Wallis J, Glasscock J, Wilson RK, Reily AD, Duckworth J, Ventura M, Hardy J, Warren WC, Eichler EE. Evolutionary toggling of the MAPT 17 q21.31 inversion region. Nature Genetics. 2008;40:1076–1083. doi: 10.1038/ng.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Emi Y, Ikushiro S, Iyanagi T. Xenobiotic responsive element-mediated transcriptional activation in the UDP-glucuronosyltransferase family 1 gene complex. J Biol Chem. 1996;271:3952–3958. doi: 10.1074/jbc.271.7.3952. [DOI] [PubMed] [Google Scholar]
- (19).Favreau L, Pickett C. Transcriptional regulation of the rat NAD(P)H:quinone reductase gene. Identification of regulatory elements controlling basal level expression and inducible expression by planar aromatic compounds and phenolic antioxidants. J Biol Chem. 1991;266:4556–4561. [PubMed] [Google Scholar]
- (20).Fujisawa-Sehara A, Sogawa K, Yamane M, Fujii-Kuriyama Y. Characterization of xenobiotic responsive elements upstream from the drug-metabolizing cytochrome P-450c gene: a similarity to glucocorticoid regulatory elements. Nucleic Acids Res. 1987;15:4179–4191. doi: 10.1093/nar/15.10.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Pimental RA, Liang B, Yee GK, Wilhelmsson A, Poellinger L, Paulson KE. Dioxin receptor and C/EBP regulate the function of the glutathione S-transferase Ya gene xenobiotic response element. Mol Cell Biol. 1993;13:4365–4373. doi: 10.1128/mcb.13.7.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Yoo HY, Chang MS, Rho HM. Xenobiotic-responsive element for the transcriptional activation of the rat Cu/Zn superoxide dismutase gene. Biochem Biophys Res Commun. 1999;256:133–137. doi: 10.1006/bbrc.1999.0299. [DOI] [PubMed] [Google Scholar]
- (23).Zhang L, Savas U, Alexander DL, Jefcoate CR. Characterization of the mouse Cyp1B1 gene. Identification of an enhancer region that directs aryl hydrocarbon receptor-mediated constitutive and induced expression. J Biol Chem. 1998;273:5174–5183. doi: 10.1074/jbc.273.9.5174. [DOI] [PubMed] [Google Scholar]
- (24).Reisdorph R, Lindahl R. Constitutive and 3-methylcholanthrene-induced rat ALDH3A1 expression is mediated by multiple xenobiotic response elements. Drug Metab Dispos. 2007;35:386–393. doi: 10.1124/dmd.106.012393. [DOI] [PubMed] [Google Scholar]
- (25).Rhead B, Karolchik D, Kuhn R, Hinrichs A, Zweig A, Fujita P, Diekhans M, Smith K, Rosenbloom K, Raney B, Pohl A, Pheasant M, Meyer L, Learned K, Hsu F, Hillman-Jackson J, Harte R, Giardine B, Dreszer T, Clawson H, Barber G, Haussler D, Kent W. The UCSC Genome Browser database: update 2010. Nucleic Acids Research. 2010;38:D613. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Boverhof DR, Burgoon LD, Tashiro C, Chittim B, Harkema JR, Jump DB, Zacharewski TR. Temporal and dose-dependent hepatic gene expression patterns in mice provide new insights into TCDD-Mediated hepatotoxicity. Toxicol Sci. 2005;85:1048–1063. doi: 10.1093/toxsci/kfi162. [DOI] [PubMed] [Google Scholar]
- (27).Eckel J, Gennings C, Therneau T, Burgoon L, Boverhof D, Zacharewski T. Normalization of two-channel microarray experiments: a semiparametric approach. Bioinformatics. 2005;21:1078–1083. doi: 10.1093/bioinformatics/bti105. [DOI] [PubMed] [Google Scholar]
- (28).Eckel J, Gennings C, Chinchilli V, Burgoon L, Zacharewski T. Empirical bayes gene screening tool for time-course or dose-response microarray data. J Biopharm Stat. 2004;14:647–670. doi: 10.1081/BIP-200025656. [DOI] [PubMed] [Google Scholar]
- (29).Murray IA, Morales JL, Flaveny CA, Dinatale BC, Chiaro C, Gowdahalli K, Amin S, Perdew GH. Evidence for ligand-mediated selective modulation of aryl hydrocarbon receptor activity. Molecular Pharmacology. 2010;77:247–254. doi: 10.1124/mol.109.061788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Boutros PC, Yan R, Moffat ID, Pohjanvirta R, Okey AB. Transcriptomic responses to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in liver: comparison of rat and mouse. BMC Genomics. 2008;9:419. doi: 10.1186/1471-2164-9-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Boverhof DR, Burgoon LD, Tashiro C, Sharratt B, Chittim B, Harkema JR, Mendrick DL, Zacharewski TR. Comparative toxicogenomic analysis of the hepatotoxic effects of TCDD in Sprague Dawley rats and C57BL/6 mice. Toxicol Sci. 2006;94:398–416. doi: 10.1093/toxsci/kfl100. [DOI] [PubMed] [Google Scholar]
- (32).Fletcher N, Wahlström D, Lundberg R, Nilsson CB, Nilsson KC, Stockling K, Hellmold H, Håkansson H. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) alters the mRNA expression of critical genes associated with cholesterol metabolism, bile acid biosynthesis, and bile transport in rat liver: a microarray study. Toxicol Appl Pharmacol. 2005;207:1–24. doi: 10.1016/j.taap.2004.12.003. [DOI] [PubMed] [Google Scholar]
- (33).Hayes K, Zastrow G, Nukaya M, Pande K, Glover E, Maufort J, Liss A, Liu Y, Moran S, Vollrath A, Bradfield C. Hepatic transcriptional networks induced by exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Chem Res Toxicol. 2007;20:1573–1581. doi: 10.1021/tx7003294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Puga A, Maier A, Medvedovic M. The transcriptional signature of dioxin in human hepatoma HepG2 cells. Biochem Pharmacol. 2000;60:1129–1142. doi: 10.1016/s0006-2952(00)00403-2. [DOI] [PubMed] [Google Scholar]
- (35).Tijet N, Boutros PC, Moffat ID, Okey AB, Tuomisto J, Pohjanvirta R. Aryl hydrocarbon receptor regulates distinct dioxin-dependent and dioxin-independent gene batteries. Mol Pharmacol. 2006;69:140–153. doi: 10.1124/mol.105.018705. [DOI] [PubMed] [Google Scholar]
- (36).Ovando BJ, Vezina CM, McGarrigle BP, Olson JR. Hepatic gene downregulation following acute and subchronic exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol Sci. 2006;94:428–438. doi: 10.1093/toxsci/kfl111. [DOI] [PubMed] [Google Scholar]
- (37).Yauk CL, Jackson K, Malowany M, Williams A. Lack of change in microRNA expression in adult mouse liver following treatment with benzo(a)pyrene despite robust mRNA transcriptional response. Mutation research. 2010 doi: 10.1016/j.mrgentox.2010.02.012. [DOI] [PubMed] [Google Scholar]
- (38).Bourdeau V, Deschênes J, Métivier R, Nagai Y, Nguyen D, Bretschneider N, Gannon F, White JH, Mader S. Genome-wide identification of high-affinity estrogen response elements in human and mouse. Mol Endocrinol. 2004;18:1411–1427. doi: 10.1210/me.2003-0441. [DOI] [PubMed] [Google Scholar]
- (39).Lemay DG, Hwang DH. Genome-wide identification of peroxisome proliferator response elements using integrated computational genomics. J Lipid Res. 2006;47:1583–1587. doi: 10.1194/jlr.M500504-JLR200. [DOI] [PubMed] [Google Scholar]
- (40).Menendez D, Inga A, Resnick MA. Estrogen receptor acting in cis enhances WT and mutant p53 transactivation at canonical and noncanonical p53 target sequences. Proc Natl Acad Sci USA. 2010;107:1500–1505. doi: 10.1073/pnas.0909129107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Nukaya M, Moran S, Bradfield CA. The role of the dioxin-responsive element cluster between the Cyp1a1 and Cyp1a2 loci in aryl hydrocarbon receptor biology. Proc Natl Acad Sci USA. 2009;106:4923–4928. doi: 10.1073/pnas.0809613106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Ortiz-Barahona A, Villar D, Pescador N, Amigo J, Del Peso L. Genome-wide identification of hypoxia-inducible factor binding sites and target genes by a probabilistic model integrating transcription-profiling data and in silico binding site prediction. Nucleic acids research. 2010 doi: 10.1093/nar/gkp1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).van Batenburg MF, Li H, Polman JA, Lachize S, Datson NA, Bussemaker HJ, Meijer OC. Paired hormone response elements predict caveolin-1 as a glucocorticoid target gene. PLoS ONE. 2010;5:e8839. doi: 10.1371/journal.pone.0008839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Xue Y, Sun D, Daly A, Yang F, Zhou X, Zhao M, Huang N, Zerjal T, Lee C, Carter NP, Hurles ME, Tyler-Smith C. Adaptive evolution of UGT2B17 copy-number variation. Am J Hum Genet. 2008;83:337–346. doi: 10.1016/j.ajhg.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Farnham PJ. Insights from genomic profiling of transcription factors. Nat Rev Genet. 2009;10:605–616. doi: 10.1038/nrg2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Li Q, Barkess G, Qian H. Chromatin looping and the probability of transcription. Trends Genet. 2006;22:197–202. doi: 10.1016/j.tig.2006.02.004. [DOI] [PubMed] [Google Scholar]
- (47).Long X, Miano JM. Remote control of gene expression. J Biol Chem. 2007;282:15941–15945. doi: 10.1074/jbc.R700010200. [DOI] [PubMed] [Google Scholar]
- (48).Silkworth JB, Carlson EA, McCulloch C, Illouz K, Goodwin S, Sutter TR. Toxicogenomic analysis of gender, chemical, and dose effects in livers of TCDD-or aroclor 1254-exposed rats using a multifactor linear model. Toxicol Sci. 2008;102:291–309. doi: 10.1093/toxsci/kfm313. [DOI] [PubMed] [Google Scholar]
- (49).Walker NJ, Wyde ME, Fischer LJ, Nyska A, Bucher JR. Comparison of chronic toxicity and carcinogenicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in 2-year bioassays in female Sprague-Dawley rats. Mol Nutr Food Res. 2006;50:934–944. doi: 10.1002/mnfr.200600031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Twigger SN, Pruitt KD, Fernández-Suárez XM, Karolchik D, Worley KC, Maglott DR, Brown G, Weinstock G, Gibbs RA, Kent J, Birney E, Jacob HJ. What everybody should know about the rat genome and its online resources. Nat Genet. 2008;40:523–527. doi: 10.1038/ng0508-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Nakajima M, Iwanari M, Yokoi T. Effects of histone deacetylation and DNA methylation on the constitutive and TCDD-inducible expressions of the human CYP1 family in MCF-7 and HeLa cells. Toxicol Lett. 2003;144:247–256. doi: 10.1016/s0378-4274(03)00216-9. [DOI] [PubMed] [Google Scholar]
- (53).Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, Kuznetsov H, Wang C-F, Coburn D, Newburger DE, Morris Q, Hughes TR, Bulyk ML. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Jaeger SA, Chan ET, Berger MF, Stottmann R, Hughes TR, Bulyk ML. Conservation and regulatory associations of a wide affinity range of mouse transcription factor binding sites. Genomics. 2010 doi: 10.1016/j.ygeno.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Cicatiello L, Addeo R, Sasso A, Altucci L, Petrizzi VB, Borgo R, Cancemi M, Caporali S, Caristi S, Scafoglio C, Teti D, Bresciani F, Perillo B, Weisz A. Estrogens and progesterone promote persistent CCND1 gene activation during G1 by inducing transcriptional derepression via c-Jun/c-Fos/estrogen receptor (progesterone receptor) complex assembly to a distal regulatory element and recruitment of cyclin D1 to its own gene promoter. Mol Cell Biol. 2004;24:7260–7274. doi: 10.1128/MCB.24.16.7260-7274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Owen GI, Richer JK, Tung L, Takimoto G, Horwitz KB. Progesterone regulates transcription of the p21(WAF1) cyclin-dependent kinase inhibitor gene through Sp1 and CBP/p300. J Biol Chem. 1998;273:10696–10701. doi: 10.1074/jbc.273.17.10696. [DOI] [PubMed] [Google Scholar]
- (57).Stoecklin E, Wissler M, Schaetzle D, Pfitzner E, Groner B. Interactions in the transcriptional regulation exerted by Stat5 and by members of the steroid hormone receptor family. J Steroid Biochem Mol Biol. 1999;69:195–204. doi: 10.1016/s0960-0760(99)00052-7. [DOI] [PubMed] [Google Scholar]
- (58).Ahmed S, Valen E, Sandelin A, Matthews J. Dioxin increases the interaction between aryl hydrocarbon receptor and estrogen receptor alpha at human promoters. Toxicol Sci. 2009;111:254–266. doi: 10.1093/toxsci/kfp144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Beischlag TV, Perdew GH. ER alpha-AHR-ARNT protein-protein interactions mediate estradiol-dependent transrepression of dioxin-inducible gene transcription. J Biol Chem. 2005;280:21607–21611. doi: 10.1074/jbc.C500090200. [DOI] [PubMed] [Google Scholar]
- (60).Boutros PC, Moffat ID, Franc MA, Tijet N, Tuomisto J, Pohjanvirta R, Okey AB. Dioxin-responsive AHRE-II gene battery: identification by phylogenetic footprinting. Biochem Biophys Res Commun. 2004;321:707–715. doi: 10.1016/j.bbrc.2004.06.177. [DOI] [PubMed] [Google Scholar]
- (61).Sogawa K, Numayama-Tsuruta K, Takahashi T, Matsushita N, Miura C, Nikawa J.-i., Gotoh O, Kikuchi Y, Fujii-Kuriyama Y. A novel induction mechanism of the rat CYP1A2 gene mediated by Ah receptor-Arnt heterodimer. Biochem Biophys Res Commun. 2004;318:746–755. doi: 10.1016/j.bbrc.2004.04.090. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.