Abstract
In mammals, spinal cord injury results in permanent sensory-motor loss due in part to a failure in reinitiating local neurogenesis. However, zebrafish show robust neuronal regeneration and functional recovery even after complete spinal cord transection. Postembryonic neurogenesis is dependent upon resident multipotent progenitors, which have been identified in multiple vertebrates. One candidate cell population for injury repair expresses Dbx1, which has been shown to label multipotent progenitors in mammals. In this study, we use specific markers to show that cells expressing a dbx1a:GFP reporter in the zebrafish spinal cord are radial glial progenitors that continue to generate neurons after embryogenesis. We also use a novel larval spinal cord transection assay to show that dbx1a:GFP+ cells exhibit a proliferative and neurogenic response to injury, and contribute newly-born neurons to the regenerative blastema. Together, our data indicate that dbx1a:GFP+ radial glia may be stem cells for the regeneration of interneurons following spinal cord injury in zebrafish.
Keywords: Zebrafish, larvae, spinal, injury, regeneration, neurogenesis
Introduction
During vertebrate embryogenesis, neurons of the central nervous system (CNS) are initially derived from neuroepithelial progenitors, some of which transform into radial glia (Mori et al., 2005). By the end of embryogenesis most mammalian radial glia differentiate as astrocytes (Rakic, 2003). However in anamniotes radial glia persist widely in the CNS (García-Verdugo et al., 2002; Naujoks-Manteuffel and Roth, 1989; Zupanc and Clint, 2003), and their continued presence has been implicated in the striking ability of these animals to regenerate following injury (Chernoff et al., 2003; Hui et al., 2010; Rehermann et al., 2011). Thus radial glia have been suggested to represent an endogenous neural stem cell population.
In contrast to the permanent loss of sensory and motor function after spinal cord injury observed in mammals, urodele amphibians and teleost fish regenerate lost tissue and reestablish damaged connections, restoring function to nearly pre-injury levels (Chernoff et al., 2002; Kuscha et al., 2012). In addition to axonal regrowth after spinal cord transection in adult zebrafish (Becker et al., 2004, 1997; Goldshmit et al., 2012; Schweitzer et al., 2007), spinal lesion triggers generation of motoneurons and interneurons, with pre-injury levels restored by 6–8 weeks post injury (wpi, Reimer et al., 2008). While olig2+ radial glia represent a pool of motoneuron progenitors that contribute to neurogenesis after lesion, the identity and behavior of other progenitor populations remains unknown.
In vertebrates, Dbx genes encode a family of homeodomain transcription factors expressed in the intermediate spinal cord that are necessary for spinal cord development (Jessell, 2000; Lu et al., 1992). Dbx1-expressing cells predominately produce Evx1/2+ interneurons (Pierani et al., 2001), but also generate radial glia, astrocytes and oligodendrocytes (Fogarty et al., 2005). In mouse, Dbx1 expression is not detectable beyond E16.5, suggesting that mammalian Dbx1+ progenitors terminally differentiate (Fogarty et al., 2005). Zebrafish have two Dbx1 orthologs, dbx1a and dbx1b, which are similarly expressed in the intermediate spinal cord (Gribble et al., 2007; Seo et al., 1999); however, their lineage is uncharacterized. Based on the multipotency of Dbx1+ progenitors in amniotes, and the persistence of radial glia in zebrafish, we hypothesized that Dbx1-expressing cells might represent a population that could contribute to regeneration of the spinal cord following injury.
We previously generated a dbx1a:GFP transgenic reporter line (Gribble et al., 2009), and showed that GFP expression colocalized with endogenous dbx1a expression in embryonic spinal progenitors. In this study, we characterize the identity of dbx1a:GFP expressing cells and their progeny in the embryonic and larval zebrafish, and their response to spinal cord transection. We show that dbx1a mRNA expression persists beyond embryogenesis, and that the dbx1a:GFP reporter transgene labels a neurogenic spinal progenitor population. We also show that dbx1a:GFP expressing cells are slowly dividing neural progenitors that increase their rate of neurogenesis beyond basal levels in response to transection. Together, our data suggest that dbx1a:GFP+ radial glia may represent a neural stem cell population in the postembryonic spinal cord that can be activated in response to injury.
Materials and Methods
Fish Strains and Staging
Embryos were obtained from wildtype (AB*), Tg(olig2:dsRed)vu19, Tg(elavl3:EGFP)knu3, and Tg(-3.5 dbx1a:EGFP)zd3 crosses (Gribble et al., 2009; Kucenas et al., 2008; Park et al., 2000, 2007), and staged according to Kimmel et al., 1995. Zebrafish were raised and bred according to standard procedures; experiments were approved by the University of Utah Institutional Animal Care and Use Committee.
In Situ Hybridization
Embryos were fixed in fresh 4% paraformaldehyde (PFA) overnight at room temperature, washed in PBS, decapitated and coarsely chopped. In situ hybridization was performed as described previously (Oxtoby and Jowett, 1993). For sectioning, embryos were cryoprotected in sucrose, embedded in OCT, and sectioned at 20µm thickness on a Leica CM3050 cryostat. Images were taken on an Olympus BX51WI compound microscope using an Olympus Microfire camera. Images were processed using GNU Image Manipulation Program (GIMP.org).
Immunohistochemistry
Embryos up to 48hpf (hours post fertilization) were fixed in fresh 4% PFA for 3 hours at room temperature then overnight at 4°C; embryos over 48hpf were fixed for one hour at room temperature. After fixation, embryos were washed in PBS, cryoprotected in sucrose, embedded in OCT and sectioned at 12µm or 50µm thickness on a Leica CM3050 cryostat. For BrdU antigen retrieval, thick sections were incubated at room temperature for 90 minutes in 2N HCl. For PCNA antigen retrieval, thin sections were incubated in 100°C 10mM sodium citrate buffer, pH 6.0. Primary antibodies used were: rabbit anti-GFP (1:5000, Invitrogen #A-11122), chicken anti-GFP (1:1000, Aves #GFP-1020), mouse anti-HuC/D (1:500, Invitrogen #A-21271), chicken anti-BrdU (1:500, ICL #CBDU-65A-Z), mouse anti-PCNA (1:1000, Sigma #p8825), rabbit anti-PCNA (1:100, Santa Cruz Biotechnology #F2212), rabbit anti-Sox3 (1:200, gift from Dr. Mike Klymkowsky, University of Colorado-Boulder), rabbit anti-Sox3 (1:200, Pierce Custom Antibodies and Peptides), rabbit anti-DsRed (1:200, Clontech #632496), mouse zrf-1 (1:200, Zebrafish International Resource Center #zrf-1). Secondary antibodies used were: goat anti-rabbit 488 (1:200, Invitrogen #A- 11008), goat anti-rabbit 568 (1:200, Invitrogen #A-11041), goat anti-rabbit cy3 (1:200, Jackson ImmunoResearch #111-165-003), goat anti-mouse 633 (1:200, Invitrogen #A-21050), goat anti-mouse cy3 (1:200, Jackson ImmunoResearch #115-165-003), goat anti-chicken 488 (1:200, Invitrogen #A-11039), goat anti-chicken 633 (1:200, Invitrogen #A-21103), donkey anti-chicken 488 (1:200, Jackson ImmunoResearch #703-485-155). Hoechst 33342 was added to secondary antibodies to visualize nuclei.
Confocal Microscopy
Sections were imaged using an Olympus FV-1000XY confocal microscope using a 60x oil-immersion objective. Images were processed using ImageJ (rsbweb.nih.gov/ij) and GIMP (gimp.org). Projections were generated using FluoRender (Wan et al., 2012).
Spinal Cord Transection
Tg(elavl3:EGFP)knu3 or Tg(-3.5dbx1a:EGFP)zd3 4dpf (days post fertilization) embryos were raised in E2 medium + 0.2mM Phenothiourea (Sigma) + 10mg/L Gentamycin Sulfate (Amresco), treated with 10mM BrdU (Sigma) for 24hours then immediately lesioned at 5dpf as described previously (Bhatt et al., 2004; Briona and Dorsky, 2013) and transferred back into E2-PTU-Gentamycin Sulfate media. Briefly, microinjection glass pipettes were broken, beveled and used as a scalpel. Fish were anesthetized with 0.016% Tricaine (Sigma), braced with microforceps, and lesioned by driving a glass scalpel through the spinal cord at the level of the anal pore, and moved dorsally to sever the entire spinal cord. Sham treated animals were anesthetized and braced as described, and touched with the glass scalpel on the dorsal flank at the level of the anal pore without breaking the skin.
Quantification of dbx1a+ Progeny
Five non-consecutive transverse 12µm cryosections from five Tg(-3.5dbx1a:EGFP)zd3 embryos were quantified for antibody colocalization based on MaxZ projections generated with ImageJ(rsbweb.nih.gov/ij). dbx1a:eGFP+ cells were analyzed based on single-slice verification of colocalization with nuclear staining, verifying that the contiguous area of GFP expression was at least as large as the nucleus (Supplemental Figure 1).
Quantification of Spinal Cord Regeneration
Nineteen Tg(elavl3:EGFP)knu3 animals were injured at 5dpf, and imaged every other day until end of experiment. For BrdU labeling studies, 50µm sagittal cryosections from 95 Tg(-3.5 dbx1a:EGFP)zd3 embryos were quantified for antibody colocalization at 1–9dpi (days post injury). MaxZ projections were stitched together using ImageJ and GIMP. The plane of injury was identified, the spinal cord was outlined, and the leading edge of healthy spinal cord proximal to the injury site was labeled as 0µm. Three regions in the spinal cord were characterized: a 30µm region 200 µm rostral to the leading edge of uninjured spinal cord was identified as “rostral,” a 30µm region immediately adjacent to the rostral stump was identified as “proximal”, and the region adjacent to the leading edge of healthy spinal cord at the injury site was characterized as “blastema.” In sham-treated animals, spinal cord at the level of the anal pore was labeled as 0µm, and the rostral and proximal regions were identified relative to this point as described previously.
Statistical Analysis
All results were expressed as mean ± SEM. Two-tailed two-sample equal variance Student t-tests were calculated using Excel (Microsoft Corporation) or R (GNU S). Differences were considered significant at p<0.05.
Results
dbx1a mRNA and reporter expression persists beyond embryogenesis
Previous research in our laboratory using whole mount in situ hybridization showed that dbx1a was expressed in the intermediate spinal cord by 15 hours post fertilization (hpf), but was no longer detectable by 72hpf (Gribble et al., 2007). However, probe penetration becomes difficult beyond 48hpf due to mesenchymal condensation around the spinal cord (Bader et al., 2009; Simmons and Appel, 2012; Thisse et al., 2004) and cartilage formation at later stages (Sisson and Topczewski, 2009; Williams et al., 2000). Using coarsely chopped wildtype trunk sections we re-examined dbx1a mRNA expression from 3 days post fertilization (dpf) through 14dpf, and at all timepoints we observed expression in the intermediate spinal cord (Figure 1A–C). These data suggested that dbx1a-expressing cells may have a role beyond embryonic neurogenesis.
Figure 1.
Expression of dbx1a mRNA and dbx1a:GFP reporter. (A–C) In situ hybridization shows that dbx1a mRNA is expressed in the intermediate spinal cord through 14dpf. (D–F) In 3dpf dbx1a:GFP embryos, GFP protein expression is observed in lateral cells negative for GFP mRNA (arrows), suggesting that perdurance of protein can be used to trace the lineage of reporter-expressing cells. Scalebars = 10µm.
To determine the identity and ultimate fate of dbx1a-expressing cells, we used the Tg(-3.5dbx1a:EGFP)zd3 transgenic line previously characterized in our laboratory (Gribble et al., 2009). In order to test whether GFP protein perdurance in this line could be used as a marker for lineage tracing experiments, we examined the expression of GFP mRNA and GFP protein in 3dpf spinal sections. We found that mRNA expression was strongest in cells of the intermediate spinal cord immediately adjacent to the central canal and weak or absent in the lateral spinal cord where postmitotic motor neurons reside (Figure 1D). In the same sections, we observed GFP protein overlapping with mRNA expression, but also more laterally in cells negative for GFP mRNA (Figure 1E, F). Thus, we conclude that expression of GFP protein in postmitotic neurons is due to perdurance of the protein after cessation of mRNA expression, and that GFP protein expression marks both dbx1a-expressing progenitors and their immediate progeny.
dbx1a:GFP+ radial glial progenitors are neurogenic beyond embryogenesis
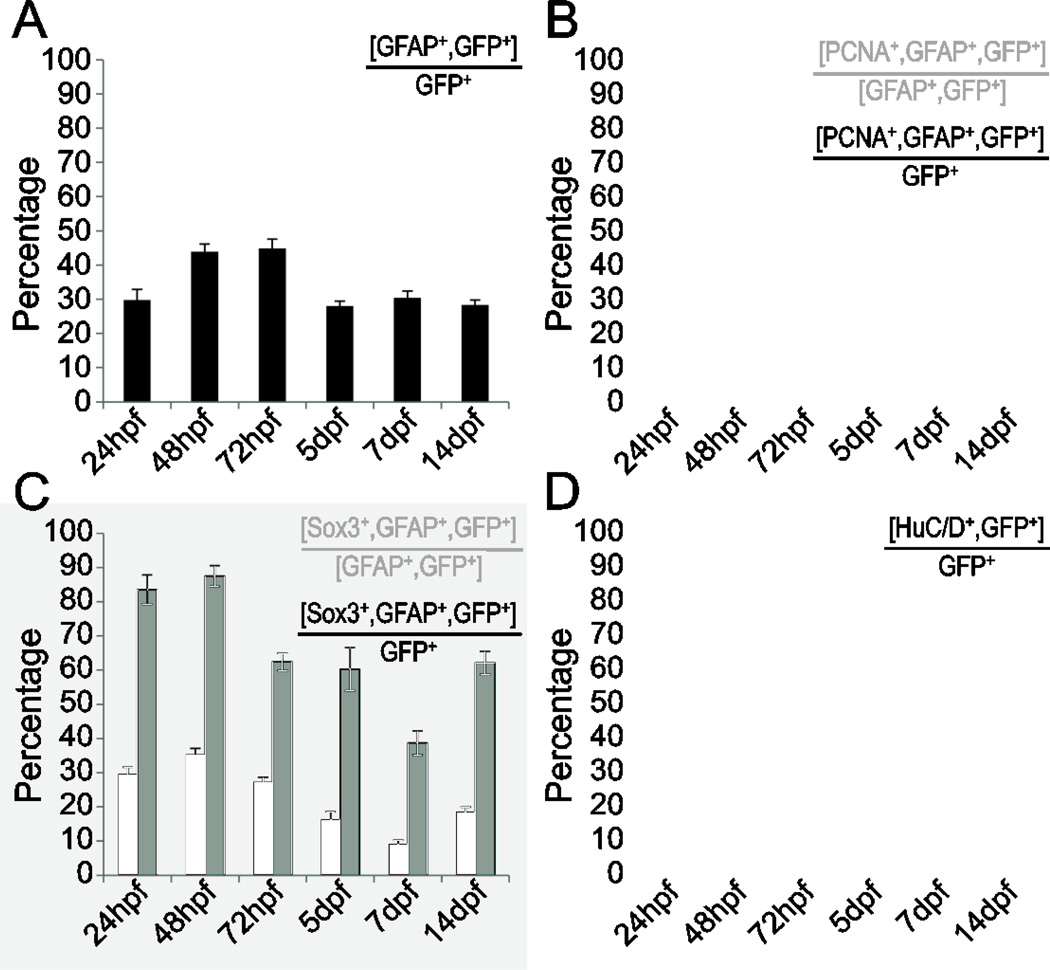
To determine which cell types express dbx1a:GFP during embryonic and larval stages, we examined the coexpression of GFP with cell type specific markers in transgenic fish (Supplemental Figure 1). Zebrafish embryogenesis is considered complete by 5dpf, as this marks the developmental timepoint when feeding begins.(Westerfield, 2000) At 24hpf, 30% of GFP+ cells were co-labeled by the zrf-1 antibody that recognizes GFAP, a marker of radial glia (Trevarrow et al., 1990). Because GFAP is a cytoskeletal intermediate filament while GFP is nuclear and cytoplasmic, the two antigens do not exhibit identical cellular expression. (Barresi et al., 2010). To confirm colocalization of GFP and GFAP, individual cells were examined for a cortical ring of GFAP expression around a GFP+ nucleus, as well as axonal extensions that were both GFAP+ and GFP+ (Figure 2). The peak colocalization of GFP and GFAP occurred at 48hpf and was maintained at 44% until 72hpf. At 5dpf, levels of GFP and GFAP coexpression returned to 30%, and remained steady until 14dpf (Figure 3A–C; Figure 4A). To further characterize this population, we examined colocalization of GFAP, GFP, and PCNA to identify proliferating radial glia, and GFAP, GFP, and Sox3 to identify neural progenitors (Goldman, 2003; Kim and Dorsky, 2011; Malatesta et al., 2000; Wang et al., 2006). At 24hpf, 90% of GFP+ radial glia were also PCNA positive. This percentage dropped over time to 18% at 5dpf, after which the percentage of GFP+ radial glia that were also PCNA+ returned to 40% at both 7dpf and 14dpf (Figure 3D–G’; Figure 4B). We also found several GFP+, GFAP+ radial glia positive for Sox3 expression at the central canal starting at 24hpf, and at all timepoints examined (Figure 3H–K’; Figure 4C). Together, these data suggest that dbx1a:GFP+ cells persist as proliferating neural progenitors beyond embryogenesis.
Figure 2.
GFP and GFAP colocalization. (A–D) Colocalization of dbx1a:GFP with GFAP at 48hpf in a single confocal slice. Arrow denotes a double-labeled cell based on colocalization of GFP with nuclear staining and cortical GFAP around the nucleus, and colocalization of GFP and GFAP in the process. White dashed box in (D) denotes region of interest in (E–P). (E–H) Maximum Z-projection showing colocalization of GFP and GFAP. (I–P) Rotated projection views showing colocalization of GFP and GFAP. Scale bar = 10µm.
Figure 3.
dbx1a:GFP+ progenitors persist beyond embryogenesis. (A–C) dbx1a:GFP+ cells contribute to a GFAP+ glial population. Arrow marks a double-labeled cell. (D–G) dbx1a:GFP+ glia remain proliferative beyond embryogenesis. Arrowheads mark triple-labeled cells. White box marks region shown in D’–G’. (HK) dbx1a:GFP+ glia persist as Sox3+ neural progenitors. Arrowheads mark triple-labeled cells. White box marks region shown in H’–K’. (L–N) dbx1a:GFP+ cells contribute to an intermediate HuC/D+ neuronal population. Arrows mark double-labeled cells. Scalebars = 10µm; all images are single optical sections.
Figure 4.
A stable population of dbx1a:GFP+ progenitors persists in the spinal cord. (A) Percentage of dbx1a:GFP+ progenitors that are GFAP+. (B) Percentage of PCNA+ cells in the overall dbx1a:GFP+ population (black), and among the GFAP+ subpopulation (grey). (C) Percentage of Sox3+ neural progenitors in the overall dbx1a:GFP+ population (black), and among the GFAP+ subpopulation (grey). (D) Percentage of HuC/D+ neurons in the dbx1a:GFP+ population. n=25 at each timepoint; error bars = SEM. Scalebar = 10µm.
We next asked whether dbx1a:GFP-expressing progenitors contribute to the neuronal population by examining HuC/D coexpression with GFP (Figure 3L–N). At 24hpf, 42% of the GFP+ cells were also HuC/D+ (Figure 4D). This coexpression expanded by 5dpf to a maximum of 73% of GFP+ cells, and returned to baseline levels at 7dpf, with 43% of GFP+ cells also HuC/D+. Consistent with our observations that GFP protein expression perdures in the progeny of dbx1a:gfp+ radial glia, we found that a 2-hour pulse of BrdU did not immediately label any GFP+ neurons, while an additional 24 hour chase allowed us to label these cells (data not shown). Taken together, these data show that dbx1a:GFP+ progenitors continue to divide and produce neurons beyond the end of embryogenesis, suggesting that they represent a potential source of regenerative potential following injury.
dbx1a:GFP+ progenitors are distinct from the olig2+ population
Progenitors in the ventral spinal cord expressing olig2 have been proposed as neural stem cells in postembryonic zebrafish (Park et al., 2007; Reimer et al., 2008). To confirm that dbx1a:GFP expressing cells are separate from the olig2+ population, we crossed Tg(olig2:dsRed)vu19 and Tg(-3.5 dbx1a:EGFP)zd3 fish and examined transgene colocalization in offspring. At earlier timepoints we did not observe any dsRed, GFP double positive cells (data not shown), and even at 5dpf, the peak of neurogenesis for dbx1a:GFP+ cells, dsRed did not colocalize with GFP (Figure 5D–F), suggesting that dbx1a:GFP+ cells are not a subset of olig2:dsRed+ progenitors. To determine the relative contributions of both lineages to neural progenitors in the developing spinal cord, we labeled 5dpf olig2:dsRed;dbx1a:GFP larvae with the neural progenitor marker Sox3. Colabeling showed that at 5dpf, 29.46%±1.93 of Sox3+ cells were GFP+ while 11.69%±1.41 of Sox3+ cells were dsRed+. These data suggest that while both populations contribute to the neural progenitor population, dbx1a:GFP+ cells represent a comparatively larger progenitor source.
Figure 5.
dbx1a:GFP+ and olig2:dsRed+ mark two independent progenitor populations. (A–E) At 5dpf, the dbx1a:GFP+ and olig2:dsRed+ populations do not overlap, suggesting they constitute separate lineages. Both populations contain Sox3+ neural progenitors (arrowhead and arrow). (F) At 5dpf, more dbx1a:GFP+ cells than olig2:dsRed+ cells express Sox3. N=25; error bars = SEM, scalebar = 10µm.
Larval zebrafish rapidly regenerate their spinal cord following transection
We next wanted to determine whether dbx1a:GFP progenitors in the post-embryonic spinal cord were able to regenerate lost neurons post-injury. While the adult zebrafish is a well-established model for studying spinal cord regeneration following transection, crush injury, or laser ablation of cells (Becker et al., 2004; Goldshmit et al., 2012; Hui et al., 2010), we chose to study spinal cord regeneration in larval zebrafish, which are transparent and thus allow an in vivo analysis of the regenerative process (Hale et al., 2001). The larval stage begins at 5dpf, which coincides with a decrease in spinal progenitor proliferation and differentiation to a level that remains stable for weeks (Figure 4 and Park et al., 2007). Therefore, neurogenesis after injury in larvae beyond this timepoint should require reactivation of progenitors into a regenerative program. While anatomical and functional recovery following spinal cord transection in the adult zebrafish takes 6–8 weeks (Reimer et al., 2008), we hypothesized that regeneration after transection would occur faster in the larva, based on maturity and size (Navarro et al., 1988; Sun et al., 2005).
Using a broken beveled micropipette as a glass scalpel, we completely transected the spinal cord of anesthetized 5dpf larvae at the level of the anal pore. Complete transection was verified visually by confirming a complete gap between rostral and caudal cord stumps (Figure 6A), and physiologically by lack of touch response caudal to the injury site. Sham animals were treated the same as experimental, except that they were only touched with the glass scalpel; no incision was made. Over 95% of transected fish survived the surgery until 7 days post injury (7dpi), at which point about 50% of the survivors died, likely due to the inability to feed as a result of failure to inflate the swimbladder (Haffter et al., 1996). Over 95% of sham treated animals survived until the end of the experiment.
Figure 6.
Larval zebrafish recover rapidly from spinal cord transection. (A–C) Compound microscope images of regeneration from a single live larva, using elavl3:EGFP to label postmitotic neurons. Spinal cord is marked by dashed line. (A) At 1dpi there is a clear gap between rostral and caudal stump ends after transection. (B) Processes projecting from the rostral stump into injury site are visible by 7dpi. (C) By 11dpi, the rostral and caudal stumps are contiguous across injury site. (D–I) Confocal images of regeneration from a single live larva, using elavl3:EGFP to label postmitotic neurons. Spinal cord is marked by dashed line. (D) At 1dpi there is a gap between the rostral and caudal stumps after injury; brightfield overlay shows location of wound site and scar tissue. (E, F) By 5dpi, rostral processes toward the pial surface can be seen (E, inset, arrows), but processes are absent from the caudal end (F, inset, arrow). (G) By 7dpi, numerous processes are visible at the rostral severed end (arrow), while the recovery response of the caudal end (H) resembles the rostral end at 5dpi (arrow). (I) By 9dpi, GFP+ soma are present in the injury site (arrows). All confocal images are single slices except (D), which is a Max-Z projection. Dorsal (D) and Rostral (R) are marked in panel (A). Scalebar = 50µm, YE = yolk extension. (J–O): Functional recovery following spinal cord transections. C-bends and S-bends are inducible at 3dpi (J,M). Magnitude and frequency of movement increases at 5dpi (K, N) and 9dpi (L, O).
Using elavl3:GFP larvae to visualize postmitotic neurons of the spinal cord, we first examined regeneration in live fish using compound fluorescence microscopy (Figure 6A–C). At 1dpi (6dpf) no regeneration was visible (Figure 5A). By 7dpi, neuronal processes from the rostral end of the spinal cord were visible, and appeared to be projecting towards the dorsal pial surface (Figure 6B). By 11dpi, the severed spinal cord was once again contiguous across the injury site (Figure 6C). To examine the regeneration process more closely, we visualized injured elavl3:GFP larvae in vivo using confocal microscopy (Figure 6D–I). At 1dpi, no regeneration was visible (Figure 6D). By 5dpi, rostral neuronal processes were visible (Figure 6E, arrows); however, the regenerative response was delayed on the caudal side of the injury, as no processes were visible at this time (Figure 6F). At 7dpi, rostral neuronal processes had become more numerous, and caudal processes were visible as well (Figure 6G, H, arrows). By 9dpi, elavl3:gfp+ soma were present in the injury site (Figure 6F, arrows).
We also examined the recovery of spinal cord function following transection. At 1dpi, all injured fish were nonresponsive to touch on their tail caudal to the injury site with a tungsten needle probe; however, touch rostral to injury site did elicit a startle response (Supplemental Movie 1). By 2dpi, 11/165 fish showed sporadic response to touch caudal to injury site (Supplemental Movie 2). At 3dpi, 28.5% exhibited a sporadic C-bend response to touch caudal to injury site, while 5.1% consistently responded to touch on both sides of the tail caudal to injury site, at all levels tested (Figure 6J, M; Supplemental Movie 3). Since the C-bend startle response is dependent upon reticulospinal neurons (Burgess and Granato, 2007), we used acetylated tubulin to label spinal axons. At 1dpi, there were no axons crossing the injury site, but by 5dpi, numerous axons crossed the injury gap (Supplemental Figure 2). By 5dpi, all injured fish still alive exhibited either sporadic (58.31%) or consistent response to touch, with some having resumed voluntary swimming (Figure 6K, N; Supplemental Movie 4). By 9dpi, all surviving fish showed robust swimming using both pectoral and caudal fins and a consistent startle response to touch caudal to injury site (Figure 6L, O; Supplemental Movie 5). Together these data show that larval zebrafish are capable of regenerating a transected spinal cord with regenerated axons crossing the injury gap and neuronal soma present at the injury site by 9dpi, reestablishing both sensory and motor function within 9dpi.
dbx1a:GFP+ progenitors contribute to neurogenesis after spinal cord transection
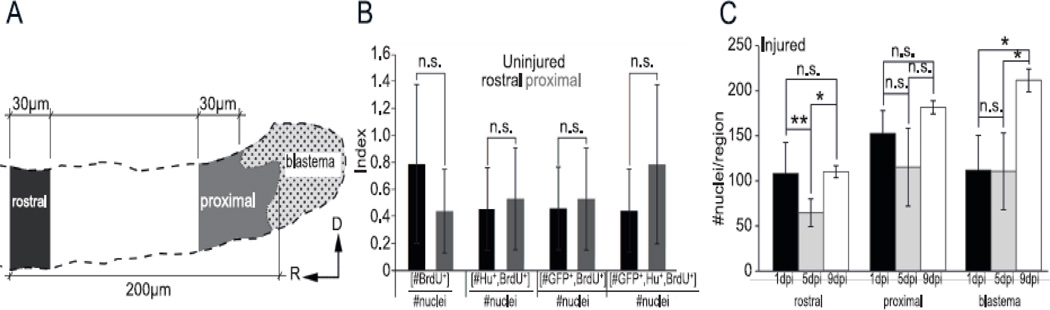
To determine whether dbx1a:GFP-expressing progenitors undergo a proliferative and neurogenic response following injury, we examined BrdU incorporation and HuC/D expression in dbx1a:GFP fish during recovery from spinal cord transection. To avoid potential complications from differences in BrdU accessibility following injury, we chose to label both transected and control larvae via incubation in BrdU for 24 hours immediately before injury. To facilitate characterization of the recovery process, we defined three zones of examination. The blastema was defined as the active recovery zone closest to the transection with its rostral edge located where neurons became discontinuous (Figure 7A). The neighboring proximal zone was defined as 30µm rostral to the blastema, and a rostral zone was defined as 170–200µm away from the rostral edge of the blastema. We hypothesized that the blastema zone would have the highest rate of sustained neurogenesis after injury, while the proximal zone would have the earliest proliferative response due to a high concentration of reactive progenitors. We also hypothesized that the regenerative response would be local to the injury site, and thus the rostral zone would reflect rates of proliferation and neurogenesis similar to that observed in sham treated animals. Our analysis was limited to the spinal cord rostral to the injury site to focus on the earliest-regenerating cells identified in our initial characterization.
Figure 7.
Definition of responsive zones following transection. (A) Schematic of zone identification in transected rostral spinal cord. The blastema was defined as the area containing non-contiguous HuC/D+ neurons. Rostral and proximal zones were defined by their position relative to the blastema. (B) Rostral (black) and proximal (grey) zones in the uninjured spinal cord are similar in proliferation and rate of neurogenesis, suggesting that the larval spinal cord primarily exhibits homeostatic maintenance beyond 5dpf. (C) The number of nuclei in rostral and proximal zones remain relatively unchanged following transection; however, there is a significant increase in the number of nuclei in the blastema by 9dpi. n=5–7 fish per condition, per timepoint. Error bars=SEM; *p<0.05, **p<0.01, ***p<0.001.
To establish baseline rates of proliferation and neurogenesis in sham-treated animals, 5–8 larvae at 1dpi- 9dpi timepoints were examined. No day-to-day differences between proximal and rostral zones (as defined relative to the transection site in injured animals) were observed in comparing the following indices: proliferation (%BrdU+ nuclei), proliferation of dbx1a:GFP+ cells (%BrdU+,GFP+ nuclei), rate of neurogenesis (%BrdU+,Hu+ nuclei), and rate of neurogenesis of dbx1a:GFP+ cells (%BrdU+,GFP+,Hu+ nuclei) (Figure 7B). When data were averaged across all nine days and compared, no significant differences between proximal and rostral regions were observed. Thus, both the rates of proliferation and neurogenesis in uninjured animals are stable between 5dpf-14dpf, and could be aggregated together as sham data without regard for position analyzed. To characterize proliferation after injury, we counted the number of nuclei present in each region at 1, 5, and 9dpi (Figure 7C). In the rostral zone, there was a transient decrease in the number of nuclei at 5dpi. The number of nuclei remained relatively consistent during regeneration in the proximal zone; however, there was a significant increase in the number of nuclei in the blastema by 9dpi.
Representative maximum intensity Z-projections of the neurogenic process are shown in Figure 8, and colocalization of markers was confirmed using single slices (Supplemental Figure 3). Many BrdU+ cells were detectable outside of the spinal cord following injury; these are likely to be part of the immune response and dermal and myogenic repair (Suzuki et al., 2005). However, our analysis was limited to the spinal cord. When we examined neurogenesis after injury in dbx1a:GFP fish, we observed no newly born neurons or GFP+ cells in the blastema at 1dpi (Figure 8A–D; A’–D’). At 5dpi GFP+, Hu+, BrdU+ cells were observed at the proximal zone-blastema transition area (arrows) as well as in the rostral zone (Figure 8EH, E’–H’). By 9dpi, multiple neurons were present in the blastema, both GFP+, Hu+, BrdU+ (arrows) and GFP+, Hu+, BrdU+ (arrowheads, Figure 8I–L, I’–L’).
Figure 8.
dbx1a:GFP+ cells exhibit a proliferative and neurogenic response to injury. (A–D) At 1dpi, [GFP+, BrdU+, Hu−] cells (red arrowheads) are present in healthy spinal cord (yellow dashed line), as are [GFP+, Hu+, BrdU−] cells (yellow arrowheads); however, no GFP+ cells are present in the blastema (marked by solid yellow line). White box marks region shown in A’–D’. (E–H) At 5dpi, [GFP+, Hu+, BrdU−] cells (arrows) representing neurons arising from the dbx1a:GFP+ lineage born after injury are present in proximal and rostral zones; a few triple positive cells are present in the blastema as well. [GFP+, Hu+, BrdU−] cells (arrowheads) are present in proximal zone. White box marks region shown in E;-H’. (I–L) At 9dpi, multiple [GFP+, Hu+, BrdU+] (arrows) and [GFP+, Hu+, BrdU−] (arrowheads) cells are present in the growing blastema; triple positive cells observed at proximal zone-blastema boundary but not in the rostral zone. White box marks region shown in I’–L’. Scalebar = 50µm. All figures are Max-Z projections.
We found there was an increase in BrdU+ cells in all regions following injury (Figure 9A–C), and normalizing to the number of nuclei, we were able to conclude that proliferation occurred at all three regions by 5dpi, continuing until 9dpi (Figure 9D–F). To specifically determine if dbx1a:GFP+ cells proliferate in response to injury, we examined them as a subset of total BrdU+ cells (Figure 9G–I). In the rostral zone, there was a significant increase in the index of BrdU+,GFP+ cells at 1dpi, continuing through 9dpi. In the proximal zone, the index of BrdU+,GFP+ cells showed a trend of increase by 5dpi, with a significant increase at 9dpi. In the blastema, there was a brief reduction in the index of BrdU+,GFP+ cells compared to baseline at 1dpi, which is consistent with the cell death and debris clearing associated with spinal cord transection (Hui et al., 2010). The index of BrdU+,GFP+ cells increased significantly by 5dpi, and remained elevated through 9dpi. These data show that dbx1a:GFP+ cells proliferate in response to injury, and that the increase in proliferation is also systemic.
Figure 9.
Time course of proliferative and neurogenic responses following transection. (A–C) The number of BrdU+ cells following injury remains relatively unchanged in the rostral region during recovery (A), but a steady increase is observed in proximal (B) and blastema (C) zones. (D–F) Significant increases in the BrdU labeling index are observed in proximal and blastema zones by 5dpi. (G–I) The index of BrdU+,dbx1a:GFP+ cells also increases following injury, with a large number of BrdU+, GFP+ cells appearing in the blastema by 9dpi. (J–L) Significant accumulations of newly-born (BrdU+) neurons are observed in proximal and blastema zones by 9dpi. (M–O) A significant increase in newly-born GFP+ neurons is observed in the proximal zone by 5dpi, and in the blastema by 9dpi. (P–R) Percentage of newly born neurons arising from dbx1a:GFP+ population. Dashed line represents sham data, black lines are transected animals. *p<0.05, **p<0.01, ***p<0.001; n=5–7 animals per condition, per timepoint for each set of experiments; error bars=SEM.
We also found that neurogenesis was increased in response to injury (Figure 9J–L). In the rostral zone, the number of newly born (BrdU+) neurons in injured fish did not significantly vary from basal levels at all timepoints examined, suggesting that BrdU+ progenitor cells observed in this area do not differentiate in this location. In the proximal zone, a significant increase in the number of neurons born after injury was detected at 5dpi, continuing through 9dpi. Interestingly, a significant increase in the index of HuC/D+,BrdU+ cells was not observed in the blastema until 9dpi, suggesting that the neurons born shortly after injury in the proximal zone may migrate into this region. To determine whether proliferating dbx1a:GFP+ cells specifically contribute to de novo neurogenesis after injury, we examined them as a subset of total HuC/D+,BrdU+ cells (Figure 9M–O). A maximum of 2% of BrdU+ cells in the rostral zone were neurons arising from the dbx1a:GFP+ lineage, and levels in the rostral zone beyond 1dpi were not significantly different than controls. However, the neurogenic index of dbx1a:GFP+ cells in the proximal zone was significantly greater than controls at 5dpi and 9dpi. In the blastema, there was no significant neurogenesis from dbx1a:GFP+ cells until 9dpi. To determine the neurogenic contribution of dbx1a:GFP+ cells to injury, we examined the percentage of newly born neurons arising from the dbx1a:GFP population. In the proximal zone, 74.40% ± 18.9% of newly born neurons were GFP+ by 5dpi; at 9dpi this percentage was reduced to 38.38% ± 12.64%. Interestingly, 63.04% ± 9.11% of newly born neurons in the blastema were GFP+ at 9dpi (Figure 9P–R). Together these data demonstrate that dbx1a:GFP+ cells respond to injury by first rapidly expanding their progenitor pool throughout the rostral spinal cord with a subset of these new progenitors differentiating as neurons beginning at 5dpi, and that dbx1a:GFP+ cells represent a neural progenitor population with a robust neurogenic response to spinal cord injury.
Discussion
dbx1a+ cells are neural progenitors
In the embryonic mammalian spinal cord, Dbx1 expression labels a multipotent progenitor population that gives rise to neurons, astrocytes and oligodendrocytes (Fogarty et al., 2005). Here, our data show that dbx1a expression identifies a progenitor population in the zebrafish spinal cord that persist as radial glia and give rise to neurons during embryogenesis. This suggests an evolutionary conservation of Dbx1 expression between mammals and teleosts identifying neural progenitors of the intermediate spinal cord. We have also shown that at 5dpf, dbx1a:GFP+ cells are separate from olig2:dsRed+ cells, suggesting that these are two different progenitor populations. Previous analysis has shown that Dbx1+ cells contribute to the V0 and V1 interneuron populations during amniote development (Pierani et al., 2001). However, in the absence of immunohistochemical markers to identify these interneurons, we have not yet been able to determine which neuronal subtypes dbx1a:GFP+ cells produce during zebrafish development.
dbx1a:GFP+ cells persist as progenitors beyond embryogenesis
In the mammalian spinal cord, Dbx1+ cells are no longer detectable by E16.5 (Fogarty et al., 2005), indicating a loss of a multipotent progenitor population consistent with the observation that mammals cannot reinitiate neurogenesis following spinal cord injury. In contrast to mammals, dbx1a expression persists beyond embryogenesis until at least 14dpf in zebrafish, and dbx1a:GFP expression labels a slowly dividing neural progenitor population as evidenced by coexpression of GFAP, PCNA, and Sox3 with GFP. It remains to be shown whether dbx1a:GFP+ progenitor cells persist into adulthood, and whether this marker identifies a neural stem cell population in the intermediate spinal cord at that time.
Larval zebrafish exhibit rapid and robust spinal cord regeneration
The adult zebrafish has been effectively established as a model for studying spinal cord regeneration (Becker et al., 2004; Goldshmit et al., 2012; Hui et al., 2010). Zebrafish take 2–4 months to reach sexual maturity, and coupled with a recovery time from spinal injury of 6–8 wpi, we sought to establish a novel spinal cord injury model in the larval zebrafish that might represent shorter lead and recovery times and could also take advantage of genetic tools not available in the adult. We chose to transect the spinal cord at 5dpf because our data indicate that the peak of neurogenesis by dbx1a:GFP+ cells occurs at this time, and beyond that point the progenitors become more quiescent. While there was variability in recovery from injury, we have demonstrated that larval zebrafish injured at 5dpf have neuronal soma in the injury site by 9dpi, compared to 4wpi in the adult (Hui et al., 2010). Additionally, sensory and motor function recovery in the larval zebrafish is complete by 9dpi, instead of the 6–8wpi observed in the adult zebrafish (Reimer et al., 2008). We therefore propose that the larval zebrafish is an effective model for studying spinal cord regeneration. While our data do not conclusively prove that functional recovery in larvae requires neurogenesis or axon regrowth, the timing of these three events is well correlated.
dbx1a+ cells contribute to a proliferative and neurogenic response during spinal cord regeneration
By using BrdU to label proliferative cells before spinal cord transection, we were able to track equivalent populations in injured and control animals, and monitor their proliferation. Our data show that the proliferative response to injury is systemically present by 5dpi, but is more pronounced in the region proximal to the injury site and in the regenerating blastema. When we examined dbx1a:GFP+ cells as a subset of the BrdU+ population, we observed similar dynamics with a higher relative increase in proliferation compared to uninjured controls. We therefore conclude that spinal cord transection results in a rapid proliferative response from existing mitotic progenitors, including dbx1a:GFP+ cells.
BrdU labeling before injury also allowed us to trace the differentiation of dividing progenitors into neurons. When examining all BrdU+ cells, we found that the significant neurogenic response to injury was restricted to the region proximal to the injury site and the blastema, unlike the more widespread proliferative response. These data suggest that more rostral progenitors which proliferate following injury may die, remain undifferentiated, or migrate prior to differentiation. Significant levels of newly born neurons did not accumulate until after 5dpi, showing evidence of a delay between the proliferative and neurogenic responses, and the subset of dbx1a:GFP+ cells labeled by BrdU showed similar dynamics, indicating that the transgene marks neural progenitors contributing to repair following injury. Furthermore, our analysis suggests that most neurons born after injury arise from the dbx1a:GFP+ progenitor population. Our experiments do not allow us to distinguish between the possibilities that progenitors differentiate directly in the blastema, or that newly born dbx1a:GFP+ neurons migrate to the blastema from uninjured spinal cord. Regardless, it is likely that migration of either progenitors or neurons occurs because the blastema region is almost entirely devoid of cells following transection.
In conclusion, we have shown that dbx1a:GFP expression marks a pool of neurogenic spinal radial glial progenitors that persist beyond the end of embryogenesis, and contribute to the regenerative response by proliferating and subsequently differentiating as neurons. Based on their multipotency, longevity, slow rate of proliferation in the absence of injury and their rapid regenerative response, our data indicate that dbx1a:GFP+ radial glia are likely stem cells for the regeneration of interneurons following spinal cord injury in zebrafish.
Supplementary Material
Highlights.
A dbx1a:GFP reporter transgene labels radial glial progenitors beyond embryogenesis
Zebrafish larvae rapidly regenerate lost neurons and regain sensory and motor function after complete spinal transection
dbx1a:GFP+ cells proliferate and undergo neurogenesis after spinal cord transection
Acknowledgements
We are indebted to M. Klymkowsky for providing us with Sox3 antibody, and the University of Utah zebrafish facility for animal husbandry. We thank F.E. Poulain and J.A. Gaynes for helpful discussion. R.I.D. was supported by NIH R56NS053897, and L.K.B. was a predoctoral trainee supported by the HHMI Med-Into-Grad initiative.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Lisa K. Briona, Email: Lisa.Briona@neuro.utah.edu.
Richard I. Dorsky, Email: Richard.Dorsky@neuro.utah.edu.
References
- Bader HL, Keene DR, Charvet B, Veit G, Driever W, Koch M, Ruggiero F. Zebrafish collagen XII is present in embryonic connective tissue sheaths (fascia) and basement membranes. Matrix Biol. 2009;28:32–43. doi: 10.1016/j.matbio.2008.09.580. [DOI] [PubMed] [Google Scholar]
- Barresi MJF, Burton S, Dipietrantonio K, Amsterdam A, Hopkins N, Karlstrom RO. Essential genes for astroglial development and axon pathfinding during zebrafish embryogenesis. Dev. Dyn. 2010;239:2603–2618. doi: 10.1002/dvdy.22393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker CG, Lieberoth BC, Morellini F, Feldner J, Becker T, Schachner M. L1.1 is involved in spinal cord regeneration in adult zebrafish. J. Neurosci. 2004;24:7837–7842. doi: 10.1523/JNEUROSCI.2420-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Wullimann MF, Becker CG, Bernhardt RR, Schachner M. Axonal regrowth after spinal cord transection in adult zebrafish. J. Comp. Neurol. 1997;377:577–595. doi: 10.1002/(sici)1096-9861(19970127)377:4<577::aid-cne8>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Bhatt DH, Otto SJ, Depoister B, Fetcho JR. Cyclic AMP-induced repair of zebrafish spinal circuits. Science. 2004;305:254–258. doi: 10.1126/science.1098439. [DOI] [PubMed] [Google Scholar]
- Briona LK, Dorsky RI. Spinal cord transection in the larval zebrafish. JoVE. 2013 doi: 10.3791/51479. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess HA, Granato M. Sensorimotor gating in larval zebrafish. J. Neurosci. 2007;27:4984–4994. doi: 10.1523/JNEUROSCI.0615-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff EAG, Sato K, Corn A, Karcavich RE. Spinal cord regeneration: intrinsic properties and emerging mechanisms. Semin. Cell Dev. Biol. 2002;13:361–368. doi: 10.1016/s1084952102000927. [DOI] [PubMed] [Google Scholar]
- Chernoff EAG, Stocum DL, Nye HLD, Cameron JA. Urodele spinal cord regeneration and related processes. Dev. Dyn. 2003;226:295–307. doi: 10.1002/dvdy.10240. [DOI] [PubMed] [Google Scholar]
- Fogarty M, Richardson WD, Kessaris N. A subset of oligodendrocytes generated from radial glia in the dorsal spinal cord. Development. 2005;132:1951–1959. doi: 10.1242/dev.01777. [DOI] [PubMed] [Google Scholar]
- García-Verdugo JM, Ferrón S, Flames N, Collado L, Desfilis E, Font E. The proliferative ventricular zone in adult vertebrates: a comparative study using reptiles, birds, and mammals. Brain Res. Bull. 2002;57:765–775. doi: 10.1016/s0361-9230(01)00769-9. [DOI] [PubMed] [Google Scholar]
- Goldman S. Glia as neural progenitor cells. Trends Neurosci. 2003;26:590–596. doi: 10.1016/j.tins.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Goldshmit Y, Sztal TE, Jusuf PR, Hall TE, Nguyen-Chi M, Currie PD. Fgf-dependent glial cell bridges facilitate spinal cord regeneration in zebrafish. J. Neurosci. 2012;32:7477–7492. doi: 10.1523/JNEUROSCI.0758-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribble SL, Nikolaus OB, Carver MS, Hoshijima K, Dorsky RI. Chromosomal position mediates spinal cord expression of a dbx1a enhancer. Dev. Dyn. 2009;238:2929–2935. doi: 10.1002/dvdy.22122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribble SL, Nikolaus OB, Dorsky RI. Regulation and function of Dbx genes in the zebrafish spinal cord. Dev. Dyn. 2007;236:3472–3483. doi: 10.1002/dvdy.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang YJ, Heisenberg CP, Kelsh RN, Furutani-Seiki M, Vogelsang E, Beuchle D, Schach U, Fabian C, Nüsslein-Volhard C. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- Hale ME, Ritter DA, Fetcho JR. A confocal study of spinal interneurons in living larval zebrafish. J. Comp. Neurol. 2001;437:1–16. doi: 10.1002/cne.1266. [DOI] [PubMed] [Google Scholar]
- Hui SP, Dutta A, Ghosh S. Cellular response after crush injury in adult zebrafish spinal cord. Dev. Dyn. 2010;239:2962–2979. doi: 10.1002/dvdy.22438. [DOI] [PubMed] [Google Scholar]
- Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Kim H-S, Dorsky RI. Tcf7l1 is required for spinal cord progenitor maintenance. Dev. Dyn. 2011;240:2256–2264. doi: 10.1002/dvdy.22716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kucenas S, Takada N, Park H-C, Woodruff E, Broadie K, Appel B. CNS-derived glia ensheath peripheral nerves and mediate motor root development. Nature Neuroscience. 2008;11:143–151. doi: 10.1038/nn2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuscha V, Barreiro-Iglesias A, Becker CG, Becker T. Plasticity of tyrosine hydroxylase and serotonergic systems in the regenerating spinal cord of adult zebrafish. J. Comp. Neurol. 2012;520:933–951. doi: 10.1002/cne.22739. [DOI] [PubMed] [Google Scholar]
- Lu S, Bogarad LD, Murtha MT, Ruddle FH. Expression pattern of a murine homeobox gene, Dbx, displays extreme spatial restriction in embryonic forebrain and spinal cord. Proc. Natl. Acad. SciU.S.A. 1992;89:8053–8057. doi: 10.1073/pnas.89.17.8053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malatesta P, Hartfuss E, Götz M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development. 2000;127:5253–5263. doi: 10.1242/dev.127.24.5253. [DOI] [PubMed] [Google Scholar]
- Mori T, Buffo A, Götz M. The novel roles of glial cells revisited: the contribution of radial glia and astrocytes to neurogenesis. Curr. Top. Dev. Biol. 2005;69:67–99. doi: 10.1016/S0070-2153(05)69004-7. [DOI] [PubMed] [Google Scholar]
- Naujoks-Manteuffel C, Roth G. Astroglial cells in a salamander brain (Salamandra salamandra) as compared to mammals: a glial fibrillary acidic protein immunohistochemistry study. Brain Res. 1989;487:397–401. doi: 10.1016/0006-8993(89)90849-4. [DOI] [PubMed] [Google Scholar]
- Navarro X, Kamei H, Kennedy WR. Effect of age and maturation on sudomotor nerve regeneration in mice. Brain Research. 1988;447:133–140. doi: 10.1016/0006-8993(88)90973-0. [DOI] [PubMed] [Google Scholar]
- Oxtoby E, Jowett T. Cloning of the zebrafish krox-20 gene (krx-20) and its expression during hindbrain development. Nucleic Acids Res. 1993;21:1087–1095. doi: 10.1093/nar/21.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HC, Kim CH, Bae YK, Yeo SY, Kim SH, Hong SK, Shin J, Yoo KW, Hibi M, Hirano T, Miki N, Chitnis AB, Huh TL. Analysis of upstream elements in the HuC promoter leads to the establishment of transgenic zebrafish with fluorescent neurons. Dev. Biol. 2000;227:279–293. doi: 10.1006/dbio.2000.9898. [DOI] [PubMed] [Google Scholar]
- Park H-C, Shin J, Roberts RK, Appel B. An olig2 reporter gene marks oligodendrocyte precursors in the postembryonic spinal cord of zebrafish. Dev. Dyn. 2007;236:3402–3407. doi: 10.1002/dvdy.21365. [DOI] [PubMed] [Google Scholar]
- Pierani A, Moran-Rivard L, Sunshine MJ, Littman DR, Goulding M, Jessell TM. Control of interneuron fate in the developing spinal cord by the progenitor homeodomain protein Dbx1. Neuron. 2001;29:367–384. doi: 10.1016/s0896-6273(01)00212-4. [DOI] [PubMed] [Google Scholar]
- Rakic P. Elusive radial glial cells: historical and evolutionary perspective. Glia. 2003;43:19–32. doi: 10.1002/glia.10244. [DOI] [PubMed] [Google Scholar]
- Rehermann MI, Santiñaque FF, López-Carro B, Russo RE, Trujillo-Cenóz O. Cell proliferation and cytoarchitectural remodeling during spinal cord reconnection in the fresh-water turtle Trachemys dorbignyi. Cell Tissue Res. 2011;344:415–433. doi: 10.1007/s00441-011-1173-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer MM, Sörensen I, Kuscha V, Frank RE, Liu C, Becker CG, Becker T. Motor neuron regeneration in adult zebrafish. J. Neurosci. 2008;28:8510–8516. doi: 10.1523/JNEUROSCI.1189-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer J, Gimnopoulos D, Lieberoth BC, Pogoda H-M, Feldner J, Ebert A, Schachner M, Becker T, Becker CG. Contactin1a expression is associated with oligodendrocyte differentiation and axonal regeneration in the central nervous system of zebrafish. Mol. Cell. Neurosci. 2007;35:194–207. doi: 10.1016/j.mcn.2007.02.018. [DOI] [PubMed] [Google Scholar]
- Seo HC, Nilsen F, Fjose A. Three structurally and functionally conserved Hlx genes in zebrafish. Biochim. Biophys. Acta. 1999;1489:323–335. doi: 10.1016/s0167-4781(99)00187-6. [DOI] [PubMed] [Google Scholar]
- Simmons T, Appel B. Mutation of pescadillo disrupts oligodendrocyte formation in zebrafish. PLoS ONE. 2012;7:e32317. doi: 10.1371/journal.pone.0032317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisson BE, Topczewski J. Expression of five frizzleds during zebrafish craniofacial development. Gene Expr. Patterns. 2009;9:520–527. doi: 10.1016/j.gep.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Colello RJ, Daugherty WP, Kwon TH, McGinn MJ, Harvey HB, Bullock MR. Cell Proliferation and Neuronal Differentiation in the Dentate Gyrus in Juvenile and Adult Rats following Traumatic Brain Injury. Journal of Neurotrauma. 2005;22:95–105. doi: 10.1089/neu.2005.22.95. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Satoh A, Ide H, Tamura K. Nerve-dependent and -independent events in blastema formation during Xenopus froglet limb regeneration. Dev. Biol. 2005;286:361–375. doi: 10.1016/j.ydbio.2005.08.021. [DOI] [PubMed] [Google Scholar]
- Thisse B, Heyer V, Lux A, Alunni V, Degrave A, Seiliez I, Kirchner J, Parkhill J-P, Thisse C. Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol. 2004;77:505–519. doi: 10.1016/s0091-679x(04)77027-2. [DOI] [PubMed] [Google Scholar]
- Trevarrow B, Marks DL, Kimmel CB. Organization of hindbrain segments in the zebrafish embryo. Neuron. 1990;4:669–679. doi: 10.1016/0896-6273(90)90194-k. [DOI] [PubMed] [Google Scholar]
- Wan Y, Otsuna H, Chien C-B, Hansen C. FluoRender: An Application of 2D Image Space Methods for 3D and 4D Confocal Microscopy Data Visualization in Neurobiology Research. IEEE Pac Vis Symp. 2012:201–208. doi: 10.1109/pacificvis.2012.6183592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T-W, Stromberg GP, Whitney JT, Brower NW, Klymkowsky MW, Parent JM. Sox3 expression identifies neural progenitors in persistent neonatal and adult mouse forebrain germinative zones. J. Comp. Neurol. 2006;497:88–100. doi: 10.1002/cne.20984. [DOI] [PubMed] [Google Scholar]
- Westerfield M. A guide for the laboratory use of zebrafish (Danio rerio) 4th ed. Eugene, OR: University of Oregon Press; 2000. The zebrafish book. [Google Scholar]
- Williams JA, Barrios A, Gatchalian C, Rubin L, Wilson SW, Holder N. Programmed cell death in zebrafish rohon beard neurons is influenced by TrkC1/NT-3 signaling. Dev. Biol. 2000;226:220–230. doi: 10.1006/dbio.2000.9860. [DOI] [PubMed] [Google Scholar]
- Zupanc GKH, Clint SC. Potential role of radial glia in adult neurogenesis of teleost fish. Glia. 2003;43:77–86. doi: 10.1002/glia.10236. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.