Abstract
Spore germination in Bacillus species represents an excellent model system with which to study the molecular mechanisms underlying the nutritional control of growth and development. Binding of specific chemical nutrients to their cognate receptors located in the spore inner membrane triggers the germination process that leads to a resumption of metabolism in spore outgrowth. Recent studies suggest that the inner membrane GerD lipoprotein plays a critical role in the receptor-mediated activation of downstream germination events. The 121-residue core polypeptide of GerD (GerD60-180) from Geobacillus stearothermophilus forms a stable α-helical trimer in aqueous solution. The 2.3-Å-resolution crystal structure of the trimer reveals a neatly twisted superhelical rope, with unusual supercoiling induced by parallel triple-helix interactions. The overall geometry comprises three interleaved hydrophobic screws of interacting helices linked by short turns that have not been seen before. Using complementation analysis in a series of Bacillus subtilis gerD mutants, we demonstrated that alterations in the GerD trimer structure have profound effects on nutrient germination. This important structure–function relationship of trimeric GerD is supported by our identification of a dominant negative gerD mutation in B. subtilis. These results and those of others lead us to propose that GerD mediates clustering of germination proteins in the inner membrane of dormant spores and thus promotes the rapid and cooperative germination response to nutrients.
Keywords: Bacillus, spores, spore germination, germinant receptors, GerD
Introduction
One of the most characteristic features of the Firmicute phylum, which includes various Bacillales and Clostridiales species, is the ability of some of these species to form endospores in sporulation, a process induced in response to adverse growth conditions [1,2]. These spores are extremely resistant to all manner of environmental insults, properties that allow spores to exist in their metabolically dormant state indefinitely and remain viable for hundreds of years without water or nutrients [3,4]. However, during their long period of dormancy, spores are constantly sensing the environment, and when favorable conditions return, spores can return to active metabolism within minutes through the process of spore germination followed by outgrowth to generate growing cells [1,3,5,6]. As such, spores are ubiquitous throughout our environment, and pathogenic varieties have had a significant impact on human health and disease [6]. For example, Clostridium botulinum, Clostridium perfringens and Bacillus cereus are major agents of food spoilage and food-borne disease, while Bacillus anthracis spores cause anthrax in animals and man and can be used as a biological weapon. Thus, a detailed understanding of the mechanisms of sporulation and germination has both basic and applied interests.
A major signal that triggers spore germination is the presence of specific nutrients called germinants in spores’ environments. These nutrient germinants are typically amino acids, purine nucleosides or sugars that are recognized in a stereospecific manner by cognate germinant receptors (GRs) located in the inner membrane of the spore. Three functional GRs are found in Bacillus subtilis spores, each encoded by the homologous tricistronic gerA, gerB and gerK operons [4,7]. The GerA GR responds to l-alanine or l-valine, while the GerB and GerK GRs cooperate to respond to an amino acid and sugar combination of l-asparagine, d-glucose, d-fructose and potassium ions (AGFK). Specific germinant–GR interaction results in transduction of a signal that leads to the release of the large depot (~10% of the spores dry weight) of pyridine-2,6-dicarboxylic acid [dipicolinic acid (DPA)] and associated cations, predominantly Ca2+ (CaDPA) from the spore core, likely via a channel composed at least in part of SpoVA proteins [8,9]. DPA release then triggers degradation of the spore’s peptidoglycan cortex by cortex-lytic enzyme, eventually leading to resumption of metabolism, macromolecular synthesis and vegetative growth.
While a number of the physical changes accompanying spore germination and the proteins involved in this process have been identified, there is as yet no understanding of how signals are transduced from GRs to other spore components to initiate the physiological route to germination. The fact that spores can integrate and amplify signals from multiple GRs in determining rates of commitment and germination [10] suggests that there is an additional protein involved in GR-dependent signal transduction. In Bacillus species, an obvious candidate for an intermediate role in this signal transduction pathway is the GerD protein. The gerD-null mutation greatly decreases rates of GR-dependent germination in response to nutrient germinants with spores of B. subtilis and Bacillus megaterium but does not affect spore germination induced by agents that do not act through GRs (Gupta, S. and Christie, G., personal communication) [11]. Moreover, recent work demonstrates that, in B. subtilis spores, GRs and GerD colocalize in a small cluster termed the germinosome in the spore’s inner membrane and that GerD is essential for this GR clustering [12]. Interestingly, in Bacillus spores, both the SpoVA proteins and the germination-specific cortex-lytic enzyme SleB are also located in the spore inner membrane [9,13]. It thus appears that GerD likely plays a role in mediating the rapid transduction of signals from the germinant–receptor complex to downstream effectors.
A single gerD gene is present in all spore-forming Bacillales species and its expression takes place only in the forespore under the control of forespore-specific RNA polymerase sigma factor σG, as is the case for transcription of the GR genes [1,14]. It is worth noting that there is no obvious GerD homolog in the more distantly related Clostridiales genus, suggesting that either the function of GerD is not required in spores of this genus or its function is fulfilled by an alternative undefined protein [1]. In keeping with its inner membrane location in spores, GerD is likely to be a lipoprotein that has a diacylglycerol anchor linked to an amino-terminal cysteine residue [15,16] and appears to be on the outer surface of the inner membrane [17]. Although the amino acid sequences of the GerD proteins are well conserved throughout the Bacilli, they exhibit no significant sequence homology to proteins of known function.
In an effort to understand the role of GerD in promoting spore germination, we have determined the crystal structure of the 121-residue core domain of the Geobacillus stearothermophilus GerD protein. The 2.3-Å-resolution structure reveals that GerD possesses a previously uncharacterized rod-like fold consisting of three parallel helical polypeptide chains wrapping into a supercoiled rope. Both secondary-structure prediction and structure-based alignment suggest that this fold represents the prototype of the GerD proteins from different species. We showed that single-point mutations or helix truncation mutations that affect the overall topology and helical fold of GerD exert deleterious effects on nutrient germination. Using an independent genetic screen, we also identified a dominant negative mutation in gerD, reinforcing the importance of GerD in nutrient germination in addition to GRs. Our structural and functional analyses support a model in which GerD serves a scaffolding function in formation of a GR-containing complex that may facilitate the signal transmission from GRs to downstream components of the germination pathways.
Results
Expression and structure determination
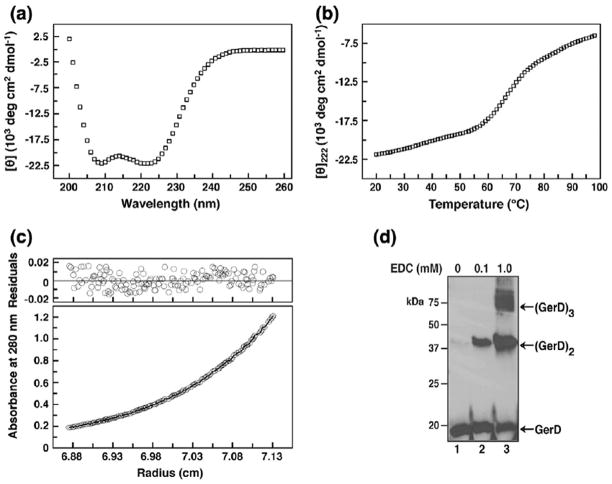
We crystallized a 121-residue fragment of the G. stearothermophilus GerD protein (GerD60-180). GerD60-180 lacks the N-terminal signal and lipobox sequences (residues 1–28) and the predicted H01 and H02 helices (residues 29–59), as well as the C-terminal acidic tail (residues 181–195). The recombinant GerD60-180 protein was expressed in Escherichia coli and purified to homogeneity by anion-exchange and gel-filtration chromatography. The truncated G. stearothermophilus GerD protein was chosen for the current study because we were unable to obtain diffraction-quality crystals of the full-length protein and the GerD homologs from other Bacillus species. Circular dichroism (CD) measurements showed that GerD60-180 is ~80% helical at 20 °C (Fig. 1a) and displays a cooperative thermal unfolding transition with a melting temperature of ~68 °C (Fig. 1b). Sedimentation equilibrium experiments indicated that GerD60-180 forms a clean trimer and exhibits no systematic dependence of molecular mass on protein concentration between 30 and 500 μM (Fig. 1c). Thus, GerD60-180 forms a stable, well-ordered three-helix bundle in solution. Moreover, cross-linking of the total lysate fractions of B. subtilis spores using a zero-length cross-linking agent produces multiple higher-molecular-weight species corresponding to the dimer, trimer and even higher-order oligomeric forms of GerD (Fig. 1d), suggesting that GerD can indeed self-associate to form oligomers in spores. The crystal structure of GerD60-180 was determined at 2.3 Å resolution by the single-wavelength anomalous dispersion method using data from a selenomethionine (SeMet)-substituted crystal. Iterative model building and crystallographic refinement resulted in a model with an R factor of 20.7% and a Rfree value of 25.6% (Table 1). The final model of the GerD60-180 structure contains two trimers.
Fig. 1.
GerD forms a stable α-helical trimer in solution. (a) CD spectrum of G. stearothermophilus GerD60-180 (25 μM) at 20 °C in TBS (pH 8.0). (b) Thermal melt monitored by CD at 222 nm with 35 μM protein. (c) Representative sedimentation equilibrium data for GerD60-180 (35 μM) at 20 °C and 14,000 rpm in TBS (pH 7.6). The data fit closely to a trimeric complex. (Upper) The deviation in the data from the linear fit for a trimeric model is plotted. (d) Cross-linking of total lysates of the wild-type B. subtilis PS533 spores by varying concentrations of EDC, which cross-links adjacent carboxyl groups to primary amines. Equal amounts of the resulting lysates were analyzed by SDS-PAGE\Western blotting using anti-B. subtilis GerD serum. Similar results were obtained when a lysate from spores lacking the GerA, GerB and GerK GRs were cross-linked with EDC (data not shown). The black arrows on the right side of the blot indicate the predicted molecular weights for GerD monomer, dimer and trimer.
Table 1.
Summary of crystallographic analysis.
| Native | SeMet (peak) | |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 0.9600 | 0.9792 |
| Space group | P212121 | P212121 |
| Cell dimensions (Å) | ||
| a, b, c (Å) | 59.7, 98.5, 127.5 | 142.4, 142.4, 187.7 |
| Resolution (Å) | 50.0–2.30 (2.38–2.30) | 50.0–2.70 (2.80–2.70) |
| Rsym (%) | 8.9 (46.3) | 22.2 (92.5) |
| I/σI | 30.5 (6.3) | 12.9 (3.8) |
| Completeness (%) | 99.9 (99.9) | 100.0 (99.9) |
| Redundancy | 14.4 (14.4) | 13.9 (11.8) |
| SAD Analysis | ||
| rms FH/e | 0.83 | |
| Mean figure of merit before DM | 0.43 | |
| Mean figure of merit after DM | 0.54 | |
| Refinement | ||
| Resolution (Å) | 50.0–2.30 | |
| No. of reflections (|F|> 0σ) | 32,570 | |
| R-factor/Rfree | 20.7/25.6 | |
| Total protein atoms | 5410 | |
| Water molecules | 159 | |
| B-factors (Å2) | ||
| Protein | 48.2 | |
| Water | 48.6 | |
| rmsd | ||
| Bond lengths (Å) | 0.011 | |
| Bond angles (°) | 1.144 | |
| Ramachandran (%) | ||
| Within favored | 97.7 | |
| Within allowed | 100.0 | |
| Outliers | 0 |
Values in parentheses are for the highest-resolution shell.
Structure of the GerD60-180 trimer
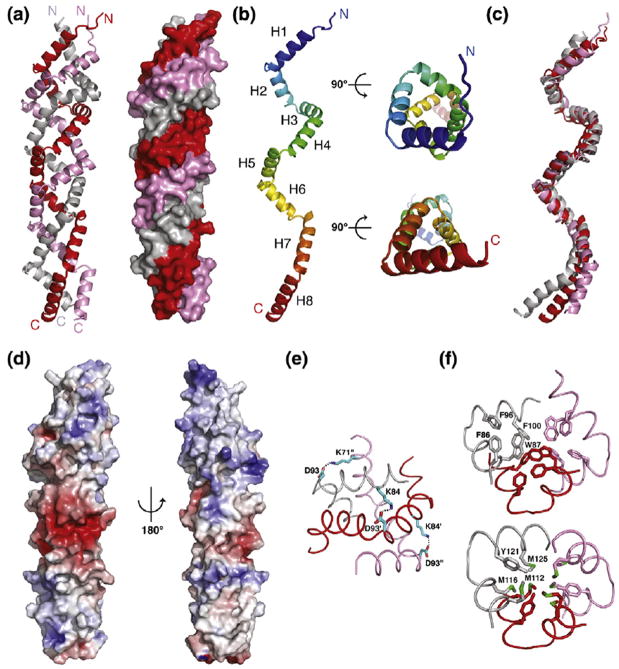
The GerD60-180 trimer consists of three parallel polypeptide chains twisted into a superhelical, right-handed rope with several novel and unique features (Fig. 2a). The superhelical rope includes approximately 110–120 residues from each chain; a few N- and C-terminal tail residues are not well defined in the electron density maps. This triple-stranded rope has a straight supercoil axis and forms a cylinder ~110 Å in length and ~22 Å in diameter. The individual polypeptide chains of GerD60-180 form eight consecutive helices (H1 to H8) linked by short turns (Figs. 2b and 3) and can be superimposed on each other with root-mean-square (rms) deviations for α carbon atoms of 2.6–2.8 Å and with poor superposition of the loosely packed H8 helices (Fig. 2c). One hundred residues (83%) of the GerD60-180 molecule are in an α-helical conformation, consistent with the CD analysis results (Fig. 1a).
Fig. 2.
Crystal structure of G. stearothermophilus GerD60-180. (a) Ribbon diagram (left) and molecular surface (right) of the GerD60-180 trimer. (b) Side (left) and axial (right) views of the GerD60-180 monomer. (c) Superposition of three GerD60-180 protomers. (d) Molecular surface representation of the GerD60-180 trimer, colored according to the local electrostatic potential ranging from −6 kT/e in deep red (most negative) to +6 kT/e in dark blue (most positive). The local electrostatic potential was calculated using the program ABPS [18]. (e) Close-up view of the salt-bridge network (indicated by black dots) formed between Lys71, Lys84 and Asp93 of the GerD60-180 trimer. (f) Cross-sections of the superhelical GerD60-180 trimer showing the hydrophobic packing involving either aromatic (upper) or methionine (lower) residues. Side chains of residues are shown as stick representation and the three monomers are shown as Cα traces.
Fig. 3.
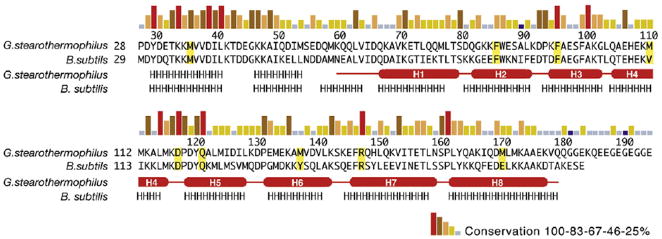
Sequence and structure conservation among GerD orthologs of Bacillales species. Sequence conservation of GerD is shown as a bar graph, with red bars indicating identity among the 24 GerD orthologs in Bacillales species. Secondary-structure assignments of GerD60-180 from the structure are shown as red cylinders (α-helices) and lines (turns or loops). Predicted secondary-structure elements (http://www.predicprotein.org) for B. subtilis GerD and the N-terminal region of G. stearothermophilus GerD are indicated by letters (H, α-helix).
Helices H1–H7 in each GerD60-180 chain twist around the central axis to form two complete turns of a right-handed supercoil with a pitch of ~44 Å. Interestingly, the N-terminal supercoil turn containing helices H1–H4 (total 51 residues) is packed and organized in positions similar to those of four residues in an α-helix turn, while the C-terminal turn containing helices H6–H8 (45 residues) is similar to those of three residues in a canonical 310 helix (Fig. 2b). The individual eight helices of GerD60-180 bend in a right-handed curl, allowing them to specifically associate with neighboring helices over the entire length of the superhelical rope. The curvature of the GerD60-180 trimer is associated with shorter main-chain hydrogen bonds in the interface compared to the outside of the helices, a feature that has been suggested to induce supercoiling of the α-helical coiled coil [19]. The CD spectrum of the GerD60-180 trimer has a ratio of 1.05 for the minima at 222 and 208 nm (Fig. 1a), consistent with typical spectra observed for interacting α-helices rather than a single-stranded α-helix [20].
Serendipitously, the three parallel polypeptide chains in the structure of the GerD60-180 trimer are offset from each other by approximately one-third of a turn of the α-helix, causing a shift in register between the adjacent helices. Consequently, surface residues of the interacting helices make unique side-to-side contacts, leading to the formation of distinctive screws of triple-helical bundles to efficiently sequester the hydrophobic interface from solvent (Fig. 2a). Approximately 4600 Å2 of accessible surface area (43% of the total accessible surface area of each isolated monomer) is buried in the GerD60-180 trimer. So far as we know, these interleaved superhelical screws have not been seen before in reported protein structures.
The GerD60-180 helices are largely amphipathic, with polar and charged residues on the surface of the triple-stranded rope, while nonpolar side chains are buried on the inside to form the highly packed hydrophobic core of the rope (Fig. 2d). A distinct network of interhelical and intrahelical salt bridges and hydrogen bonds coats the surface of the GerD60-180 trimer. For example, salt bridges between the Lys71, Lys84 and Asp93 side chains are involved in the close packing and stabilization of interhelical contacts (Fig. 2e). Clustering of Asp127, Asp131, Glu133, Glu135, Asp140 and Glu146 in the middle of the superhelix forms a large negatively charged patch on its surface (Fig. 2d). In contrast to the surface of the GerD60-180 trimer, its central core is lined with hydrophobic side chains that span the superhelix (Fig. 2f). The Phe86, Trp87, Phe96 and Phe100 residues in helices H2 and H3 are stacked with each other similar to a twisted ladder. Similarly, a group of methionine residues in helices H4 and H5 pack tightly in the middle of the superhelix through interchain van der Waals interactions (Fig. 2f). The results suggest that both optimal networks of surface electrostatic interactions and internal hydrophobic packing interactions play a crucial role in determining the structure and stability of the GerD60-180 trimer.
Structural conservation among GerD homologs
A broad BLAST search using the B. subtilis GerD protein as the query sequence identified 24 homologs of GerD in spore-forming Bacillales genomes, with one gerD gene per genome. These homologs share approximately 30–73% pairwise sequence identities with their B. subtilis counterparts. With the structure of G. stearothermophilus GerD in hand, we can now elaborate on several features relating to sequence and structure conservation between GerD homologs. First, other GerD proteins are likely to have a three-dimensional structure similar to that of G. stearothermophilus GerD. Secondary-structure predictions by the programs PredictProtein [21] and Jpred [22] suggest that all GerD homologs exhibit predominantly α-helical topology essentially identical with that of G. stearothermophilus GerD, including the N-terminal region (residues 29–59; predicted helices H01 and H02) absent in our GerD60-180 structure (Fig. 3). It is worth noting that, unlike B. subtilis GerD, most GerD proteins identified in Bacillaceae, including G. stearothermophilus GerD, contain a flexible C-terminal acidic tail (Fig. 3). The length and exact composition of the acidic tail varies among different species and its function remains to be understood. We should emphasize that G. stearothermophilus GerD without the C-terminal acidic tail can fully complement the poor spore germination of B. subtilis spores lacking GerD (see below). Second, the investigation of GerD homologous sequence conservation in the structural context of the GerD60-180 trimer reveals that invariant or highly conserved residues are located throughout the entire GerD molecule, although residues in the H01–H02 and H4–H5 clusters are more conserved than the rest of the protein (Fig. 3).
Effects of gerD mutations on germination
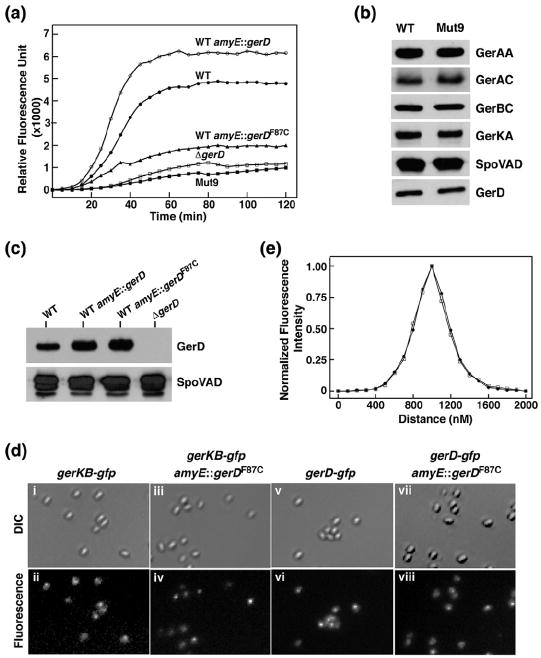
To characterize the structure–function relationship of GerD, we produced a series of G. stearothermophilus gerD mutants in which various helical coding segments were either truncated or disrupted. We then replaced the gerD gene in B. subtilis with G. stearothermophilus gerD variants and assessed the germination activities of the mutant spores by monitoring DPA release upon addition of the germinant l-valine. All mutant strains showed sporulation efficiency comparable to that of the wild-type B. subtilis strain (data not shown). The initial germination rate of spores from the B. subtilis mutant carrying the G. stearothermophilus gerD1-180 (with a deletion of the C-terminal acidic tail) is essentially identical with that of spores carrying the wild-type B. subtilis gerD gene (Fig. 4a; Table 2). In contrast, nutrient germination was essentially abolished in spores containing a deletion of the N-terminal H01 and H02 helices of GerD. While the truncation of the C-terminal H8 helix of GerD led to a 20% reduction in the initial germination rate, the effects of deleting additional C-terminal helices were much more deleterious, with 26%, 7% and 14% of initial germination rates for ΔH7-8, ΔH6-8 and ΔH5-8 variants, respectively. Next, we used site-directed mutagenesis to individually change the aforementioned Met36, Phe86, Met111, Met138 and Met171 residues involved in the GerD60-180 trimer interactions to proline to determine whether the specific side chains are crucial for GerD function (Fig. 4b). Proline was selected because proline is an α-helix breaker due to steric hindrance arising from its cyclic side chain that blocks the main-chain NH group [23]. As summarized in Table 2, the spores of the M36P, F86P, M138P and M171P mutants had 9–14% of the initial germination rate, essentially lower than that of the gerD-null spores, while the M111P mutation in the middle of the GerD superhelix resulted in a 50% reduction in the initial germination rate. Finally, we created B. subtilis strains carrying alanine point mutations of four invariant or highly conserved residues in GerD (F96A, D118A, Q122A and R148A). All these mutant spores were unable to germinate in vitro (Fig. 4c; Table 2). Taken together, these data suggest that the overall topology and precise packing of the triple-stranded superhelical structure of GerD plays an important role in mediating its function in nutrient-induced spore germination.
Fig. 4.
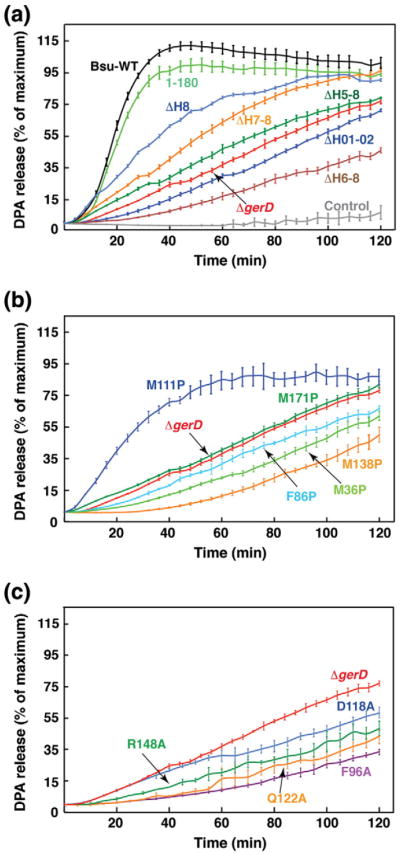
Effects of gerD mutations on germination of spores. Germinations of spores of the wild-type (a) (B. subtilis gerD and G. stearothermophilus gerD1-180) and mutant [(a) helix deletion mutations, (b) proline mutations, (c) conserved residue mutations] gerD strains with 10 mM l-valine are shown. The percentage of DPA release of spores of each strain was normalized against the maximum RFU obtained using B. subtilis spores with the wild-type G. stearothermophilus gerD gene (gerD1-180) as previously described. The germination curve of the spores of the B. subtilis strain with the B. subtilis wild-type gerD gene and the kanamycin resistance cassette is shown. The fluorescence measurements of the spores of this latter strain in the absence of germinants are labeled as Control. The germination curve of the B. subtilis ΔgerD spores is also shown. Data represent means ± standard deviations from at least three independent measurements.
Table 2.
Kinetic analysis of germination of wild-type and mutant gerD spores with l-valine
| gerD strains | Relative rates (min−1)a | C50 (mM)b | Vmax (min−1) |
|---|---|---|---|
| 1-180 | 100 | 2.2 ± 0.2 | 186 ± 5 |
| 1-185 (B. subtilis) | 113 | 2.4 ± 0.5 | 230 ± 17 |
| ΔgerD | 15 | 4.2 ± 1.2 | 38 ± 4 |
| Helix truncation | |||
| 1-28, 55-18 (ΔH01-02)c | 13 | 5.1 ± 1.5 | 29 ± 4 |
| 1-163 (ΔH8) | 80 | 2.9 ± 0.3 | 85 ± 3 |
| 1-146 (ΔH7-8) | 26 | 4.4 ± 1.3 | 61 ± 7 |
| 1-133 (ΔH6-8) | 7 | 6.2 ± 1.2 | 26 ± 8 |
| 1-120 (ΔH5-8) | 14 | 3.9 ± 1.2 | 56 ± 6 |
| Helix disruption | |||
| M36P (H01) | 9 | 4.4 ± 1.5 | 18 ± 4 |
| F86P (H2) | 11 | 6.2 ± 1.7 | 40 ± 5 |
| M111P (H4) | 50 | 3.0 ± 0.6 | 104 ± 7 |
| M138P (H6) | 9 | 4.0 ± 1.3 | 20 ± 4 |
| M171P (H8) | 14 | 5.1 ± 1.5 | 29 ± 4 |
| Conservation | |||
| F96A (H3) | 4 | 7.4 ± 2.0 | 16 ± 2 |
| D118A (between H4 and H5) | 10 | 4.0 ± 0.4 | 27 ± 1 |
| Q122A (H5) | 7 | 9.7 ± 1.7 | 18 ± 2 |
| R148A (H7) | 7 | 7.5 ± 2.4 | 20 ± 3 |
The initial rate of the germination of the GerD1-180 spores was set at 100.
C50, germinant concentration that stimulates a half-maximal initial rate of germination.
Values in parentheses indicate the location of the mutation.
We further examined the effects of the above gerD truncating and point mutations on the germination of the wild-type and mutant spores with varying concentrations of l-valine. The relative maximum initial germination rates (Vmax) and the l-valine concentrations needed to achieve 50% of Vmax (C50) for the wild-type and mutant spores were obtained from hyperbolic curves of germination rates with l-valine (Table 2). Based on these analysis, the gerD1-163 (ΔH8) and gerDM111P mutant spores had a C50 value close to that of the wild-type spore and a Vmax of about half of the wild-type one, in keeping with their relatively good germination with l-valine (Fig. 5a and b). All other mutant spores had values of C50 and Vmax similar to those of the gerD-null strain, indicating the loss of GerD function in those mutant strains.
Fig. 5.
F87C is a dominant negative mutation in B. subtilis GerD. (a) Effects of the F87C mutation on germination of spores with 10 mM l-valine. The symbols for the strains used to generate spores are as follows: (●) PS832 (wild-type); (○) PS832 amyE∷gerD; (▲) PS832 amyE∷gerDF87C; (□) FB62 (ΔgerD); (■) Mut9. (b) Levels of germination proteins in the total lysates of spores of strains PS832 and Mut9. Equal amounts of lysate proteins extracted from decoated spores from both strains were analyzed by SDS-PAGE\Western blotting using polyclonal antisera against B. subtilis proteins. (c) Levels of GerD in spores of various strains. Equivalent amounts of total lysates from spores of strains PS832 (wild type), PS832 amyE∷gerD, PS832 amyE∷gerDF87C and FB62 (ΔgerD) were analyzed by SDS-PAGE\Western blotting using antiserum against B. subtilis GerD. The blot was also probed for SpoVAD as an additional loading control. (d) The F87C mutation in GerD does not disrupt germinosome formation in spores. Panels show fluorescence images of spores expressing (ii) GerKB-GFP (KGB0412), (iv) GerKB-GFP and GerDF87C, (vi) GerD-GFP (KGB7312) and (viii) GerD-GFP and GerDF87C. Shown in (i), (iii), (v) and (vii) are differential interference contrast images of the same spores in each column. Each image in this figure was scaled independently in order to show the foci. (e) Plots show the normalized averaged distribution of the fluorescence signal along the long axis of spores of strains GerD-GFP [KGB73; (●) 20 spores] and GerD-GFP/GerDF87C [(□) 18 spores]. The full width at half-maximal intensity for the GerD-GFP signal was ~300 nm, similar to that found previously [12], and considerably smaller than the signal from soluble GFP in the spore core that was not present in discrete foci.
Since some severe germination defects were observed for the gerD mutant spores, an obvious question is whether these mutations affect GerD protein solubility, stability and/or protein level in spores. To this end, we expressed and purified several G. stearothermophilus GerD variants with the truncations and single-point mutations (ΔH01-02, ΔH6-8, ΔH5-8, M36P, F86P, M111P, M138P, D118A, Q122A and R148A). Both gel-filtration chromatography and dynamic light scattering showed that all the GerD variants maintained a trimeric configuration in solution (data not shown). CD spectroscopy and thermal melting analyses further indicated that these proteins retained much of the helical character of GerD60-180 and generally had only slightly decreased Tm values (data not shown). These relatively normal biophysical properties of the purified G. stearothermophilus GerD variants suggest that they should be stable in spores. Unfortunately, we did not have an antiserum against the G. stearothermophilus GerD protein in order to assess the levels of the GerD variants in spores by Western blot analysis, and our anti-B. subtilis GerD serum did not cross-react with G. stearothermophilus GerD (data not shown).
A dominant negative gerD mutation in B. subtilis
While GerD and GRs are essential for nutrient-mediated germination, it is not clear whether other proteins are also involved. To this end, we performed a genetic screen to identify additional B. subtilis genes required for GR-mediated spore germination and isolated nine mutants whose spores did not germinate appreciably with nutrients. Complementation and DNA sequencing established that six of these mutants have short deletions in gerD, two have mutations in the gerA operon and these mutations were responsible for the slow spore germination phenotype.
Spores of the final mutant (Mut9) exhibited very slow germination with all nutrients, similar to that of ΔgerD spores (Fig. 5a). However, this defect was not complemented by wild-type gerD, the gerA operon or genes encoding transcription factors that promote genes encoding GRs (data not shown). In addition, levels of GerD, GR subunits and the SpoVA protein SpoVAD in spore lysates were essentially identical in Mut9 and wild-type spores (Fig. 5b). Sequencing of the genomic DNA of Mut9 and the parental strain revealed a single T-to-G mutation in Mut9 that changes the Phe87 residue to a cysteine in GerD. To confirm that this change alone was responsible for Mut9 spores’ slow GR-dependent germination, we inserted B. subtilis gerD and the gene with the F87C mutation (gerDF87C) at the amyE locus of the parental strain. The nutrient germination of spores with gerDF87C at amyE was essentially identical with that of Mut9 spores, as well as that of ΔgerD spores (Fig. 5a), indicating that the F87C change in GerD is sufficient to almost eliminate GR-dependent spore germination. Importantly, the new gerDF87C strain is a merodiploid with both wild-type and mutant forms of GerD, suggesting that gerDF87C is interfering with the function of the wild-type protein and F87C is a dominant negative mutation. This result indicates that functional GerD is oligomeric, in agreement with the structural finding and the cross-linking results in this study. As expected, spores with an extra copy of gerD at amyE germinated slightly faster than wild-type spores with l-valine (Fig. 5a). Western blot analysis further showed that levels of GerD in spores with gerD or gerDF87C at amyE were roughly twice that of the parental spores (Fig. 5c), indicating that GerDF87C does not affect protein stability. It is notable that the effect of GerDF87C on B. subtilis spore germination was essentially identical with that of the G. stearothermophilus GerD variant in which the corresponding Phe residue was changed to Pro (F86P) (compare Figs. 4b and 5a), consistent with G. stearothermophilus GerDF86P also likely being a dominant negative variant.
GerD and GR subunits have recently been found to colocalize primarily in a single focus, the germinosome, in the inner membrane of dormant spores, and this colocalization is GerD dependent [12]. To determine whether GerDF87C disrupt germinosome formation, we inserted gerDF87C at the amyE locus of B. subtilis strains that express functional GerKB-GFP or GerD-GFP from the normal chromosomal locus [12]. Following a previously developed protocol [12], we observed similar discrete fluorescent foci in both GerDF87C-containing strains (Fig. 5d). The distribution and intensity of the foci in spores of both mutant strains were also similar to those in parental spores (Fig. 5e), suggesting that GerDF87C did not disrupt germinosome formation.
In the crystal structure of the G. stearothermophilus GerD60-180 trimer, the side chain of Phe86 (Phe87 in B. subtilis GerD) packs into the hydrophobic core of the superhelix and stacks with several other aromatic residues (Trp87, Phe96 and Phe100) from all three monomers (Fig. 2f). The importance of these hydrophobic residues in GerD is underscored by their conservation among GerD homologs; only 5 out of 24 GerD homologs have used different amino acid residues in these positions (Fig. 3). It thus appears that the Phe-to-Cys mutant of GerD interferes with the ability of the wild-type GerD to form the functional superhelix rope, thereby giving rise to the observed dominant negative phenotype. In addition, since we did not identify additional genes essential for GR-dependent germination in our genetic screen, this is further evidence that GerD is an indispensable component in this process.
Discussion
How signals are integrated and transferred from GRs to downstream signaling molecules in the process of germination of spores of Bacillus species remains an unresolved issue. Previous studies have demonstrated that loss of GerD dramatically slows GR-dependent germination of Bacillus spores and further that GRs depend on GerD for clustering into the germinosome in the spore’s inner membrane [11,12]. Thus, GerD likely plays a role in assembling the germinosome, thereby facilitating GR-dependent spore germination. The goal of this study was to elucidate the structural and functional properties of GerD at the atomic level. Our results support a model in which GerD acts as a scaffold protein to promote germinosome assembly and perhaps to mediate the signal transmission events from GRs to downstream components of the germination pathway.
Our crystallographic analysis reveals that the central core domain of GerD from a thermophilic Bacillus forms a previously uncharacterized type of a trimeric rod-shaped structure, with an unusual α-helical register shift between adjacent staggered helices. Both secondary structure and primary sequence alignment suggest that this novel right-handed superhelical fold is well conserved among GerD homologs in spores of Bacillus species. Structure-guided site-directed mutagenesis coupled with analysis of spore germination induced by the nutrient l-valine demonstrates the critical relationship between GerD structure and function. Identification of the dominant negative F87C mutation in B. subtilis GerD also strongly suggests that GerD functions as an oligomer in vivo, as nonfunctional molecules result when even one or two mutated chains are incorporated in the GerD trimer. We also noted that the association between three polypeptide parallel GerD chains is somewhat reminiscent of the triple-helix conformation largely associated with collagen and other fibrous structural proteins within the extracellular matrix [24,25]. Collagen is composed of a right-handed bundle of three parallel, left-handed polyproline-II-type helices, and its molecular conformation confers strict amino acid sequence constraints, including glycine as every third residue and a high content of imino acids [25,26]. In striking contrast, the molecular architecture of the trimeric GerD superhelix is determined by repeating secondary-structure motifs. Consequently, there may be a stronger requirement for regularity of the GerD trimer packing because such extensive interdigitation of topologically arranged helices with interruptions can only occur at optimal packing angles.
The crystal structure of GerD60-180 displays a distinctive packing arrangement in which the two trimers in the asymmetric unit associate laterally along their superhelical axes (Fig. 6). The intertrimer association found in the crystalline state is largely mediated by multiple noncovalent interactions, such as salt bridges, hydrogen bonds and hydrophobic interactions, among backbone and side-chain atoms and solvent molecules. This compact arrangement makes it possible to create sufficient intertrimer contacts to stabilize the crystal lattice. We have also attempted to use chemical cross-linking reagents to analyze the interaction network of GerD in spores. Our preliminary data using a zero-length cross-linking reagent showed that GerD likely forms dimer, trimer and even higher-order oligomeric complexes in spores. As GerD is likely to be held on the inner membrane periphery through an N-terminal diacylglycerol anchor attached to an amino-terminal cysteine residue, the GerD triplex may self-associate to form higher-order structures, consistent with the observation that GerD-GFP by itself localizes to a single focus in the spore’s inner membrane [12]. It seems entirely possible that the GerD trimer can assemble into a large mechanically rigid structure that serves as a binding platform for various germination proteins to facilitate their interaction and communication. Nevertheless, in vitro affinity pull-down experiments with purified GerD lacking the lipobox anchor show that GerD does not associate strongly with either the GerBC [27] subunit of the GerB GR or the SpoVAD protein [8] that is a part of the SpoVA protein channel for CaDPA. One possible explanation for the latter observation is that GerD and its binding partners associate only in the milieu of the spore inner membrane, where all the GRs and most downstream germination proteins are localized. Indeed, recent studies have shown that many signal transduction processes utilize colocalization and clustering of sequentially acting signaling proteins for the selective activation of downstream functions [28]. It is also possible that combining multiple weak interactions may be generally beneficial when the assembly of membrane-associated signaling protein complexes is dynamic and/or regulated, while each individual interaction is too weak to allow successful pull-downs. Indeed, it has been suggested that several large multiprotein complexes in bacteria including the spore coat [29] in B. subtilis and the DNA replisome [30] in E. coli are assembled using a large number of weak interactions. In addition, GerD may change its oligomerization state and conformation in response to changes or signals in its environment, although the nature of the changes or signals is unknown. Clearly, further study is required to determine the precise role of GerD in GR-dependent germination.
Fig. 6.
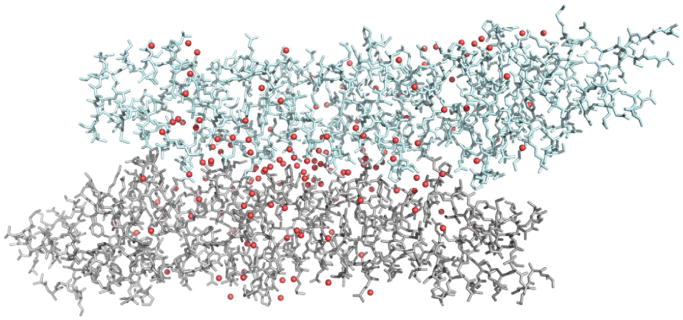
GerD may form a higher-order structure. Side view showing lateral assembly of two GerD60-180 trimers in the asymmetry unit. Water molecules are depicted as red spheres.
Materials and Methods
Protein expression and purification
The G. stearothermophilus gerD gene was amplified by PCR from chromosomal DNA of G. stearothermophilus strain PS3001. Mutations were introduced into the gerD gene by PCR or by overlapping PCR to piece together two PCR fragments when necessary. The gerD variants were cloned into a modified pGEX vector containing an N-terminal glutathione S-transferase tag. The GerD proteins were expressed in E. coli and purified by G4B affinity chromatography followed by tobacco etch virus protease cleavage of the glutathione S-transferase tag and by anion-exchange and gel-filtration chromatography. For crystallization, the G. stearothermophilus GerD60-180 protein (residues 60–180) was concentrated to 26 mg/ml by ultrafiltration in 20 mM Tris (pH 7.6) and 150 mM NaCl (TBS). SeMet-substituted GerD was produced following established procedures [31] and purified and concentrated as described above.
CD spectroscopy
CD measurements were carried out on an Aviv 410 CD spectrophotometer (Aviv Biomedical Inc., Lakewood, NJ) at 20 °C in TBS (pH 8.0) with 25 μM protein. Spectra were recorded as the average of five scans using a 5-s integration time at 1.0-nm wavelength increments. Spectra were baseline corrected against the cuvette with buffer alone. A [θ]222 value of −33,000 degrees cm2 dmol−1 was taken to correspond to 100% α-helix [32]. Thermal stability was assessed by monitoring [θ]222 as a function of temperature. Melting profiles were reversible, as > 90% of the CD signal was regained upon cooling. Tm values were estimated by nonlinear regression fitting to the Boltzmann equation of the OriginPro 7.5 software program (OriginLab Corporation, Northampton, MA).
Analytical ultracentrifugation analysis
Sedimentation equilibrium measurements were carried out on a Beckman XL-A analytical ultracentrifuge (Beckman Coulter Inc., Brea, CA) using an An-60 Ti rotor. Protein samples were dialyzed overnight against TBS (pH 7.6); loaded at initial concentrations of 35, 150 and 500 μM and analyzed at rotor speeds of 14,000 and 17,000 rpm at 20 °C. Data were acquired at two wavelengths per rotor speed setting and processed globally for the best fit to a single-species model of absorbance versus radial distance by using Origin provided by the manufacturer. Solvent density and protein partial specific volume were calculated according to solvent and protein composition, respectively [33]. Apparent molecular masses were within 10% of those calculated for an ideal trimer, with no systematic deviation of the residuals.
Western blot and chemical cross-linking analyses of the spore total lysates
Total lysates of spores of various B. subtilis strains were prepared as described previously [34]. For Western blot experiments, equal amounts of the total lysates were run on SDS-PAGE, and the levels of the subunits of GRs, the SpoVAD subunit of the SpoVA channel and the GerD protein were detected using specific rabbit polyclonal antisera [35]. For cross-linking experiments, the total lysate prepared from spores of strain PS533 was dialyzed at 4 °C overnight using a Slide-A-Lyzer MINI Dialysis device (10,000 molecular weight cutoff; Thermo Scientific, Rockford, IL) against a buffer containing 50 mM Hepes (pH 7.4), 5 mM ethylenediaminetetraacetic acid and 0.1 mM phenylmethanesulfonyl fluoride. Equal amounts of the dialyzed total lysate were then cross-linked with varying concentrations (0.1–10 mM) of 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC; Thermo Scientific) at room temperature for 30 min in the dark. The cross-linking reactions were quenched with 20 mM of 2-mercaptoethanol at room temperature for 15 min, and equal amounts of the treated total lysates were run on SDS-PAGE and the GerD protein was detected by Western blotting.
Crystallization and structure determination
The native and SeMet-substituted GerD60-180 proteins were crystallized from a solution consisting of 15% ethyl alcohol and 0.1 M Tris–HCl (pH 8.5) by the hanging-drop vapor diffusion method at 4 °C. Crystals were flash frozen in crystallization solution containing 35% (w/v) polyethylene glycol 400. Data collection on the native and SeMet GerD crystals was performed at the X29A beamline of the National Synchrotron Light Source (Brookhaven National Laboratories). Data were processed using the HKL2000 suite [36]. The crystals contain six GerD60-180 molecules in the asymmetric unit. The data collected at the selenium peak wavelength were selected to search for SeMet sites using the program SOLVE [37] as implemented in PHENIX [38]. About 50 sites were identified and initial phases calculated from these sites were improved by density modification using RESOLVE/PHENIX. The resulting electron density map was readily interpretable and used to build two-thirds of the molecule using the program Coot [39]. Iterative cycles of refinement in REFMAC with TLS [40] followed by manual rebuilding in Coot were carried out until no further improvement of the Rfree factor was observed. The final model contains six GerD molecules (chain A, residues 69–178; chain B, 63–179; chain C, 64–176; chain D, 60–179; chain E, 61–175; chain F, 66–180). Ramachandran statistics were calculated using MolProbity [41]. Molecular graphics were rendered using PyMOL (Delano Scientific LLC).
BLAST search and sequence conservation analysis
The B. subtilis GerD amino acid sequence was used as query sequence for the initial homology sequence search of spore-forming members of the Bacillales on the National Center for Biotechnology Information genomic BLAST server†. All the top hits (E-value < e-45 and query coverage > 90%) were confirmed to have the recognition sequence for addition of a diacylglycerol anchor near the N-termini of the protein. A single gerD gene was found for each species. A ClustalW alignment of GerD homologs was performed using DNASTAR Lasergene Suite 8 (DNASTAR Inc., Madison, WI).
Preparation of gerD mutant strains
Wild-type and mutagenized G. stearothermophilus gerD PCR products were cloned into a modified pBluescript II KS(–) vector containing a kanamycin-resistance cassette separated by two inserts from the B. subtilis gerD region. The first insert consisted of 510 bp within the upstream region of the B. subtilis gerD gene followed by G. stearothermophilus gerD variants. Downstream from this insert is the kanamycin-resistance cassette, followed by 542 bp of sequence downstream of B. subtilis gerD, to provide homology for a double crossover with B. subtilis chromosomal DNA. The insertion of the G. stearothermophilus gerD gene and the mutation sites in the plasmids were confirmed by DNA sequencing. The wild-type or mutagenized plasmids were used to transform competent cells of B. subtilis strain PS832 (a laboratory derivative of strain 168) with selection for kanamycin resistance (10 μg/ml). Transformants in which the G. stearothermophilus gerD gene had integrated into the chromosome with replacement of B. subtilis gerD were identified by PCR, and the PCR-amplified regions were sequenced to confirm the presence of the mutations. The wild-type B. subtilis gerD gene was also cloned and transformed in the same way and used as a positive control.
Spore preparation, purification and germination
Spores of various gerD strains were prepared at 37 °C on double-strength Schaeffer’s-glucose (2×SG) agar plates, and spores were harvested, purified and stored as previously described [42,43]. All spores used in this work were free (>98%) of growing or sporulating cells, germinated spores and cell debris as determined by phase-contrast microscopy.
Spores were heat shocked at 70 °C for 30 min and then cooled on ice prior to germination. Germination experiments were performed with spores at an optical density at 600 nm (OD600) of 0.5 in 200 μl of 25 mM K–Hepes buffer (pH 7.4) at 37 °C containing 10 mM l-valine and 50 μM TbCl3. Germinations were initiated by addition of spores, and the process of germination was followed by monitoring real-time DPA release by Tb-DPA fluorescence in an automated fluorescence plate reader [10]. Each reaction was tested in quadruplicate, and the fluorescence detected at zero time was subtracted as the background. The maximum number of RFUs (relative fluorescence units) recorded using the spores with the wild-type G. stearothermophilus gerD gene was set at 100%, and the percentage of DPA released at each time point of spores of other strains was calculated against this maximum number. The germination reactions of all spores in the absence of the germinant were used as negative controls. The percentages of spores that had germinated by the end of reaction incubations were also examined by phase-contrast microscopy. These measurements invariably agreed with those determined from RFU values. Germination rates (percentage of DPA release, min−1) were calculated as the slope of the linear segment of DPA release that followed the lag phase immediately after addition of spores.
To determine kinetic parameters of spores’ germination with l-valine, we performed germination in the presence of varying l-valine concentrations. All curves generated by plotting the germinant concentration versus germination rate were fitted using nonlinear regression to the Hill equation (n = 1) of OriginPro 7.5 to determine the maximum initial rate of germination (defined as Vmax) and the germinant concentration needed to achieve 50% of maximal rate (defined as C50).
Mutagenesis and mutant enrichment of B. subtilis spores
A 2-ml aliquot of spores of B. subtilis strain PS533, a derivative of the wild-type strain PS832 that carries plasmid pUB110 encoding kanamycin resistance (10 μg/ml) [44], at an OD600 of 2 (~6 × 108 spores) was mutagenized by irradiating with ultraviolet light (254 nm) from a UVG-1 short wave UV lamp (UVP Inc., San Gabriel, CA) in a petri dish for 10 min. The percentage of spores killed by irradiation was determined by counting the colonies formed when appropriate dilutions were spotted on Luria Broth (LB) medium plates before and after UV treatment. Mutagenized spores were germinated with a mixture of 10 mM l-valine and AGFK (10 mM each of l-asparagine, d-glucose, d-fructose and KCl) in LB medium for 2 h at 37 °C, and the mix was then transferred into 2×SG liquid medium for sporulation at 37 °C with good aeration for 3 days. Spores were purified and germinated in liquid by the 10 mM nutrient mixture described above in LB medium at 37 °C for 1 h, and the mix was heated at 70 °C for 1 h to kill any germinated spores and then pelleted by centrifugation, washed with water and incubated in 60 mM CaDPA at 25 °C for 2 h to germinate remaining dormant spores. The mix was washed several times by centrifugation with water to remove CaDPA and transferred into 2×SG liquid medium for sporulation at 37 °C for 3 days. The cycle of elimination of spores that germinated with nutrients and recovery of spores that germinated with CaDPA was repeated three times to enrich for spores that do not germinate with nutrient germinants but do germinate with the GR-independent germinant CaDPA. After the final enrichment, the spore mix was germinated with CaDPA and spread on LB plates. Approximately 2000 individual colonies were screened by the plate assay (below).
Tetrazolium plate assay
The abovementioned mutagenized B. subtilis spores were screened by a tetrazolium overlay technique developed previously [45,46]. Briefly, individual colonies from CaDPA-germinated mutagenized spores were picked onto 2×SG agar plates (~50/plate) and plates were incubated at 37 °C for at least 48 h. Colonies from the wild-type PS533 strain and the strain lack all functional GR genes (FB72) [47] were picked onto these plates as positive and negative controls, respectively. The wellsporulated colonies were lifted onto Whatman 3MM filter paper disks that were then baked at 70 °C for 3 h to kill any remaining vegetative or growing cells. After cooling, the filters were soaked in a nutrient germination mixture [10 mM l-valine, 12.5 mM each of AGFK and 25 mM Tris–HCl (pH 7.4)] plus 2,3,5-triphenyltetrazolium chloride (1 mg/ml) and left in a covered petri dish at 37 °C for 2–3 h. The colonies that generate germinated spores turn red in this assay because they reduce the tetrazolium dye, while colonies with spores that remain predominantly or completely dormant are brown.
Whole-genome sequencing
The genomic DNA of the parental wild-type strain and the Mut9 strain isolated from the abovementioned mutagenesis experiments were prepared and sequenced at the Tufts University Core Facility genomic core at the Tufts University School of Medicine using the Hiseq 2500 system powered by Illumina Sequencing Technology. A DNA library consisting of 500-bp inserts was constructed for each genome and sequenced with the paired-end strategy resulting in about 100-fold coverage. The data were analyzed using CLC Genomics Workbench Package by the same facility.
gerD complementation studies
The B. subtilis gerD transcription unit was PCR amplified from PS832 chromosomal DNA with primers designed to include the promoter (upstream primer began 203 bp upstream of the translation start site), the ORF (open reading frame) and the terminator sequence (downstream primer began 144 bp downstream of the translation stop codon) of gerD. The F87C mutation was introduced into the gerD gene using the QuikChange II XL Site-Directed Mutagenesis kit (Agilent Technologies, Santa Clara, CA). Both the wild-type and mutant gerD genes were cloned into plasmid pDG364 that is used to integrate genes at the B. subtilis amyE locus by a double-crossover event [48], and the insertions in plasmids were confirmed by DNA sequencing. These plasmids were used to transform B. subtilis strains PS832 and FB62 (ΔgerD) [49] to an amyE chloramphenicol-resistant phenotype, and the presence of the wild-type or mutant gerD gene in the resultant chloramphenicol-resistant strains was confirmed by PCR and DNA sequencing.
Microscopic analysis
The pDG364 plasmids that contained the wild-type or mutant (F87C) gerD gene were used to transform B. subtilis strains KGB04 (gerKB-gfp)12 and KGB73 (gerD-gfp)12 to an amyE chloramphenicol-resistant phenotype as described above. Detection of the fluorescence signals from the spores of those strains was performed as described previously [12]. In each experiment, images used in subsequent analysis were generated by averaging 100 consecutive images in the acquisition sequence followed by subtraction of the background signal present in cell-free areas. To compensate for the autofluorescence of spores, we assigned a threshold value for the GFP channel based on the maximum fluorescence intensity of the parental spores without a GFP fusion.
Supplementary Material
Acknowledgments
We thank W. Shi at the National Synchrotron Light Source for support at beamline X29A and L. Luo at Malvern Instruments Ltd. for dynamic light-scattering measurement and analysis using a Zetasizer ZS system. This work was supported by a Department of Defense Multi-University Research Initiative award to P.S. and B.H. through the U.S. Army Research Laboratory and the U.S. Army Research Office under contract number W911NF-09-1-0286 and by National Institutes of Health Grants GM099948 to B.H. and AI094599 to M.L. This publication was made possible by the Center for Synchrotron Biosciences Grant P30-EB-009998 from the National Institute of Biomedical Imaging and Bioengineering.
Abbreviations used
- GR
germinant receptor
- DPA
dipicolinic acid
- SeMet
selenomethionine
Footnotes
Accession numbers
Coordinates and structure factors have been deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank under accession number 4O8W.
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jmb.2014.02.004.
References
- 1.Paredes-Sabja D, Setlow P, Sarker MR. Germination of spores of Bacillales and Clostridiales species: mechanisms and proteins involved. Trends Microbiol. 2010;19:85–94. doi: 10.1016/j.tim.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 2.Setlow P. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J Appl Microbiol. 2006;101:514–25. doi: 10.1111/j.1365-2672.2005.02736.x. [DOI] [PubMed] [Google Scholar]
- 3.Setlow P. Spore germination. Curr Opin Microbiol. 2003;6:550–6. doi: 10.1016/j.mib.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Moir A. How do spores germinate? J Appl Microbiol. 2006;101:526–30. doi: 10.1111/j.1365-2672.2006.02885.x. [DOI] [PubMed] [Google Scholar]
- 5.Ross C, Abel-Santos E. The Ger receptor family from sporulating bacteria. Curr Issues Mol Biol. 2010;12:147–58. [PMC free article] [PubMed] [Google Scholar]
- 6.Setlow P, Johnson EA. Spores and their significance. In: Doyle MP, Buchanan RL, editors. Food microbiology: fundamentals and frontiers. 4. Washington, DC: ASM Press; 2012. pp. 45–79. [Google Scholar]
- 7.Paidhungat M, Setlow P. Spore germination and outgrowth. In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillus subtilis and its relative: from genes to cells. Washington, DC: ASM Press; 2002. pp. 537–48. [Google Scholar]
- 8.Li Y, Davis A, Korza G, Zhang P, Li YQ, Setlow B. Role of a SpoVA protein in dipicolinic acid uptake into developing spores of Bacillus subtilis. J Bacteriol. 2012;194:1875–84. doi: 10.1128/JB.00062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vepachedu VR, Setlow P. Role of SpoVA proteins in release of dipicolinic acid during germination of Bacillus subtilis spores triggered by dodecylamine or lysozyme. J Bacteriol. 2007;189:1565–72. doi: 10.1128/JB.01613-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yi X, Setlow P. Studies of the commitment step in the germination of spores of Bacillus species. J Bacteriol. 2010;192:3424–33. doi: 10.1128/JB.00326-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pelczar PL, Igarashi T, Setlow B, Setlow P. Role of GerD in germination of Bacillus subtilis spores. J Bacteriol. 2007;189:1090–8. doi: 10.1128/JB.01606-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffiths KK, Zhang J, Cowan AE, Yu J, Setlow P. Germination proteins in the inner membrane of dormant Bacillus subtilis spores colocalize in a discrete cluster. Mol Microbiol. 2011;81:1061–77. doi: 10.1111/j.1365-2958.2011.07753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masayama A, Fukuoka H, Kato S, Yoshimura T, Moriyama M, Moriyama R. Subcellular localization of a germination-specific cortex-lytic enzyme, SleB, of Bacilli during sporulation. Genes Genet Syst. 2006;81:163–9. doi: 10.1266/ggs.81.163. [DOI] [PubMed] [Google Scholar]
- 14.Kemp EH, Sammons RL, Moir A, Sun D, Setlow P. Analysis of transcriptional control of the gerD spore germination gene of Bacillus subtilis 168. J Bacteriol. 1991;173:4646–52. doi: 10.1128/jb.173.15.4646-4652.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mongkolthanaruk W, Robinson C, Moir A. Localization of the GerD spore germination protein in the Bacillus subtilis spore. Microbiology. 2009;155:1146–51. doi: 10.1099/mic.0.023853-0. [DOI] [PubMed] [Google Scholar]
- 16.Pelczar PL, Setlow P. Localization of the germination protein GerD to the inner membrane in Bacillus subtilis spores. J Bacteriol. 2008;190:5635–41. doi: 10.1128/JB.00670-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korza G, Setlow P. Topology and accessibility of germination proteins in the Bacillus subtilis spore inner membrane. J Bacteriol. 2013;195:1484–91. doi: 10.1128/JB.02262-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–41. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pauling L, Corey RB. Compound helical configurations of polypeptide chains: structure of proteins of the alpha-keratin type. Nature. 1953;171:59–61. doi: 10.1038/171059a0. [DOI] [PubMed] [Google Scholar]
- 20.Zhou NE, Zhu BY, Kay CM, Hodges RS. The two-stranded alpha-helical coiled-coil is an ideal model for studying protein stability and subunit interactions. Biopolymers. 1992;32:419–26. doi: 10.1002/bip.360320419. [DOI] [PubMed] [Google Scholar]
- 21.Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32:W321–6. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barlow DJ, Thornton JM. Helix geometry in proteins. J Mol Biol. 1988;201:601–19. doi: 10.1016/0022-2836(88)90641-9. [DOI] [PubMed] [Google Scholar]
- 24.Bella J, Eaton M, Brodsky B, Berman HM. Crystal and molecular structure of a collagen-like peptide at 1.9 Å resolution. Science. 1994;266:75–81. doi: 10.1126/science.7695699. [DOI] [PubMed] [Google Scholar]
- 25.Brodsky B, Ramshaw JA. The collagen triple-helix structure. Matrix Biol. 1997;15:545–54. doi: 10.1016/s0945-053x(97)90030-5. [DOI] [PubMed] [Google Scholar]
- 26.Brondijk TH, Bihan D, Farndale RW, Huizinga EG. Implications for collagen I chain registry from the structure of the collagen von Willebrand factor A3 domain complex. Proc Natl Acad Sci U S A. 2012;109:5253–8. doi: 10.1073/pnas.1112388109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Setlow B, Setlow P, Hao B. Crystal structure of the GerBC component of a Bacillus subtilis spore germinant receptor. J Mol Biol. 2010;402:8–16. doi: 10.1016/j.jmb.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teruel MN, Meyer T. Translocation and reversible localization of signaling proteins: a dynamic future for signal transduction. Cell. 2000;103:181–4. doi: 10.1016/s0092-8674(00)00109-4. [DOI] [PubMed] [Google Scholar]
- 29.Kim H, Hahn M, Grabowski P, McPherson DC, Otte MM, Wang R, et al. The Bacillus subtilis spore coat protein interaction network. Mol Microbiol. 2006;59:487–502. doi: 10.1111/j.1365-2958.2005.04968.x. [DOI] [PubMed] [Google Scholar]
- 30.Johansson E, Dixon N. Replicative DNA polymerases. Cold Spring Harbor Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a012799. http://dx.doi.org/10.1101/cshperspect.a012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229:105–24. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 32.Chen YH, Yang JT, Chau KH. Determination of the helix and beta form of proteins in aqueous solution by circular dichroism. Biochemistry. 1974;13:3350–9. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- 33.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding S, Rowe A, Horton J, editors. Analytical ultracentrifugation in biochemistry and polymer science. Cambridge, UK: Royal Society of Chemistry; 1992. pp. 90–125. [Google Scholar]
- 34.Stewart KA, Setlow P. Numbers of individual nutrient germinant receptors and other germination proteins in spores of Bacillus subtilis. J Bacteriol. 2013;195:3575–82. doi: 10.1128/JB.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghosh S, Scotland M, Setlow P. Levels of germination proteins in dormant and superdormant spores of Bacillus subtilis. J Bacteriol. 2012;194:2221–7. doi: 10.1128/JB.00151-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–26. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 37.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr Sect D Biol Crystallogr. 1999;55:849–61. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr Sect D Biol Crystallogr. 2002;58:1948–54. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–21. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 41.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicholson WL, Setlow P. Sporulation, germination and outgrowth. In: Harwood CR, Cutting SM, editors. Molecular biological methods for Bacillus. Chichester, UK: John Wiley and Sons; 1990. pp. 391–450. [Google Scholar]
- 43.Paidhungat M, Ragkousi K, Setlow P. Genetic requirements for induction of germination of spores of Bacillus subtilis by Ca2+-dipicolinate. J Bacteriol. 2001;183:4886–93. doi: 10.1128/JB.183.16.4886-4893.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Setlow B, Setlow P. Role of DNA repair in Bacillus subtilis spore resistance. J Bacteriol. 1996;178:3486–95. doi: 10.1128/jb.178.12.3486-3495.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moir A, Lafferty E, Smith DA. Genetics analysis of spore germination mutants of Bacillus subtilis 168: the correlation of phenotype with map location. J Gen Microbiol. 1979;111:165–80. doi: 10.1099/00221287-111-1-165. [DOI] [PubMed] [Google Scholar]
- 46.Ragkousi K, Eichenberger P, van Ooij C, Setlow P. Identification of a new gene essential for germination of Bacillus subtilis spores with Ca2+-dipicolinate. J Bacteriol. 2003;185:2315–29. doi: 10.1128/JB.185.7.2315-2329.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paidhungat M, Setlow P. Role of ger proteins in nutrient and nonnutrient triggering of spore germination in Bacillus subtilis. J Bacteriol. 2000;182:2513–9. doi: 10.1128/jb.182.9.2513-2519.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cutting SM, Vander Horn PB. Genetic analysis. In: Harwood CR, Cutting SM, editors. Molecular biological methods for Bacillus. Chichester, UK: John Wiley and Sons; 1990. pp. 22–74. [Google Scholar]
- 49.Igarashi T, Setlow B, Paidhungat M, Setlow P. Effects of a gerF (lgt) mutation on the germination of spores of Bacillus subtilis. J Bacteriol. 2004;186:2984–91. doi: 10.1128/JB.186.10.2984-2991.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.