Abstract
Glutaredoxins (Grxs) have been shown to be critical in maintaining redox homeostasis in living cells. Recently, an emerging subgroup of Grxs with one cysteine residue in the putative active motif (monothiol Grxs) has been identified. However, the biological and physiological functions of this group of proteins have not been well characterized. Here, we characterize a mammalian monothiol Grx (Grx3, also termed TXNL2 / PICOT) with high similarity to yeast ScGrx3 / ScGrx4. In yeast expression assays, mammalian Grx3s were localized to the nuclei and able to rescue growth defects of grx3grx4 cells. Furthermore, Grx3 inhibited iron accumulation in yeast grx3gxr4 cells and suppressed the sensitivity of mutant cells to exogenous oxidants. In mice, Grx3 mRNA was ubiquitously expressed in developing embryos, adult tissues and organs, and was induced during oxidative stress. Mouse embryos absent of Grx3 grew smaller with morphological defects and eventually died at 12.5 days of gestation. Analysis in mouse embryonic fibroblasts revealed that Grx3−/− cells had impaired growth and cell cycle progression at the G2/M phase, whereas the DNA replication during the S phase was not affected by Grx3 deletion. Furthermore, Grx3-knockdown HeLa cells displayed a significant delay in mitotic exit and had a higher percentage of binucleated cells. Therefore, our findings suggest that the mammalian Grx3 has conserved functions in protecting cells against oxidative stress and deletion of Grx3 in mice causes early embryonic lethality which could be due to defective cell cycle progression during late mitosis.
Keywords: cell cycle, embryogenesis, glutaredoxin, mouse, oxidative stress
Introduction
Reactive oxygen species (ROS) can be formed as by-products in all oxygenic organisms during aerobic metabolism [1,2]. Cells also actively generate ROS as signals through activation of various oxidases and peroxidases in response to internal developmental cues and external stresses [3]. ROS-dependent signals are vital for normal growth and developmental processes, like blastocyst cleavage, neuronal differentiation, digit formation, immune response and hormone action [4-8]. However, because of the cytotoxic and extremely reactive nature of ROS, excess ROS, namely oxidative stress, can cause a wide range of damage to macromolecules, which are often associated with pathogenesis [9-12].
To overcome such oxidative damage and control signaling events, cells have orchestrated an elaborate antioxidant network [1,13,14]. Of these antioxidant systems, glutaredoxins (Grxs) appear to be involved in many cellular processes and play an important role in protecting cells against oxidative stress [15]. Grxs are ubiquitous, small heat-stable disulfide oxidoreductases which are conserved in both prokaryotes and eukaryotes [16]. Biochemical analyses of Grxs (dithiol Grx) from various organisms reveal that this group of proteins can catalyze the reduction of protein disulfides and glutathione–protein mixed disulfides via a dithiol or monothiol mechanism [17,18]. Recently, a group of Grxs have been identified that contain a single cysteine residue in the putative motif, ‘CGFS’, and have been termed monothiol Grxs [19]. Monothiol Grxs were initially identified in yeast (ScGrx3, −4 and −5), and subsequently found in all types of living cells [19]. There is a growing body of evidence that monothiol Grxs may have multiple functions in biogenesis of iron–sulfur clusters, iron homeostasis, protection of protein oxidation, cell growth and proliferation [19].
In yeast, ScGrx3 and ScGrx4 have a conserved thioredoxin homology domain (Trx-HD) and Grx-HD [19,20]. ScGrx3 and ScGrx4 have been shown to be critical in regulating iron homeostasis through interactions with a transcriptional factor, Aft1 and protecting cells against oxidative stress [21,22]. Furthermore, ScGrx4 interacts with a p53-related protein kinase, piD261/Bud32, and is proposed to have a critical function in cell proliferation [23]. Interestingly, a recent study suggests that ScGrx3 and ScGrx4 appeared to modulate the mitochondrial iron–sulfur cluster synthesis [24,25].
In mammalian cells, there are two monothiol Grxs identified [26,27]. Mammalian Grx5, similar to yeast and zebrafish Grx5s, is a mitochondrial Grx and plays a critical role in iron–sulfur cluster biogenesis and heme synthesis in red blood cells [28-32]. Grx3, also termed thioredoxin-like 2 (Txnl2) or PICOT, was originally identified through a yeast two-hybrid screening, in which Grx3 physically interacts with the protein kinase C theta isoform [27]. Transient expression of Grx3 positively regulates calcineurin–NFAT activation in rat basophilic leukemia cells (RBL-2H3) [33]. Furthermore, forced expression of Grx3 in transgenic mice (heart) enhances cardiomyocyte contractility and inhibits calcineurin–NFAT-mediated signaling in the progression of pressure-overload-induced heart hypertrophy [34,35]. Other studies show that a single Grx3 allele deletion augments cardiac hypertrophy in transgenic mice under pressure overload [36]. Our previous work indicates that Grx3 plays a critical role in regulating human beast cancer cell growth and metastasis via redox homeostasis and NF-κB signaling [37]. However, the physiological functions of mammalian Grx3 in oxidative stress and ROS-mediated signaling remain to be explored.
In this study, we analyzed the functions of mammalian Grx3s by heterologous expression of mouse Grx3 (MmGrx3) or human Grx3 (HsGrx3) in yeast grx3grx4 mutants. We examined the expression pattern of MmGrx3 mRNA in mouse tissues and its response to oxidative stress in myoblast cells. We generated Grx3-deficient mice and characterized the vital role of MmGrx3 in embryo development. We also generated HsGrx3 knockdown (KD) HeLa cells and examined the function of HsGrx3 in cell cycle progression. Taken together, these findings suggest that mammalian Grx3s have important roles in controlling cell cycle progression and growth.
Results
Grx3 is able to complement the growth defects of yeast grx3grx4 double mutant
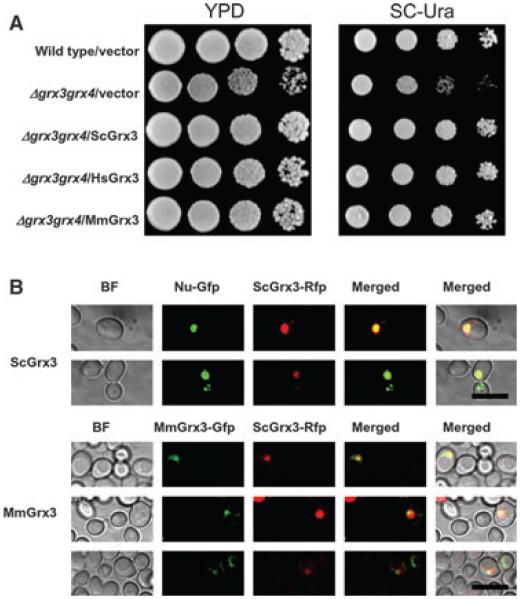
HsGrx3 is 95% identical to MmGrx3 at the amino acid level (data not shown) and both Grxs have a conserved Trx-HD and two tandem Grx-HDs, which are similar to yeast monothiol Grxs, ScGrx3 and ScGrx4, whereas ScGrx3 and ScGrx4 have only one Grx-HD at their C-termini [19]. ScGrx3 and ScGrx4 appear to have redundant functions in cell growth [21,22]. Neither ScGrx3 nor ScGrx4 deletion affects yeast cell growth, however deletion of both ScGrx3 and ScGrx4 reduced cell growth in both nutrient rich medium (YPD) and minimal medium (Fig. 1A and Fig. S1) [28]. The impaired growth was rescued by the overexpression of ScGrx3 and ScGrx4 (Fig. 1A and Fig. S1).
Fig. 1.
Mammalian Grx3s can rescue the growth defects of yeast grx3grx4 cells. (A) Vector-expressing wild-type cells, and vector-, ScGrx3-, HsGrx3- and MmGrx3-expressing grx3grx4 cells were grown on nutrient rich YPD and SC-Ura media for 48 h at 30 °C. (B) Subcellular localization of ScGrx3–RFP (upper) and MmGrx3–GFP (lower) in yeast cells. Scale bars = 10 μm.
To examine whether mammalian Grx3 could complement ScGrx3 and ScGrx4 function in grx3grx4 cells, HsGrx3 and MmGrx3 were expressed in the yeast mutant strain. As shown in Fig. 1A and Fig. S1, both mammalian Grx3s rescued mutant cell growth in a manner similar to ScGrx3 and ScGrx4. ScGrx3 targeted to nuclei when ScGrx3–RFP was expressed in grx3grx4 mutant cells (Fig. 1B) and this nuclear localization has been shown to be a prerequisite for ScGrx3 function [23,38]. To determine whether MmGrx3 could target to nuclei in yeast cells, MmGrx3 was fused with green fluorescent protein (GFP) and expressed in grx3grx4 cells. The MmGrx3–GFP appeared functional because the fusion protein rescued the growth defects of yeast mutant cells in a manner similar to MmGrx3 expression (data not shown). When coexpressed with ScGrx3–RFP in yeast, MmGrx3–GFP colocalized with ScGrx3 in the nuclei (Fig. 1B). These heterologous expression studies suggest that mammalian Grx3 functions in cell growth.
Grx3 suppresses the sensitivity of grx3grx4 mutants to oxidative stress
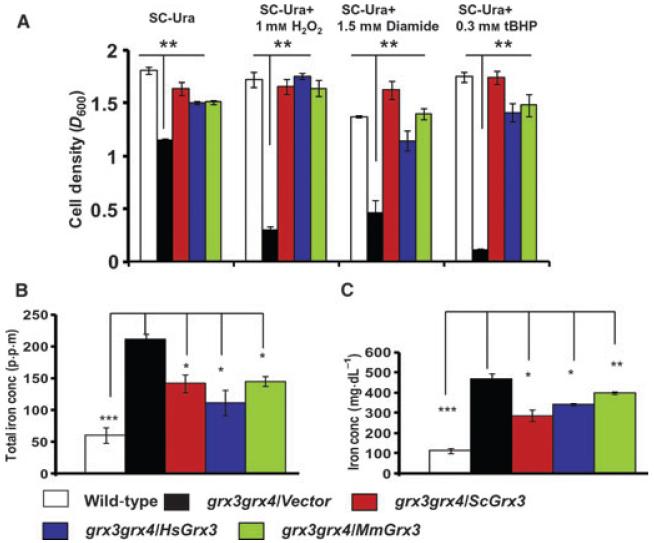
Previous studies indicate that yeast ScGrx3 and ScGrx4 are required for cell survival under oxidative stress [21]. To determine whether mammalian Grx3 could suppress the sensitivity of grx3grx4 cells to external oxidants, both human and mouse Grx3s were expressed in mutant cells and grown in media with or without oxidants. Yeast grx3grx4 cells grew more slowly than wild-type cells and mutant cells expressing ScGrx3, ScGrx4 or two mammalian Grx3s in synthetic media (Fig. 2A and Fig. S1B). The growth of yeast mutant cells was significantly inhibited when exposed to exogenous oxidants, whereas both ScGrx3 and ScGrx4 were able to restore the growth of mutant cells (Fig. 2A and Fig. S1B). Furthermore, mammalian Grx3s were also able to rescue the growth of mutant cells as did ScGrx3 or ScGrx4 (Fig. 2A and Fig. S1B).
Fig. 2.
Mammalian Grx3s are able to suppress the sensitivity of grx3grx4 cells to oxidants and iron accumulation. (A) Yeast grx3grx4 cells expressing plasmids as indicated were grown in SC-Ura liquid media and the same media supplemented with 1.0 mM H2O2, 1.5 mM diamide, 0.3 mM tBHP, respectively. Cell density was measured at A600 after growth for 24 h at 30 °C. Shown is one representative experiment from four independent experiments conducted. The bars indicate the standard deviation (n = 3). (B) Whole-cell iron contents were measured using inductively coupled plasma mass spectrometry (ICP-MS). All results shown here are the means of three independent experiments, and the bars indicate the standard deviation. (C) Intracellular iron levels were measured by a QuantiChron™ Iron Assay Kit. Shown is one representative experiment of four independent experiments. The bars represent standard deviations (n = 3). Student’s t-test, *P < 0.01; **P < 0.001; ***P < 0.0001.
Previous studies have shown that disruption of iron regulation and/or intracellular iron accumulation, which subsequently causes a Fenton reaction, may account for the sensitivity of mutant cells to excess oxidants [21,22,39]. Human and mouse Grx3s, like ScGrx3, could partially inhibit both intracellular and total iron accumulation in grx3grx4 cells (Fig. 2B,C). Together, these findings demonstrate that monothiol Grxs have a conserved function in protecting cells against oxidative stress.
Grx3 expression in tissues and embryos
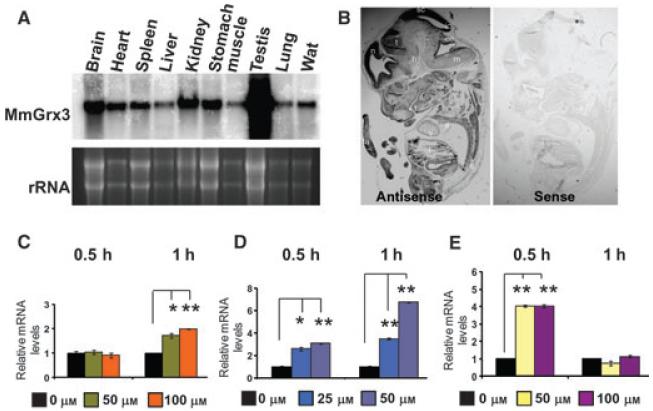
The tissue distribution of HsGrx3 was previously determined by semiquantitative RT-PCR and the expression of HsGrx3 mRNA is ubiquitous [27]. Similarly, RNA blotting analysis revealed that MmGrx3 mRNA was expressed ubiquitously in the tissues and organs tested (Fig. 3A). In contrast to human tissues, mouse Grx3 expression appeared to be more robust in testis, brain, kidney and stomach (Fig. 3A). In addition, there was no significant difference in MmGrx3 mRNA levels and tissue distribution between male and female mice (data not shown). To determine the spatial expression of MmGrx3 in mouse embryos, we performed in situ hybridization using E10.5 embryo and MmGrx3-specific probes. As shown in Fig. 3B, similar to adult mouse, MmGrx3 was ubiquitously expressed in most tissues and developing organs, with stronger signals in the brain (Fig. 3B). This ubiquitous distribution and expression implies that Grx3 may play a role in various tissues and organs.
Fig. 3.
MmGrx3 expression is in tissues and embryos and induced by oxidative stress. (A) Ten micrograms of total RNA isolated from brain, heart, spleen, liver, kidney, stomach, muscle, testis, lung and fat tissues were prepared, blotted and probed for MmGrx3. Ethidium bromide-stained rRNAs are shown as a loading control. (B) In situ hybridization. Embryo specimens at E10.5 were hybridized with digoxygenin-labeled antisense RNA probe (left) and sense RNA probe (right). Magnification of images is 7.5×. a, heart atrium; ce, cerebellum; h, hypothalamus; ic, infecrior colliculus; l, liver; m, medulla; n, neocortex; s, spinal cord; sc, superior colliculus; t, tongue; ta, tail; th, thalamus; v, heart ventricle. (C–E) Quantitative real-time PCR analysis of MmGrx3 mRNA levels in C2C12 cells treated with various concentrations of H2O2 (C), diamide (D) and tBHP (E) for different time points as indicated. The data shown are relative mRNA levels (fold change) as compared with C2C12 cells without treatments. The housekeeping gene cyclophilin was used to normalize Grx3 expression. All values are means ± SD. Student’s t-test, *P < 0.05; **P < 0.01.
Grx3 expression in response to oxidative stress
To understand how MmGrx3 expression is regulated by oxidative stress, mouse myoblast cells (C2C12 cells) were treated with various oxidants. To determine the effect of exogenous oxidants on cell viability, C2C12 cells were treated with 100 μm H2O2, 50 μm diamide or 100 μm tert-butylhydroperoxide (tBHP) for 1 h, and then stained with a Trypan Blue stain. The staining patterns indicated that the various treatments did not reduce cell viability (Fig. S2). Quantitative RT-PCR of MmGrx3 mRNA levels revealed that MmGrx3 was induced by all tested oxidants (Fig. 3C-E). Interestingly, the responsiveness (duration and amplitude) of MmGrx3 to exogenous oxidants appeared to be different among the tested conditions. For example, MmGrx3 expression was not induced when cells were treated with H2O2 for 0.5 h, but was enhanced at 1 h (Fig. 3C). In contrast, when cells were treated with tBHP, MmGrx3 expression was rapidly increased at 0.5 h, but reset to the resting levels when cells were treated for 1 h (Fig. 3E). Among the three oxidants, only diamide treatment resulted in induction of MmGrx3 expression at both 0.5 and 1.0 h (Fig. 3D). These results suggest Grx3 may function in response to oxidative stress.
Disruption of Grx3 in mice results in embryonic lethality
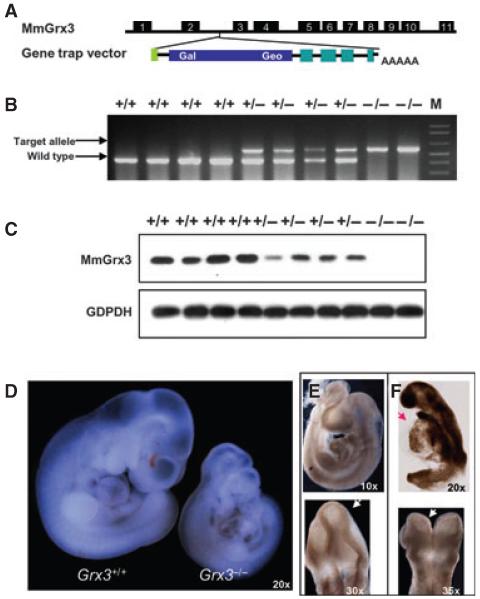
To delineate the function of Grx3 in vivo, we generated Grx3-deficient mice (Fig. 4). The Grx3 gene trap 129/SvEv embryonic stem cells contained a beta-galactosidase neomycin insertion within the second intron of Grx3 at chromosome 7 (Fig. 4A). This target allele could be detected by diagnostic PCR (Fig. 4B). After splicing, the third exon of Grx3 was fused to the insertion to generate a truncated Grx3 transcript, but it did not produce the full-length Grx3 transcript (data not shown). Thus, the Grx3 protein was not produced (Fig. 4C). With the single Grx3 allele (heterozygote), the protein levels of Grx3 were reduced by ~50% compared with wild-type embryos (Fig. 4C). Genetic analysis of F2 transgenic mice generated by sibling crossing revealed that 79 of 120 F2 mice had the target allele, but were all heterozygous (Table 1), the ratio of heterozygous to wild-type mice was ~2:1. No homozygous F2 mice were found at weaning age (Table 1), although homozygous embryos could be identified from early embryos at E12.5 (Fig. 4D-F and Table 1). The majority of homozygous embryos appeared to be morphologically normal, but smaller than heterozygous and wild-type embryos (Fig. 4D). However, some homozygous embryos displayed growth defects, such as open anterior neural tubes and pericardial effusion (Fig. 4E,F). Heterozygous and wild-type embryos were morphologically indistinguishable, but heterozygous embryos may have minor pericardial effusion as well (data not shown). These observations indicate that Grx3 is required for mouse embryo development.
Fig. 4.
Disruption of Grx3 in mice. (A) Shown is the schematic diagram of MmGrx3 genomic DNA structure and the gene trap vector. (B) Genotyping of embryos dissected at E10.5 from F2 sibling-crossing female by PCR using a combination of gene-specific and target vector-specific primers. The large PCR fragments indicated target alleles, whereas the smaller bands indicated wild-type alleles. (C) Western blot analysis of the lysates from the same group of embryos shown in (B). Shown are MmGrx3 protein levels in wild-type, reduced levels in heterozygous alleles and absence of MmGrx3 in homozygous alleles. (D) Shown are examples for wild-type and homozygous embryos at E10.5. Magnification is 20×. (E) Wild-type embryo showing closed neural tube (arrowhead). (F) Homozygous embryo showing open neural tube and pericardial effusion (arrowhead).
Table 1.
Offspring genotypes from heterozygous matings.
| No. of progeny with genotype |
No. of resorbing embryos |
Total no. of zygotes |
|||
|---|---|---|---|---|---|
| Age | +/+ | +/− | −/− | ||
| 21 days | 41 | 79 | 0 | 120 | |
| Embryos | |||||
| 9.5 dpc | 17 | 24 | 13 | 1 | 55 |
| 10.5 dpc | 13 | 22 | 16 | 1 | 52 |
| 12.5 dpc | 12 | 19 | 5 | 9 | 45 |
| 13.5 dpc | 6 | 10 | 0 | 11 | 27 |
Grx3−/− mouse embryonic fibroblasts (MEF) have impaired cell proliferation
In yeast, the deletion of ScGrx3 and ScGrx4 impairs cell proliferation, particularly under oxidative stress [21]. Mammalian Grx3 is able to complement ScGrx3 or ScGrx4 function (Fig. 1 and Fig. S1). Grx3 null allelic mice displayed early embryonic lethality (Fig. 4). These findings that Grx3-deficient embryos could not survive beyond E12.5 indicate that Grx3 is critical for cell viability.
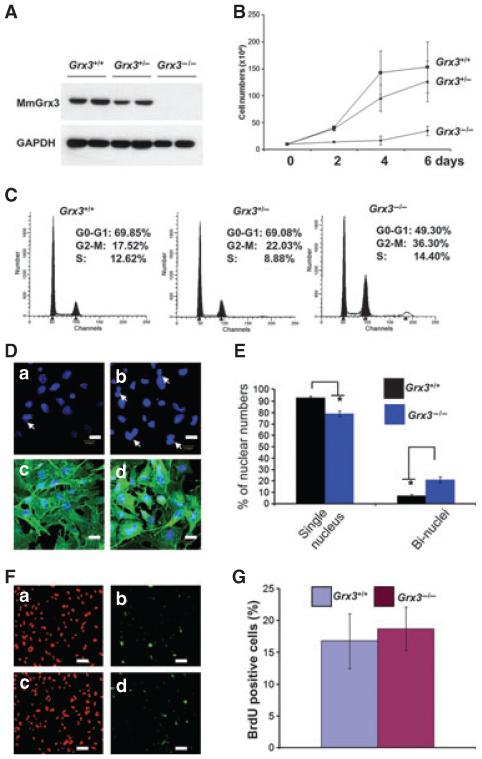
To investigate the role of Grx3 in cell proliferation, mouse embryonic fibroblasts (MEFs) were derived from Grx3+/+, Grx3+/− and Grx3−/− embryos. Similarly, Grx3 was not produced in Grx3−/− MEFs and the protein levels of Grx3 were reduced in Grx3+/− MEFs in comparison with that of Grx3+/+ MEFs (Fig. 5A). Cell proliferation was impaired in Grx3−/− MEFs during the 6-day growth period (Fig. 5B). The growth of Grx3+/− MEFs was slightly slower than that of Grx3+/+ MEFs (Fig. 5B). Notably, Grx3−/− MEFs could not survive beyond passage 4 under our culture conditions, whereas Grx3+/+ and Grx3+/− MEFs grew normally at passage 4 (data not shown).
Fig. 5.
Grx3 null allele MEFs display impaired cell proliferation and are defective in cell-cycle progression. (A) Cell lysates from Grx3+/+, Grx3+/−, and Grx3−/− MEFs were subjected to western blot analysis of MmGrx3 (1:1000). Monoclonal antibody against Gapdh (1: 1000) was used as a loading control. (B) MEF proliferation (Passage 2) was examined during a 6-day period. (C) Cell-cycle profiles of MEFs (P2) were conducted by flow cytometry (FACScan, Coulter). Cells were grown in 10% fetal bovine serum Dulbecco’s modified Eagle’s medium media for 72 h, fixed then stained with propidium iodide. (D) DAPI staining of nuclei of Grx3+/+ (a) and Grx3−/− (b) MEFs showed Grx3−/− MEFs accumulated binucleated cells (arrow). Grx3+/+ (c) and Grx3−/− (d) MEFs were counterstained with β-actin (1:200). Scale bars = 10 μm. (E) Quantification of binucleated cells in Grx3+/+ and Grx3−/− MEFs. Total 615 cells counted for Grx3+/+ and 527 cells counted for Grx3−/−. The bars represent means ± SD. Student’s t-test *P < 0.0001. (F,G) Grx3+/+ and Grx3−/− MEFs were pulse-labeled with BrdU for 5 h before being harvested. Cells were first stained with anti-BrdU-Alex 688 and then counterstained with Sytox orange. Results in (D) show the same field of cells stained with Sytox orange (a and c) or anti-BrdU-Alex 688 (b and d). (a,b) Grx3+/+ MEFs; (c,d) Grx3−/− MEFs. Scale bars = 30 μm. The percentage of BrdU-positive cells in (E) was calculated by counting the BrdU-positive cells in six independent fields and dividing by the total number of Sytox orange-stained cells (~ 200 cells counted in each field). The bars represent means ± SD.
Analysis of cell cycle distribution in asynchronously growing MEFs revealed that there was an increase in the proportion of Grx3−/− cells at the G2/M phase compared with Grx3+/+ and Grx3+/− cells (Fig. 5C). DAPI staining of nuclei revealed that Grx3−/− MEFs consisted of more binucleated cells (21.1 ± 2.62%) compared with wild-type MEFs (7.1 ± 1.31%) (Fig. 5D,E). To directly determine whether deletion of Grx3 affects the G1-to-S progression, DNA replication in MEFs was examined by measuring the percentage of incorporation of BrdU. As shown in Fig. 5(F,G) Grx3−/− cells consist of 18 ± 3.4% BrdU-positive cells, which is comparable with Grx3+/+ (16 ± 4.3% of BrdU positive cells), indicating that deletion of Grx3 did not alter the G1/S phase progression.
Grx3 is required for efficient mitotic exit during cell cycle progression
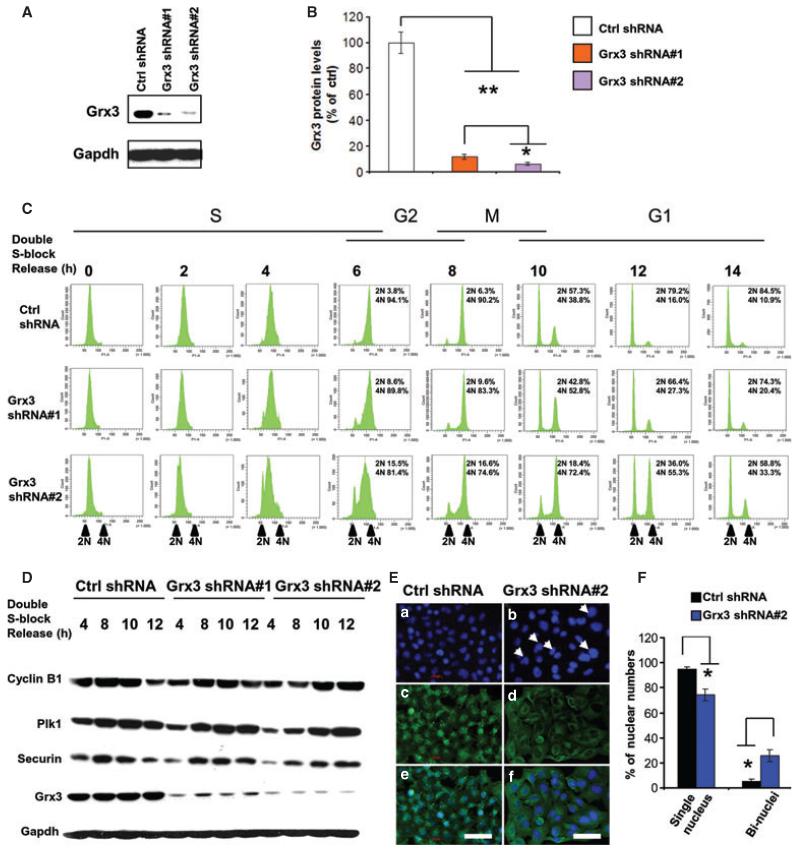
To understand the underlying mechanism that Grx3−/− MEFs accumulated at the G2/M phase, we generated stable Grx3-KD HeLa cells using two Grx3-specific small hairpin RNAs shRNAs [37]. In comparison with control cells, Grx3 shRNA1- and 2-KD cells had significantly reduced Grx3 protein levels (12.1 ± 2.0% of control levels in shRNA#1 cells and 6.97 ± 1.19% of control levels in shRNA#2 cells) (Fig. 6A,B). These Grx3-KD Hela cells also proliferated at a slower rate (data not shown).
Fig. 6.
Grx3 is critical for cell-cycle progression at G2/M phase. (A) Western blot analysis of Grx3 expression in Grx3-KD HeLa cells in comparison with control cells. (B) Quantification of Grx3 protein levels in Grx3-KD cells compared with control cells. The bars represent standard deviations (n = 3). Student’s t-test *P < 0.05, **P < 0.01. (C) Grx3-KD and control HeLa cells were synchronized using double thymidine block, then released, and progressed through S, G2 and M phases to finish cell cycle back to G1 phase. Cells were harvested at 2-h intervals. Half of cells were stained with propidium iodide, and analyzed by FACS. Another half of cells were prepared for cell lysate. (D) Analysis of key cell-cycle regulators in Grx3-KD and control cells Cell lysates from control, Grx3-KD shRNA#1, Grx3-KD shRNA#2 cells were prepared as described above and subjected to western blotting for cyclin B1, Plk1, Securin, Grx3 and Gapdh. (E) DAPI staining of nuclei of control (a) and Grx3-KD (b) cells showed Grx3-KD cells accumulated binucleated cells (arrowhead). Control (c) and Grx3-KD (d) were counterstained with β-actin (1: 200). (e,f) Mergered images of control (e) and Grx3-KD (f) staining cells. Scale bars = 200 μm. (F) Quantification of binucleated cells in control and Grx3-KD cells. Total 600 cells counted for control cells and 360 cells counted for Grx3-KD cells. The bars represent means ± SD. Student’s t-test *P < 0.0001.
To closely monitor cell cycle progression in Grx3-KD cells, we synchronized the cells using a double thymidine block (DTB) method [40]. More than 95% of the cells proceeded through S phase synchronously after being released from DTB and no difference was observed among control and Grx3-KD cells (Fig. 6C). Like control cells, 6–8 h after DTB release, most of Grx3-KD cells progressed into the G2/M phase (Fig. 6C). However, Grx3-KD cells displayed a significant delay in mitotic exit at 10–12 h after DTB release, whereas the control cells completed mitosis and entered the next G1 phase (Fig. 6C). Accordingly, protein levels of Plk1, cyclin B1 and Securin, which are important mitotic regulators, remained high in Grx3-KD cells compared with control cells at 10–12 h after DTB release (Fig. 6D). Furthermore, similar to Grx3−/− MEFs, Grx3-KD HeLa cells had a higher percentage of binucleated cells (15.7 ± 3.8% in shRNA#1 cells and 25.9 ± 4.7% in shRNA#2 cells) than control cells (5.2 ± 1.6%) at 16 h after DTB release (Fig. 6E,F), indicating that more cytokinesis failure occurred in Grx3-KD cells. Taken together, our results suggest that Grx3 may be involved in the regulation of mitotic progression, particularly at the later stages of mitosis.
Discussion
In this study, we demonstrate that a mammalian monothiol glutaredoxin, Grx3, has conserved functions in the protection of cells against oxidative stress and complete loss of Grx3 causes mouse embryonic lethality likely due to cell cycle defects at the G2/M progression.
The ability of Grx3 to complement yeast ScGrx3/ScGrx4 function in mutant cells suggests that this group of Grxs may have conserved protective roles in response to oxidative stress (Figs 1 and 2A and Fig. S1). Deletion of ScGrx3 and ScGrx4 in the strain (CML235) used in this study results in mild growth retardation under normal growth conditions, but mutant cells are sensitive to oxidative stress (Fig. 1 and Fig. S1). Our results are consistent with the original report [28] and a publication from another group [21]. Notably, recent studies report severe growth defects in some yeast strains when ScGrx3 and ScGrx4 are deleted [22,25]. We speculate that this phenotypic change could be due to the genetic background. For example, Wanat et al. [41] reported that mlh1 alleles (DNA mismatch repair gene MLH1) in both S288c and SK1 strains displayed difference in mismatch repair efficiency that is strain dependent. In yeast, ScGrx3 and ScGrx4 contain one Trx-HD and one Grx-HD, but all mammalian Grx3 have two repeated Grx-HDs (data not shown; [19]). Computational analysis indicated that multiple Grx-HDs have also been identified in Grx3 from plants, nematodes and fish, but not from prokaryotes, fungi and insects (data not shown). The conserved biological function among various Grxs suggests that one Grx-HD may be sufficient for Grx activity [42]. For example, yeast ScGrx5, a mitochondrial monothiol Grx, is able to suppress grx3grx4 mutant phenotypes, when the mitochondrial targeting sequence is removed [22]. In addition, yeast ScGrx3, a nucleocytoplasmic Grx, can restore Grx5 function in grx5 cells when ScGrx3 is targeting to mitochondria [38]. Most interestingly, a single Grx-HD from a poplar monothiol Grx, GrxS17, which has three Grx-HDs at its C-terminus, can fully suppress yeast grx5 mutant phenotypes [43]. However, the interchangeability among mammalian Grx3 Grx-HD and Grx5 has not been determined. In comparison with the C-terminal Grx-HD, the N-terminal Trx-HD is more diverse (data not shown). It has been proposed that the Trx-HD is involved in protein–protein interactions instead of modulating oxidative stress response [26]. For example, the yeast Grx4 N-terminal region physically interacts with piDB26, a p53-related protein kinase [23], and a human Grx3 interacts with protein kinase C theta isoform through the N-terminal Trx-HD [26]. A recent report indicates that Grx3 binds two bridging [2Fe–2S] clusters in a homodimeric complex with the active site Cys residue of its two Grx-HDs, suggesting that this unique structure could act as a redox sensor [44]. Future studies are needed to determine how Grx3 and its functional domains regulate cellular redox homeostasis and antioxidative processes in vivo.
There is growing evidence that ScGrx3/ScGrx4 play essential roles in iron sensing, trafficking and homeostasis in yeast [21,22,24,25,45-47]. Mammalian Grx3s are able to inhibit free iron accumulation in grx3grx4 mutant cells (Fig. 2B,C). Notably, similar to ScGrx3, Grx3 could only partially rescue double-mutant phenotypes in terms of iron accumulation (Fig. 2B,C). This partial restoration has been seen in several previous studies, in which single mutant cells (either grx3 or grx4) accumulate significantly more free iron and/or less efficiently incorporate Fe into some Fe-containing proteins; this is most likely caused by the nonredundant functions of ScGrx3 and ScGrx4 or gene-dosage effects [21-23,25,48]. Nevertheless, our findings suggest that Grx3s may have similar functions to yeast ScGrx3/ScGrx4 in regulating iron homeostasis in mammalian cells (Fig. 2B,C).
Ubiquitous expression of Grx3 in both mouse embryos and tissues indicates that Grx3 is required for cell growth, organ development and normal metabolism during growth and development (Fig. 3). Grx3 expression was induced by exogenous oxidants (Fig. 3). Interestingly, the induction of Grx3 expression was differentially modulated by oxidants. We speculate that each oxidant may preferentially activate different stress-responsive signaling pathways and/or transcriptional machinery. For example, H2O2 (100 μm) activates AP-1 in cultured chondrocytes, whereas tBHP triggers increased expression of hypoxia-inducible factor-1alpha (HIF-1α) [49,50]. Furthermore, diamide, a thiol oxidant, activates NF-κB activity and tumor necrosis factor-α-induced gene expression [51]. Further studies will be required to clarify factors modulating Grx3 expression in response to oxidative stress and identify downstream targets.
Mice lacking Grx3 are unable to survive beyond E12.5 (Fig. 4) [36]. The embryonic lethality of Grx3 null mice indicates that Grx3 is essential for embryogenesis. Although Grx3−/− embryos could survive up to E12.5, the effects of Grx3 dysfunction on embryo development might take place at early stages because we noticed smaller Grx3−/− embryos compared with Grx3+/− and Grx3+/+ embryos, and an increased number of resorbing embryos at E9.5–E11.5 (Fig. 4D and Table 1). We did not observe any morphological change in embryos among three genotypes at or before E8.5, or any difference in decidua at early stages, although molecular and biochemical changes might occur before the phenotypes of whole embryo could be distinguished (data not shown). It is worth noting that Grx3−/− embryos survive to E12.5 at a variable frequency that may depend on the genetic background. For example, most E12.5 Grx3−/− embryos were dissected from pregnant female mice in a mixed background (50% 129Sv and 50% C57BL6) and most E9.5 Grx3−/− embryos were dissected from C57BL6 mice (after being backcrossed to C57BL6 for 10 generations). The majority of embryos with severe growth defects were found in C57BL6 mice (data not shown). Interestingly, this observation has been reported in Igf-1 mutant mice, where the survival rate of homozygous mutants depends on the genetic background [52]. In young adulthood, Grx3+/+ and Grx3+/− mice (both male and female) did not appear to have any distinguishable phenotypic differences. However, in a recent report, a single Grx3 allele deletion augments cardiac hypertrophy in transgenic mice under pressure overload [36]. The results suggest that reduction of Grx3 expression may also be critical in the pathogenesis of human diseases.
Oxidative stress can cause cell cycle arrest and subsequently embryonic death [6,53-55]. Reduced levels of Grx3 causes increased ROS production in cells [37], suggesting that Grx3 may have a protective role in counteracting oxidative stress during cell cycle progression. In support of this notion, Grx3−/− MEFs display impaired cell proliferation and G2/M progression (Fig. 5). Furthermore, Grx3-KD HeLa cells showed similar cell cycle defects during mitotic exit (Fig. 6). Grx3-KD Hela cells have a higher percentage of binucleated cells than that of control cells, indicating an increased failure of cytokinesis in the absence of Grx3. This is also the case in Grx3−/− MEFs (Fig. 5D,E). Binucleated mammalian cells are prevented from entering the next cell cycle through a p53-dependent tetraploidy checkpoint [56]. This at least partially explains why Grx3−/− MEF cells accumulated at the G2/M stage (Fig. 5) and the early death phenotype in Grx3−/− embryos (Fig. 4 and Table 1). Therefore, it will be interesting to determine whether inactivation of p53 can rescue Grx3−/− MEF growth and Grx3−/− embryo development. Given the complexity of cell cycle regulation and the numerous intracellular components responding to oxidative stress, it is not surprising that Grx3-mediated cell cycle control may be dependent on multiple regulatory pathways and the underlying mechanisms remain unresolved.
Conclusion
The characterization of mammalian Grx3 reported here is particularly noteworthy in that a comprehensive in vivo function of mammalian monothiol Grxs has not been previously defined. Grx3 appears to be evolutionarily conserved across species and the capability of mammalian Grx3s to rescue yeast Scgrx3/Scgrx4 deficiency phenotypes suggests a conserved biochemical mechanism among monothiol Grxs. The Grx3 null mice demonstrate that this protein is essential for embryogenesis. Specifically, mammalian Grx3 is required for efficient cell cycle progression. Thus, the characterization of Grx3 provides insights into the molecular mechanism of redox regulation in cell growth and organ development in mammals.
Materials and methods
Antibodies
Cyclin B1 (sc-752) and Plk1 (sc-17783) were purchased from Santa Cruz Biotechnology (Santa Cruz Biotechnology, Santa Cruz, CA, USA); Securin (Pds1) was ordered from (MS-1511-P1) Neomarkers (Lab Vision, Fremont, CA, USA). Cdc2 (#9112), phosphor-cdc2 Tyr15 (#9111), and cleaved caspase 3 antiobdy (#9664), goat against mouse IgG1, and rabbit IgG conjugated with horseradish peroxidase were purchased from Cell Signaling Technology (Cell Signaling Technology, Beverly, MA, USA). Alexa Fluor 488 conjugated goat anti-mouse IgG1 and antibromodeoxyuridine conjugated with Alexa Fluor 680 antibodies were purchased from Invitrogen (Invitrogen, Carlsbad, CA, USA). Monoclonal antibody against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was ordered from Chemicon (Chemicon, San Diego, CA, USA). Proteinase inhibitor cocktail tablets were purchased from Roche Diagnostics (Roche Diagnostics, Indianapolis, IN, USA). Monoclonal antibody against β actin IgG1 and all chemicals were purchased from Sigma-Aldrich (Sigma-Aldrich, St. Louis, MO, USA). Monoclonal antibody against human Grx3 (full length) was generated in the Baculovirus/Monoclonal Antibody Core Facility at Baylor College of Medicine.
Yeast strains, DNA constructs, and growth assays
Saccharomyces cerevisiae wild-type strain CML235 (MATa ura3-52 leu2Δ1 his3Δ200) and grx3grx4 (MATa ura3-52 leu2Δ1 his3Δ200 MATa grx3::kanMX4 grx4::kanMX4) were generously gifted by Enrique Herrero (Universitat de Lleida, Lleida, Spain) [28] and were used in all yeast experiments. Yeast ScGrx3 and ScGrx4 were amplified by PCR using gene-specific primers. For ScGrx3, we used the forward primer 5′-GGCTCTAGAATGTGTTCTTTTCAGGTTCCAT-3′ and the reverse primer 5′-CCGGAGCTCTTAAGATTGGAGAGCATGCTG-3′. For ScGrx4, the we used the forward primer 5′-GCCGGATCCATGACTGTGGTTGAAATAAAAAG-3′ and the reverse primer 5′-CCGGAGCTCTTACTGTAGAGCATGTTGGAAATA-3′. Full-length cDNA of HsGrx3 and MmGrx3 were amplified by PCR using gene-specific primers. For HsGrx3, the we used forward primer 5′-GCCGGATCCATGGCGGCGGGGGCGGCTGAGGCA-3′ and the reverse primer 5′-GGCGTCGACCCGCGGTTAATTTTCTCCTCTCAGTAT-3′; and for MmGrx3, we used the forward primer 5′-GGGCTCGAGAGATCTGCGATGGCGGCGGGGGCGGCCGA-3′ and the reverse primer 5′-GGCCCGCGGCTATAGGATCCCATTTTCTCCTTTCAGTATAGG-3′. The PCR products were cloned into pGEM-T Easy (Promega, Madison, WI, USA). The fidelity of all clones was confirmed by sequencing. Yeast ScGrx3 and mammalian Grx3s were subcloned into a yeast expression vector, piUGpd [57]. Yeast cells were transformed using the LiOAc method [20]. All yeast strains were assayed on YPD medium (yeast peptone dextrose, rich media) and SC (synthetic complete) medium with or without various concentrations of H2O2, diamide, and tBHP [20]. Measurement of iron concentration was conducted as described previously [20].
Localization of MmGrx3–GFP fusion in yeast
Full-length yeast ScGrx3 and MmGrx3 were fused to the N-terminus of red fluorescent protein (Clontech Laboratories, Inc., Mountain View, CA, USA) and green fluorescent protein (GFP), respectively, using a procedure described previously [58]. The fluorescent protein constructs were subcloned into yeast vectors as described previously [58]. The subcellular localization of the fused proteins was imaged in comparison with nuclear–GFP markers as described previously [58].The fluorescence signals were detected at 510 nm (excitation at 488 nm) for GFP, at 582 nm (excitation at 543 nm) for red fluorescent protein as previously described [58].
RNA gel-blotting analysis
Total RNA was extracted from mouse (C57BL6) tissues of both 12-week-old male and female mice using TRIzol reagents (Invitrogen). To analyze gene expression, 10 μg of total RNA was loaded onto a formaldehyde-containing 1.0% agarose gel, blotted onto nylon membrane (Amersham Biosciences, Little Chalfont, UK), and subjected to hybridization with a 32P-labeled gene-specific probe [59].
Quantitative reverse transcription-PCR of Grx3 expression
Mouse myocytes (C2C12) were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal bovine serum and 1% antibiotics (Pen-Strep from Invitrogen). Cells were cultured at 37 °C in a humidified atmosphere of 5% CO2 until 85–90% confluence, then treated with or without 50 and 100 μm H2O2, 25 and 50 μm diamide, 50 and 100 μm tBHP for 0.5 or 1 h, respectively, prior to be harvested for RNA extraction. To check cell viability after oxidant treatments, C2C12 cells were treated with 100 μm H2O2, 50 μm diamide and 100 μm tBHP for 1 h, respectively, in triplicate for each oxidant. Cells were trypsinized and suspended in 0.5 mL of Dulbecco’s modified Eagle’s medium media without serum, then mixed thoroughly with 0.1 mL of 0.4% Trypan Blue stain (EMD Chemicals Inc., Gibbestown, NJ, USA). After incubation at room temperature for 5 min, dead cells, which were stained blue, and viable cells excluding the stain were counted with a hemocytometer. Cell viability was calculated as percentage of viable cells against the total number of cells counted. RNA extraction was conducted as described above. Quantitative RT-PCR was performed using a TaqMan system as described previously [60]. Forward primer (Grx3-58F): 5′-TCTGCCCAGCAGTTTGAAGA-3′; reverse primer (Grx3-131R): 5′-CATGGTGCCCAGAAATGAAC-3′; probe (Grx3-79T): 5′-CTACTGCGCCTCAAAACCAAGTCACTCCT-3′. The housekeeping gene cyclophilin was used to normalize the gene expression data.
Generation of Grx3-deficient mice
A mouse 129/SvEv embryonic stem cell clone (RRF094) harboring gene trap vector within Grx3 gene at chromosome 7 was obtained from Baygenomics (http://www.genetrap.org/) [61]. The Grx3+/− heterozygous embryonic stem cell was injected into C57BL/6 albino blastocytes to generate chimeric founder mice in the Darwin Transgenic Mouse Core at Baylor College of Medicine (https://www.bcm.edu/labs/darwin_core/). Those male chimeric founder mice were bred to C57BL/6 or C57BL/6 albino mice, and germline transmission was achieved. By using invert PCR, the insertion loci of gene trap vector was determined to locate within the second intron of Grx3 gene. Heterozygous F1 transgenic mice were determined by PCR using a combination of gene-specific and vector-specific primers. A wild-type allele could be identified by amplification of 450 bp of PCR product using a Grx3 forward primer: 5′-CCTAGAAGGTAACCCTAAAATGTC-3′ and a Grx3 reverse primer: 5′-CCATCACTGCGTTACTCCAGA-3′. A mutant allele could be detected by amplification of 550 bp of PCR product with the Grx3 forward primer and a vector-specific primer: 5′-GCTACCGGCTAAAACTTGAGA-3′. All animal protocols were approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine.
Embryo dissection and in situ hybridization
Grx3+/− male and female mice were mated overnight and vaginal plugs were checked the following morning (E0.5). Pregnant mice were sacrificed by cervical dislocation and embryos at various somite stages were dissected from decidua in NaCl/Pi medium. Maternal tissues and Reichert membrane were removed. The yolk sac was detached from the embryo properly and used for genotyping by PCR. The morphological appearance of whole embryos was first evaluated at various somite stages. For in situ hybridization, embryos at E10.5 were fixed in 4% paraformaldehyde in NaCl/Pi overnight at 4 °C, rinsed with NaCl/Pi, dehydrated in a series of ethanol solutions (50%, 60%, 70%, 80%, 90% and 100%), and embedded in wax. Fixed embryos were sectioned at 6 μm and specimens were subjected to hybridization using digoxygenin-labeled antisense and sense riboprobes (nonradioactive) generated by in vitro transcription from Grx3 DNA templates using T7 (sense) or Sp6 (antisense) bacteriophage RNA polymerases. Prehybridization, hybridization, stringency washes, incubation with antidigoxygenin antibody coupled to peroxidase, tyramin–biotin amplification reaction, alkaline phosphatase–streptavidin detection of covalently attached biotin, colorimetric detection of phosphatase moiety using BCPI/NBT reagents were sequentially carried out by the GenePaint robot in an automated manner in the Core laboratory at Baylor College of Medicine [62].
Mouse embryonic fibroblasts
Dissected individual embryo was minced and incubated in trypsin (0.05%; Invitrogen) for 10 min at 37 °C. Cells were pipetted up and down, and washed with NaCl/Pi once. Cells were plated in high-glucose Dulbecco’s modified Eagle’s medium medium with 10% fetal bovine serum, l-glutamine (2 μm), and 1% Penstrep, then maintained at 37 °C in 5%CO2.
shRNA and HeLa cell stable transfectants
shRNA constructs in pLKO.1-puro specifically targeting human Grx3 sequences were purchased from Sigma-Aldrich. The human Grx3 shRNA1 sequence is 5′-CCGGGCTCTTTATGAAAGGAAACAACTCGAGTTGTTTCCTTTCATAAAGAGCTTTTTG-3′. The human Grx3 shRNA2 sequence is 5′-CCGGGAACGAAGTTATGGCAGAGTTCTCGAGAACTCTGCCATAACTTCGTTCTTTTTG-3′. Control cells were transfected with a control shRNA that does not match any known human coding cDNA. Stable knockdown clones of HeLa cells were pooled and used for experiments [37]. To quantify the Grx3 protein levels in Grx3-KD HeLa cells, 20 μg of protein lysates from control, shRNA#1 and shRNA#2 cells were subjected to western blot analysis using antibody against Grx3 IgG1 and antibody against GAPDH IgG1 as controls. The blots were analyzed using Molecular Imager Chemi Doc™ XRS System (Bio-Rad Laboratories, Hercules, CA, USA). The signal intensity was measured by using a quantity one (v. 4.6.5) software following the manufacture instruction.
Cell proliferation, flow cytometry, BrdU-labeling and immunolabeling
MEFs at passage 2 (105) were plated in six-well plates and cell proliferation was measured every 2 days by cell numeration of triplicates of indicated samples (Grx3+/+, Grx3+/− and Grx3−/−) [63]. Parallel samples were collected for analysis of cell-cycle profile by staining with propidium iodide, followed by flow cytometry (FACScan, Beckman Coulter, Fullerton, CA, USA) [64]. For anti-BrdU staining, cells were maintained in medium containing 0.1% fetal bovine serum for 2 days, replenishing with 10% fetal bovine serum for 12 h. Cells were pulsed-chased for 5 h with 25 μm BrdU (Sigma). Cells were fixed with 70% ethanol and then treated with 2 m HCl (in 0.5% Triton-X 100) for 10 min. After washing with NaCl/Pi, the cells were incubated with a 1: 50 dilution of anti-BrdU conjugated with Alexa Fluor 680 antibody for 1 h. Cells were washed with NaCl/Pi and counterstained with Sytox orange for 5 min. The images were captured by confocal microscope and the numbers of positive stained cells were counted from six different fields with each field ~200 cells were recorded. To synchronize cells, Grx3-KD and control HeLa cells were arrested by double thymidine block [40]. In brief, cells were blocked for 18 h with 2 mm thymidine (final concentration), relased for 9 h by washing out the thymidine using fresh media, and then blocked with 2 mm thymidine for additional 17 h. The cells were released by washing out the thymidine with fresh media. Synchronized Grx3-KD and control cells were harvested every 2 h after release from thymidine and subjected to flow cytometry analysis as described above. Cell lysates were prepared in the protein extraction buffer (0.5% nonide P-40, 0.5% sodium deoxycholate, 50 mm NaCl, 1 mm EDTA, 5 mm Na3VO4, 20 mm NaF, 20 mm Na4O7P2, 50 mm Hepes pH 7.5, 1 mm phenylmethanesulfonyl fluoride, one tablet complete protease inhibitors (Roche). Extracted proteins were then subjected to western blot analysis using the antibodies as indicated in the figure legends. For immunolabeling, Grx3+/+ and Grx3−/− MEFs and unsynchronized Grx3-KD and control HeLa cells were plated on LabTek chambered coverslips (Nunc) and grown for 24 h at 37 °C. The cells were rinsed three times in 37 °C NaCl/Pi and fixed in absolute methanol at −20 °C for 5 min. The cells were washed three times with NaCl/Pi for 3 min each, and then blocked in 10% normal goat serum for 1 h at room temperature. The cells were incubated in mAb against β-actin (1: 100) for 1 h at room temperature, and then washed five times with NaCl/Pi for 5 min each, followed by a secondary antibody (goat against mouse IgG conjugated with fluorescein isothiocyanate) at 1: 250 for 30 min. The cells were washed five times with NaCl/Pi for 5 min each, and then stained with 4′,6-diamidino-2-phenylindole (300 nm) for 5 min. The images were captured by confocal microscope.
Supplementary Material
Fig. S1. Yeast ScGrx3 and ScGrx4 can rescue the growth defects of yeast grx3grx4 and suppress the sensitivity of grx3grx4 cells to oxidants.
Fig. S2. Effect of exogenous oxidants on C2C12 cell viability.
Acknowledgements
We thank Dr Enrique Herrero for wild-type and yeast grx strains. Bolanle A. Bukoye and Annette Frank, participants in the Baylor SMART program, were involved in the initial construction of Grx3 plasmids and the initial yeast assays. Jianping Jin is a Pew Scholar and supported by a grant (AU-1711) from the Welch Foundation. This work is supported by the United States Department of Agriculture/Agricultural Research Service under Cooperation Agreement 6250-51000-055 (N.-H. Cheng).
Abbreviations
- DTB
double thymidine block
- GFP
green fluorescent protein
- Grx
glutaredoxin
- HD
homology domain
- HsGrx3
Homo sapiens glutaredoxin 3
- KD
knock-down
- MEF
mouse embryonic fibroblast
- MmGrx3
Mus musculus glutaredoxin 3
- ROS
reactive oxygen species
- ScGrx3
Saccharomyces cerevisiae glutaredoxin 3
- ScGrx4
Saccharomyces cerevisiae glutaredoxin 4
- shRNA
small hairpin RNA
- tBHP
tert-butylhydroperoxide
- Trx
thioredoxin
- Txnl2
thioredoxin-like 2
Footnotes
The following supplementary material is available:
This supplementary material can be found in the online version of this article.
Please note: As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- 1.Droge W. Free radicals in the physiological control of cell function. Physiol Rev. 2002;82:47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 2.Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 4.Finkel T. Oxidant signals and oxidative stress. Curr Opin Cell Biol. 2003;15:247–254. doi: 10.1016/s0955-0674(03)00002-4. [DOI] [PubMed] [Google Scholar]
- 5.Goldstein BJ, Mahadev K, Wu X, Zhu L, Motoshima H. Role of insulin-induced reactive oxygen species in the insulin signaling pathway. Antioxid Redox Signal. 2005;7:1021–1031. doi: 10.1089/ars.2005.7.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dennery PA. Effects of oxidative stress on embryonic development. Birth Defects Res C Embryo Today. 2007;81:155–162. doi: 10.1002/bdrc.20098. [DOI] [PubMed] [Google Scholar]
- 7.Trachootham D, Lu W, Ogasawara MA, Nilsa RD, Huang P. Redox regulation of cell survival. Antioxid Redox Signal. 2008;10:1343–1374. doi: 10.1089/ars.2007.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ushio-Fukai M, Nakamura Y. Reactive oxygen species and angiogenesis: NADPH oxidase as target for cancer therapy. Cancer Lett. 2008;266:37–52. doi: 10.1016/j.canlet.2008.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finkel T. Radical medicine: treating ageing to cure disease. Nat Rev Mol Cell Biol. 2005;6:971–976. doi: 10.1038/nrm1763. [DOI] [PubMed] [Google Scholar]
- 10.Halliwell B. Oxidative stress and cancer: have we moved forward? Biochem J. 2007;401:1–11. doi: 10.1042/BJ20061131. [DOI] [PubMed] [Google Scholar]
- 11.Betts DH, Madan P. Permanent embryo arrest: molecular and cellular concepts. Mol Hum Reprod. 2008;14:445–453. doi: 10.1093/molehr/gan035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leung PS, Chan YC. Role of oxidative stress in pancreatic inflammation. Antioxid Redox Signal. 2009;11:135–165. doi: 10.1089/ars.2008.2109. [DOI] [PubMed] [Google Scholar]
- 13.Kondo N, Nakamura H, Masutani H, Yodoi J. Redox regulation of human thioredoxin network. Antioxid Redox Signal. 2006;8:1881–1890. doi: 10.1089/ars.2006.8.1881. [DOI] [PubMed] [Google Scholar]
- 14.Berndt C, Lillig CH, Holmgren A. Thiol-based mechanisms of the thioredoxin and glutaredoxin systems: implications for diseases in the cardiovascular system. Am J Physiol Heart Circ Physiol. 2007;292:H1227–H1236. doi: 10.1152/ajpheart.01162.2006. [DOI] [PubMed] [Google Scholar]
- 15.Lillig CH, Berndt C, Holmgren A. Glutaredoxin systems. Biochim Biophys Acta. 2008;1780:1304–1317. doi: 10.1016/j.bbagen.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 16.Fernandes AP, Holmgren A. Glutaredoxins: glutathione-dependent redox enzymes with functions far beyond a simple thioredoxin backup system. Antioxid Redox Signal. 2004;6:63–74. doi: 10.1089/152308604771978354. [DOI] [PubMed] [Google Scholar]
- 17.Holmgren A. Thioredoxin and glutaredoxin systems. J Biol Chem. 1989;264:13963–13966. [PubMed] [Google Scholar]
- 18.Bushweller JH, Aslund F, Wuthrich K, Holmgren A. Structural and functional characterization of the mutant Escherichia coli glutaredoxin (C14S) and its mixed disulfide with glutathione. Biochemistry. 1992;31:9288–9293. doi: 10.1021/bi00153a023. [DOI] [PubMed] [Google Scholar]
- 19.Herrero E, de la Torre-Ruiz MA. Monothiol glutaredoxins: a common domain for multiple functions. Cell Mol Life Sci. 2007;64:1518–1530. doi: 10.1007/s00018-007-6554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng NH, Liu JZ, Brock A, Nelson RS, Hirschi KD. AtGRXcp, an Arabidopsis chloroplastic glutaredoxin, is critical for protection against protein oxidative damage. J Biol Chem. 2006;281:26280–26288. doi: 10.1074/jbc.M601354200. [DOI] [PubMed] [Google Scholar]
- 21.Pujol-Carrion N, Belli G, Herrero E, Nogues A, de la Torre-Ruiz MA. Glutaredoxins Grx3 and Grx4 regulate nuclear localisation of Aft1 and the oxidative stress response in Saccharomyces cerevisiae. J Cell Sci. 2006;119:4554–4564. doi: 10.1242/jcs.03229. [DOI] [PubMed] [Google Scholar]
- 22.Ojeda L, Keller G, Muhlenhoff U, Rutherford JC, Lill R, Winge DR. Role of glutaredoxin-3 and glutaredoxin-4 in the iron regulation of the Aft1 transcriptional activator in Saccharomyces cerevisiae. J Biol Chem. 2006;281:17661–17669. doi: 10.1074/jbc.M602165200. [DOI] [PubMed] [Google Scholar]
- 23.Lopreiato R, Facchin S, Sartori G, Arrigoni G, Casonato S, Ruzzene M, Pinna LA, Carignani G. Analysis of the interaction between piD261/Bud32, an evolutionarily conserved protein kinase of Saccharomyces cerevisiae, and the Grx4 glutaredoxin. Biochem J. 2004;377:395–405. doi: 10.1042/BJ20030638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumanovics A, Chen OS, Li L, Bagley D, Adkins EM, Lin H, Dingra NN, Outten CE, Keller G, Winge D, et al. Identification of FRA1 and FRA2 as genes involved in regulating the yeast iron regulon in response to decreased mitochondrial iron–sulfur cluster synthesis. J Biol Chem. 2008;283:10276–10286. doi: 10.1074/jbc.M801160200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mühlenhoff U, Molik S, Godoy JR, Uzarska MA, Richter N, Seubert A, Zhang Y, Stubbe J, Pierrel F, Herrero E, et al. Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron–sulfur cluster. Cell Metab. 2010;12:373–385. doi: 10.1016/j.cmet.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isakov N, Witte S, Altman A. PICOT-HD: a highly conserved protein domain that is often associated with thioredoxin and glutaredoxin modules. Trends Biochem Sci. 2000;25:537–539. doi: 10.1016/s0968-0004(00)01685-6. [DOI] [PubMed] [Google Scholar]
- 27.Witte S, Villalba M, Bi K, Liu Y, Isakov N, Altman A. Inhibition of the c-Jun N-terminal kinase/AP-1 and NF-kappaB pathways by PICOT, a novel protein kinase C-interacting protein with a thioredoxin homology domain. J Biol Chem. 2000;275:1902–1909. doi: 10.1074/jbc.275.3.1902. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez-Manzaneque MT, Ros J, Cabiscol E, Sorribas A, Herrero E. Grx5 glutaredoxin plays a central role in protection against protein oxidative damage in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:8180–8190. doi: 10.1128/mcb.19.12.8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wingert RA, Galloway JL, Barut B, Foott H, Fraenkel P, Axe JL, Weber GJ, Dooley K, Davidson AJ, Schmid B, et al. Deficiency of glutaredoxin 5 reveals Fe–S clusters are required for vertebrate haem synthesis. Nature. 2005;436:1035–1039. doi: 10.1038/nature03887. [DOI] [PubMed] [Google Scholar]
- 30.Camaschella C, Campanella A, De Falco L, Boschetto L, Merlini R, Silvestri L, Levi S, Iolascon A. The human counterpart of zebrafish shiraz shows side-roblastic-like microcytic anemia and iron overload. Blood. 2007;110:1353–1358. doi: 10.1182/blood-2007-02-072520. [DOI] [PubMed] [Google Scholar]
- 31.Linares GR, Xing W, Govoni KE, Chen ST, Mohan S. Glutaredoxin 5 regulates osteoblast apoptosis by protecting against oxidative stress. Bone. 2009;44:795–804. doi: 10.1016/j.bone.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye H, Jeong SY, Ghosh MC, Kovtunovych G, Silvestri L, Ortillo D, Uchida N, Tisdale J, Camaschella C, Rouault TA. Glutaredoxin 5 deficiency causes sideroblastic anemia by specifically impairing heme biosynthesis and depleting cytosolic iron in human erythroblasts. J Clin Invest. 2010;120:1749–1761. doi: 10.1172/JCI40372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kato N, Motohashi S, Okada T, Ozawa T, Mashima K. PICOT, protein kinase C theta-interacting protein, is a novel regulator of FcepsilonRI-mediated mast cell activation. Cell Immunol. 2008;251:62–67. doi: 10.1016/j.cellimm.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 34.Jeong D, Cha H, Kim E, Kang M, Yang DK, Kim JM, Yoon PO, Oh JG, Bernecker OY, Sakata S, et al. PICOT inhibits cardiac hypertrophy and enhances ventricular function and cardiomyocyte contractility. Circ Res. 2006;99:307–314. doi: 10.1161/01.RES.0000234780.06115.2c. [DOI] [PubMed] [Google Scholar]
- 35.Jeong D, Kim JM, Cha H, Oh JG, Park J, Yun SH, Ju ES, Jeon ES, Hajjar RJ, Park WJ. PICOT attenuates cardiac hypertrophy by disrupting calcineurin–NFAT signaling. Circ Res. 2008;102:711–719. doi: 10.1161/CIRCRESAHA.107.165985. [DOI] [PubMed] [Google Scholar]
- 36.Cha H, Kim JM, Oh JG, Jeong MH, Park CS, Park J, Jeong HJ, Park BK, Lee YH, Jeong D, et al. PICOT is a critical regulator of cardiac hypertrophy and cardiomyocyte contractility. J Mol Cell Cardiol. 2008;45:796–803. doi: 10.1016/j.yjmcc.2008.09.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qu Y, Wang J, Ray PS, Guo H, Huang J, Shin-Sim M, Bukoye BA, Liu B, Lee AV, Lin X, et al. Thioredoxin-like 2 regulates human cancer cell growth and metastasis via redox homeostasis and NF-κB signaling. J Clin Invest. 2011;121:212–225. doi: 10.1172/JCI43144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Molina MM, Belli G, de la Torre MA, Rodriguez-Manzaneque MT, Herrero E. Nuclear monothiol glutaredoxins of Saccharomyces cerevisiae can function as mitochondrial glutaredoxins. J Biol Chem. 2004;279:51923–51930. doi: 10.1074/jbc.M410219200. [DOI] [PubMed] [Google Scholar]
- 39.Halliwell B. Biochemistry of oxidative stress. Biochem Soc Trans. 2007;35:1147–1150. doi: 10.1042/BST0351147. [DOI] [PubMed] [Google Scholar]
- 40.Whitfield ML, Zheng LX, Baldwin A, Ohta T, Hurt MM, Marzluff WF. Stem–loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol Cell Biol. 2000;20:4188–4198. doi: 10.1128/mcb.20.12.4188-4198.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wanat JJ, Singh N, Alani E. The effect of genetic background on the function of Saccharomyces cerevisiae mlh1 alleles that correspond to HNPCC missense mutations. Human Mol Genet. 2007;16:445–452. doi: 10.1093/hmg/ddl479. [DOI] [PubMed] [Google Scholar]
- 42.Molina-Navarro MM, Casas C, Piedrafita L, Belli G, Herrero E. Prokaryotic and eukaryotic monothiol glutaredoxins are able to perform the functions of Grx5 in the biogenesis of Fe/S clusters in yeast mitochondria. FEBS Lett. 2006;580:2273–2280. doi: 10.1016/j.febslet.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 43.Bandyopadhyay S, Gama F, Molina-Navarro MM, Gualberto JM, Claxton R, Naik SG, Huynh BH, Herrero E, Jacquot JP, Johnson MK, et al. Chloroplast monothiol glutaredoxins as scaffold proteins for the assembly and delivery of [2Fe–2S] clusters. EMBO J. 2008;27:1122–1133. doi: 10.1038/emboj.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haunhorst P, Berndt C, Eitner S, Godoy JR, Lillig CH. Characterization of the human monothiol glutaredoxin 3 (PICOT) as iron-sulfur protein. Biochem Biophys Res Commun. 2010;394:372–376. doi: 10.1016/j.bbrc.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 45.Li H, Mapolelo DT, Dingra NN, Naik SG, Lees NS, Hoffman BM, Riggs-Gelasco PJ, Huynh BH, Johnson MK, Outten CE. The yeast iron regulatory proteins Grx3/4 and Fra2 form heterodimeric complexes containing a [2Fe–2S] cluster with cysteinyl and histidyl ligation. Biochemistry. 2009;48:9569–9581. doi: 10.1021/bi901182w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mercier A, Labbé S. Both Php4 function and subcellular localization are regulated by iron via a multistep mechanism involving the glutaredoxin Grx4 and the exportin Crm1. J Biol Chem. 2009;284:20249–20262. doi: 10.1074/jbc.M109.009563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Mapolelo DT, Dingra NN, Keller G, Riggs-Gelasco PJ, Winge DR, Johnson MK, Outten CE. Histidine 103 in Fra2 is an iron–sulfur cluster ligand in the [2Fe–2S] Fra2–Grx3 complex and is required for in vivo iron signaling in yeast. J Biol Chem. 2011;286:867–876. doi: 10.1074/jbc.M110.184176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jablonowski D, Butler AR, Fichtner L, Gardiner D, Schaffrath R, Stark MJ. Sit4p protein phosphatase is required for sensitivity of Saccharomyces cerevisiae to Kluyveromyces lactis zymocin. Genetics. 2001;159:1479–1489. doi: 10.1093/genetics/159.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin G, Andriamanalijaona R, Mathy-Hartert M, Henrotin Y, Pujol JP. Comparative effects of IL-1[beta] and hydrogen peroxide (H2O2) on catabolic and anabolic gene expression in juvenile bovine chondrocytes. Osteoarthr Cartilage. 2005;13:915–924. doi: 10.1016/j.joca.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 50.Ramakrishnan P, Hecht BA, Pedersen DR, Lavery MR, Maynard J, Buckwalter JA, Martin JA. Oxidant conditioning protects cartilage from mechanically induced damage. J Orthopaedic Res. 2010;28:914–920. doi: 10.1002/jor.21072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ozsoy HZ, Sivasubramanian N, Wieder ED, Pedersen S, Mann DL. Oxidative stress promotes ligand-independent and enhanced ligand-dependent tumor necrosis factor receptor signaling. J Biol Chem. 2008;283:23419–23428. doi: 10.1074/jbc.M802967200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu JP, Baker J, Perkins AS, Robertson EJ, Efstratiadis A. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r) Cell. 1993;75:59–72. [PubMed] [Google Scholar]
- 53.Shackelford RE, Kaufmann WK, Paules RS. Oxidative stress and cell cycle checkpoint function. Free Radic Biol Med. 2000;28:1387–1404. doi: 10.1016/s0891-5849(00)00224-0. [DOI] [PubMed] [Google Scholar]
- 54.Covarrubias L, Hernandez-Garcia D, Schnabel D, Salas-Vidal E, Castro-Bregon S. Function of reactive oxygen species during animal development: passive or active? Dev Biol. 2008;320:1–11. doi: 10.1016/j.ydbio.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 55.Nonn L, Williams RR, Erickson RP, Powis G. The absence of mitochondrial thioredoxin 2 causes massive apoptosis, exencephaly, and early embryonic lethality in homozygous mice. Mol Cell Biol. 2003;23:916–922. doi: 10.1128/MCB.23.3.916-922.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andreassen PR, Lohez OD, Lacroix FB, Margolis RL. Tetraploid state induces p53-dependent arrest of nontransformed mammalian cells in G1. Mol Biol Cell. 2001;12:1315–1328. doi: 10.1091/mbc.12.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nathan DF, Vos MH, Lindquist S. Identification of SSF1, CNS1, and HCH1 as multicopy suppressors of a Saccharomyces cerevisiae Hsp90 loss-of-function mutation. Proc Natl Acad Sci USA. 1999;96:1409–1414. doi: 10.1073/pnas.96.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng NH, Liu JZ, Nelson RS, Hirschi KD. Characterization of CXIP4, a novel Arabidopsis protein that activates the H+/Ca2+ antiporter, CAX1. FEBS Lett. 2004;559:99–106. doi: 10.1016/S0014-5793(04)00036-5. [DOI] [PubMed] [Google Scholar]
- 59.Cheng NH, Hirschi KD. Cloning and characterization of CXIP1, a novel PICOT domain-containing Arabidopsis protein that associates with CAX1. J Biol Chem. 2003;278:6503–6509. doi: 10.1074/jbc.M210883200. [DOI] [PubMed] [Google Scholar]
- 60.Durgan DJ, Trexler NA, Egbejimi O, McElfresh TA, Suk HY, Petterson LE, Shaw CA, Hardin PE, Bray MS, Chandler MP, et al. The circadian clock within the cardiomyocyte is essential for responsiveness of the heart to fatty acids. J Biol Chem. 2006;281:24254–24269. doi: 10.1074/jbc.M601704200. [DOI] [PubMed] [Google Scholar]
- 61.Stryke D, Kawamoto M, Huang CC, Johns SJ, King LA, Harper CA, Meng EC, Lee RE, Yee A, L’Italien L, et al. BayGenomics: a resource of insertional mutations in mouse embryonic stem cells. Nucleic Acids Res. 2003;31:278–281. doi: 10.1093/nar/gkg064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Visel A, Thaller C, Eichele G. GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 2004;32:D552–D556. doi: 10.1093/nar/gkh029. GenePaint.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Louie MC, Revenko AS, Zou JX, Yao J, Chen HW. Direct control of cell cycle gene expression by proto-oncogene product ACTR, and its autoregulation underlies its transforming activity. Mol Cell Biol. 2006;26:3810–3823. doi: 10.1128/MCB.26.10.3810-3823.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ormerod MG, Imrie PR, Loverock P, Ter Haar G. A flow cytometric study of the effect of heat on the kinetics of cell proliferation of Chinese hamster V-79 cells. Cell Prolif. 1992;25:41–51. doi: 10.1111/j.1365-2184.1992.tb01436.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Yeast ScGrx3 and ScGrx4 can rescue the growth defects of yeast grx3grx4 and suppress the sensitivity of grx3grx4 cells to oxidants.
Fig. S2. Effect of exogenous oxidants on C2C12 cell viability.