Abstract
The rapid encoding of contextual memory requires the CA3 region of hippocampus, but the necessary genetic pathways remain unclear. We found that the activity-dependent transcription factor Npas4 regulates a transcriptional program in CA3 that is required for contextual memory formation. Npas4 was specifically expressed in CA3 after contextual learning. Global knockout or selective deletion of Npas4 in CA3 both resulted in impaired contextual memory, and restoration of Npas4 in CA3 was sufficient to reverse the deficit in global knockout mice. By recruiting RNA polymerase II to promoters and enhancers of target genes, Npas4 regulates a learning-specific transcriptional program in CA3 that includes many well-known activity-regulated genes, suggesting that Npas4 is a master regulator of activity-regulated gene programs and is central to memory formation.
The ability to form a long-term memory after a single experience is essential for the survival of higher organisms. In rodents and humans, memory of places or contexts can be formed after a single brief exposure to a novel environment, and this process requires the hippocampus (1, 2). It has been suggested that hippocampal area CA3 is required for rapid encoding of contextual memory (3-6). However, CA3-specific molecular pathways underlying contextual memory formation remain uncharacterized.
The formation and maintenance of long-term memories requires new gene and protein synthesis (7, 8). Learning-induced expression of activity-regulated genes, especially immediate early genes (IEGs), provides a link between behavioral experience and the molecular events required to encode memory (9, 10). Genetic perturbations of IEGs or transcription factors that control activity-regulated gene expression thus often lead to deficits in neuronal plasticity and memory (11-15). However, most IEGs can be induced by a wide range of stimuli and are involved in processes essential to normal cellular function and survival (16, 17), suggesting that their function may not be specific to learning-related neuronal activity. Therefore, identifying IEGs whose function is selectively correlated with both synaptic activity and learning may help to reveal the genetic programs required for memory encoding.
The expression of the activity-dependent transcription factor Npas4 (neuronal PAS domain protein 4) was previously shown to selectively coupled to neuronal activity (18). We therefore investigated whether it regulates a learning-specific transcriptional program underlying the formation of contextual memories.
Npas4 expression is selectively induced by neuronal activity and contextual learning
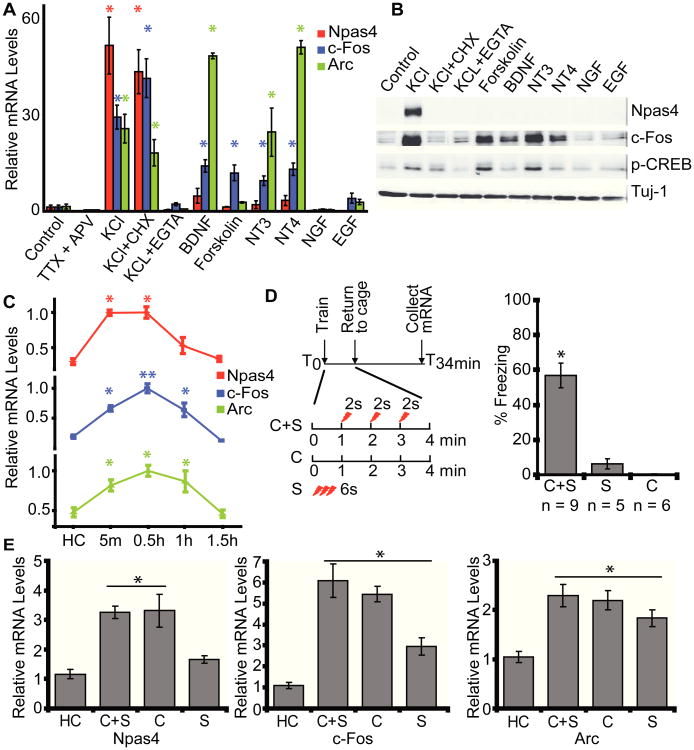
We first characterized the induction of Npas4, together with several other IEGs, in cultured mouse hippocampal neurons. Membrane depolarization resulted in robust protein synthesis-independent expression of Npas4 mRNA, suggesting that Npas4 is an IEG (Fig. 1A). Npas4 was selectively induced by depolarization and Ca2+ influx, but not by activators of several other signaling pathways that induce IEGs, such as c-Fos, Arc (activity-regulated cytoskeleton-associated protein) and Zif268 (Fig. 1A and 1B), similar to previous observations in dissociated rat neurons (18).
Fig. 1. Npas4 expression is selectively induced by neuronal activity in vitro and by learning in vivo.
(A, B) Npas4 mRNA (A) and protein (B) in cultured hippocampal neurons is selectively induced by membrane depolarization and dependent on Ca2+ influx. Induction of Npas4 mRNA does not require new protein synthesis. n = 4 cultures.
(C) Npas4 is rapidly induced after contextual fear conditioning (CFC). Separate groups of mice were sacrificed 5min (n = 8), 30min (n = 9-11), 1hr (n = 6), or 4.5hr (n = 5) after CFC and compared to naive home cage mice (HC, n = 10). Values are plotted relative to peak timepoint.
(D) Experimental scheme and behavioral outcomes of CFC and control conditions. C+S: context + shock; C: context exposure; S: immediate shock; HC: home cage.
(E) Npas4 mRNA is selectively induced by context learning. C+S: n = 8-10; C: n = 8; S: n = 8 and HC: n = 10. All groups were sacrificed 30min after training. *p < 0.001.
To examine experience-induced expression of Npas4, we trained mice in a hippocampus-dependent contextual fear conditioning (CFC) paradigm, which is thought to be dependent on new gene and protein synthesis (8), and examined Npas4 mRNA expression in dorsal hippocampus (DH). We focused on DH on the basis of extensive work showing that DH is required for CFC (19).
Mice were sacrificed at various time points following CFC to measure expression of Npas4, c-Fos, and Arc mRNA using quantitative PCR (qPCR; Fig. 1C). Npas4 mRNA reached its peak level 5 minutes after training and returned to baseline levels 4.5 hours later. c-Fos and Arc reached their peak levels by 30 minutes and returned to baseline levels 4.5 hours after training (Fig. 1C).
Next we trained mice under CFC conditions that provided both context learning and shock association (C+S), or under conditions that involved just context learning (C) or shock (S) alone (Fig. 1D). Both C+S and C represent learning conditions, because the hippocampus forms contextual representations independent of shock delivery (20, 21), but only C+S provides a behavioral readout of learning (Fig. 1D). Immediate shock (S) fails to induce long-term contextual memories, as the context exposure is not long enough for the hippocampus to form a representation (Fig 1D) (20, 22). Therefore, this served as a control condition, allowing us to distinguish IEG induction specific to context learning from induction due to the shock.
Gene expression analysis in mice sacrificed 30 minutes after training indicated that, compared to naive subjects, Npas4 was induced in the C+S and C groups, but not in the S group. In contrast, c-Fos and Arc were significantly induced in all behavioral conditions (Fig. 1E).
Learning and memory deficits in Npas4 global knockout mice
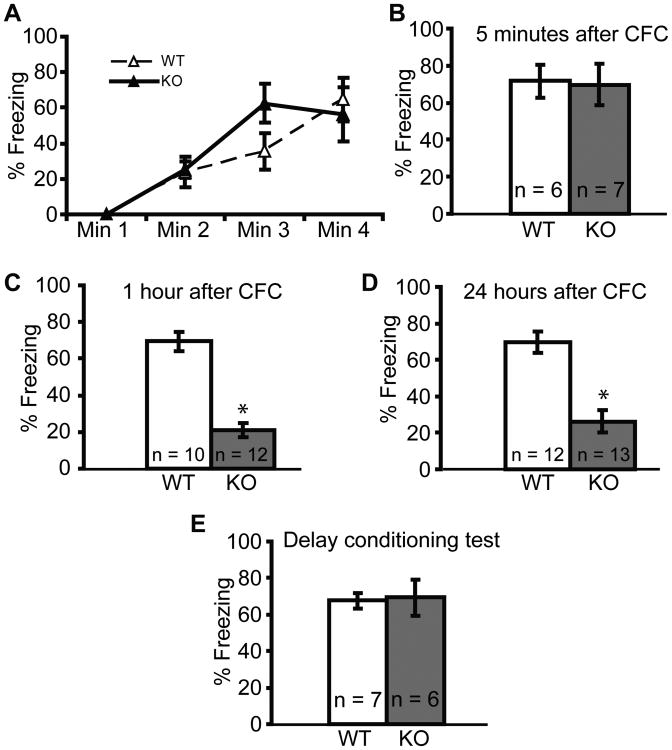
We next determined whether CFC was impaired in Npas4 knockout (Npas4-/-) mice. During the training session and the memory test 5 minutes later, we observed robust freezing behavior in both Npas4-/- and wildtype (Npas4+/+) littermates, suggesting that the ability to acquire CFC is normal in Npas4-/- mice (Fig. 2A and 2B). Furthermore, locomotor activity, anxiety levels, footshock sensitivity, and hippocampal morphology were similar across genotypes (figs. S1 and S2). However, despite having intact memories five minutes after training, freezing elicited by the context 1 hour and 24 hours after training was significantly reduced in Npas4-/- mice (Fig. 2C and 2D), suggesting that both short-term memory (STM) and long-term memory (LTM) are impaired.
Fig. 2. Npas4 global knockout mice exhibit impaired hippocampal-dependent STM and LTM.
(A,B) Npas4-/- and Npas4+/+ littermates exhibit similar freezing during the training session (A) and 5 min after training (B). p = 0.879.
(C,D) 1h (C) and 24h (D) after CFC Npas4-/- mice freeze at a significantly lower level than Npas4+/+ littermates. *p ≤ 0.001.
(E) 24h after auditory delay conditioning, Npas4-/- and Npas4+/+ mice exhibit similar freezing during a tone memory test. p = 0.859.
There is now a general consensus that the amygdala is required for all forms of fear conditioning, while only a subset of fear conditioning paradigms (including CFC) rely on hippocampal integrity (23, 24). We therefore tested whether Npas4-/- mice were deficient in auditory delay conditioning, a form of fear conditioning known to depend on the amygdala but not the hippocampus (24). We saw no difference between Npas4-/- and wildtype mice when tone-induced freezing was measured 24 hours after training, confirming that sensory detection and fear memory acquisition are normal in Npas4-/- mice and suggesting that the impairment we observed in CFC was likely due to a deficit in the hippocampus, and not the amygdala (Fig. 2E).
Selective deletion of Npas4 from CA3, but not CA1, impairs long-term contextual memory
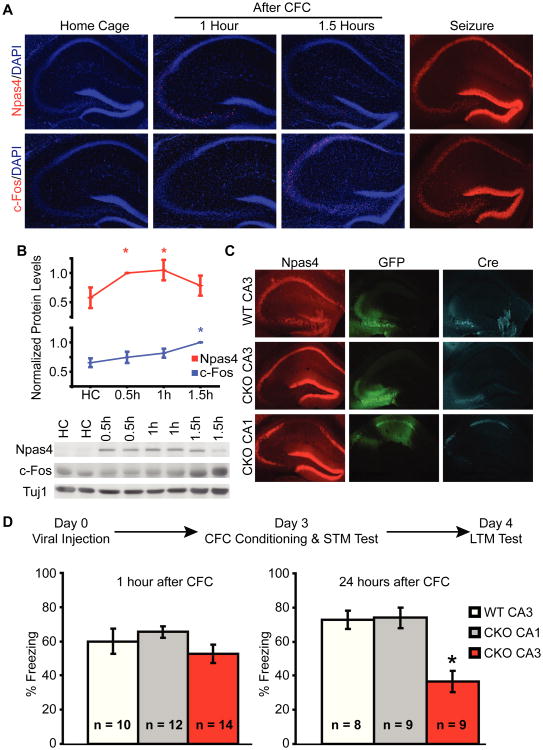
We hypothesized that the memory impairment observed in the global knockout was due to a loss of learning-induced Npas4 expression in DH, based on its selective expression after context learning (Fig. 1E). Because the different subregions within DH may play dissociable roles in contextual memory formation (3), we examined whether CFC resulted in a regionally-selective expression of Npas4. While Npas4 was expressed broadly in several brain regions after CFC, including amygdala and entorhinal cortex (fig. S3), within the hippocampus Npas4 expression after CFC was largely restricted to the CA3 subregion (Fig. 3A), with higher expression in dorsal CA3 than in ventral CA3 (fig. S4). In contrast, c-Fos was robustly expressed in both CA1 and CA3 (Fig. 3A) and similar patterns of induction have been reported for Arc and Zif268 (25, 26). We also noted that the highest level of Npas4 was observed 30 minutes after CFC, 1 hour before the peak expression of c-Fos (Fig. 3B). This observation suggests that pathways activating Npas4 may be distinct from those for other IEGs. Importantly, localized induction of Npas4 in CA3 appears to be specific to contextual learning, because Npas4 was induced in all regions of hippocampus following kainic acid-stimulated seizures (Fig. 3A). While the CA3 region is known to be required for rapid contextual learning, these data are the first to show the selective induction of a specific IEG within CA3 by this form of learning.
Fig. 3. Npas4 expression in CA3 is required for contextual fear conditioning.
(A) Upper panel: Npas4 expression is increased only in CA3 and to a lesser extent in dentate gyrus after CFC. Lower panel: c-Fos expression is induced in all subregions of DH after CFC. Right hand column: seizure induces Npas4 and c-Fos in all subregions of hippocampus.
(B) Western blot analysis of Npas4 and c-Fos expression in DH at various times after CFC. n = 5 mice/condition. Values are plotted relative to peak timepoint. *p < 0.04.
(C) Selective deletion of Npas4 in CA3 or CA1 three days after injecting HSV-Cre.
(D) Selective deletion of Npas4 in CA3 impairs long term CFC memory formation. *p < 0.001.
If induction of Npas4 in CA3 is required for contextual memory, then deleting Npas4 in CA3 should replicate the memory impairments seen in the global knockout (Fig. 2). We acutely deleted Npas4 by stereotaxically injecting a herpes simplex virus (HSV) expressing Cre recombinase (HSV-Cre) into the CA3 region of Npas4 conditional knockout (Npas4flx/flx) mice (Fig. 3C). HSV is naturally neurotropic and reaches peak expression within three days of delivery (27, 28). In another group of mice, we used an equivalent amount of virus to delete Npas4 from a similar volume of cells in dorsal CA1, where we see no activation of Npas4 after CFC (Fig. 3C). To control for any effects of expressing Cre recombinase we injected HSV-Cre into CA3 of wild type mice. Mice were injected with HSV-Cre 3 days before CFC and tested 1 hour and 24 hours after training (Fig. 3D). All animals showed similar freezing during the 1 hour context test. However, 24 hours after training animals with Npas4 deletions in CA3 had attenuated freezing responses compared to animals with Npas4 deletions in CA1 or wild type animals injected with HSV-Cre in CA3 (Fig. 3D).
Npas4 regulates an activity-dependent genetic program that includes several IEGs
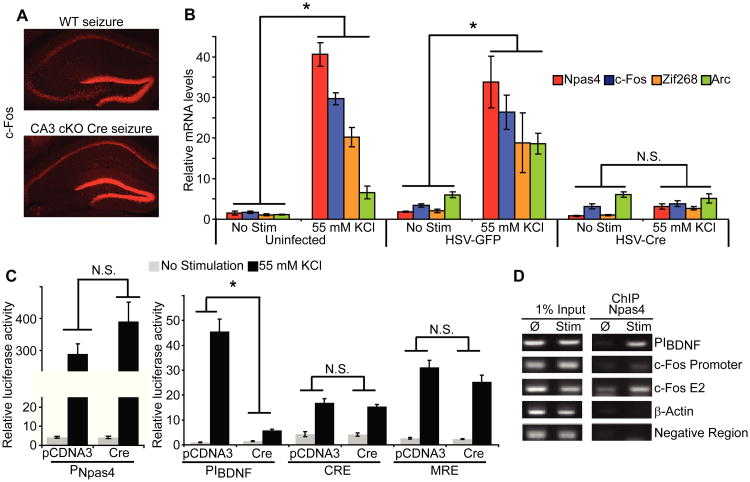
As an activity-dependent transcription factor, Npas4 likely regulates a genetic program that is required for CA3-dependent encoding of contextual memory. Npas4 expression peaks prior to that of several other IEGs (Fig 1C and 3B), and its acute deletion abolished expression of c-Fos (Fig 4A); together these data suggest that Npas4 may regulate the activity-dependent expression of other IEGs. To explore this possibility, we acutely deleted Npas4 in a high percentage of cultured Npas4flx/flx hippocampal neurons by infecting them with HSV-Cre and assayed the mRNA expression of several IEGs following membrane depolarization. Compared to uninfected and HSV-GFP infected controls, deletion of Npas4 abolished depolarization-induced expression of Arc, c-Fos and Zif268 mRNA (Fig. 4B). Expression of the housekeeping gene GAPDH (glyceraldehyde 3-phosphate dehydrogenase) was not altered.
Fig. 4. Npas4 regulates the expression of several IEGs.
(A) Conditional deletion of Npas4 in CA3 results in loss of c-Fos expression.
(B) Dramatic loss of depolarization-induced IEG expression by acute deletion of Npas4 in cultured mouse neurons. n = 3 cultures. *p < 0.001.
(C) The activity of PIBDNF reporter is abolished in the absence of Npas4, while PNpas4, CRE, and MRE reporters show similar induction in the presence or absence of Npas4. n = 4 cultures. *p < 0.001.
(D) ChIP experiments showing that under depolarized conditions Npas4 binds to PIBDNF and enhancer II of c-Fos (c-Fos E2). No binding is observed at the c-Fos promoter, the β-actin promoter, or a negative control region.
Deletion of Npas4 could affect expression of activity-regulated IEGs indirectly, for example by generally disrupting the cellular response to neuronal activity. To examine this possibility, we designed a series of luciferase reporter assays to determine whether other activity-dependent transcriptional pathways function normally in the absence of Npas4. We first characterized transcription from the promoter of Npas4 (PNpas4-Luc), to look at pathways upstream of Npas4. This reporter was induced in response to KCl depolarization but not to activators of other signaling pathways, similar to endogenous Npas4 (fig. S5; compare Fig. 1A and 1B). When Npas4 was acutely deleted by the expression of Cre recombinase in cultured hippocampal neurons generated from Npas4flx/flx mice, activity of PNpas4-Luc in response to KCl depolarization was unchanged (Fig. 4C). We also examined the activity of the transcription factors CREB (cAMP responsive element binding protein) and MEF2 (myocyte enhancer factor-2). Unlike Npas4, these proteins are constitutively expressed, and are activated by post-translational modifications in response to depolarization (29, 30). Reporters expressing luciferase under the control of CREB and MEF2 response elements (CRE and MRE) were unaffected by acute deletion of Npas4 (Fig. 4C).
We then directly determined whether Npas4 binds to the genomic DNA of two activity-regulated genes, BDNF and c-Fos, using chromatin immunoprecipitation (ChIP). These genes are dependent on Npas4 for their expression in response to neuronal activity (Fig. 4A-C and (18)), have well characterized genomic structures (31-35), and have been implicated in learning and memory (14, 36, 37). We examined one of the activity-regulated promoters of BDNF, promoter I (PIBDNF), the proximal promoter region of c-Fos and one of its upstream enhancer regions, E2 (38). After depolarization, Npas4 bound to PIBDNF and c-Fos E2, but not to the c-Fos proximal promoter (Fig. 4D).
Npas4 is required for recruitment of RNA polymerase II to regulatory regions of its target genes
Genome-wide ChIP-sequencing has revealed that Npas4 co-localizes with RNA polymerase II (Pol II) at enhancer and promoter sites of many activity-regulated genes, including BDNF and c-Fos (38). However, it is not known whether this co-localization plays an important role in regulating transcription of these genes. We hypothesized that Npas4 is required for activity-dependent recruitment of Pol II to promoter and enhancer regions of its targets, in order to activate their transcription.
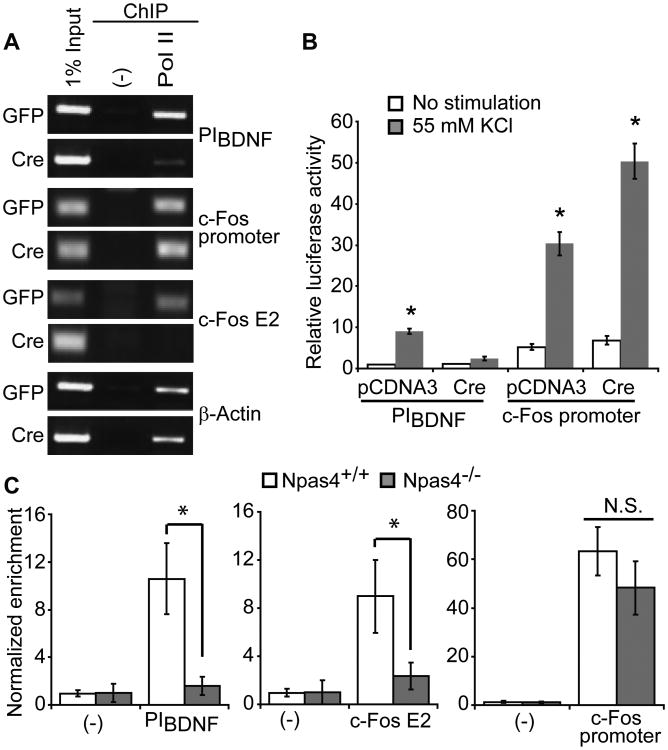
We acutely deleted Npas4 using HSV-Cre in a high percentage of cultured Npas4flx/flx cortical neurons and then performed ChIP for Pol II after 2 hours of membrane depolarization. In control neurons infected with HSV-GFP, Pol II localized to PIBDNF, the c-Fos enhancer E2, the c-Fos promoter region, and the β-actin promoter after depolarization (Fig. 5A). When Npas4 was deleted by HSV-Cre, localization of Pol II to PIBDNF and c-Fos E2 was impaired (Fig. 5A). As we described above, Npas4 binds to both of these regions. Pol II binding to the promoter regions of c-Fos and β-actin, where we did not observe Npas4 binding (Fig. 4D), was not affected by deletion of Npas4. To confirm that the Npas4-dependent binding of Pol II is important for gene expression, we compared luciferase reporters driven by PIBDNF and the c-Fos promoter and found that expression from PIBDNF was abolished by deletion of Npas4, while expression from the c-Fos promoter was not attenuated (Fig. 5B).
Fig. 5. Npas4 is required for the recruitment of RNA polymerase II to enhancer and promoter regions of activity-regulated genes.
(A) Localization of Pol II to PIBDNF and c-Fos enhancer region E2 is dependent on Npas4. No change is observed in Pol II binding at the c-Fos or β-actin promoter.
(B) Depolarization-induced activity of PIBDNF reporter is abolished in the absence of Npas4, but the c-Fos promoter reporter is unaffected. n = 4 cultures. *p < 0.001.
(C) qPCR analysis of ChIP samples from seized Npas4-/- and Npas4+/+ littermates. Pol II binding to PIBDNF (*p ≤ 0.008, n=7/genotype) and c-Fos E2 (*p ≤ 0.048, n=6/genotype) is diminished in Npas4-/- mice relative to Npas4+/+ littermates. No change is observed in Pol II binding at the c-Fos promoter (p = 0.333, n = 6/genotype).
To confirm our findings in vivo, we performed ChIP for Pol II from hippocampal tissue extracted from adult Npas4+/+ and Npas4-/- littermates. Npas4 is expressed only in a sparse population of neurons following CFC (Fig. 3A), making it difficult to detect Pol II binding in these cells. We therefore used kainic acid-induced seizures to activate all neurons in order to determine the genomic localization of Pol II in vivo. Seizure has been shown to robustly induce activity-regulated genes, many of which have been implicated in memory formation, and under certain conditions can induce potentiation similar to LTP (39). In line with our in vitro observations, localization of Pol II to PIBDNF and c-Fos E2 was impaired in Npas4-/- mice compared to Npas4+/+ littermates, while Pol II binding to the promoter regions of c-Fos and β-actin was similar across genotypes (Fig. 5C and fig. S6).
Expression of Npas4 in CA3 rescues transcription and memory formation in global knockouts
We next investigated whether re-expressing Npas4 in CA3 of Npas4-/- mice leads to expression of its genetic program and consequently rescues memory formation.
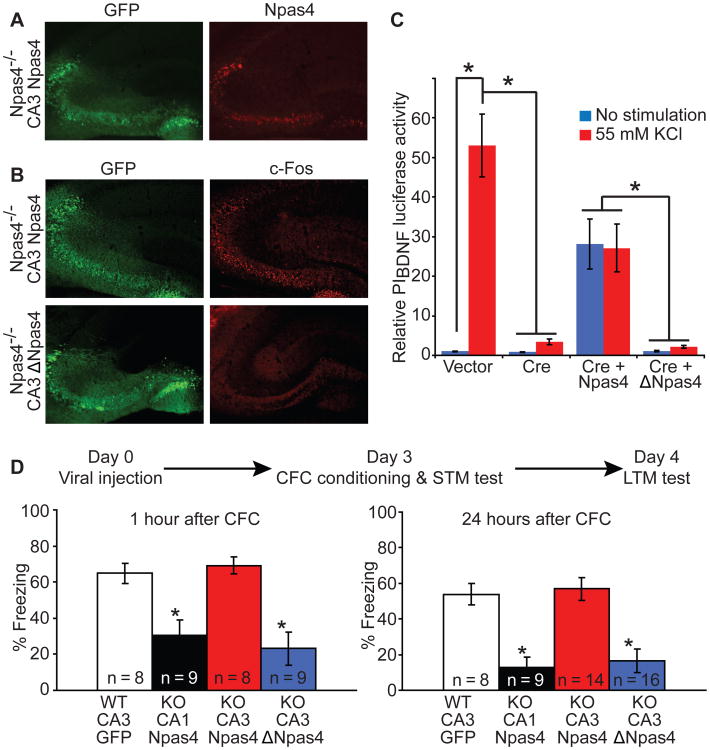
The CA3 region of Npas4-/- mice was infected with HSV expressing Npas4 (HSV-Npas4) (Fig. 6A) and activation of Npas4 gene targets was examined using immunostaining. HSV-Npas4 induced the expression of c-Fos (Fig. 6B), but a transcriptionally inactive version of Npas4 (ΔNpas4) did not, confirming that the transcription activation ability of Npas4 is required. We also tested whether expression of Npas4 is sufficient to induce BDNF by measuring the activity of a PIBDNF reporter construct in vitro. We transfected Cre into Npas4flx/flx neurons and found that activity of the PIBDNF reporter was abolished. Co-transfecting Npas4, but not ΔNpas4, rescued the activity of the PIBDNF reporter (Fig. 6C).
Fig. 6. Acute expression of Npas4 in CA3 reverses short-term and long-term memory deficits observed in Npas4-/- mice.
(A) Expression of Npas4 in CA3 of Npas4-/- mice using HSV-Npas4.
(B) Npas4, but not ΔNpas4, restores expression of c-Fos in vivo.
(C) Expression of Npas4, but not ΔNpas4, drives activity of PIBDNF independent of KCl depolarization. n = 4 cultures. *p ≤ 0.001.
(D) Npas4-/- mice with Npas4 injected into CA3 freeze at similar levels to Npas4+/+ mice injected with GFP 1hr and 24hrs after training. CA1 injection of Npas4 or CA3 injection of ΔNpas4 did not reverse the memory deficit. *p < 0.001.
We then determined whether expressing Npas4 in CA3 of the global knockout mice is sufficient to restore the ability to form long-term contextual memories. The use of HSV allowed us to acutely express Npas4, with a peak expression 3 days after injection (27, 28). Mice were injected with virus, trained three days after injection, and tested 1 hour and 24 hours after training (Fig. 6D). Expressing Npas4 in CA3 completely reversed both the short-term and long-term contextual memory deficits observed in the global knockouts, since Npas4 knockout mice with HSV-Npas4 injected into CA3 showed similar freezing behavior to wild type control animals injected with GFP. Global knockouts with HSV-Npas4 delivered to CA1 showed no such recovery (Fig. 6D). Expressing ΔNpas4 in CA3 failed to overcome the memory deficits in Npas4-/- mice, confirming that activation of the genetic program regulated by Npas4 is required for rescue of memory formation.
Discussion
We have identified a genetic pathway in CA3 required for rapid encoding of hippocampal-dependent contextual memory. While several studies have identified CA3 function and output as essential to the encoding of contextual information (3-6), very little is known about the molecular mechanisms underlying this process. We found that acute deletion of Npas4 from CA3 resulted in a dramatic reduction in IEG expression and impaired contextual memory formation, and that expression of transcriptionally active Npas4 in CA3 was sufficient to restore both IEG expression and memory formation in the global knockout. Additionally, we found that expression of Npas4 in CA1 is neither necessary nor sufficient for contextual memory formation. While our viral strategy cannot target all of CA1, these findings are in line with other studies using transgenic mouse lines targeting CREB in CA1 (40, 41), but see (42)). Our data indicate that regulation of a transcriptional program by Npas4 is a mechanism through which CA3 supports the rapid acquisition and consolidation of contextual information.
Activity-dependent gene expression is thought to be required for LTM, but not for STM (8). We observed a STM deficit in the Npas4 global knockout mice, but not in the conditional CA3 knockout (Fig. 3C). Although the STM impairment could be due to a developmental deficit caused by germline deletion of Npas4 in the global knockout, the rescue by acute expression of Npas4 argues against this explanation. It is possible that basal levels of neuronal activity maintain a low level of Npas4, which in turn provides a moderate level of the downstream molecules required for STM. Then, perhaps, while acute deletion of Npas4 does not reduce the level of those genes below that required for STM, chronic deletion in the global knockout results in insufficient levels to support STM.
It is intriguing that Npas4 global knockout mice function normally in auditory delay conditioning, which is hippocampus-independent but amygdala-dependent, because long-term memory formation in the amygdala is thought to be dependent on activity-regulated gene expression. We observed that the expression of Npas4 gene targets is attenuated in the Npas4 global knockout, but not to the degree that was observed in the conditional deletion (fig. S7), suggesting that compensatory pathways may result in some expression of target IEGs. Conceivably these pathways are sufficient to support memory formation in the amygdala, but IEG expression fails to reach a level sufficient to support the hippocampal learning required for CFC. Alternatively, or additionally, the activity-regulated genetic program induced through compensating pathways independent of Npas4, although including certain IEGs such as c-Fos and BDNF, may not contain all the components necessary for CFC. It seems likely that acute deletion of Npas4 in the amygdala will result in impairment of auditory delay conditioning.
Our findings suggest the possibility of a hierarchical genetic program in which Npas4 is upstream of several activity-regulated genes. However, Npas4 itself is regulated by activity at the mRNA level and though it reaches peak expression slightly earlier than other rapidly responding IEGs (Fig 1C and Fig. 3B), it is unclear whether Npas4 protein is synthesized quickly enough to initiate the first wave of IEG expression. It seems more likely that Npas4, through the recruitment of Pol II, only enhances and sustains IEG expression at later time points, as suggested recently for Npas4-dependent regulation of BDNF transcripts (43).
The mechanism by which Npas4 affects Pol II recruitment to its target genes is not immediately obvious. It could directly recruit Pol II to genomic regions in a manner similar to CBP, or it could be indirectly involved through interactions with other proteins, such as CREB (44, 45).
Our previous work identified a role for Npas4 in the activity-dependent regulation of inhibitory synapse development (18). Thus the genetic program controlled by Npas4 may be involved in contextual memory formation, at least in part, through the modulation of inhibitory synapses in the hippocampal circuit. Consistent with this idea, learning-induced increases in inhibitory synaptic transmission have recently been reported in the hippocampus (46, 47).
We have focused here on the role of Npas4 in hippocampus-dependent contextual learning, but the genetic program regulated by this transcription factor likely contributes to several other experience-dependent processes. We hope to leverage the function of Npas4 in order to dissect specific neural circuits actively engaged in information processing in order to better understand the molecular and cellular mechanisms underlying learning and memory.
Supplementary Material
Acknowledgments
We thank C. M. Fletcher, A. Heynen, C. Jennings, R. Desimone, M. Baratta, J. Biedenkapp, R. Froemke, J. Czerniawski, and D. Bambah-Mukku for critical reading of the manuscript. We also thank G. Hale and M. Wilson for help characterizing the Npas4-/- mice and S. Ramirez for help with the open field assay. Y.L. acknowledges the generous support of the McGovern Institute for Brain Research at MIT. This work was supported by the MIT Presidential Marcus Fellowship to Honor Norman B. Leventhal (K.R.), a postdoctoral fellowship from the MIT Simons Initiative on Autism and the Brain (G.M.B), NSF Grant IOS 0919159 (T.O.), and by a Whitehall Foundation research grant, an Anonymous Foundation research grant, the John Merck Scholar Program, and NIH grant MH091220-01 (Y.L.).
References and Notes
- 1.Neves G, Cooke SF, Bliss TV. Nat Rev Neurosci. 2008;9:65. doi: 10.1038/nrn2303. [DOI] [PubMed] [Google Scholar]
- 2.Scoville WB, Milner B. J Neuropsychiatry Clin Neurosci. 2000;12:103. doi: 10.1176/jnp.12.1.103. [DOI] [PubMed] [Google Scholar]
- 3.Kesner RP. Learn Mem. 2007;14:771. doi: 10.1101/lm.688207. [DOI] [PubMed] [Google Scholar]
- 4.Nakashiba T, Young JZ, McHugh TJ, Buhl DL, Tonegawa S. Science. 2008;319:1260. doi: 10.1126/science.1151120. [DOI] [PubMed] [Google Scholar]
- 5.Nakazawa K, et al. Neuron. 2003;38:305. doi: 10.1016/s0896-6273(03)00165-x. [DOI] [PubMed] [Google Scholar]
- 6.Lee I, Kesner RP. Hippocampus. 2004;14:301. doi: 10.1002/hipo.10177. [DOI] [PubMed] [Google Scholar]
- 7.Davis HP, Squire LR. Psychol Bull. 1984;96:518. [PubMed] [Google Scholar]
- 8.Alberini CM. Physiol Rev. 2009;89:121. doi: 10.1152/physrev.00017.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tischmeyer W, Grimm R. Cell Mol Life Sci. 1999;55:564. doi: 10.1007/s000180050315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kubik S, Miyashita T, Guzowski JF. Learn Mem. 2007;14:758. doi: 10.1101/lm.698107. [DOI] [PubMed] [Google Scholar]
- 11.Guzowski JF. Hippocampus. 2002;12:86. doi: 10.1002/hipo.10010. [DOI] [PubMed] [Google Scholar]
- 12.Jones MW, et al. Nat Neurosci. 2001;4:289. doi: 10.1038/85138. [DOI] [PubMed] [Google Scholar]
- 13.Plath N, et al. Neuron. 2006;52:437. doi: 10.1016/j.neuron.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 14.Fleischmann A, et al. J Neurosci. 2003;23:9116. doi: 10.1523/JNEUROSCI.23-27-09116.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kida S, et al. Nat Neurosci. 2002;5:348. doi: 10.1038/nn819. [DOI] [PubMed] [Google Scholar]
- 16.Lonze BE, Riccio A, Cohen S, Ginty DD. Neuron. 2002;34:371. doi: 10.1016/s0896-6273(02)00686-4. [DOI] [PubMed] [Google Scholar]
- 17.Ramanan N, et al. Nat Neurosci. 2005;8:759. doi: 10.1038/nn1462. [DOI] [PubMed] [Google Scholar]
- 18.Lin Y, et al. Nature. 2008;455:1198. doi: 10.1038/nature07319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fanselow MS, Dong HW. Neuron. 2010;65:7. doi: 10.1016/j.neuron.2009.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rudy JW, O'Reilly RC. Behav Neurosci. 1999;113:867. doi: 10.1037//0735-7044.113.5.867. [DOI] [PubMed] [Google Scholar]
- 21.Fanselow MS. Behav Brain Res. 2000;110:73. doi: 10.1016/s0166-4328(99)00186-2. [DOI] [PubMed] [Google Scholar]
- 22.Landeira-Fernandez J, DeCola JP, Kim JJ, Fanselow MS. Behav Neurosci. 2006;120:873. doi: 10.1037/0735-7044.120.4.873. [DOI] [PubMed] [Google Scholar]
- 23.Maren S. Eur J Neurosci. 2008;28:1661. doi: 10.1111/j.1460-9568.2008.06485.x. [DOI] [PubMed] [Google Scholar]
- 24.Anagnostaras SG, Gale GD, Fanselow MS. Hippocampus. 2001;11:8. doi: 10.1002/hipo.1051. [DOI] [PubMed] [Google Scholar]
- 25.Guzowski JF, McNaughton BL, Barnes CA, Worley PF. Nat Neurosci. 1999;2:1120. doi: 10.1038/16046. [DOI] [PubMed] [Google Scholar]
- 26.Lonergan ME, Gafford GM, Jarome TJ, Helmstetter FJ. Neural Plast. 2010;2010:139891. doi: 10.1155/2010/139891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrot M, et al. Proc Natl Acad Sci U S A. 2002;99:11435. doi: 10.1073/pnas.172091899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han JH, et al. Science. 2007;316:457. doi: 10.1126/science.1139438. [DOI] [PubMed] [Google Scholar]
- 29.Kornhauser JM, et al. Neuron. 2002;34:221. doi: 10.1016/s0896-6273(02)00655-4. [DOI] [PubMed] [Google Scholar]
- 30.Mao Z, Bonni A, Xia F, Nadal-Vicens M, Greenberg ME. Science. 1999;286:785. doi: 10.1126/science.286.5440.785. [DOI] [PubMed] [Google Scholar]
- 31.Coulon V, Chebli K, Cavelier P, Blanchard JM. PLoS One. 2010;5:e11235. doi: 10.1371/journal.pone.0011235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheng M, Dougan ST, McFadden G, Greenberg ME. Mol Cell Biol. 1988;8:2787. doi: 10.1128/mcb.8.7.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Treisman R. Cell. 1985;42:889. doi: 10.1016/0092-8674(85)90285-5. [DOI] [PubMed] [Google Scholar]
- 34.Treisman R. Cell. 1986;46:567. doi: 10.1016/0092-8674(86)90882-2. [DOI] [PubMed] [Google Scholar]
- 35.Wagner BJ, Hayes TE, Hoban CJ, Cochran BH. Embo J. 1990;9:4477. doi: 10.1002/j.1460-2075.1990.tb07898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minichiello L. Nat Rev Neurosci. 2009;10:850. doi: 10.1038/nrn2738. [DOI] [PubMed] [Google Scholar]
- 37.Lu Y, Christian K, Lu B. Neurobiol Learn Mem. 2008;89:312. doi: 10.1016/j.nlm.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim TK, et al. Nature. 2010;465:182. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ben-Ari Y, Represa A. Trends Neurosci. 1990;13:312. doi: 10.1016/0166-2236(90)90135-w. [DOI] [PubMed] [Google Scholar]
- 40.Balschun D, et al. J Neurosci. 2003;23:6304. doi: 10.1523/JNEUROSCI.23-15-06304.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pittenger C, et al. Neuron. 2002;34:447. doi: 10.1016/s0896-6273(02)00684-0. [DOI] [PubMed] [Google Scholar]
- 42.Athos J, Impey S, Pineda VV, Chen X, Storm DR. Nat Neurosci. 2002;5:1119. doi: 10.1038/nn951. [DOI] [PubMed] [Google Scholar]
- 43.Pruunsild P, Sepp M, Orav E, Koppel I, Timmusk T. J Neurosci. 2011;31:3295. doi: 10.1523/JNEUROSCI.4540-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakajima T, et al. Cell. 1997;90:1107. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 45.Kwok RP, et al. Nature. 1994;370:223. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 46.Cui Y, et al. Cell. 2008;135:549. doi: 10.1016/j.cell.2008.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruediger S, et al. Nature. 2011;473:514. doi: 10.1038/nature09946. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.