SUMMARY
Long noncoding RNAs (lncRNAs) are thought to be prevalent regulators of gene expression, but the consequences of lncRNA inactivation in vivo are mostly unknown. Here we show that targeted deletion of mouse Hotair lncRNA leads to de-repression of hundreds of genes, resulting in homeotic transformation of the spine and malformation of metacarpal-carpal bones. RNA-seq and conditional inactivation reveal an ongoing requirement of Hotair to repress HoxD genes and several imprinted loci such as Dlk1-Meg3 and Igf2-H19, without affecting imprinting choice. Hotair binds to both Polycomb repressive complex 2 that methylates histone H3 at lysine 27 (H3K27) and Lsd1 complex that demethylates histone H3 at lysine 4 (H3K4) in vivo. Hotair inactivation causes H3K4me3 gain and, to a lesser extent, H3K27me3 loss at target genes. These results reveal the function and mechanisms of Hotair lncRNA to enforce silent chromatin state at Hox and additional genes.
INTRODUCTION
Long noncoding RNAs (lncRNAs) are pervasively transcribed in mammalian genomes [reviewed by (Rinn and Chang, 2012)]. Thousands of lncRNA species have been reported, but their in vivo functions are mostly unknown. Some lncRNAs act at the interface between the genome and chromatin modification machinery, such as Xist and Air that recruit repressive chromatin modifications to silence nearby genes in cis for dosage compensation and imprinting (Lee, 2009; Nagano et al., 2008). Other lncRNAs, such as HOTAIR and linc-p21, act in trans to guide silencing complexes to sites throughout the genome (Huarte et al., 2010; Rinn et al., 2007). Histone H3 lysine 27 trimethylation (H3K27me3) mediates developmental silencing while histone H3 lysine 4 trimethylation (H3K4me3) is associated with transcriptional activation. Human HOTAIR, a 2.2 kb RNA transcribed from the HOXC locus, binds both Polycomb repressive complex 2 (PRC2) and LSD1 complexes, and recruits them to hundreds of genomic sites to promote coordinated H3K27 methylation and H3K4 demethylation, respectively, for gene silencing (Chu et al., 2011; Rinn et al., 2007; Tsai et al., 2010). HOTAIR silences human HOXD genes, a function that is believed to contribute to cell positional identity (Rinn et al., 2007), and overexpression of HOTAIR in several types of human cancers have been linked to metastasis and cancer progression (Gupta et al., 2010; Kim et al., 2012; Kogo et al., 2011). HOTAIR has been considered a prototype of lncRNA-guided chromatin modification that typifies a large class of lncRNAs associated with PRC2 and other chromatin modification complexes (Khalil et al., 2009; Zhao et al., 2010).
The evolutionary conservation of lncRNA sequence and function is potentially distinct from that of protein coding genes (Derrien et al., 2012; Ulitsky et al., 2011). While lncRNAs show greater sequence conservation than introns, even functionally redundant lncRNAs exhibit only limited sequence identity (e.g. roX RNAs in Drosophila). LncRNAs show greater conservation of genomic synteny than sequence identity (Ulitsky et al., 2011). These findings raise the possibility that functional lncRNAs may quickly arise in evolution, but also suggest that understanding lncRNA function across evolution likely requires direct experimental analysis. Mouse Hotair (hereafter Hotair) is a lncRNA transcribed from the syntenic location in the HoxC locus, and is expressed in posterior or distal anatomic sites (Rinn et al., 2007; Schorderet and Duboule, 2011). Analysis of a large deletion of mouse HoxC locus [HoxCΔ (Suemori and Noguchi, 2000), which includes Hotair] found little change in HoxD gene expression or chromatin state, which led to the interpretation that mouse and human Hotair are functionally distinct (Schorderet and Duboule, 2011). However, because HoxCΔ also removes eight HoxC genes, two microRNAs, and additional lncRNAs, its use to assign function to Hotair may be less than ideal. Here, we generate and analyze the targeted deletion of Hotair, and discover its function in modulating the chromatin state and gene expression of HoxD and imprinted genes in vivo.
RESULTS
Hotair knockout causes homeotic transformation and skeletal malformation
We first examined Hotair expression in developing embryos at embryonic day 11.5, 12.5 and 13.5 stages by in situ hybridization. Hotair is specifically expressed in the posterior trunk and distal limb bud (Figure S1A), as previously described (Rinn et al., 2007); (Schorderet and Duboule, 2011). Hotair is expressed from somites 33–34 onward posteriorly, corresponding to the developing lumbosacral anatomical region. Hotair is also expressed in specific mesenchymal cells and condensates in E11.5 and E15.5 forelimb including wrist and digital condensates (Figures S1B and S1C). The site-specific expression pattern of Hotair suggests potential roles in vertebrae and wrist morphogenesis during development.
To understand the functions of mouse Hotair, we generated conditional and constitutive knockout alleles of the Hotair locus (Figure 1A, Methods). We introduced LoxP sites to flank exons 1 and 2, which comprise the entirety of the known Hotair transcript; crossing to HPRT-Cre mice (activating cre in female germline) yielded targeted deletion of the locus (hereafter “Hotair KO”), and resulted in no detectable Hotair RNA expression (Figures 1B, S1D). Hotair KO allele was backcrossed to C57/BL6 background for six generations, yielding over 99% C57/BL6 background as confirmed by strain- specific SNP array analysis (Methods). Heterozygous intercrosses generated Hotair wild type and KO littermates for comparison.
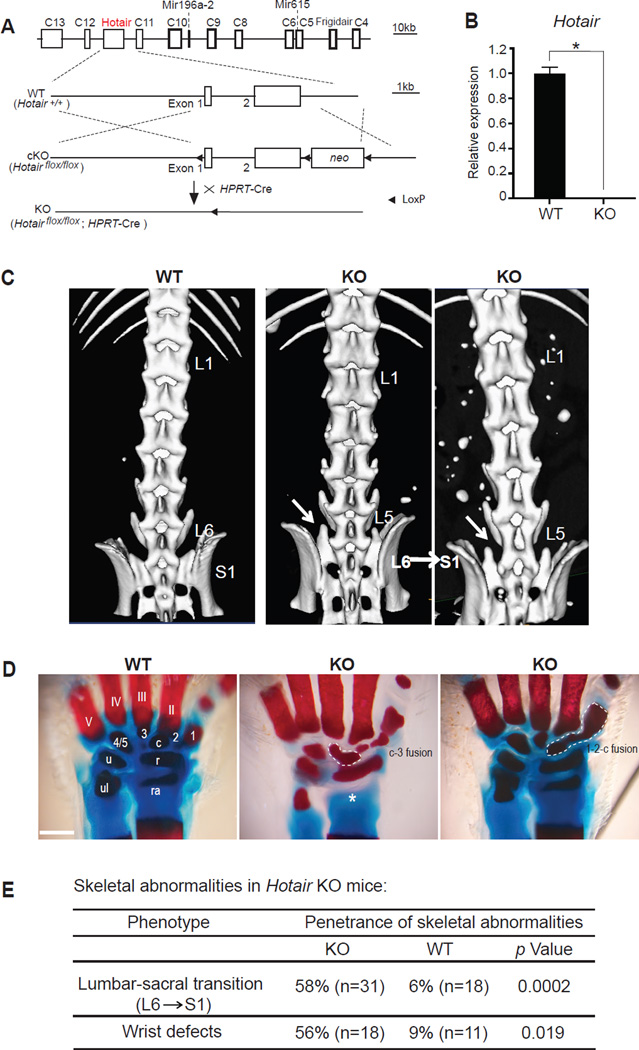
Figure 1. Hotair knockout causes homeotic transformations.
(A) Schematic of Hotair conditional knockout (cKO) allele. Arrowhead, Loxp site.
(B) qRT-PCR confirms loss of Hotair expression in KO TTF.
(C) Micro-CT scans showing “L6a→S1” homeotic transformation of the lumbar vertebrae, which resulted in losing the 6th lumbar and having structurally deformed 1st sacral vertebrae (arrow) in Hotair KO mice. L, lumbar vertebrae; S, sacral vertebrae.
(D) Alizarin Red-Alcian Blue staining showing the deformed wrist bones in KO mice. Digits (II-V), carpal elements (1, 2, 3, 4/5), central element (c), radiale (r), ulnare (u), radius (ra) and ulna (ul). Note the fusion of carpal elements c-3, and 1-2-c (circled area), and missing radius (asterisk) in KOs. 4/5 are always naturally fused in WT wrist. (Scale bar: 0.5mm)
(E) Summary of skeletal abnormalities in Hotair KO mice. Phenotype penetrance, number (n) of animals examined, and p-values (Fisher’s exact test) are indicated.
See also Figure S1.
Homozygous Hotair KO animals were viable and fertile, but showed three notable skeletal phenotypes (Figures 1C–1E, Figures S1E and S1F). First, in the C57/BL6 genetic background, wild type littermates possess six lumbar vertebrae (L1 to L6); whereas 58% of Hotair KO mice have five lumbar vertebrae (p=0.0002, Fisher’s exact test). Micro-scale computed tomography revealed that the sacral 1(S1) vertebrae in Hotair KO still had the lateral processes typical of L6 vertebrae, indicative of a L6 to S1 transformation (Figure 1C). These micro-CT findings were confirmed by Alcian blue staining of the vertebral skeleton (Figure S1E). Second, detailed examination of the limb skeleton also revealed a majority of KOs (56% vs 9% in WT) with abnormalities in the metacarpal and carpal bones, including fusions and missing bony elements (Figures 1D and 1E). The spine and wrist abnormalities do not necessarily co-occur in individual animals; hence up to 78% of KO animals exhibit one or more of these abnormalities. Third, Hotair KO animals exhibited a subtle but fully penetrant transformation of the caudal 4 vertebrae (Figures 1E and S1F). These phenotypes were robust through all the backcrosses, and we never saw the phenotype segregate independently from the Hotair allele. These results suggest that Hotair, first identified in the context of adult skin positional identity (Rinn et al., 2007), is also important for embryonic patterning of the skeletal system in vivo.
Gene de-repression in Hotair knockout cells and embryos
We analyzed gene expression patterns in Hotair KO to gain insights into the molecular basis for the observed phenotypes. While Hotair expression is segmentspecific and heterogeneous in embryos and mouse embryonic fibroblasts (MEFs), we found that primary tail tip fibroblasts (TTFs, derived from a posterior site where Hotair is highly expressed) maintained position-specific and consistent Hotair expression (Figures S2A and S2B). Single cell analysis shows that the vast majority of individual TTF cells (83%) express Hotair while only 28% of MEF cells express Hotair (Figure S2C). The enrichment of a relatively pure population of Hotair+ cells is ideal to address the impact of Hotair KO at the molecular level. Previous studies with human HOTAIR used RNA interference, which were not able to fully deplete HOTAIR (Rinn et al., 2007; Tsai et al., 2010). Hence, the consequences of targeted and complete Hotair inactivation on gene expression are not known. RNA-seq of TTFs derived from wild type, heterozygous, and Hotair KO mice revealed significant differences in expression (Figure 2A). Validaton by microarray analysis and qRT-PCR of TTFs from independent animals yielded genes with consistent expression changes in multiple platforms that we consider Hotair-dependent target genes (Figure S2D).
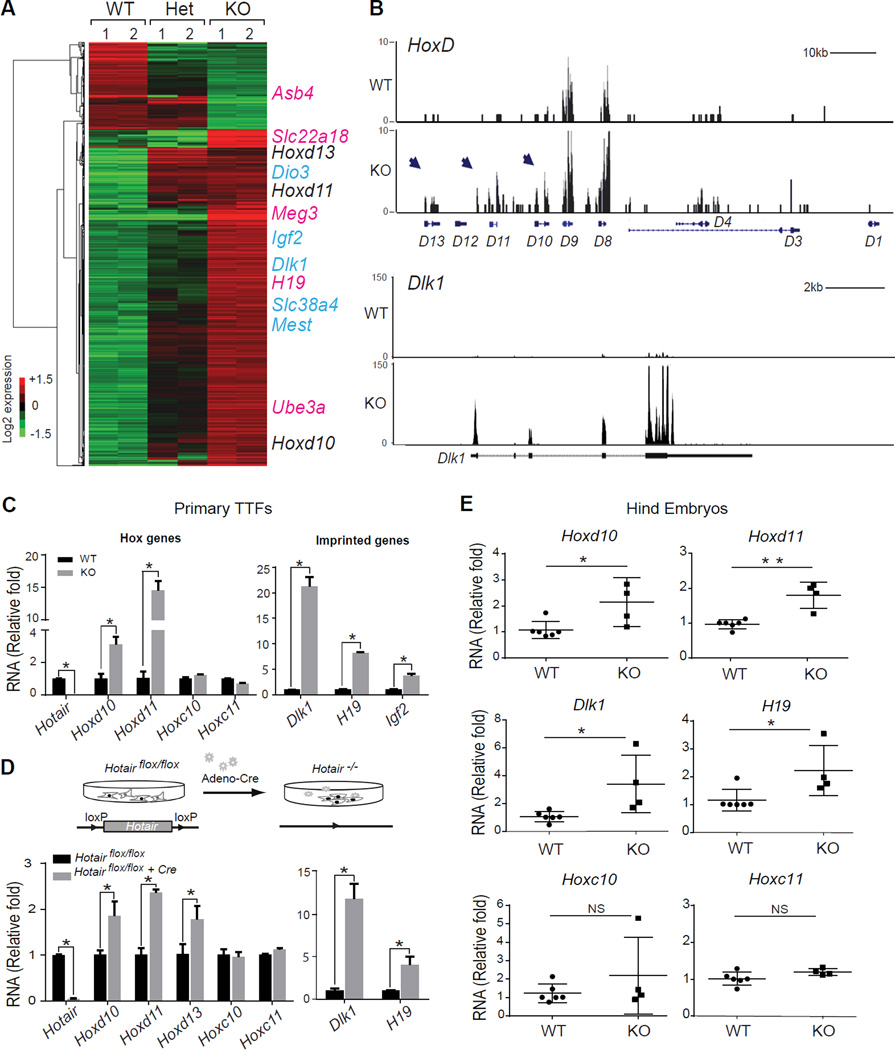
Figure 2. Hotair knockout de-represses HoxD and imprinted genes.
(A) Differential gene expression by RNA-seq in Hotair WT, heterozygous (Het), and KO TTF. Each row is a transcript; each column is a sample. De-repressed HoxD (black) and imprinted genes (maternally expressed, pink; paternally expressed, blue) are indicated.
(B) RNA-seq data of HoxD and Dlk1 loci. Distal HoxD genes (D13, D11 and D10, arrow) and Dlk1 were derepressed in KO cells. x-axis showing the genomic coordinate; y-axis showing the normalized RNA-seq signals. Box represents known mRNA exons.
(C) qRT-PCR of indicated genes in Hotair KO cells or (D) after acute Hotair deletion in cKO cells. Top: schematic of acute deletion assay.
(E) qRT-PCR of indicated genes in Hotair KO embryos. The hind portions of E13.5 embryos from the same litters were analyzed (n>3). Mean± s.d are shown for all panels;
*, p<0.05; **, p<0.01 by student's t-test (n>3). NS, not significant.
See also Figure S2.
Hotair KO resulted in predominantly de-repression of gene expression. Approximately 80% of the gene expression changes are increased in the KO cells, suggesting that Hotair functions primarily as a repressor, either directly or indirectly. Prominent among the de-repressed genes are several HoxD genes, including Hoxd10, Hoxd11, and Hoxd13 (Figures 2A and 2B). Multiple HoxC genes are expressed and well detected, but no significant difference in HoxC expression level was observed despite the fact that Hotair is embedded in the HoxC locus (Figure S2F). In addition, gene expression from HoxA and HoxB loci was not significantly affected (Figure S2F). Thus, similar to human HOTAIR, mouse Hotair appears to be a trans-acting regulator of gene expression.
Moreover, Hotair KO increased the expression of approximately 30 genes from imprinted loci. These include paternally expressed genes Dlk1, Dio3, Igf2, Mest, Slc38a4 (Figure 2A, blue) and maternally expressed genes H19, Meg3 (also known as Gtl2), (Figure 2A, pink). Genes surrounding the imprinted loci were not affected (Figure 2B, Figure S2F). Although only a minority of all known imprinted genes was de-repressed, we noted that the affected genes tend to be clustered in chromosomal loci (Figure S2F). For example, Dlk1, Meg3, and Dio3 reside in the same imprinted locus on mouse chromosome 12 (da Rocha et al., 2009; Takahashi et al., 2009). Notably, partial de-repression of HoxD genes are observed in Hotair +/− cells, whereas derepression of imprinted genes was only observed in the Hotair KO cells. This result suggests that the effect of Hotair on HoxD genes may be dose-dependent, and potentially explains why prior studies knocking down HOTAIR did not observe effects on imprinted genes.
Genes with altered expression had significant enrichment for Gene Ontology terms related to transcriptional regulation, cell proliferation, and development (p<0.05 for each, FDR < 0.05, Figure S2E). Quantitative reverse transcription-PCR (qRT-PCR) of independent TTF cells from WT and Hotair KO mice confirmed the de-repression of multiple HoxD and imprinted genes, but no significant changes in Hoxc10 and Hoxc11 (Figure 2C). Hoxd11 is induced over 10-fold and is the most strongly de-repressed gene among the HoxD genes. Dlk1, H19, and Igf2 are also de-repressed from 5- to over 20- fold in Hotair KO tissue. Fluorescence-activated cell sorting and immunofluorescence confirmed the increased synthesis of Dlk1 and Igf2 proteins in Hotair KO cells (Figures S2G and S2H).
To determine whether ongoing Hotair function is required for proper gene expression, we studied the consequences of inducible acute deletion of Hotair. Introduction of cre-expressing adenovirus into Hotairflox/flox TTFs led to high efficiency deletion and silencing of Hotair expression (Figure 2D). Introduction of control adenovirus served as negative control. Acute deletion of Hotair led to de-repression of HoxD and imprinted genes after 3–5 passages, albeit with lower-fold effect than the constitutive Hotair KO; while expression of Hoxc10, Hoxc11 were not affected (Figure 2D). The acute genetic deletion in isogenic cells rules out potential background effects, and suggests an ongoing requirement of Hotair for proper expression of its target genes in trans. Quantitative RT-PCR analysis of posterior and distal embryonic tissues, where endogenous Hotair is normally expressed confirmed de-repression of HoxD and imprinted genes in Hotair KO embryos without significant changes in HoxC genes (Figure 2E).
Hotair knockout alters spatial pattern of gene expression in vivo
Hotair KO alters both the spatial pattern as well as the quantitative levels of gene expression. Hox genes are expressed along the anterior-posterior and proximal-distal axis in a nested, segmental fashion related to their positions on the chromosomes (Krumlauf, 1994). Hence, 5’ HoxD genes are expressed posteriorly and distally, a pattern that requires proper chromatin-based silencing mechanisms such as Polycomb (Soshnikova and Duboule, 2009). Hotair KO embryos showed anterior expansion of the Hoxd10, and Hoxd11 expression domain in the trunk compared to wild type littermates, an effect that is recognizable starting at E12.5 and confirmed by examination of E13.5 embryos (Figure 3A and 3B). The anterior boundary of Hoxd10 expression is shifted domain from somites 31–32 (WT) to somites 28–29 (KO), and Hoxd11 domain from somites 34–35 (WT) to somites 31–32 (KO), which are the precursors of the developing lumbar-sacral vertebraes (Figure 3A and 3B) (Burke et al., 1995). In addition, the level of Hoxd11 expression in the posterior trunk and distal limb buds was consistently elevated in the Hotair KO (Figure 3B). In contrast, HoxC genes did not show anteriorization or increased intensity of expression (Figure S3). We also analyzed the expression pattern of the imprinted gene Dlk1. Intriguingly, while Dlk1 is under imprinted control in all cells (da Rocha et al., 2008), Hotair KO led to ectopic Dlk1 expression in the posterior trunk and in the distal limbs, which corresponds to the anatomic sites of endogenous Hotair expression (Figure 3C).
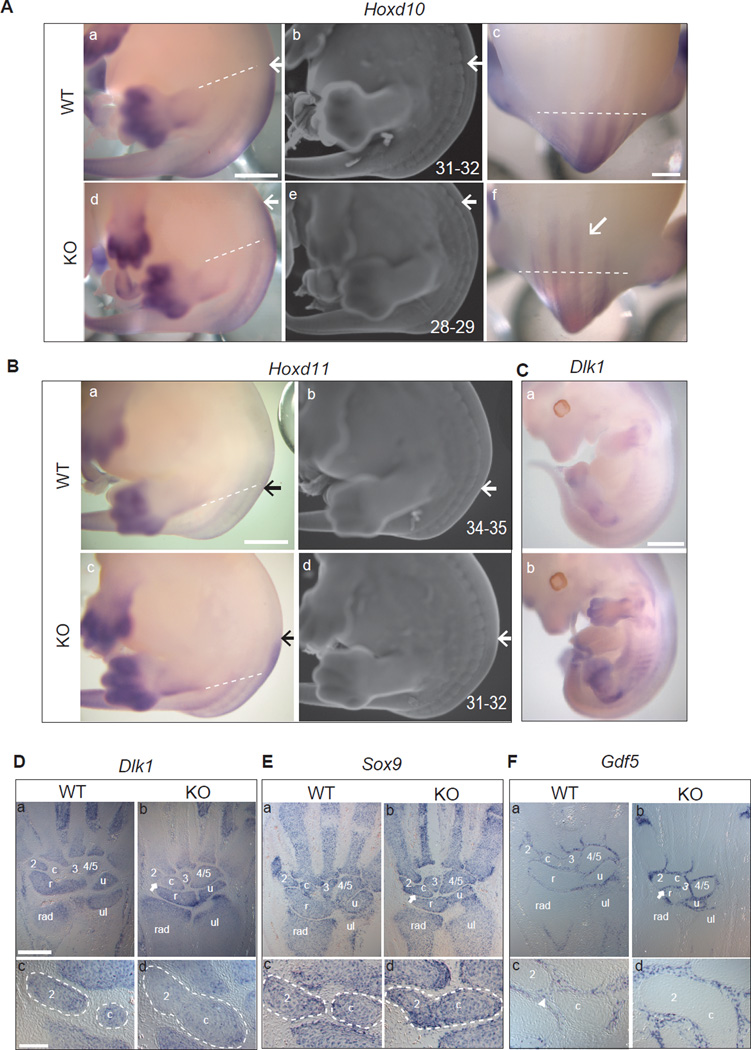
Figure 3. Spatial and temporal gene expression patterns in Hotair KO mice.
(A-B) Whole mount in situ hybridization (WISH) of Hoxd10 (A-a/c/d/f) and Hoxd11 (Ba/ c) of E13.5 embryos (n>3 for each genotype). KO embryos showed increased intensity and anterior shift of the expression domains of HoxD genes (highlight with arrows, the dotted lines across hind limbs are used as anatomical limit). Same embryos were co-stained with ethidium bromide, and the somite position of the anterior expression domain were numbered and marked with arrows (A-b/e; B-b/d). Scale bar: 1mm for A-a/b/d/e, B-a/b/c/d; 600µm for A-c/f.
(C) WISH of Dlk1 on E12.5 embryos showing ectopic expression in Hotair KO embryos. (WT, n=4; KO, n=5; scale bar: 1mm).
(D, E, F) Altered Dlk1 expression and mesenchymal cell fates in Hotair KO wrists. Dlk1 (D), Sox9 (E) and Gdf5 (F) expression in E15.5 wrist sections (n>3 for each genotype). Arrows indicate the joint regions in KO. Dotted circles marked the carpal element 2 and central element c. Arrowhead indicates the intervening Gdf5-positive domain in WT. Note 2-c fusion in KO wrist, showing continuous Dlk1 and Sox9 expression in the junction area of 2 and c; and loss of Gdf5 signal as well. (Scale bar: 300µm for D-a/b, E-a/b, F-a/b; 100µm for D-c/d, E-c/d, F-c/d)
See also Figure S3.
Detailed examination of the limb suggested a connection between Dlk1 de-repression and wrist skeletal element abnormalities. Dlk1 functions as a delta-like ligand of the Notch pathway, and has been implicated in osteogenesis (Abdallah et al., 2004). At E15.5, Dlk1 expression is normally confined to the mesenchymal condensation of the wrist bones (Figure 3D). These condensations are also marked by Sox9 expression and demarcated by Gdf5, which is expressed in cells of the perichondrium that later form cortical bone (Bi et al., 1999; Francis-West et al., 1999). We found that Hotair KO animals showed expansion of the Dlk1 expression domain, such that multiple Sox9- positive condensations become contiguous and the intervening Gdf5-positive domains are lost (Figure 3D–3F). These results suggest a cell fate switch where a subset of perichondrial cells--destined to form bone-- can become cartilage-producing chondrocytes, analogous to mesenchymal fate changes seen with perturbed Wnt or Shh signaling (Day et al., 2005; Niedermaier et al., 2005). The domain of increased HoxD expression is much broader in the Hotair KO than the site of wrist bone abnormalities; on the other hand, the alteration in Dlk1 expression tracks closely with the wrist phenotype. Thus, the localized phenotype suggests the involvement of either Dlk1 alone, or both Dlk1 and HoxD genes. Collectively, these results suggest that Hotair is required to silence genes for proper pattern of gene expression in vivo.
Hotair regulates histone modification at select loci
Biochemical and functional studies indicate that Hotair regulates histone modification patterns genome-wide (Figure 4). Because protein partners of mouse Hotair are not fully characterized, we examined its association with key chromatin modification complexes. Immunoprecipitation (IP) of PRC2 subunits Ezh2 or Suz12, and separately the histone demethylase LSD1, from posterior E11.5 embryos specifically retrieved Hotair but not Malat1 nor U1 RNA, while NF-κB subunit p65 (also known as RelA) and control IgG retrieved neither (Figure 4A). This result suggests that Hotair binds both H3K27 methylase and H3K4 demethylase complexes, similar to its human counterpart. Hotair KO led to loss of H3K27me3 and gain of H3K4me3 at HoxD (including Hoxd1, Hoxd3, Hoxd11, Hoxd13) and imprinted gene loci (such as Dlk1, H19, Igf2, Plag1, and Dcn), as shown by chromatin IP (ChIP) followed by qPCR in TTFs (Figure 4B). ChIP signal at Fgf4 was not changed in KO and served as negative control. We also found that PRC2 occupancy at HoxD and imprinted genes are significantly reduced in the Hotair KO (Figure S4A).
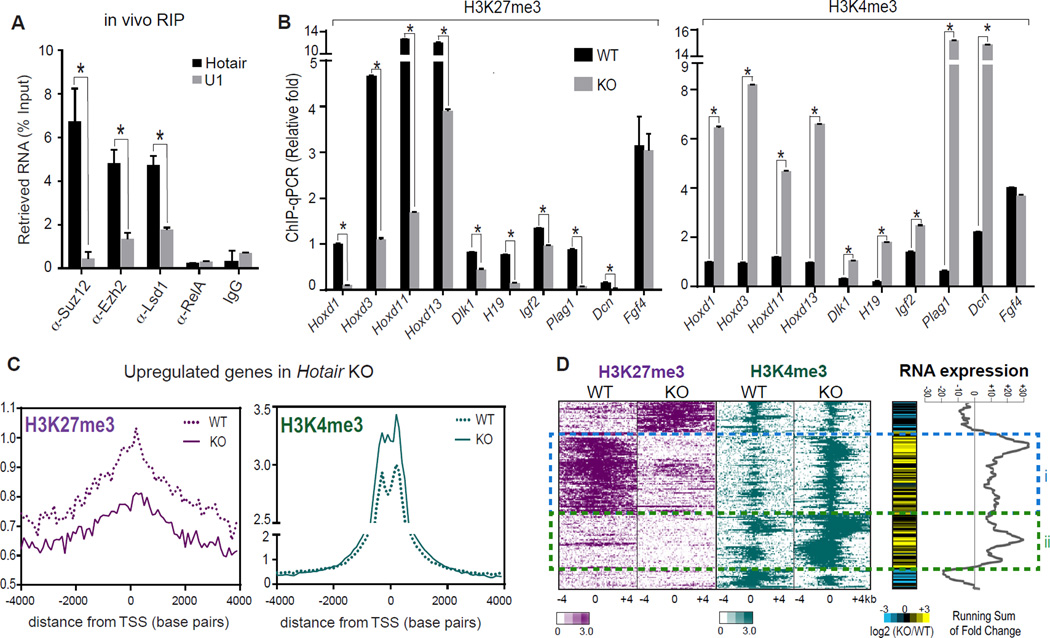
Figure 4. Hotair regulates silent chromatin state genome-wide.
(A) Hotair binds PRC2 and LSD1 complexes in vivo. RIP of E11.5 embryos with the indicated antibodies was followed by qRT-PCR of Hotair and control RNAs (U1, Malat1) and normalized with 1% input performed in parallel (n=3). Mean±s.d. is shown for all panels; *, p<0.05 student's t-test.
(B) ChIP-qPCR in Hotair KO vs. WT cells of H3K27me3 (left panel) and H3K4me3 (right panel) of the indicated genes. Fgf4 was not changed in KO and served as a negative control. (n=3).
(C) Average H3K27me3 (left panel) and H3K4me3 (right panel) ChIP-seq signal across the transcription start site (TSS) of upregulated genes in Hotiar KO cells is shown.
(D) Relationship of histone modifications to gene de-repression in Hotair WT and KO cells. Heat map zoom-in of ChIP-seq signal in 8-kb bins centered on peak summits after unsupervised hierarchical clustering (left). RNA expression changes (KO/WT) and running sums across clusters are shown (right). Gene activation is seen in cluster i (75 loci) with both H3K27me3 loss and H3K4me3 gain; cluster ii shows 70 loci with only H3K4me3 gain in Hotair KO.
See also Figure S4.
We next analyzed the global pattern of H3K27me3 and H3K4me3 by ChIP-seq. Genes that are de-repressed by Hotair KO showed, on average, broad decrease of H3K27me3 occupancy and focal gain of H3K4me3 centered around their transcriptional start sites (Figure 4C). Conversely, promoters with Hotair-dependent loss of H3K27me3 or gain of H3K4me3 are significantly enriched for de-repressed genes in Hotair KO (p < 0.0001, GSEA for concordance between each chromatin change and gene induction). Next, we organized the H3K27me3 and H3K4me3 ChIP-seq data by unsupervised hierarchical clustering, and displayed the RNA level changes in KO vs WT in parallel (Figure 4D). This analysis revealed two main patterns of histone modification change in association with gene de-repression: One cluster of loci (termed cluster i) showed coordinate broad loss of H3K27me3 and focal gain of H3K4me3 in Hotair KO; another cluster of loci (termed cluster ii) showed only H3K4me3 gain but lacked H3K27me3 in either WT or KO cells. The two clusters demonstrate comparable levels of gene derepression in Hotair KO, but cluster i has lower level of H3K4me3 in WT cells, consistent with their co-occupancy with H3K27me3. These results suggest that Hotair can regulate coordinated H3K4 and H3K27 methylation at some loci (via both PRC2 and LSD1) and solely H3K4 methylation at other loci (via LSD1). While the emergence of H3K4me3 signal may be due to the well-known association of H3K4me3 with active promoters or secondary effects, the fact that not all loci with H3K4me3 gain showed RNA increase suggests that H3K4me3 gain in KO cells is not simply a consequence of increased transcription. Additional genes de-repressed in Hotair KO are associated with different and heterogeneous chromatin patterns, which may occur through alternative or indirect mechanisms.
Because DNA methylation is a well-studied regulator of imprinting status (Abramowitz and Bartolomei, 2011), we tested whether Hotair may also influence DNA cytosine methylation. Bisulfite conversion and sequencing of the intergenic differentially methylated region (IG-DMR) from Dlk1-Gtl2 locus showed that deletion of Hotair had no significant impact on DNA methylation at this locus (Figure S4B). These results suggest that Hotair affects the Dlk1 locus principally through control of histone methylation genome-wide.
DISCUSSION
LncRNAs are increasingly recognized as potential mediators of gene regulation and pathogenic loci in human diseases (Rinn and Chang, 2012). Hence there is an important need to understand their physiological functions in model organisms. The targeted and conditional knockout of Hotair provides a model to analyze lncRNA functions in vivo for development and cancer. Human HOTAIR was the first lncRNA reported to silence genes in trans, notably HOXD genes (Rinn et al., 2007). Hotair KO is now shown to cause derepression at multiple genes, including Hoxd10 and Hoxd11 that are important for patterning of lumbar-sacral junction and of metacarpal and carpal bones in the limbs (Favier et al., 1995; Gerard et al., 1996). Hotair KO causes increased expression and anterior expansion of Hoxd10 and d11 domains, which increases the dosage of Hoxd genes in posterior embryo. Notably, L6->S1 transformation is the same phenotype that is observed when ectopic copies of the HoxD locus are introduced into mouse genome (Spitz et al., 2001), or when endogenous Hoxd10 and Hoxd11 expression domains are anteriorized by deletion of cis repressor element (Gerard et al., 1996). These findings suggest that de-repression of Hoxd10 and Hoxd11 likely contribute to the homeotic axial transformation in Hotair KO mice. In silico analyses suggest that Hotair is conserved predating the eutherian-marsupial split, but Hotair is conserved in gene synteny and RNA structure rather than primary sequence (He et al., 2011; Yu et al., 2012). The similar roles of human and mouse Hotair on HoxD provides another example of conservation of lncRNA function at syntenic locations despite limited sequence conservation (Ulitsky et al., 2011). This knowledge sets the stage for potential in silico analyses to highlight conserved RNA domains for Hotair function, and phenotypic rescue of Hotair KO with human HOTAIR and mutants should provide definitive structure-function studies.
Because some homeotic transformations occur with different frequency in different genetic backgrounds, care and proper controls are important to interpret this result. After extensive backcrossing to homogeneous genetic background, Hotair KO demonstrated significantly increased (~10 fold) L6->S1 transformation compared to WT littermates. Moreover, inducible deletion of Hotair in isogenic cells also de-repressed Hoxd and Dlk1, suggesting that these are direct effects of Hotair removal. However, the frequency of the L6 transition in Hotair KO (58%) is less than that observed in HoxD transgene (>80%) or derepression in cis (~100%) (Gerard et al., 1996; Spitz et al., 2001). These differences may be due to less potent regulation in cis vs. in trans, potential redundancy of in recruitment mechanisms of silencing complexes, or different genetic backgrounds.
An intriguing question is why HoxCΔ did not reveal a more drastic phenotype. Schoderet and Duboule also observed that Hoxd8, d9, and d10 are derepressed by approximately two-fold in HoxCΔ compared to wildtype (Schorderet and Duboule, 2011), but these changes were not apparently sufficient to cause skeletal transformations. Re-analysis of the published RNA-seq data from HoxCΔ (Schorderet and Duboule, 2011) revealed modest but consistent up-regulation of all three remaining Hox loci, such that the total dosage of Hox transcripts is maintained in HoxCΔ tissue (Figure S4C and S4D). This finding may also explain why HoxCΔ has a milder phenotype than deletions of individual Hoxc genes (Suemori and Noguchi, 2000). Alternatively, HoxCΔ may remove genes with functions antagonistic to Hotair, which are preserved in Hotair KO. Although not detected in the tissues and time points examined, we cannot rule out the possibility that Hotair affects one or more Hoxc genes in cis at other times in development, which would be absent in HoxCΔ. The comparison between HoxCΔ and targeted Hotair KO also demonstrate the value of multiple and fine-scale manipulations to define lncRNA function in vivo.
Hotair KO reveals an unexpected role for Hotair in transcriptional repression of several imprinted gene loci. Imprinting involves the selective expression of genes between two nearly identical copies, the paternal vs. maternal alleles; similarly, developmental Hox expression involves the selective expression from among highly homologous copies of homoeodomain genes, from the Hox loci. Beyond these conceptual parallels, our results suggest a direct cross regulation between Hox and some imprinted loci. The diversity of imprinting mechanisms potentially explains why Hotair KO only affects the expression of a small subset of imprinted genes. Hotair is likely to be involved in the maintenance of proper gene expression levels rather than the initial imprint choice because (i) Hotair is not expressed in early zygotes when imprinted alleles are marked by DNA methylation, and (ii) DNA methylation of the imprinted loci (which reflect imprinting choice) at not altered in Hotair KO. Consistently, Hotair KOs do not demonstrate phenotypes of complete imprinting loss, such as postnatal lethality from altered dosages of the Dlk1 locus (da Rocha et al., 2009; Takahashi et al., 2009). The role of Hotair may be similar to that of Bmi1, a Polycomb protein that control the expression level, but not the imprinting choice, of multiple imprinted genes to regulate self-renewal of adult stem cells (Zacharek et al., 2011). In an analogous fashion, Hotair controls Polycomb- and Lsd1-related histone modification state to repress several imprinted genes; alteration of imprinted genes and consequently stem cell self-renewal provide potentially new insights for human cancers that overexpress HOTAIR (Gupta et al., 2010; Kim et al., 2012; Kogo et al., 2011).
The set of genes that show altered expression or chromatin state may represent direct or indirect effects of Hotair KO. Delineating the direct targets of Hotair, such as by ChIRP (Chu et al., 2011), will be an important future direction. The ability of Hotair to affect the chromatin state may arise from direct regulation, or due to the known physical clustering and mutual influence of the epigenetic states of some imprinted gene loci (Sandhu et al., 2009). The generation of Hotair cKO allows these and other potential mechanisms to be dissected in future studies.
Experimental Procedures
Detailed experimental and analysis methods can be found in Extended Experimental Procedures.
Animals
Hotair conditional KO mice were generated by homologous recombination and were crossed to HPRT-Cre mice to yield ubiquitous deletion of the Hotair Locus. All the mice were bred in the Stanford University Research Animal Facility in accordance with the guidelines (see details in Extended Experimental Procedures).
RNA-seq
Poly-A selected RNA was isolated from the fibroblast of Hotair WT, heterozygous and KO mice. The libraries were prepared with the dUTP protocol and sequenced using the Illumina Genome Analyzer IIX platform with 36bp reads. Raw reads were aligned to the mouse reference sequences NCBI Build 37/mm9 with the TopHat (v1.1.3) algorithm. Expression levels of RefSeq annotated genes were calculated in unit of reads per kilobase of exon model per million mapped fragments (RPKM). Detailed analysis is presented in Extended Experimental Procedures.
ChIP-seq and ChIP-qPCR
ChIP-qPCR and ChIP-seq were performed as described (Tsai et al., 2010). Sequencing libraries were made following Illumina’s protocol. qPCR analysis were performed with Roche’s Lightcycler. Sequencing reads (36bp) were generated on Illumina GAIIX Genome Analyzer and were uniquely mapped to mouse reference genome (NCBI37/mm9) using bowtie (version 0.12.6). Peaks for each sample were called using MACS algorithm (version 1.4.2). Detailed analysis is presented in Extended Experimental Procedures.
Supplementary Material
ACKNOWLEDGEMENT
We thank D. Frendewey and V. Lai for communicating unpublished observations of the C4 vertebrae (Lai et al, in preparation), members of Chang lab and A. Oro for discussion, Stanford Small Animal Imaging Facility for micro CT analysis, P. Schoderet and D. Duboule for HoxCΔ RNA-seq data and reagents, E. Zelzer for reagents, and P. Grote for in vivo RIP protocol. Supported by NIH (R01-CA118750 to H.Y.C), National Science Foundation (O.L.W.), and Susan G. Komen Foundation (M.C.T.). H.Y.C. is an Early Career Scientist of the Howard Hughes Medical Institute.
Footnotes
ACCESSION NUMBERS
The GEO accession number for all genomic data herein is GSE48007. Available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=hpgxpigmqmyaula&acc=GSE48007
References
- Abdallah BM, Jensen CH, Gutierrez G, Leslie RG, Jensen TG, Kassem M. Regulation of human skeletal stem cells differentiation by Dlk1/Pref-1. J Bone Miner Res. 2004;19:841–852. doi: 10.1359/JBMR.040118. [DOI] [PubMed] [Google Scholar]
- Abramowitz LK, Bartolomei MS. Genomic imprinting: recognition and marking of imprinted loci. Curr Opin Genet Dev. 2011;22:72–78. doi: 10.1016/j.gde.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Sox9 is required for cartilage formation. Nat Genet. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- Burke AC, Nelson CE, Morgan BA, Tabin C. Hox genes and the evolution of vertebrate axial morphology. Development. 1995;121:333–346. doi: 10.1242/dev.121.2.333. [DOI] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic Maps of Long Noncoding RNA Occupancy Reveal Principles of RNA-Chromatin Interactions. Mol Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rocha ST, Charalambous M, Lin SP, Gutteridge I, Ito Y, Gray D, Dean W, Ferguson-Smith AC. Gene dosage effects of the imprinted delta-like homologue 1 (dlk1/pref1) in development: implications for the evolution of imprinting. PLoS Genet. 2009;5:e1000392. doi: 10.1371/journal.pgen.1000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rocha ST, Edwards CA, Ito M, Ogata T, Ferguson-Smith AC. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 2008;24:306–316. doi: 10.1016/j.tig.2008.03.011. [DOI] [PubMed] [Google Scholar]
- Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;8:739–750. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favier B, Le Meur M, Chambon P, Dolle P. Axial skeleton homeosis and forelimb malformations in Hoxd-11 mutant mice. Proc Natl Acad Sci U S A. 1995;92:310–314. doi: 10.1073/pnas.92.1.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis-West PH, Abdelfattah A, Chen P, Allen C, Parish J, Ladher R, Allen S, MacPherson S, Luyten FP, Archer CW. Mechanisms of GDF-5 action during skeletal development. Development. 1999;126:1305–1315. doi: 10.1242/dev.126.6.1305. [DOI] [PubMed] [Google Scholar]
- Gerard M, Chen JY, Gronemeyer H, Chambon P, Duboule D, Zakany J. In vivo targeted mutagenesis of a regulatory element required for positioning the Hoxd-11 and Hoxd-10 expression boundaries. Genes Dev. 1996;10:2326–2334. doi: 10.1101/gad.10.18.2326. [DOI] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Liu S, Zhu H. The sequence, structure and evolutionary features of HOTAIR in mammals. BMC Evol Biol. 2011;11:102. doi: 10.1186/1471-2148-11-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits prooncogenic activity in pancreatic cancer. Oncogene. 2012 doi: 10.1038/onc.2012.193. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- Lee JT. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23:1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- Niedermaier M, Schwabe GC, Fees S, Helmrich A, Brieske N, Seemann P, Hecht J, Seitz V, Stricker S, Leschik G, et al. An inversion involving the mouse Shh locus results in brachydactyly through dysregulation of Shh expression. J Clin Invest. 2005;115:900–909. doi: 10.1172/JCI200523675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu KS, Shi C, Sjolinder M, Zhao Z, Gondor A, Liu L, Tiwari VK, Guibert S, Emilsson L, Imreh MP, et al. Nonallelic transvection of multiple imprinted loci is organized by the H19 imprinting control region during germline development. Genes Dev. 2009;23:2598–2603. doi: 10.1101/gad.552109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorderet P, Duboule D. Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS Genet. 2011;7:e1002071. doi: 10.1371/journal.pgen.1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soshnikova N, Duboule D. Epigenetic temporal control of mouse Hox genes in vivo. Science. 2009;324:1320–1323. doi: 10.1126/science.1171468. [DOI] [PubMed] [Google Scholar]
- Spitz F, Gonzalez F, Peichel C, Vogt TF, Duboule D, Zakany J. Large scale transgenic and cluster deletion analysis of the HoxD complex separate an ancestral regulatory module from evolutionary innovations. Genes Dev. 2001;15:2209–2214. doi: 10.1101/gad.205701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suemori H, Noguchi S. Hox C cluster genes are dispensable for overall body plan of mouse embryonic development. Dev Biol. 2000;220:333–342. doi: 10.1006/dbio.2000.9651. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Okamoto A, Kobayashi R, Shirai M, Obata Y, Ogawa H, Sotomaru Y, Kono T. Deletion of Gtl2, imprinted non-coding RNA, with its differentially methylated region induces lethal parent-origin-dependent defects in mice. Hum Mol Genet. 2009;18:1879–1888. doi: 10.1093/hmg/ddp108. [DOI] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Lindsay J, Feng ZP, Frankenberg S, Hu Y, Carone D, Shaw G, Pask AJ, O'Neill R, Papenfuss AT, et al. Evolution of coding and non-coding genes in HOX clusters of a marsupial. BMC Genomics. 2012;13:251. doi: 10.1186/1471-2164-13-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharek SJ, Fillmore CM, Lau AN, Gludish DW, Chou A, Ho JW, Zamponi R, Gazit R, Bock C, Jager N, et al. Lung stem cell self-renewal relies on BMI1-dependent control of expression at imprinted loci. Cell Stem Cell. 2011;9:272–281. doi: 10.1016/j.stem.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.