Abstract
Biomphalaria glabrata susceptibility to Schistosoma mansoni has a strong genetic component, offering the possibility for investigating host–parasite interactions at the molecular level, perhaps leading to novel control approaches. The identification, mapping and molecular characterisation of genes that influence the outcome of parasitic infection in the intermediate snail host is, therefore, seen as fundamental to the control of schistosomiasis. To better understand the evolutionary processes driving disease resistance/susceptibility phenotypes, we previously identified polymorphic random amplification of polymorphic DNA and genomic simple sequence repeats from B. glabrata. In the present study we identified and characterised polymorphic expressed simple sequence repeats markers (Bg-eSSR) from existing B. glabrata expressed sequence tags. Using these markers, and with previously identified genomic simple sequence repeats, genetic linkage mapping for parasite refractory and susceptibility phenotypes, the first known for B. glabrata, was initiated. Data mining of 54,309 expressed sequence tag, produced 660 expressed simple sequence repeats of which dinucleotide motifs (TA)n were the most common (37.88%), followed by trinucleotide (29.55%), mononucleotide (18.64%) and tetranucleotide (10.15%). Penta- and hexanucleotide motifs represented <3% of the Bg-eSSRs identified. While the majority (71%) of Bg-eSSRs were monomorphic between resistant and susceptible snails, several were, however, useful for the construction of a genetic linkage map based on their inheritance in segregating F2 progeny snails derived from crossing juvenile BS-90 and NMRI snails. Polymorphic Bg-eSSRs assorted into six linkage groups at a logarithm of odds score of 3. Interestingly, the heritability of four markers (Prim1_910, Prim1_771, Prim6_1024 and Prim7_823) with juvenile snail resistance were, by t-test, significant (P < 0.05) while an allelic marker, Prim24_524, showed linkage with the juvenile snail susceptibility phenotype. On the basis of our results it is possible that the gene(s) controlling juvenile resistance and susceptibility to S. mansoni infection in B. glabrata are not only on the same linkage group but lie within a short distance (42 cM) of each other.
Keywords: Expressed sequence tags, Genetic linkage mapping, Resistance, Susceptibility, Biomphalaria glabrata, Schistosoma mansoni
1. Introduction
The freshwater snail, Biomphalaria glabrata, is an intermediate host of the parasitic trematode, Schistosoma mansoni. This parasite causes schistosomiasis in the developing world of the tropics and sub-tropics (Chitsulo et al., 2004; Gryseels et al., 2006). Sustaining control of the disease for the long-term remains a challenge due to the intractable problems of re-infection frequently experienced in the human population even after targeted chemotherapy has been successful. There is also the concern that parasites that are resistant to praziquantel, the best drug available to treat this disease, might be surfacing in certain parts of the globe (Mattos et al., 2007). To address these problems and to improve the public health of people adversely affected by this disease, new tools that will help reduce transmission for the long term are being developed (Bologna et al., 2003). While new schistosome vaccines are being developed, it is also recognised that the development of an alternative method to control schistosomiasis aimed at blocking transmission during the intramolluscan phase of its development would complement existing methods to combat schistosomiasis.
While this transmission blocking strategy is not new (it was first proposed in the 1950s by Hubendick (1958)) it has gained momentum in the past few years with the belief that using resistant snails to control transmission will reduce the use of molluscicides, the mainstay traditional method for controlling schistosomiasis. The impact of molluscicide use on the environment and the economic cost of its long-term application remains inconclusive. However, the present consensus of opinion remains that the use of resistant snails to combat schistosomiasis, as a form of biological control, will be a more environmentally friendly alternative for sustaining the long-term reduction of this disease.
Towards this end, a variety of molecular tools helping to elucidate the molecular basis of the snail schistosome interaction are being developed. The snail is also seen as a model organism for studies involving host/pathogen issues, especially in topics related to invertebrate innate defense. From these studies we are beginning to appreciate the complexities of the molecular basis of the snail–host schistosome relationship. Lately, the importance of several genes in the ability of snails to either sustain or reject parasitic infection has been recognised. For example, anti-oxidants (Bender et al., 2005; Humphries and Yoshino, 2008), proteolytic enzymes (Myers et al., 2008), stress genes (Ittiprasert et al., 2009; Ittiprasert and Knight, 2012; Lockyer et al., 2012) and the expression of certain lectins (Adema et al., 1997; Zhang et al., 2008; Hanington et al., 2010) all appear to be important in the snail host/parasite relationship. Differential gene expression studies have also shown that the temporal and spatial expression of some of these genes in the anti-parasite response may be important in whether a snail becomes infected or not (Moon et al., 1990; Bouchut et al., 2006; Hanington et al., 2010) with results that clearly point to an involvement of parasite mediated transcription regulatory mechanisms in controlling gene expression of resistant and susceptible snails. The non-random spatial repositioning of gene loci of the snail host by the parasite has also been identified (Knight et al., 2011a), offering evidence for the first time of possible epigenetic markings of the snail host genome by invading schistosomes.
While significant advances have been made in deciphering the nature of genes whose expression might be relevant in the susceptibility status of the snail to a given strain of parasite, there are no robust biomarkers for distinguishing resistant and susceptible snails from each other. In a previous study, although we were able to show the segregation of two random amplification of polymorphic DNA (RAPD) markers for resistance, further attempts to characterise the genes associated with these markers was challenging due to significant sequence redundancy of these markers in the snail genome (Knight et al., 1999). It is anticipated that the entire 1.0 Gb sequence of B. glabrata will be soon realised (Pat Minx, personal communication), with the view that more sophisticated genotyping tools (e.g. single nucleotide polymorphisms [SNPs]) that facilitate high density mapping will soon be developed.
Juvenile resistance and susceptibility of B. glabrata to S. mansoni are complex traits (Richards and Merritt, 1972; Richards and Shade, 1987). Both of these characteristics are governed in snails at this young age by at least four to five genes, each with multiple alleles. To identify the genes involved in juvenile resistance and susceptibility, we adopted a reverse genetic approach using linkage analysis of polymorphic expressed sequence tags (ESTs) – expressed simple sequence repeats (eSSRs) and previously identified bi-allelic microsatellite markers, genomic (g)SSRs (Wake and Vredenburg, 2008) that were capable of distinguishing between our representative laboratory maintained resistant (BS-90) and susceptible (NMRI) snail stocks.
The rationale for using eSSRs for genotying was to avoid the pitfall that was previously encountered; identifying genetic variant markers with excessive repeats (Knight et al., 1999). Furthermore, there are also a number of advantages for using expressed genes compared with anonymous genomic sequences as genetic markers (Hunter et al., 1961; Hoffmann et al., 1998; Imasheva et al., 1999). First, if the eSSR marker is found to be genetically associated with a trait of interest, it may be possible to directly identify the gene affecting the trait (Chen et al., 2001; Thiel et al., 2003). For this reason, these markers provide opportunities for gene discovery and enhance the role of genetic markers by assaying variations in transcribed genes, allowing correlations to be drawn between genetic variations and known gene function. Second, these markers are likely to be highly conserved and, therefore, may be more transferable between species than anonymous sequence-derived markers (Patz et al., 1996; Ansaldo et al., 2009; Chu and Guo, 2009). Third, eSSR markers that share homology to candidate genes can be specifically targeted for genetic mapping and can be helpful for genome alignment and linkage across distantly related species for comparative analysis studies (Aisemberg et al., 2005). Therefore, generating eSSRs is seen as an attractive alternative approach complementing existing gSSR collections for genetic mapping studies.
SSRs, also known as microsatellites, have increasingly become the marker of choice for population genetic analyses. Unfortunately, the development of these traditional ‘anonymous’ SSRs from genomic DNA (gSSR) is costly and time-consuming. These problems are further compounded by a paucity of resources in some taxa that lack clear research importance. However, the advent of the genomics age has resulted in the production of vast amounts of publicly available DNA sequence data, including large collections of ESTs. These are, potentially, a rich source of SSRs (eSSRs) that help to reveal polymorphisms not only within the source taxon but in related taxa as well.
In this study, the cost-saving marker development option, data mining of B. glabrata ESTs (Bg-eSSR) in GenBank combined with Simple Sequence Repeat Identification Tool (SSRIT) and RepeatMasker was utilised to identify potential functional markers for differentiating between resistant and susceptible snails. We used three different types of primers; 33 primer pairs flanking Bg-eSSR motifs, six inter-SSR (ISSR) primers designed according to high frequency motifs from Bg-eSSRs and 46 primers designed from gSSRs. By tracking the inheritance of polymorphic eSSRs and gSSRs (previously identified microsatellites) in 206 segregating F2 progeny populations (from two families, A and B) derived by crossing juvenile resistant (BS-90) and juvenile susceptible (NMRI) snails, these markers assorted into six linkage groups (LGs). The logarithm of odds (LOD) score (≥3) obtained was statistically significant for four markers (Prim1_910, Prim1_771, Prim6_1024 and Prim7_823) and an allelic marker, Prim24_524, for B. glabrata resistance and susceptibility to S. mansoni, respectively. On the basis of our results it is possible that the gene(s) controlling juvenile resistance and susceptibility to S. mansoni infection in B. glabrata are not only on the same LG but lie within a short distance 42 centiMorgans (cM) of each other.
2. Materials and methods
2.1. EST-SSR of B. glabrata (Bg-eSSR) data mining
The total number of 54,309 non-redundant B. glabrata EST sequences available at http://www.ncbi.nlm.nih.gov/sites/entrez including our subtraction suppressive hybridisation (SSH)-ESTs as previously described (Ittiprasert et al., 2010) were used for SSR analysis by SSRIT (http://www.gramene.org/db/searches/ssrtool) (Temnykh et al., 2001; Lockyer et al., 2012). The eSSRs were found to contain motifs of two to six nucleotides in size. The minimum repeats unit was defined as five for dinucleotides and four for the higher order motifs including tri-, tetra-, penta- and hexa-nucleotides. The sequences that contained SSRs were further analysed to screen out the low complexity/simple repeats using RepeatMasker (http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker) (Bajpai et al., 2007; van Die and Cummings, 2010). Also, the identical and poly A tail sequences were eliminated by Vector NTI v.10 (Invitrogen Life Science, USA).
2.2. Primer design
We attempted to design primers flanking SSRs for all of the Bg-eSSRs using Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome). However, it proved to be difficult or impossible to design primers for most of the Bg-eSSRs since potential priming sites were inconveniently located either at the border of the EST or surrounded by excessive AT-rich sequences. Another two groups of primers were included: (i) genomic microsatellite primers that were shown previously to differentiate between resistant and susceptible snails (Wake and Vredenburg, 2008) and (ii) and ISSR primers previously designed and identified with high frequency from Bg-eSSRs (Geleta and Bryngelsson, 2009). The major parameters for Bg-eSSR primer design were set as follows: expected size of PCR product, between 100 and 350 bp; optimum annealing temperature between 57 and 60 °C so all PCR amplifications could be conveniently achieved at the same annealing temperature. Also, the designed primers were checked for species specificity between Biomphalaria (taxid: 6525) and Schistosomatoidea (taxid: 31244) to confirm that they would amplify snail-specific genes and not cross hybridise with Schistosoma spp. DNA. This was important as S. mansoni infection would be used to determine heritability of the susceptibility phenotype. All of the primer pairs were used to ascertain polymorphisms between BS-90 (paternal), NMRI (maternal), individual F1 DNA and pooled F2 DNAs from two inbred families as described previously (Knight et al., 1999). The primers that amplified codominant markers in hybrid F1 and were polymorphic in F2 populations were used for fragment analysis in individual snails and for mapping. Forward primers were labelled either with the flurochromes, hexachloro-fluorescein (HEX) or 6-fluorescein (6FAM). Bg-eSSRs were named according to the following convention: primer name and fragment size, respectively.
2.3. Snail progeny and phenotype analysis
DNA from 206 F2 progeny snails derived by self-fertilisation of individual (17) F1 progeny snails generated from two families (A and B), produced by cross-fertilization of paternal resistant (BS-90) and maternal susceptible (NMRI) B. glabrata were analysed. Inheritance of the susceptibility phenotype in F2 progeny snails was determined by infecting juvenile snails individually with S. mansoni miracidia (10 miracidia/snail) as previously described (Knight et al., 1999). At 5–6 weeks p.i., snails were examined individually for cercarial shedding. The susceptible phenotype was recorded for progeny snails that were positive for cercarial shedding, while progeny snails that remained negative for cercarial shedding for up to 16 weeks post-exposure were scored for inheriting the resistant phenotype. The animal maintenance, S. mansoni exposure and subsequent harvesting of the eggs from mouse livers were done according to approved IACUC protocol number 09-03.
2.4. DNA extraction, PCR amplification and fragment analysis
Genomic DNAs from parental snails, F1 progenies and segregating F2 populations were extracted (from individual snails) as previously described (Knight et al., 1999). DNA concentrations were determined by UV absorption (260 nm) using the NanoDrop 2000C (ThermoScientific, USA). PCR amplifications in a final volume of 10 μl contained 40 ng of template, 0.1 μM of each primer, 2 mM MgCl2, 0.3 mM dNTPs, 1× PCR buffer, and 0.5 units of Taq DNA polymerase (Promega, Madison, WI, USA) in a thermal cycler. All cycling began with an initial denaturation at 95 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 60 °C for 30 s and 72 °C for 30 s. Two microliters of PCR product was run on either Tris–Boric acid–EDTA (TBE) and 1.5% agarose gels or 4% Nusieve gels to determine the outcome of the PCR and for fragment size analysis. The PCR product was also diluted 1:40 and run on the ABI3100 capillary sequencer (Applied Biosystems, USA) together with the GeneScan 500® (ROX) as a standard. Base calling of florescent labelled-fragments were calculated by GeneMapper Software v3.5 (Applied Biosystems).
2.5. Genetic linkage mapping
The mapping population comprised 206 segregating F2 individual progeny snails generated from two families resulting from cross-fertilisation of the resistant and susceptible (BS-90 × NMRI) snails. Linkage analysis was performed using the software JoinMap v3.0 (Kucuktas et al., 2009). Segregating loci in F2 populations were scored by the presence/absence of amplicons and the Bg-eSSR- and gSSR-based linkage map was constructed with molecular markers obtained from 14 primer pairs (10 Bg-eSSR and four gSSR). LGs were determined using LOD thresholds ranging from 4.0 to 8.0. Map distances were calculated with the Kosambi mapping function (Blouin et al., 2013).
3. Results and discussion
3.1. Characteristics of eSSRs derived from the B. glabrata snail
A total of 54,309 ESTs of B. glabrata including specific ESTs from previously described SSH cDNA libraries (Ittiprasert et al., 2010) were analysed to develop the eSSR markers utilised in this study. Using SSRIT, SSRs were identified from 2,474 ESTs (4.56%). For further data mining, the RepeatMasker software was used to screen out low complexity sequences. This analysis revealed 1,814 Bg-eSSRs (73.32% of identified SSRs) were of low complexity, A rich (125 ESTs), T rich (264 ESTs), AT rich (1,307 ESTs), CT rich (17 ESTs), GA rich (83 ESTs) and polypurines (18 ESTs). The remaining 660 (26.67% of identified SSRs) were high complexity microsatellites that were used for primer design. The most common repeat types were dinucleotides (37.88%) followed by trinucleotides (29.55%), mononucleotides (18.64%), tetranucleotides (10.15%), pentanucleotides (2.27%) and peptanucleotides (1.52%) (Fig. 1). As shown in Supplementary Table S1, the motif with highest frequency was TA (17.58%) followed by the motifs TG (5.76%), GA (4.85%), TTA (4.70%), CA (4.55%), CAT (4.39%), TAA (3.79%), TTG (2.58%) and TAGA (1.97%).
Fig. 1.
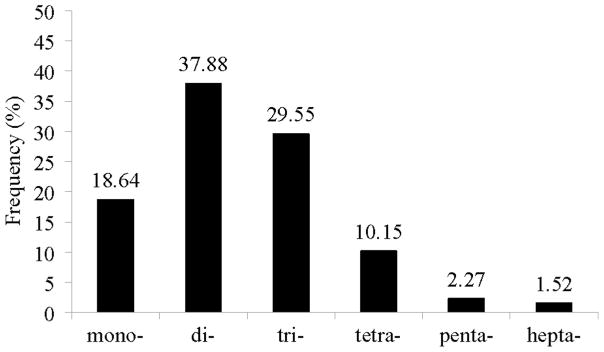
Distribution of expressed sequence tag-derived simple sequence repeats based on motif size; mono-, di-, tri-, tetra-, penta- and hepta- (X-axis). Percentage of F2 frequency shown on the Y-axis.
To determine the putative identities of the identified eSSR markers, the 660 Bg-eSSRs (after poly A tail and identical sequence elimination by Vector NTI v.10 as described in Section 2.1) were compared with sequences in the GenBank database using BLASTX with a cut-off E-value ≤10−4. Listed in Table 1 are 33 transcripts (from 56 ESTs) that showed significant sequence homology to known genes in GenBank. Twenty-two sequences corresponded to ribosomal protein, two showed significant homologies to a hypothetical protein and the majority (601) showed no hits to any existing sequences in the public domain. The biological function of the transcripts were deduced by Gene Ontology (GO) analysis (Carbon et al., 2009) with criteria as previously described (Moon et al., 1990). Results from this analysis showed that most transcripts (seven Bg-eSSRs) were in the category for DNA/RNA processing, including RNA splicing. The second most significant deduced function (five Bg-eSSRs) was in the catabolic/metabolic process category followed by (four Bg-eSSRs) the category for adhesion/receptor proteins. Two of the Bg-eSSRs were associated with categories for transcription/translation, biosynthesis process and enzymes, and one Bg-eSSR was deduced as having a functional role in signal transduction while four Bg-eSSRs had unknown function. Some important functional genes identified included B. glabrata lectins (fibrinogen related proteins, mucin), cystatin (cysteine protease inhibitor), heat shock protein (Hsp) 96, ornithine decarboxylase and protein kinase (Table 1). The involvement of all these genes in the B. glabrata snail host and S. mansoni infection has previously been described (Lockyer et al., 2008; Ittiprasert et al., 2009).
Table 1.
Expressed sequence tags of Biomphalaria glabrata protein homology and primers.
| SSR contained-ESTs homology | Motif | Primer name | Sequence of primer (5′–3′) | Biological function (GO number) |
|---|---|---|---|---|
| cAMP-specific 3′, 5′-cyclic | (tca)6 | Prim13-f Prim13-r |
ACCACCATTGCTCACACGTA AATTGATGATTCCGTTTCGC |
Signal transduction (GO:0007165) |
| CG11266-PB (Drosophila melanogaster) | (ta)5, (tg)6 | Prim22-f Prim22-f |
CTCAACCAGCAGCTACTCCC GTGTCTAGCTGTCCTTGCCC |
nd |
| Chitin-binding protein | (aca)5 | Prim26-f Prim26-r |
CTATCTCCAATTCGACGGGA TAATGGCAGATGACGGAACA |
Chitin catabolic process (GO:0066032) |
| Coagulation factor-like protein 2 | (cac)5 | Prim23-f Prim23-r |
GGGCTCTGTTATGCGTTGTT TGTAGTCGAGTTCGCTGTGG |
Cell adhesion (GO:000715) |
| Cystatin (antimicrobial activity) | (tat)10 | Could not design | Positive regulation of cell proliferation (GO:0008284) | |
| Dorsal switch protein 1-fruit fly | (gat)5 | Could not design | Negative regulation of antimicrobial humoral response (Kim et al., 2007) | |
| Eukaryotic translation elongation factor | (gt)5 | Could not design | Cellular metabolic process (GO:0044237) | |
| ENSANGP00000015826 (Anopheles gambiae) | (aga)5, (gat)5 | Prim17-f Prim17-r |
CAGAGCCCATGGCTAGGATA GGGGAGATCATCAGAATCCA |
nd |
| Fibrinogen-related proteins (FREP) | (ttta)5 | Could not design | Innate defense system (Zhang et al., 2008) | |
| G-box binding factor (GBF) | (gac)6 | Prim12-f Prim12-r |
AACAAAACCTGTCGCCAAAC GAGTTGTTGCCGGTGATTTT |
Regulation of transcription (GO:0045449) |
| Glycogen debranching enzyme | (gca)7, (ctg)11 | Prim27-f Prim27-r |
AAATTTTGTTGCTGCTGGCT TATGCAGGTACCAGGTGCAG |
Glycogen catabolic process (GO:0005980) |
| Glucosamine-6-phosphate isomerase | (att)7, (at)6 | Prim19-f Prim19-r |
GAGGATGCCACAATGGAACT ATGCCCTGATTTCGTTTGAA |
Glucosamine catabolic process (GO:0006043) |
| 94 kDa glucose regulated-protein | (aga)5 | Could not design | Glucose metabolic process (GO:0006006), glucose catabolic process (GO:0006007) | |
| Glycoprotein 93 | (aga)5 | Prim18-f Prim18-r |
CAGAGCCCATGGCTAGGATA TCCACAACACCTCGAATGAA |
nd |
| Heat shock protein gp96 | (aga)5 | Could not design | Mycelium development (GO:0043581) | |
| High mobility group B1 protein | (tca)5 | Prim24-f Prim24-r |
CCACCACCACTGCGATATAA GGGGAAGAAATGGGAAACAT |
Base-excision repair, DNA ligation (GO:0006288) |
| HM21-human high-mobility group protein 2-like 1 | (gat)6 | Prim9-f Prim9-r |
AAAACTGAATCCCTGTCCCC GCCACATCTCCCCAAGTTTA |
Base-excision repair, DNA ligation (GO:0006288) |
| Importin, moleskin | (gat)5 | Prim15-f Prim15-r |
AGCCAGCAGTATTTGGAACG TTCCTCAGCTCCATCATCTG |
Growth factor receptor signaling pathway (Lorenzen et al., 2001) |
| Integrin | (gt)6 | Could not design | Integrin biosynthetic process (GO:0045112) | |
| Intraflagellar transport 74 | (tta)9 | Could not design | Cell adhesion (GO:0007155) | |
| Mitochondrial ATP synthase subunit 9 precursor | (ca)5 | ssh5365ms-f ssh536ms-r |
CCCACCCGTTGACAAATTAC GGCCTTCCTGCTATTGTTTG |
Translation (GO:0006412) |
| Mitochondrial, mRNA | (ca)8 | Could not design | Mitochondrial RNA processing (GO:0000963), mitochondrial DNA metabolic process (GO:0032042) | |
| Mucin | (aca)5 | Could not design | Response to extracellular stimuli (GO:0009991) | |
| Mus musculas BAC clone RP24-80G20 | (tct)5 | Could not design | nd | |
| Myosin | (aca)5 | Prim10-f Prim10-r |
CCATGCAACAATTACCACCA TCCTTGCTGATGAGGAGGTC |
Myosin filament assembly (GO:0031034) |
| Neurotransmitter transporter | (tg)6, (tg)9 | Prim21-f Prim21-r |
AGAACAATGGGGCAACAAAG ACTGACCCATGACCAGTTCC |
Regulation of neurotransmitter transport (GO:0051588) |
| Orhithine decarboxylase | (ca)5, (gt)5, (tg)5 | ssh7214ms-f ssh7214ms-r ssh9020ms-f ssh9020ms-r |
ACTTCTTAACTACATCTCCAACA GGCAGGTACAACCCCAACT TCTGTTGAAATCATTGCTGACC CTTCTTAACTACATCTCCAACA |
Cellular biosynthetic process (GO:0044249) |
| Protein kinase A | (aga)5 | Could not design | Activation of protein kinase activity (GO:0034199) | |
| Retrotransposon-like 1 | (gaa)5, (agg)6 | Prim8-f Prim8-r |
TACTGTAGCCCAGAGCAGCA ACTCGACTCAACCTGACCGT |
Transposition, RNA-mediated (GO:0032197) |
| RNA-binding region containing 2 | (ta)5, (tg)6 | Prim25-f Prim25-r |
CTCAACCAGCAGCTACTCCC ATGGCCAACTTTCATTGGTC |
3′-UTR-mediated mRNA stabilisation (GO:0070935) |
| SET translocation | (atc)5 | CAGAGCCCATGGCTAGGATA GGGGAGATCATCAGAATCCA |
Nucleosome assembly (GO:0006334) | |
| Small androgen receptor-interacting protein | (tca)8 | Prim28-f Prim28-r |
CACCTCCAGTGTGTCTCCCT AAAAGGCAGAAATGCGAAGA |
nd |
| Splicing factor arginine/serine-rich | (ga)5 | Prim14-f Prim14-r |
AAGACGAGCCCTACCTGCTT CTTTTCATTGGGTTGGCATC |
RNA splicing (GO:0008380) |
| Testoseterone 6-beta-hydroxylase | (tta)5 | Prim16-f Prim16-r |
CACGTGCTGAGGAAGATCAA CTGGCGCACCATAGTTTTCT |
Hydrolase activity (GO:0010979) |
| Vitellogemin II precursor | (atg)6, (tga)5 | Prim11-f Prim11-r |
AGCAATGCCTCTTGAACCAC TTAATGATGCCTGCTGCTTG |
Low-density lipoprotein receptor biosynthetic process (GO:0045713) |
| Hypothetical protein | (tca)5 | Prim20-f Prim20-r |
TGAAGCGATCAGCAACTCTG TGAACGCCTGGAAGAAGACT |
|
| No hit | (ga)5 | prim1f prim1r |
TGGTTCTTTTGGTTTTGGGT GTATAAACGCGGGGTGGAG |
|
| (ac)6 | prim2f prim2r |
ACAGCCTTTGTTTCTGGTGG AAGGTTGAAGCGTAAGCGAA |
||
| (caa)6 | prim3f prim3r |
CGTCACTTGAGGAAGCAACA GCAGTTGTTGCTGACGTTGT |
||
| (cat)n | prim4f prim4r |
TGTTACAGCCTCAGCCTCAA AAGGTGGAGCAAGCAGTGAT |
||
| (tta)n | prim5f prim5r |
CTCGTTTTCAGGGCATCATT TGTGCGCCTTTTGAAATACA |
||
| (atc)5 | prim6f prim6r |
CTGAGAATTCCCCAAGCATT TGGGACTCAAGGTTTTCTGG |
||
| (atc)5 | prim7f prim7r |
CTGAGAATTCCCCAAGCATT TGGGACTCAAGGTTTTCTGG |
||
| (ata)7 | ssh6a21ms-f ssh6a21ms-r |
CATGTCGATAATCTCTCTTTAATCACA CGGCATTGCATTACAACAGT |
||
| (ac)5 | ssh6070ms-f ssh6070ms-r |
ATGGTTAGTTGGTTCAGACTTTG CCCGGGCAGGTACTATGAT |
nd, no data; SSR, simple sequence repeats; GO, Gene Ontology; UTR, untranslated region.
Primers were designed from all eSSRs except from those showing significant homology to ribosomal proteins. As stated above, it proved to be difficult to design primers from some of the sequences either because binding sites were located at the end of the EST or were flanked by AT-rich sequences. However, primers were successfully designed from 33 ESTs. Primers from these sequences, including 46 primers previously designed from gSSRs (Wake and Vredenburg, 2008), and the set of six ISSR primers (Hashizume et al., 2003; Geleta and Bryngelsson, 2009) were used for the marker transferability test.
3.2. Characterisation of parasite resistance and susceptibility traits in F1 progeny and segregating F2 populations
As in previous studies the resistant snail (BS-90) served as male while the susceptible snail (NMRI) served as female for the genetic cross to produce the F1 hybrid population (Knight et al., 1999). Because the BS-90 snail is a pigmented snail, a trait controlled by a single dominant gene that is inherited according to simple Mendelian genetics (Richards, 1975), selection of the recombinant inbred pigmented line (RIL) from the susceptible NMRI parent that has no pigmentation after cross-fertilisation was straightforward. Two independent crosses were performed (families A and B), and from these 21 F1 progeny snails from each cross were raised. All F1 progeny were, as expected, pigmented and displayed variability to S. mansoni infection. Two hundred and six F2 snails were derived from the F1 snails by self-fertilisation, and from these F2 populations 153 of the 206 (74.27%) progeny snails in this generation were also, as expected, pigmented confirming the Mendelian genetics (3:1 dominant gene ratio) with χ2 test significance (P value <0.05) of the basic pigmentation pattern in B. glabrata as described previously (Richards, 1975) (data not shown). This result showed that the distribution of F2 population could be accepted as significant for use in further molecular genetic analysis. The inheritance of the susceptibility phenotype was examined in all F2 progeny snails by individual S. mansoni infection as described above. Results showed that 123 (59.71%) snails from the F2 population failed to shed infective cercariae 16 weeks after parasite exposure and were thus classified as resistant snails. Variations in percentage susceptibility to parasite infection in resultant segregating F2 progenies ranged from 56.4–79.9% and 44.8–77.2% for cercariae shedding from families A and B, respectively, from their corresponding F1 hybrids. These results confirmed that as previously reported, juvenile resistance and susceptibility of B. glabrata to S. mansoni infection are both polygenic traits that are governed by non-dominant genes (Richards, 1973).
3.3. Primer determination for segregation markers
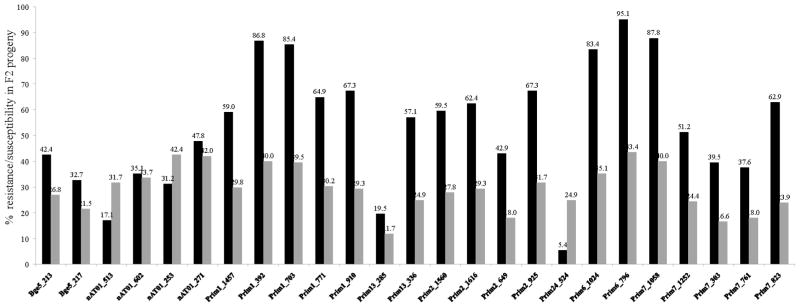
Thirty-three Bg-eSSR, 46 gSSR and six ISSR primer pairs were used to screen the parent, individual F1 population and bulks of F2 progeny. Of these, 14 primers pairs (10 from Bg-eSSR and four from gSSR) produced 35 bands that were specific either for the resistant BS-90 or susceptible NMRI parent, and consistently yielded co-dominant/segregation markers in segregating F2 snails. These primer pairs were used to amplify DNA from 206 individual F2 snails, 17 F1 snails and parents. Each marker (fluorescent labelled amplicon) was calculated after comparison with a known standard marker as described in Section 2.2. The inheritance of all markers in the F2 population were recorded and plotted for percentage resistance/susceptibility (Fig. 2). By t-test analysis, markers Prim1_910, Prim1_771, Prim7_823 and Prim6_1024, which originated from BS-90, were significantly associated with the resistant (P-value <0.05) compared with the susceptible phenotype, while Prim24_524 from the NMRI snail showed higher association with the susceptible phenotype (Fig. 2). From our study, we also found a new recombinant band at 260 bp, which amplified from the primer ‘Prim2’ that did not exist in either parent (data not shown). This result is not entirely surprising since previous studies point to the plasticity of the B. glabrata genome as in other molluscs that showed difficulties in microsatellite marker development (McInerney et al., 2010). In addition, we have shown previously in B. glabrata embryonic cell lines (isolates 1 and 2) that this snail can display extensive aneuploidy, extending the normal diploid number of 36 chromosomes (n = 18) (Odoemelam et al., 2010). Moreover, the basic chromosome number of planorbidae snails, including B. glabrata, can exhibit diploid, tetraploid, hexaploid and even octoploid levels of polyploidy (Patterson and Burch, 1978; Goldman et al., 1984; Odoemelam et al., 2009). Mechanism(s) involved in this expansion remain to be studied. However, currently available data in the public domain for the B. glabrata genome shows that aside from its inherent repetitive nature it also contains different types of mobile genetic elements. We previously characterised one of these elements, the non-long terminal repeat (LTR) retrotransposon, nimbus (Raghavan et al., 2007), and have since shown that the reverse transcriptase (RT) domain of this element is co-transcribed with the stress gene, Hsp 70, early in susceptible juvenile snails (NMRI and M-line) after S. mansoni infection (Ittiprasert et al., 2009).
Fig. 2.
Percentage of F2 snail progeny resistance and susceptibility to Schistosoma mansoni infection. Black bar = resistant, grey bar = susceptible.
3.4. Genetic mapping
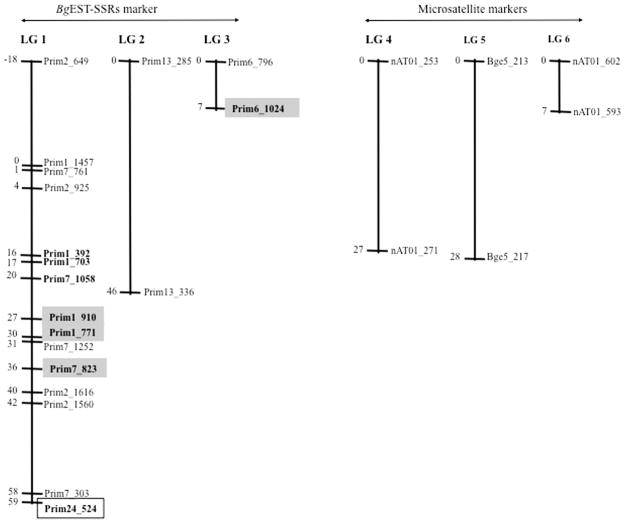
To study the linkage analysis of resistance and susceptibility of S. mansoni in B. glabrata, we constructed a genetic linkage map using 28 segregating loci identified from 10 Bg-eSSRs and four gSSRs in the F2 population. Each locus (marker) was recorded as 0 for cercarial shedding (susceptible phenotype) and 1 for no cercaria shedding (resistant phenotype). The data in Excel format was analysed by the JoinMap software to construct the LGs. The linkage map was constructed at a LOD score threshold of three or higher. From our analysis, the F2 population showed a recombination frequency, for each locus, ranging from between 0.0221–0.1591. Twenty-five loci mapped into three LGs (Fig. 3) spanning a total genetic distance of 192 cM. The length of each LG varied from 1 to 59 cM, with the closest being at 1 cM. Results showed that 19 out of 28 (67.86%) loci identified using BgEST-SSR derived markers showed significant linkage while only six loci identified using gSSR markers showed linkage. Moreover, all loci showing significant linkage to either the resistance or susceptibility traits to infection were all derived from Bg-eSSRs. We therefore show from this study that for B. glabrata, SSR based markers designed from ESTs have a higher efficiency for the construction of linkage maps compared with those designed from gSSR based markers. According to the BLASTX analysis we conducted for resistance-related markers, Prim1_910, Prim1_771 and Prim7_823, amplified using primer pair Prim1, Prim6 and Prim7, respectively, no significant hits were obtained. However, the Prim6_1024 locus (susceptibility-related) showed homology to the high mobility group B1 protein that is involved in a DNA ligation function. Interestingly, resistant and susceptible loci were located on the same LG (LG 1). On the basis of our results, it is possible that the gene(s) controlling juvenile resistance and susceptibility to S. mansoni infection in B. glabrata are not only on the same LG but lie within a short distance (42 cM) of each other (Fig. 3). From recent data showing tight linkage of gene loci associated with resistance, namely Cu–Zn superoxide dismutase (SOD), and two other genes (catalase and peroxiredoxin) of the oxidation–reduction pathway, it has been suggested there might be epistatic interactions with gene loci that are important for defense in the snail (Blouin et al., 2013).
Fig. 3.
Genetic linkage map constructed using a F2 population obtained from two families of the interspecific cross: BS-90 × NMRI snails. Linkage groups were arranged by JoinMap v.3 software. Positions of loci are given in centiMorgans (Kosambi, 1944). Fragment sizes (in bp) are given as the end of the marker names. The markers in bold within the gray and white boxes indicate markers that related to resistant and susceptible loci, respectively. BgEST-SSR, Biomphalaria glabrata expressed sequence tag simple sequence repeat.
3.5. Concluding remarks
We have identified and characterised functional polymorphic eSSR markers from existing B. glabrata ESTs in GenBank. Results showed a high frequency of transferability of these markers between parental snails and F1 and F2 progeny populations. Compared with gSSR markers, eSSRs were useful for the construction of a genetic map for this snail and are, therefore, a good alternative molecular tool for studying the genetics of B. glabrata resistance and susceptibility to parasite infection. Accordingly, 67.88% of Bg-eSSR markers helped to identify four resistant phenotype-related loci. Furthermore, because these markers were developed based on expressed sequences, we hope a novel gene silencing RNA interference (RNAi) soaking method that we have recently developed (Knight et al., 2011b) will help to reveal the function of these identified genes as linked to B. glabrata susceptibility and resistance to S. mansoni infection. In addition, using the cloned markers as probes, corresponding bacterial artificial chromosomes (BACs) have now been identified and are currently being utilised (Adema et al., 2006) for the physical mapping of ex vivo B. glabrata chromosomes (Odoemelam et al., 2009, 2010).
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Institute of Allergy and Infectious Diseases, National Institute of Health, USA, R01-AI063480 and the Intramural Research Program of the Division of Intramural Research at the National Institute of Allergy and Infectious Diseases, National Institute of Health. We are grateful to Ms Frances Barnes for technical support and to Dr. Fred Lewis for his helpful editing of the manuscript. We also thank Drs. Nithya Raghavan and Peter FitzGerald for their guidance with the determination of SSR sequences for B. glabrata in GenBank.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.ijpara.2013.03.007.
References
- Adema CM, Hertel LA, Miller RD, Loker ES. A family of fibrinogen-related proteins that precipitates parasite-derived molecules is produced by an invertebrate after infection. Proc Natl Acad Sci USA. 1997;94:8691–8696. doi: 10.1073/pnas.94.16.8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adema CM, Luo MZ, Hanelt B, Hertel LA, Marshall JJ, Zhang SM, DeJong RJ, Kim HR, Kudrna D, Wing RA, Soderlund C, Knight M, Lewis FA, Caldeira RL, Jannotti-Passos LK, Carvalho Odos S, Loker ES. A bacterial artificial chromosome library for Biomphalaria glabrata, intermediate snail host of Schistosoma mansoni. Mem Inst Oswaldo Cruz. 2006;101 (Suppl 1):167–177. doi: 10.1590/s0074-02762006000900027. [DOI] [PubMed] [Google Scholar]
- Aisemberg J, Nahabedian DE, Wider EA, Verrengia Guerrero NR. Comparative study on two freshwater invertebrates for monitoring environmental lead exposure. Toxicology. 2005;210:45–53. doi: 10.1016/j.tox.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Ansaldo M, Nahabedian DE, Di Fonzo C, Wider EA. Effect of cadmium, lead and arsenic on the oviposition, hatching and embryonic survival of Biomphalaria glabrata. Sci Total Environ. 2009;407:1923–1928. doi: 10.1016/j.scitotenv.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Bajpai A, Sridhar S, Reddy HM, Jesudasan RA. BRM-Parser: a tool for comprehensive analysis of BLAST and RepeatMasker results. In Silico Biol. 2007;7:399–403. [PubMed] [Google Scholar]
- Bender RC, Broderick EJ, Goodall CP, Bayne CJ. Respiratory burst of Biomphalaria glabrata hemocytes: Schistosoma mansoni-resistant snails produce more extracellular H2O2 than susceptible snails. J Parasitol. 2005;91:275–279. doi: 10.1645/GE-415R. [DOI] [PubMed] [Google Scholar]
- Blouin MS, Bonner KM, Cooper B, Amarasinghe V, O’Donnell RP, Bayne CJ. Three genes involved in the oxidative burst are closely linked in the genome of the snail, Biomphalaria glabrata. Int J Parasitol. 2013;43:51–55. doi: 10.1016/j.ijpara.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologna JC, Dorn G, Natt F, Weiler J. Linear polyethylenimine as a tool for comparative studies of antisense and short double-stranded RNA oligonucleotides. Nucleosides Nucleotides Nucleic Acids. 2003;22:1729–1731. doi: 10.1081/NCN-120023124. [DOI] [PubMed] [Google Scholar]
- Bouchut A, Roger E, Coustau C, Gourbal B, Mitta G. Compatibility in the Biomphalaria glabrata/Echinostoma caproni model: potential involvement of adhesion genes. Int J Parasitol. 2006;36:175–184. doi: 10.1016/j.ijpara.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Salamini F, Gebhardt C. A potato molecular function map for carbohydrate metabolism and transport. Theor Appl Genet. 2001;102:284–295. [Google Scholar]
- Chitsulo L, Loverde P, Engels D. Schistosomiasis. Nat Rev Microbiol. 2004;2:12–13. doi: 10.1038/nrmicro801. [DOI] [PubMed] [Google Scholar]
- Chu XJ, Guo JG. Impact of climate warming on schistosomiasis transmission and application of relative research techniques. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi. 2009;27:267–271. [PubMed] [Google Scholar]
- Geleta M, Bryngelsson T. Inter simple sequence repeat (ISSR) based analysis of genetic diversity of Lobelia rhynchopetalum (Campanulaceae) Hereditas. 2009;146:122–130. doi: 10.1111/j.1601-5223.2009.02111.x. [DOI] [PubMed] [Google Scholar]
- Goldman MA, LoVerde PT, Chrisman L, Franklin DA. Chromosomal evolution in planorbid snails of the genera Bulinus and Biomphalaria. Malacologia. 1984;25:427–446. [Google Scholar]
- Gryseels B, Polman K, Clerinx J, Kestens L. Human schistosomiasis. Lancet. 2006;368:1106–1118. doi: 10.1016/S0140-6736(06)69440-3. [DOI] [PubMed] [Google Scholar]
- Hanington PC, Forys MA, Dragoo JW, Zhang SM, Adema CM, Loker ES. Role for a somatically diversified lectin in resistance of an invertebrate to parasite infection. Proc Natl Acad Sci USA. 2010;107:21087–21092. doi: 10.1073/pnas.1011242107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashizume T, Shimamoto I, Hirai M. Construction of a linkage map and QTL analysis of horticultural traits for watermelon [Citrullus lanatus (THUNB) MATSUM & NAKAI] using RAPD, RFLP and ISSR markers. Theor Appl Genet. 2003;106:779–785. doi: 10.1007/s00122-002-1030-1. [DOI] [PubMed] [Google Scholar]
- Hoffmann KF, Caspar P, Cheever AW, Wynn TA. IFN-gamma, IL-12, and TNF-alpha are required to maintain reduced liver pathology in mice vaccinated with Schistosoma mansoni eggs and IL-12. J Immunol. 1998;161:4201–4210. [PubMed] [Google Scholar]
- Hubendick B. A possible method of schistosome-vector control by competition between resistant and susceptible strains. Bull WHO. 1958;18:113–116. [PMC free article] [PubMed] [Google Scholar]
- Humphries JE, Yoshino TP. Regulation of hydrogen peroxide release in circulating hemocytes of the planorbid snail Biomphalaria glabrata. Dev Comp Immunol. 2008;32:554–562. doi: 10.1016/j.dci.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter GW, Weinmann CJ, Hoffmann RG. Studies on schistosomiasis. XVII Non-reciprocal acquired resistance between Schistosoma mansoni and Schistosomatium douthitti in mice. Exp Parasitol. 1961;11:133–140. doi: 10.1016/0014-4894(61)90018-2. [DOI] [PubMed] [Google Scholar]
- Imasheva AG, Bosenko DV, Bublii OA, Lazebnyi OE. Effect of three types of ecological stress on the variability of morphological traits in Drosophila melanogaster. Genetika. 1999;35:1379–1385. [PubMed] [Google Scholar]
- Ittiprasert W, Knight M. Reversing the resistance phenotype of the Biomphalaria glabrata snail host Schistosoma mansoni infection by temperature modulation. PLoS Pathog. 2012;8:e1002677. doi: 10.1371/journal.ppat.1002677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ittiprasert W, Nene R, Miller A, Raghavan N, Lewis F, Hodgson J, Knight M. Schistosoma mansoni infection of juvenile Biomphalaria glabrata induces a differential stress response between resistant and susceptible snails. Exp Parasitol. 2009;123:203–211. doi: 10.1016/j.exppara.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ittiprasert W, Miller A, Myers J, Nene V, El-Sayed NM, Knight M. Identification of immediate response genes dominantly expressed in juvenile resistant and susceptible Biomphalaria glabrata snails upon exposure to Schistosoma mansoni. Mol Biochem Parasitol. 2010;169:27–39. doi: 10.1016/j.molbiopara.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Chae JS, Hwang SY, Yoo JY, Kim EK, Yun HJ, Cho JH, Kim J, Kim BW, Kim HC, Kang SS, Lang F, Cho SG, Choi EJ. Negative regulation of SEK1 signaling by serum-and glucocorticoid-inducible protein kinase 1. EMBO. 2007;26:3075–3085. doi: 10.1038/sj.emboj.7601755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight M, Miller AN, Patterson CN, Rowe CG, Michaels G, Carr D, Richards CS, Lewis FA. The identification of markers segregating with resistance to Schistosoma mansoni infection in the snail Biomphalaria glabrata. Proc Natl Acad Sci USA. 1999;96:1510–1515. doi: 10.1073/pnas.96.4.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight M, Ittiprasert W, Odoemelam EC, Adema CM, Miller A, Raghavan N, Bridger JM. Non-random organization of the Biomphalaria glabrata genome in interphase Bge cells and the spatial repositioning of activated genes in cells co-cultured with Schistosoma mansoni. Int J Parasitol. 2011a;41:61–70. doi: 10.1016/j.ijpara.2010.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight M, Miller A, Liu Y, Scaria P, Woodle M, Ittiprasert W. Polyethyleneimine (PEI) mediated siRNA gene silencing in the Schistosoma mansoni snail host, Biomphalaria glabrata. PLoS Negl Trop Dis. 2011b;5:e1212. doi: 10.1371/journal.pntd.0001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosambi DD. The estimation of map distance from recombination values. Annals of Eugenics. 1944;12:172–175. [Google Scholar]
- Kucuktas H, Wang S, Li P, He C, Xu P, Sha Z, Liu H, Jiang Y, Baoprasertkul P, Somridhivej B, Wang Y, Abernathy J, Guo X, Liu L, Muir W, Liu Z. Construction of genetic linkage maps and comparative genome analysis of catfish using gene-associated markers. Genetics. 2009;181:1649–1660. doi: 10.1534/genetics.108.098855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockyer AE, Spinks J, Kane RA, Hoffmann KF, Fitzpatrick JM, Rollinson D, Noble LR, Jones CS. Biomphalaria glabrata transcriptome: cDNA microarray profiling identifies resistant- and susceptible-specific gene expression in haemocytes from snail strains exposed to Schistosoma mansoni. BMC Genomics. 2008;9:634. doi: 10.1186/1471-2164-9-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockyer AE, Emery AM, Kane RA, Walker AJ, Mayer CD, Mitta G, Coustau C, Adema CM, Hanelt B, Rollinson D, Noble LR, Jones CS. Early differential gene expression in haemocytes from resistant and susceptible Biomphalaria glabrata strains in response to Schistosoma mansoni. PLoS One. 2012;7:e51102. doi: 10.1371/journal.pone.0051102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzen A, Stannek C, Lang H, Andrianov V, Kalvinsh I, Schwabe U. Characterization of a G protein-coupled receptor for nicotinic acid. Mol Pharmacol. 2001;59:349–357. doi: 10.1124/mol.59.2.349. [DOI] [PubMed] [Google Scholar]
- Mattos AC, Pereira GC, Jannotti-Passos LK, Kusel JR, Coelho PM. Evaluation of the effect of oxamniquine, praziquantel and a combination of both drugs on the intramolluscan phase of Schistosoma mansoni. Acta Trop. 2007;102:84–91. doi: 10.1016/j.actatropica.2007.04.002. [DOI] [PubMed] [Google Scholar]
- McInerney CE, Allcock AL, Johnson MP, Bailie DA, Prodohl PA. Comparative genomic analysis reveals species-dependent complexities that explain difficulties with microsatellite marker development in molluscs. Heredity. 2010;106:78–87. doi: 10.1038/hdy.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon CA, Chapman JP, Healey JC, Hannington JA. The treatment of mixed affective disorders in general practice: a comparison of trazodone and dothiepin. Curr Med Res Opin. 1990;12:34–42. doi: 10.1185/03007999009111489. [DOI] [PubMed] [Google Scholar]
- Myers J, Ittiprasert W, Raghavan N, Miller A, Knight M. Differences in cysteine protease activity in Schistosoma mansoni-resistant and -susceptible Biomphalaria glabrata and characterization of the hepatopancreas cathepsin B full-length cDNA. J Parasitol. 2008;94:659–668. doi: 10.1645/GE-1410R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odoemelam E, Raghavan N, Miller A, Bridger JM, Knight M. Revised karyotyping and gene mapping of the Biomphalaria glabrata embryonic (Bge) cell line. Int J Parasitol. 2009;39:675–681. doi: 10.1016/j.ijpara.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odoemelam EC, Raghavan N, Ittiprasert W, Miller A, Bridger JM, Knight M. FISH on chromosomes derived from the snail model organism Biomphalaria glabrata. Methods Mol Biol. 2010;659:379–388. doi: 10.1007/978-1-60761-789-1_29. [DOI] [PubMed] [Google Scholar]
- Patterson CM, Burch JB. Chromosomes of pulmonate molluscs. In: Fretter V, Peake J, editors. Pulmonates: Systematic Evolution Ecology. Academic Press; New York: 1978. pp. 171–217. [Google Scholar]
- Patz JA, Epstein PR, Burke TA, Balbus JM. Global climate change and emerging infectious diseases. JAMA. 1996;275:217–223. [PubMed] [Google Scholar]
- Raghavan N, Tettelin H, Miller A, Hostetler J, Tallon L, Knight M. Nimbus (BgI): an active non-LTR retrotransposon of the Schistosoma mansoni snail host Biomphalaria glabrata. Int J Parasitol. 2007;37:1307–1318. doi: 10.1016/j.ijpara.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards CS. Susceptibility of adult Biomphalaria glabrata to Schistosoma mansoni infection. Am J Trop Med Hyg. 1973;22:748–756. doi: 10.4269/ajtmh.1973.22.748. [DOI] [PubMed] [Google Scholar]
- Richards CS. Genetics of pigmentation in Biomphalaria straminea. Am J Trop Med Hyg. 1975;24:154–156. doi: 10.4269/ajtmh.1975.24.154. [DOI] [PubMed] [Google Scholar]
- Richards CS, Merritt JW., Jr Genetic factors in the susceptibility of juvenile Biomphalaria glabrata to Schistosoma mansoni infection. Am J Trop Med Hyg. 1972;21:425–434. doi: 10.4269/ajtmh.1972.21.425. [DOI] [PubMed] [Google Scholar]
- Richards CS, Shade PC. The genetic variation of compatibility in Biomphalaria glabrata and Schistosoma mansoni. J Parasitol. 1987;73:1146–1151. [PubMed] [Google Scholar]
- Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 2001;11:1441–1452. doi: 10.1101/gr.184001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel T, Michalek W, Varshney RK, Graner A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.) Theor Appl Genet. 2003;106:411–422. doi: 10.1007/s00122-002-1031-0. [DOI] [PubMed] [Google Scholar]
- van Die I, Cummings RD. Glycan gimmickry by parasitic helminths: a strategy for modulating the host immune response? Glycobiology. 2010;20:2–12. doi: 10.1093/glycob/cwp140. [DOI] [PubMed] [Google Scholar]
- Wake DB, Vredenburg VT. Colloquium paper: are we in the midst of the sixth mass extinction? A view from the world of amphibians. Proc Natl Acad Sci USA. 2008;105 (Suppl 1):11466–11473. doi: 10.1073/pnas.0801921105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SM, Zeng Y, Loker ES. Expression profiling and binding properties of fibrinogen-related proteins (FREPs), plasma proteins from the schistosome snail host Biomphalaria glabrata. Innate Immunity. 2008;14:175–189. doi: 10.1177/1753425908093800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.