Abstract
The Xenopus inner ear provides a useful model for studies of hearing and balance because it shares features with the mammalian inner ear, and because amphibians are capable of regenerating damaged mechanosensory hair cells. The structure and function of many proteins necessary for inner ear function have yet to be elucidated and require methods for analysis. To this end, we seek to characterize Xenopus inner ear genes outside of the animal model through heterologous expression in cell lines. As part of this effort, we aimed to optimize physical (electroporation), chemical (lipid-mediated; Lipofectamine™ 2000, Metafectene® Pro), and biological (viral-mediated; BacMam virus Cellular Lights™ Tubulin-RFP) gene delivery methods in amphibian (Xenopus; A6) cells and mammalian (Chinese hamster ovary (CHO)) cells. We successfully introduced the commercially available pEGFP-N3, pmCherry-N1, pEYFP-Tubulin, and Cellular Lights™ Tubulin-RFP fluorescent constructs to cells and evaluated their transfection or transduction efficiencies using the three gene delivery methods. In addition, we analyzed the transfection efficiency of a novel construct synthesized in our laboratory by cloning the Xenopus inner ear calcium-activated potassium channel β1 subunit, then subcloning the subunit into the pmCherry-N1 vector. Every gene delivery method was significantly more effective in CHO cells. Although results for the A6 cell line were not statistically significant, both cell lines illustrate a trend towards more efficient gene delivery using viral-mediated methods; however the cost of viral transduction is also much higher. Our findings demonstrate the need to improve gene delivery methods for amphibian cells and underscore the necessity for a greater understanding of amphibian cell biology.
Keywords: Transfection, BacMam, Electroporation, Heterologous expression, Xenopus inner ear
Introduction
Heterologous gene expression is a powerful method for characterizing the function of unknown genes. This technique requires the delivery of genes through methods developed in the last five decades (Butel et al. 1975; Scolnick and Bumgarner 1975). The success of any given gene delivery method can vary from organism to organism, as well as by cell and tissue type. We are interested in probing the function of genes expressed in the Xenopus laevis inner ear that have been identified through cDNA library construction, RACE, and microarray analysis (Varela-Ramírez et al. 1998; Serrano et al. 2001; Powers et al. 2007; Sultemeier et al. 2005). Alternative systems for genetic analysis can advance such studies because of the relatively small size and extreme inaccessibility of inner ear organs (Díaz et al. 1995; Trujillo-Provencio et al. 2009). Our aim is to characterize inner ear genes through heterologous expression in cell lines. In particular, the X. laevis (A6) kidney cell line offers advantages as a compatible expression system for these genes because it shares a species origin with the animal from which the genes were isolated and because kidney and ear tissues share a polarized epithelial arrangement and similar drug susceptibility (Knight and Serrano 2006). However, the application of traditional gene delivery methods has not been optimized for Xenopus cell lines (Kay and Peng 1991).
In order to address this problem, we evaluated the capacity of physical (electroporation), chemical (cationic lipids), or biological (viral vector), methods to provide optimal gene delivery to the A6 cell line. Cationic lipids (such as Lipofectamine™ 2000 and Metafectene® Pro) all share three structural features: a positively charged head group, a hydrophobic anchor, and a linker between these. The positively charged head group interacts with the phosphate backbone of DNA to form a liposomal structure with a positive surface charge. The positive charge of the liposomal structure facilitates its interaction with the cell membrane, and the transfection complex enters the cell by the process of endocytosis (Wyatt and Giorgio 2004). In contrast, electroporation utilizes an electrical pulse in order to create transient pores in the cell membrane, allowing the foreign DNA to enter the cell (Barry 2004). However, physical gene delivery methods frequently result in damage to cells because the integrity of the cell membrane is disrupted. All cell types are susceptible to physical and chemical gene delivery, making these methods advantageous over biological delivery techniques, which tend to be more cell specific.
Recently, the unique properties of BacMam viruses have been exploited to facilitate gene delivery. This technique uses the ability of baculoviruses, which are pathogenic only to arthropods, to enter mammalian cells through clathrin¬mediated endocytosis (Long et al. 2006). BacMam viruses are baculoviruses with DNA that has been modified to include mammalian transcriptional units. The addition of mammalian expression cassettes facilitates functional protein production in mammalian cells following transduction with the insect viruses. Because gene delivery using BacMam was designed for use in mammalian cells, we compared gene delivery efficiency in Xenopus (A6) cells to Chinese hamster ovary (CHO) cells, which are commonly used as a universal assay system (Banerjee et al. 1993; Kim et al. 2008; Ibey et al. 2009), and have been successfully transduced with BacMam (Condreay et al. 1999).
In this study, we evaluated gene delivery methods for commercially available fluorescent proteins (pmCherry-N1, pEGFP-N3). Because of our interest in mechanotransduction, the physiological task of the inner ear, we also used fluorescent constructs of proteins involved in sensory hair cell cytoskeletal organization (pEYFP-Tubulin) and electrical activity (BK β1 pmCherry-N1; see below). Furthermore, this set of experiments describes an exploratory effort to deliver genes to Xenopus cells using a commercially available virus (BacMam) that contains a fluorescent tubulin construct (Cellular Lights™ Tubulin-RFP). Tubulin is of particular relevance for inner ear research because it is a crucial structural component of the stereociliary bundles found on the apical surface of mechanosensory hair cells (Shin et al. 2005). Because of our interest in the ion channels that underlie hair cell electrical activity, we prepared a fluorescent construct with a subunit of the calcium-activated potassium channel (BK, slo1, KCa1.1, maxi-K, slowpoke, KCNMA1). The interaction between the α and β subunits of the BK channel is thought to play a role in the perception of pitch by inner ear hair cells (Fettiplace and Fuchs 1999; Oberholtzer 1999; Ramanathan et al. 1999; Duncan and Fuchs 2003; Pyott et al. 2007). The BK β subunit was cloned from X. laevis inner ear RNA and then subcloned into the pmCherry-N1 vector. Consistent with previous reports, our results show differences in the efficacy of biological, chemical, and physical gene delivery methods and suggest that amphibian kidney cells are not as amenable to techniques that have been optimized for introducing genes to mammalian cells.
Methods
Cell lines and reagents
X. laevis A6 cells, Chinese hamster ovary (CHO-K1) cells, and fetal bovine serum were obtained from American Type Culture Collection (ATCC; Manassas, VA). Fluorescent constructs pmCherry-N1, pEGFP-N3, and pEYFP-Tub as well as the SMART™ RACE cDNA Amplification Kit were purchased from Clontech Laboratories, Inc. (Mountain View, CA). Alexa Fluor® 568 phalloidin, Alexa Fluor® 488 phalloidin, 10 mg/ml Hoechst 33342 solution, SlowFade® Antifade kit, Lipofectamine™ 2000, pCR®-XL-TOPO® vector, and Cellular Lights™ Tubulin-RFP (a BacMam virus) were obtained from Invitrogen (Eugene, OR). Newborn calf serum, bovine serum albumin, formaldehyde, Triton X-100, 0.25% trypsin-EDTA, L-glutamine, F12K (Ham's nutrient modification) media, NCTC-109 media, and phosphate-buffered saline (PBS) were purchased from Sigma-Aldrich (St. Louis, MO). Eppendorf hypoosmolar electroporation buffer, 2 mm gap width electroporation cuvettes, Falcon 4¬chambered slides, and tissue culture plasticware were obtained from VWR (West Chester, PA). QiaPrep Spin Miniprep kit was purchased from Qiagen, Inc. (Valencia, CA). Metafectene® Pro was purchased from Biontex Laboratories (Planegg, Germany). BamHI, NheI, and the Quick Ligation™ kit were purchased from New England Biolabs (Ipswich, MA).
Organ extraction, RNA isolation, and quantification procedures
Dissection methods for isolation of inner ear tissue from juvenile X. laevis and the method of total RNA isolation were according to the procedures outlined in Trujillo-Provencio et al. (2009). The methods performed with X. laevis were in compliance with the Institutional Animal Care and Use Committee of New Mexico State University.
Cloning of BK beta subunit into pmCherry-N1
Primers were designed against the Xenopus tropicalis BK channel β subunit sequence obtained from the JGI X. tropicalis genome assembly 4.1 sequencing project (Joint Genome Institute 1997). The forward (sequence: GTCGTAACAACTCCGCCCC) and reverse (sequence: ATCCTCCTCGCCCTTGCT) primers were produced by Eurofins MWG Operon (Huntsville, AL). Using the primers designed against the X. tropicalis sequence, the BK β sequence from X. laevis inner ear RNA was amplified using SMART™ RACE cDNA Amplification Kit RT-PCR RACE reactions. The resultant PCR product was gel isolated and cloned initially into the pCR®-XL¬TOPO® vector (Invitrogen), following the vendor's recommendations. The BK β insert was digested from the TOPO vector with the restriction enzymes BamHI and NheI, then subcloned using a Quick Ligation™ kit (New England Biolabs) into the pmCherry-N1 vector (Clontech), which was also digested with BamHI and NheI. DNA sequencing with an ABI Prism 3100 automated sequencer verified the identity of the inner ear BK β subunit (618 base pairs; 205 amino acids) in both vectors (NCBI accession number JN679868).
Cell lines
A6 cells (an established cell line derived in 1969 from the normal kidney of an adult X. laevis male) and Chinese hamster ovary cells (CHO-K1; a subclone of the original established CHO cell line derived in 1957 from an adult female Cricetulus griseus) were obtained from the ATCC (Manassas, VA). In accordance with recommended practices for maintenance of cell line identity, A6 and CHO cells were resuspended (passage zero), then discarded following the eighth passage or after two consecutive months in culture, whichever came first. The species origin of the A6 cell line was confirmed by amplifying genomic sequence for aldolase A, fructose-bisphosphate (Aldoa) according to established procedures (Liu et al. 2004; forward primer, TGTGCCCSGTATAAGAAGGATGG; reverse primer, CCCATCAGGGAGAATTTCAGGCTCCACAA). The amplified sequence shared a 99% identity with X. laevis and 91% identity with the closely related X. tropicalis (AGTGGCGCTCCGTGCTCAAGATCTCTGAACACACCCCTCTCACCTGGCCATCATAGAGAATGCCAACGTATTGGCCCGTTACGCCAGCATCTGCCAACAG).
Propagation of A6 and CHO cells
A6 and CHO cells were cultured according to the specifications of the vendor, ATCC. X. laevis A6 kidney cells (A6, ATCC CCL-102) were grown in poly-lysine-coated T-25 flasks in a temperature (28°C) and CO2 (5%) controlled incubator. A6 cells were grown in NCTC-109 media that was supplemented with 9% ddH2O, 10% fetal bovine serum, and 2 mM L-glutamine. A6 cells were subcultured every 4 to 6 d. Chinese hamster ovary cells (CHO-K1, ATCC CRL-9618) were grown in a temperature (37°C) and CO2 (5%) controlled incubator. CHO cells were grown in poly-lysine coated T-75 culture flasks in complete Ham's F12K media supplemented with 10% fetal bovine serum, and subcultured every 2 to 4 d.
All media and solutions used to culture cells were purchased sterile or were sterilized by filtration through 0.2 μm membranes. Media were replenished for both cell lines every 48 h. All cells were subcultured by partial digestion in 0.25% trypsin-EDTA. Cells were regularly monitored for contamination and overall morphology using an inverted Nikon Diaphot TMD phase-contrast microscope.
Optimization of transfection conditions
Following the manufacturer's recommendations, transfection methods with Lipofectamine™ 2000 and Metafectene® Pro were optimized by exposing cells to varied ratios of DNA to cationic lipid. Preliminary studies tested Lipofectamine™ 2000 and Metafectene® Pro reagents on both cell lines. The ratio of DNA (in micrograms) to lipid-mediated reagent (in microliters) was varied from 1:1 to 1:5. Reagent incubation times between 6 and 48 h were tested on cells that were plated at densities that ranged from 3×104 to 4×104 cells/ cm2. For A6 cells in every condition with Lipofectamine™ 2000, the transfection efficiency was always lower than 3.7%. In contrast, a 48-h incubation with Metafectene® Pro at a DNA/lipid ratio of 1 μg: 1 μL resulted in a slightly higher efficiency of 4.0% for A6 cells in our preliminary study. Therefore, we implemented Metafectene® Pro with these parameters for our subsequent experiments. For CHO cells transfected with Metafectene® Pro, the efficiency was lower than 1.5% in every condition. In contrast, a 24¬h incubation with Lipofectamine™ 2000 using a DNA/lipid ratio of 1 μg:5 μL for our preliminary CHO cell transfection resulted in a much higher efficiency of 17.1%. This optimization procedure led to the implementation of a 24-h incubation with Lipofectamine™ 2000 using a DNA/lipid ratio of 1 μg: 5 μL for CHO cell transfection.
Transfection with lipid-mediated reagents
After trypsinization, cells were titered using a hemocytometer, centrifuged, resuspended, and plated in Falcon 4-chambered culture slides at a density of 4×104 cells/cm2. The cells were grown for 24 h prior to transfection (70–85% confluent). On the day of the transfections, the media was removed from all the chambers and 900 μL of fresh media was added to each chamber except for the autoflourescence control, which received 1 ml of fresh media. Either 5 μL of Lipofectamine™ 2000 (for CHO cells) or 1 μL Metafectene® Pro (for A6 cells) was added to 50 μL of serum-free media and incubated at room temperature for 5 min. Additionally, 1 μg of plasmid DNA from pEGFP-N3, pEYFP-Tub, pmCherry-N1, or BK β1 pmCherry-N1 was placed in separate tubes containing 50 μL serum-free media. After incubating the lipid reagents for 5 min, the DNA in media was added to the tubes containing either Lipofectamine™ 2000 or Metafectene® Pro. The lipid-DNA mixtures were incubated for 20 min at room temperature to allow formation of liposomes before adding the entire volume to the cells. Cells were incubated for 24 h (CHO) or 48 h (A6) in a temperature and CO2 controlled incubator.
Transfection with electroporation
The Eppendorf Multiporator® was used for the electroporation of the two different cell lines. The cells were trypsinized and following the manufacturer's protocol were resuspended in media with 0.5% fetal calf serum. The resuspended cells were titered using a hemocytometer, and the total amount of cells needed for the experiment was determined. Cells were centrifuged and diluted to a density of 1×106 cells/mL in hypoosmolar buffer; 450 μL of the cell suspension was transferred into a microcentrifuge tube, and 4.5 μg of plasmid DNA from either pEGFP-N3, pmCherry-N1, pEYFP-Tub, or BK β1 pmCherry-N1 were added to give a final DNA concentration of 10 μg/mL. The cell suspension containing the plasmid DNA was transferred to a 2-mm gap junction electroporation cuvette.
For both cell lines, the field strength (in volts per centimeter) of the electrical pulse was varied by applying a pulse voltage of 290, 580, 900, and 1,200 V to the cells to be electroporated. After placing the cuvette in the electroporator, the time constant was set to 40 μs and cells were exposed to one pulse of the electrical field. After the application of the current, cells were allowed to recover for 10 min at room temperature then plated in Falcon 4¬chambered culture slides at a density of approximately 1.2×105 cells/cm2. Cells in hypoosmolar solution were added to complete growth media at an approximate ratio of 1:5, then incubated for 24 h (CHO) or 48 h (A6) in a temperature and CO2 controlled incubator.
Transduction with Cellular Lights™ Tubulin-RFP, a BacMam virus
After trypsinization, cells were titered using a hemocytometer, centrifuged, resuspended, and plated in Falcon 4-chambered culture slides at a density of 4×104 cells per cm2. The cells were grown for 24 h prior to transduction. On the day of the transductions, following the manufacturer's protocol, the media were removed from all the chambers and 1 ml of the Cellular Lights™ Tubulin-RFP solution (2 ml of Cellular Lights™ Tubulin-RFP or null reagent combined with 3.5 ml of D-PBS without Ca2+/Mg2+) was added to each chamber except for a control to which 1 ml of D-PBS without Ca2+/Mg2+ was added. Slides were placed on a lab rotator and rocked gently for 2 h at room temperature in the dark. After 2 h, the Cellular Lights™ solutions and PBS were replaced with 1 ml of the enhancer solution. Slides were placed in the incubator for 2 h after which the enhancer solution was removed and 1 ml of fresh media was placed into each chamber. Slides were incubated for 24 h in a temperature and CO2-controlled incubator prior to fixation.
Cell labeling and fixation
After the allotted times for transfection or transduction, the cells were removed from the incubator and all subsequent manipulations were done at room temperature. The cells were first washed with serum-free media for 2 min and then fixed with 3.7% formaldehyde in PBS for 10 min. Following this, the cells were washed twice with 1× PBS for 2 min. Cells were permeabilized for 4 min with 0.1% Triton X-100 diluted in PBS and washed twice with 1× PBS for 2 min. Cells were then labeled for 20 min with 150 nM of either Alexa Fluor® 568 phalloidin (for cells transfected with pEGFP-N3 or pEYFP-Tub) or Alexa Fluor® 488 phalloidin (for cells transfected with pmCherry-N1, BK β1 pmCherry-N1, or Cellular Lights™ Tubulin-RFP) diluted in 1× PBS with 1% bovine serum albumin in all chambers except the auto-fluorescence control which was incubated with 1% bovine serum albumin diluted in 1× PBS. Cells were washed twice with 1× PBS for 2 min each. Nuclei were stained for ten minutes with 2 μg/mL Hoechst 33342 in 1× PBS for all samples except the autofluorescence control which was incubated with 1× PBS. Cells were then washed twice with 1× PBS for 2 min each. Following the final PBS washes, samples were equilibrated with component C from the SlowFade® Antifade kit (Invitrogen) for 15 min. Component C and the plastic slide wells were removed before adding one to two drops of solution A from the SlowFade® Antifade kit to the microscope slides, which were then cover slipped and sealed with nail polish. The microscope slides were stored for 24 h at 4°C before images were acquired.
Image acquisition, analysis, and processing
An inverted Nikon Eclipse TE-2000-S epifluorescent microscope equipped with UV-2A, B-2E/C (FITC), G-2E/C (TRITC), and Cy5 HQ filter cubes was used to capture images from the fixed and labeled cells. The combination of filters at hand were excellent for exciting and separating emission contributions from the fluorophores used in this experiment: Hoechst 33342 (UV-2A), Alexa Fluor® 568 (G-2E/ C), Alexa Fluor® 488 (B-2E/C), pEGFP-N3 (B-2E/C), RFP (G-2E/C), and mCherry (Cy5 HQ). The TE-2000-S microscope also has differential interference contrast (DIC) capabilities that was used for visual analysis of cell size, morphology, and for a representation of the field of view from which epifluorescent images were captured. Using a 20× objective, images were acquired with a CoolSNAP™ HQ CCD camera (Photometrics, Tucson, AZ) attached to the microscope and the MetaVue™ Imaging System Version 6.0r5 software (Molecular Devices Corporation, Sunnyvale, CA). Images were superimposed to ensure that the fluorescent labels corresponded to cells in the field of view and to determine true transfection or transduction events. Coloring and superimposition of images was performed with either the MetaMorph® Offline Imaging System Version 6.0r5 software (Molecular Devices Corporation) or Adobe Photoshop Version CS5 (Adobe Systems Inc., San Jose, CA).
Calculation of transfection efficiency
Captured images were analyzed using either MetaMorph® Offline Imaging System Version 6.0r5 or Adobe Photoshop Version CS5 software to count both the number of cells expressing fluorescent proteins (transfection or transduction events) and the number of nuclei. The efficiencies were calculated by dividing the number of transfection or transduction events in the field of view by the number of nuclei in the same field of view. Nuclei and transfection events from three different areas (subsamples) of each well were measured, calculated, and averaged to determine the mean efficiency of each experiment. The experiment was repeated four to six times (n=4–6) in every condition for both cell lines (see “Results”). The mean efficiency of the gene delivery method was then calculated by averaging the means derived from replicate experimental conditions (n=4–6). A one-way ANOVA was performed for each cell line to determine statistical significance between gene delivery methods. Means are shown with Gabriel comparison intervals (Gabriel 1978). Pairs of means whose Gabriel comparison intervals do not overlap are statistically different using a significance value of 0.05. Student's t tests were performed to compare conditions across cell lines; comparisons where p <0.05 were taken to be significantly different.
Results
Transfection and transduction efficiency in the A6 cell line
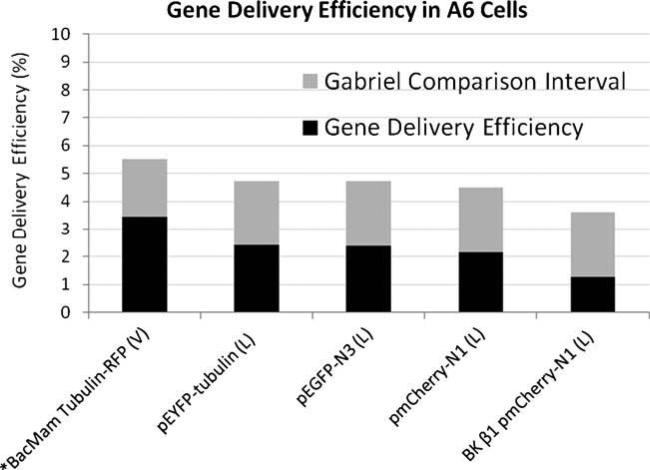
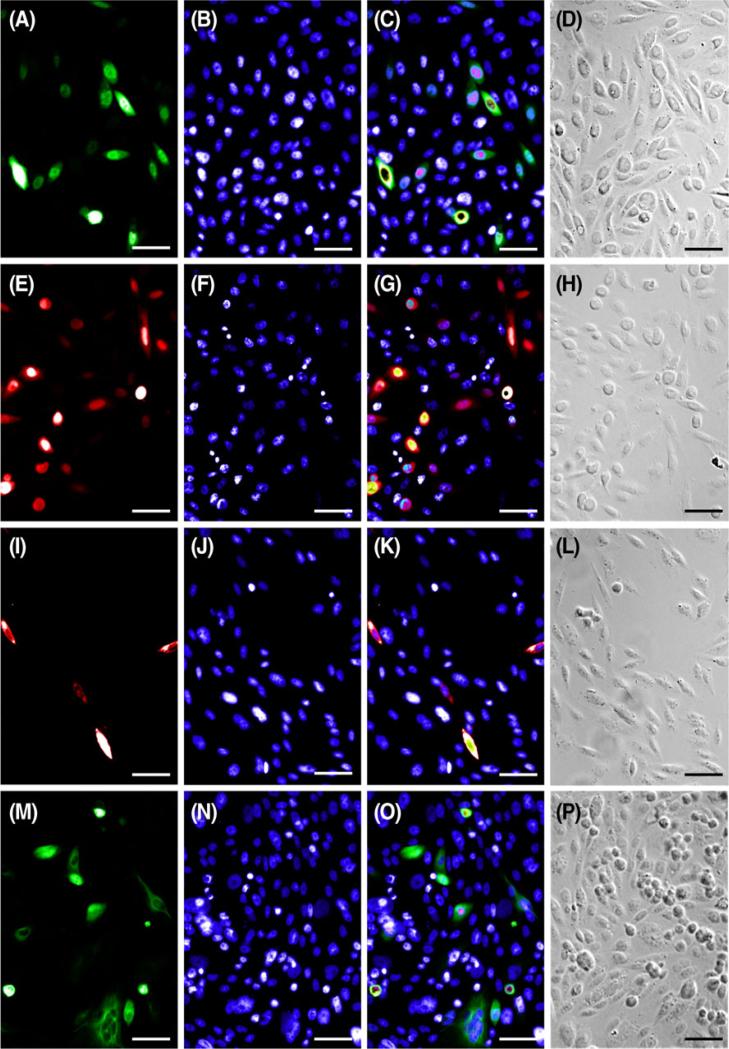
Amphibian (A6) cells were responsive to biological (viral-mediated) and chemical (lipid-mediated) delivery of commercially available fluorescent constructs as well as our novel inner ear BK β1 pmCherry-N1 construct. Gene delivery efficiency in this cell line using Metafectene® Pro was comparable regardless of the plasmid (2.38% for pEGFP-N3, Figs. 1 and 2A–D; 2.16% for pmCherry-N1, Figs. 1 and 2E–H; 1.26% for BK β1 pmCherry-N1, Figs. 1 and 2I–L; 2.41% for pEYFP-Tub, Figs. 1 and 2M–P). In every instance, the efficiency of gene delivery using the BacMam system, Cellular Lights™ Tubulin-RFP (3.43%, Figs. 1 and 3A–D) was greater than that found while using Metafectene® Pro; however, these variations were not statistically different using a significance value of 0.05 for the Gabriel comparison interval (Gabriel 1978). Electroporation was not successful in the A6 cell line, although it was attempted four to six times with each plasmid. When A6 cells were subjected to varying voltages (290–1,200 V), the cell survival rate was high but transfection events were low to nonexistent.
Figure 1. Gene delivery efficiency in A6 cells.
Mean transfection or transduction efficiencies for X. laevis kidney (A6) cells calculated from four separate experiments, except where n=5 (asterisk). Genes (listed under each column) were delivered using either Metafectene® Pro (L) or the BacMam virus (V). Pairs of means with Gabriel comparison intervals that do not overlap are significantly different (Gabriel 1978; p<0.05).
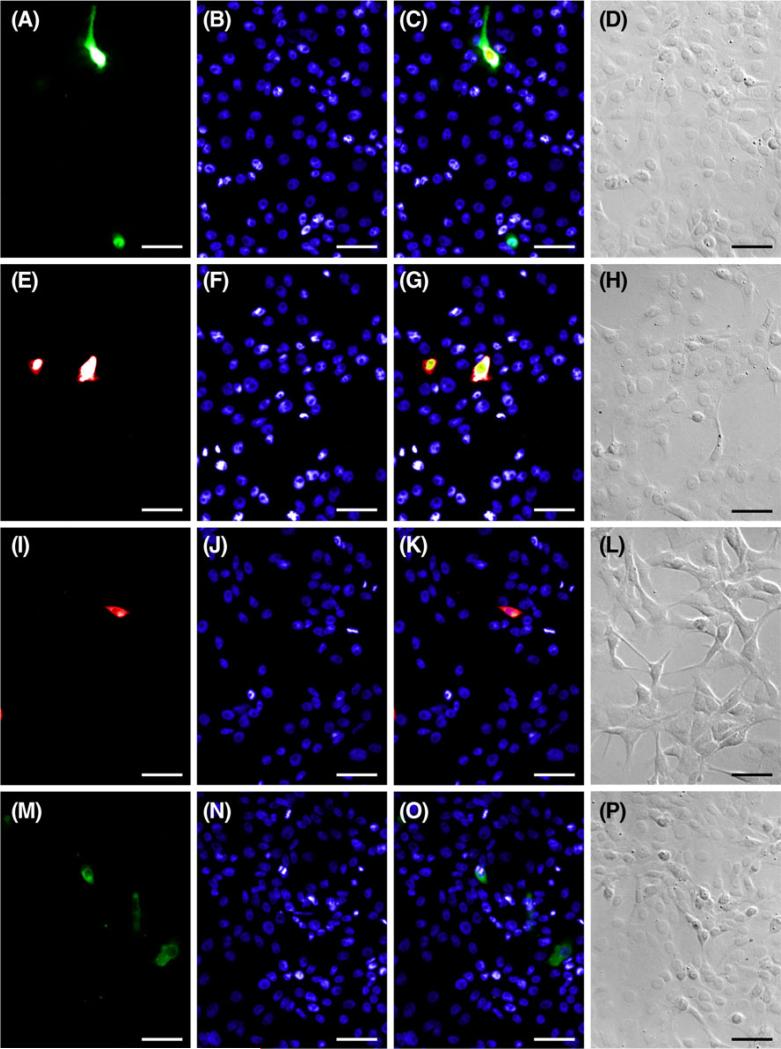
Figure 2. Lipid-mediated transfection in A6 cells.
Representative images of A6 cells used to calculate lipid-mediated transfection efficiencies. Cells transfected with pEGFP-N3 (A), pmCherry-N1 (E), pmCherry-N1 BK β1(I), and EYFP-tub (M) were counterstained with Hoechst (B, F, J, N) to calculate efficiency. Fluorescent merges (C, G, K, O) and DIC images (D, H, L, P) show that fluorescent labels localize to cells as predicted. Scale bar=50 μm.
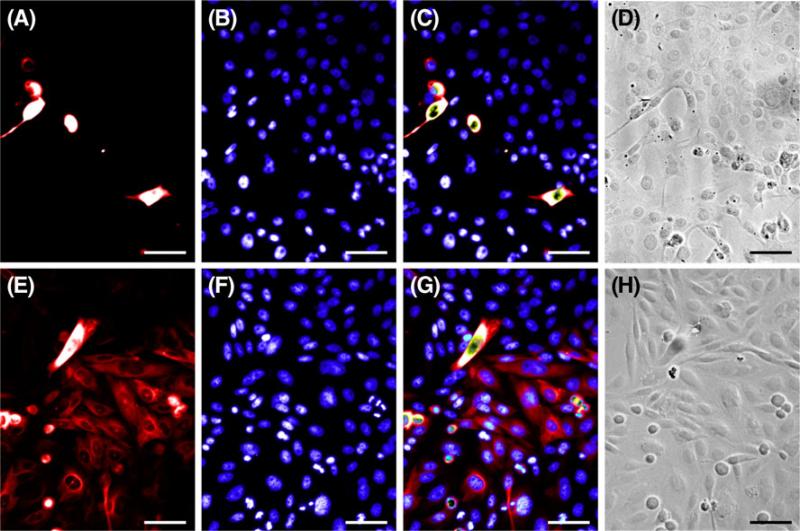
Figure 3. BacMam mediated transduction in A6 and CHO cells.
Representative images of A6 and CHO cells used to calculate viral transduction efficiencies in A6 (A–D) and CHO (E–H) cells. Cells transduced with Cellular Lights™ Tubulin-RFP (A, E) were counterstained with Hoechst (B, F) to calculate efficiency. Fluorescent merges (C, G) and DIC images (D, H) show that fluorescent labels localize to cells as predicted. Scale bar=50 μm.
Transfection and transduction efficiency in the CHO cell line
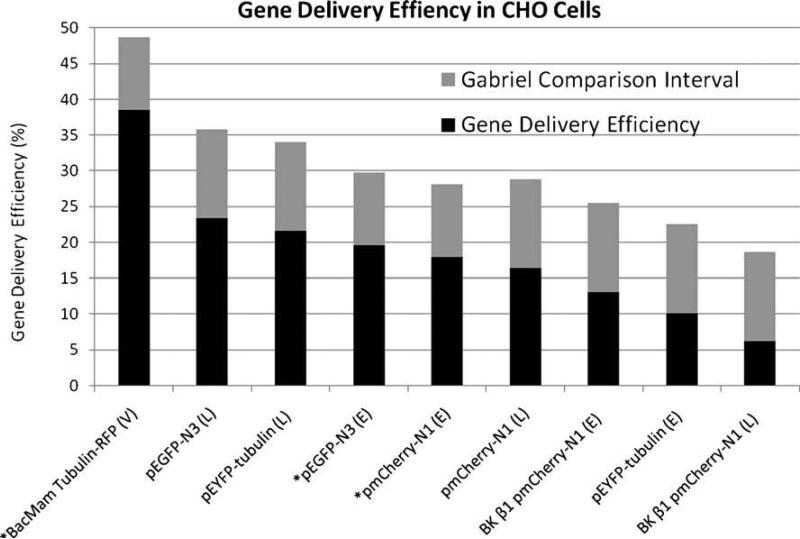
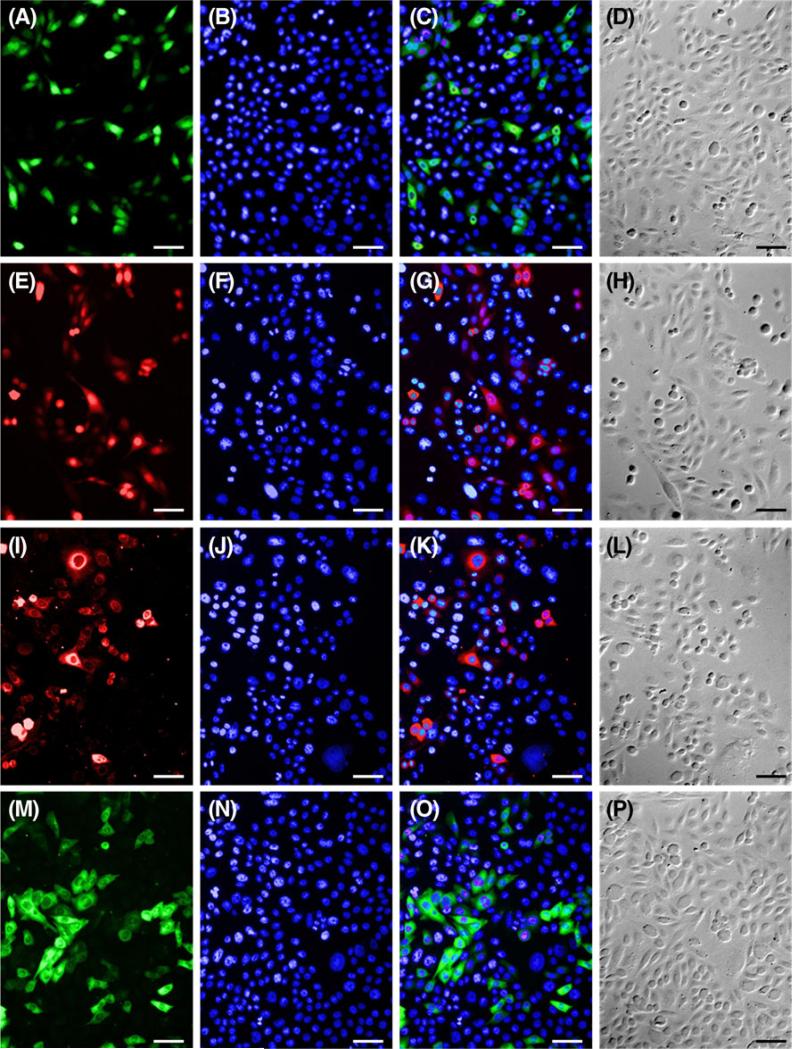
Heterologous expression of every construct was achieved in mammalian CHO cells with all three methods, including physical (electroporation). Transfection efficiencies with the chemical agent Lipofectamine™ 2000 were greater for the commercially available fluorescent constructs than for our inner ear BK β1 pmCherry-N1 construct (6.24%, Figs. 4 and 5I–L). These differences were statistically significant (p<0.05; Gabriel 1978) for pEYFP-Tub (21.6%, Figs. 4 and 5M–P) and pEGFP-N3 (23.39%, Figs. 4 and 5A–D) but not for pmCherry-N1 (16.39%, Figs. 4 and 5E–H).
Figure 4. Gene Delivery Efficiency in CHO Cells.
Mean transfection or transduction efficiencies for CHO cells where n=4, except where n=6 (asterisk). Genes (listed under each column) were delivered using Lipofectamine™ 2000 (L), electroporation (E), or BacMam virus (V). Pairs of means with Gabriel comparison intervals that do not overlap are significantly different (Gabriel 1978; p<0.05).
Figure 5. Lipid-mediated transfection in CHO cells.
Representative images of CHO cells used to calculate lipid-mediated transfection efficiencies. Cells transfected with pEGFP-N3 (A), pmCherry-N1 (E), pmCherry-N1 BK β1(I), and EYFP-tub (M) were counterstained with Hoechst (B, F, J, N) to calculate efficiency. Fluorescent merges (C, G, K, O) and DIC images (D, H, L, P) show that fluorescent labels localize to cells as predicted. Scale bar=50 μm.
A similar trend was observed in electroporation experiments. The gene delivery efficiencies of the solitary fluorescent proteins with electroporation (19.55% for pEGFP-N3, Figs. 4 and 6A–D; 17.96% for pmCherry-N1, Figs. 4 and 6E–H), were greater than the transfection efficiency of the BK β1 pmCherry-N1 fusion construct (13.09%, Figs. 4 and 6I–L). In contrast to results in the A6 cell line, the electroporation efficiency of the commercially available fluorescent fusion construct (10.16% for pEYFPTub, Figs. 4 and 6M–P) was less than that of the BK β1pmCherry-N1 fusion construct. However, none of these differences were statistically significant using a significance value of 0.05 for the Gabriel comparison interval (Gabriel 1978). Finally, the transduction efficiency with the BacMam virus, Cellular Lights™ Tubulin-RFP (38.49%, Figs. 3E–H and 4)was significantly greater than the efficiencies found with any other gene delivery method (p<0.05; Gabriel 1978).
Figure 6. Transfection with electroporation in CHO cells.
Representative images of CHO cells used to calculate transfection efficiencies for electroporation. Cells transfected with pEGFP-N3 (A), pmCherry¬N1 (E), pmCherry-N1 BK β1(I), and EYFP-tub (M) were counterstained with Hoechst (B, F,J, N) to calculate efficiency. Fluorescent merges (C, G, K, O) and DIC images (D, H, L, P) show fluorescent labels localize to cells as predicted. Scale bar=50 μm.
Comparison between the CHO and A6 cell lines
Lipid-mediated gene delivery was significantly more effective in CHO cells for every plasmid (p=0.001, pEYFP-Tub; p= 0.000006, pEGFP-N3; p=0.00004, pmCherry-N1; and p= 0.0006, BK β1 pmCherry-N1). When compared with the amphibian cells (A6), viral-mediated gene delivery using Cellular Lights™ Tubulin-RFP, a BacMam virus, was significantly more successful (p=0.008) in the mammalian (CHO) cell line. Electroporation in hypoosmolar buffer appeared to affect the A6 cell line morphology (data not shown) but not CHO cells (Fig. 6); A6 cells appeared more rounded (phase bright) and were less distended than controls. Viral-mediated (Fig. 3) and lipid-mediated (Figs. 2 and 5) gene delivery methods did not appear to affect cellular morphology for either the A6 or the CHO cell line.
Cost and exposure period versus gene delivery efficiency
BacMam had the highest gene delivery efficiency; however, this method was the most expensive with an approximate cost of $22 per experimental chamber (Table 1). The costs of gene delivery with cationic lipid reagents and electroporation were comparable. Lipid-mediated transfection was the least expensive method for each sample ($2.20 for Lipofectamine™ 2000, $0.25 for Metafectene® Pro; Table 1). Electroporation required the greatest amount of plasmid DNA (10 μg) but exposes cells to the treatment for the least amount of time. While cells require hours to days to express the fluorescent gene of interest regardless of the gene delivery method, electroporation limits the amount of time that the cellular environment is perturbed to 40 μs. In contrast, biological (viral-mediated) and chemical (lipid¬mediated) gene delivery methods require exposure to the reagent for 4–48 h. Ancillary costs such as media and trypsin are the same for all gene delivery methods and were not included in our comparison. In addition, the initial cost of the Eppendorf Multiporator® (~4,500; Institutional Pricing 2009) was not factored in to our calculations in Table 1.
Table 1. Gene delivery cost estimates.
The approximate costs per experimental treatment are provided for each gene delivery method. The cost estimates listed are based on 2011 institutional pricing with all applicable discounts
| Gene delivery method | Product Cost (including S & H) | Product packaging | Number of slide wells that can be prepared | Estimated cost of reagent per slide well |
|---|---|---|---|---|
| Biological (BacMam virus) | ||||
| Chemical (Metafectene) | 6 mL vial 1 mL vial | |||
| Chemical (Lipofectamine) | 0.75 mL vial 50 | |||
| Physical (Electroporation) | $360 $255 $330 $165 | cuvettes | 16.5 1,000 150 50 | $22 $0.25 $2.20 $3.30 |
Discussion
Xenopus affords novel opportunities for genetic investigations of inner ear structure and function due to the availability of a sequenced genome and established methods for the facile production of transgenic animals (Chesneau et al. 2008; Hellsten et al. 2010). The ability to characterize prevalent Xenopus inner ear genes through heterologous expression in cell lines expands the repertoire of available methods for genetic analysis beyond the traditional and well-established Xenopus oöcyte expression system (Terhag et al. 2010). Procedures that facilitate Xenopus gene delivery in both amphibian and mammalian cell lines are desirable for greatest experimental flexibility. Because available gene delivery methods are optimized primarily for mammalian cell lines, we hypothesized that transfection of the amphibian (Xenopus) A6 cell line might pose challenges with regard to efficiency (Kay and Peng 1991). To this end, we compared the efficiencies of physical (electroporation), chemical (lipid-mediated), and biological (viral-mediated) methods for delivering fluorescent constructs to CHO and X. laevis (kidney; A6) cells. Our experimental approach implemented many commercially available reagents with properties that may vary due to lot number and changes in production procedures.
Significantly higher fluorescent protein expression was observed in CHO cells as compared with A6 cells with all three gene delivery methods and every fluorescent construct (Figs. 1 and 4). Specific properties of CHO and A6 cells could account for the difference in gene delivery efficiency between these cell lines. For example, the X. laevis (A6) cell line is of kidney and amphibious origin. In contrast to mammalian cells, amphibian cells are adapted to a wider range of osmotic environments and are cultured in a solution that would be highly hypotonic for a mammalian cell (Balls and Worley 1973). Furthermore, A6 cells appear capable of adapting to a range of gravitational environments. A6 cells have been shown to proliferate robustly when exposed to hypergravity conditions up to 10 g (Ichigi and Asashima 2001; Tanaka et al. 2003). In our experiments, we did not observe adverse effects on A6 cell morphology in response to electrical pulses of voltages as high as 1,200 V. Cellular properties that confer these types of adaptive characteristics to the A6 cell line also could potentially reduce the efficacy of gene delivery methods.
For example, the A6 cell membrane could have a molecular composition and structure that reduces the efficacy of gene delivery, as all methods require a direct and aggressive interaction with the plasma membrane (e.g., lipid fusion or electrically induced destabilization). Methods that perturb the membrane of A6 cells in a more forceful manner may increase transfection efficiency. However, exposure to such conditions frequently results in extensive cell death and makes apoptosis an essential factor to consider when optimizing transfection methods that severely impact cell mortality (Matsuki et al. 2008).
Differences in transfection efficiency between the two cell lines could be attributable, at least to some extent, to intrinsic growth properties and culture conditions of the A6 and CHO lines. A6 cells are grown slightly above room temperature (28°C) and double approximately every 36 h (Ichigi and Asashima 2001). In contrast, the CHO cells used in this study are grown at a higher temperature (37°C) and double approximately every 12 h (Nakahara et al. 2002). Presumably, therefore, transcription and translation are more rapid in the CHO line, resulting in the ability to detect fluorescent proteins more rapidly. However, we attempted to account for this difference in our comparison of lipid-mediated transfection by allowing the A6 cells to grow for 48 h and by fixing CHO cells after only 24 h. Therefore, growth rate and temperature differences could partially account for the variation in transfection efficiency between the two cell lines used in this study. It is important to account for inherent differences between cell lines as they may account for discrepancies, not only in this study, but in all studies of cell lines.
The majority of the plasmids we used in this experiment were commercially available constructs. We also synthesized our own fusion construct by cloning an inner ear BK β1 ion channel subunit and then subcloning the sequence into the commercially available pmCherry-N1 vector. We successfully expressed our novel fusion construct, thereby laying the foundation for future studies of inner ear genes of interest. Although not statistically significant, we noted a trend in both cell lines regarding the expression of fusion constructs versus expression of the fluorescent protein tag alone. We observed that insertion of the inner ear BK β1 subunit into the pmCherry-N1 vector reduced the transfection efficiency in both cell lines (Figs. 1 and 4). These results suggest that cloning genes of interest into a viral vector for transduction may decrease the efficiency of gene expression when compared with the efficiency of expression of the fluorescent tag alone. The inner ear BK β1 insert is small (618 bp), but it is reasonable to assume that larger inserts would augment this trend.
Due to the limited commercial availability of BacMam Cellular Lights™ constructs, we compared the viral delivery of BacMam Cellular Lights™ Tubulin-RFP to the chemical and physical delivery of pEYFP-tubulin. The expressed proteins of these constructs are similarly sized and share amino acid homology and function (pEYFP-Tubulin, 688 amino acids; Cellular Lights™ Tubulin-RFP, 680 amino acids). As previously noted, BacMam viruses are designed for expression in mammalian cells (Long et al. 2006). Transduction of CHO cells with Cellular Lights™ Tubulin-RFP was almost twice as effective as gene delivery with any other method (Fig. 4). While BacMam is the most expensive method at $22 per experimental sample, the high transduction efficiency in mammalian (CHO) cells indicates that this system is suitable for heterologous expression studies of Xenopus inner ear genes. BacMam was more effective in A6 cells (3.4% transduction efficiency) than the other gene delivery methods, although this result was not statistically significant (Fig. 1).
The high gene delivery efficiency using a BacMam virus (37.49%, Fig. 4) in CHO cells offers significant advantages for experiments where co-transduction is desired. Co¬transduction studies are essential for the analysis of membrane proteins that require modulation by intracellular moieties or are comprised of several subunits that come together to form a complete protein, as with the BK channel (Orio et al. 2002). The A6 cell line would be advantageous for co-transfection studies because it shares a species origin with the inner ear genes we wish to analyze and would presumably have the species-specific cofactors necessary for protein folding. Although the BacMam virus offered a higher (though not statistically significant) gene delivery efficiency in A6 cells (3.4%), the probability of a co¬transduction event is only 0.1%, or one in every thousand A6 cells as compared with approximately one in every 15 CHO cells.
Xenopus cell lines such as A6 offer distinct advantages for our heterologous expression research because gene expression levels can be estimated using the Affymetrix GeneChip® X. laevis genome arrays. However, the low probability of a co-transfection event in the A6 cell line makes mammalian (CHO) cells a better option for our heterologous expression studies, and this is consistent with longstanding practices where genes are expressed in cell lines of different species origin. For example, insect cells have been widely employed for mammalian protein production (Smith et al. 1983; Altmann et al. 1999) and plant cells offer promise for the production of therapeutic human proteins (Desai et al. 2010). Other laboratories have used alternative methods for A6 kidney cell transfection by creating stable transformants through drug selection (Faletti et al. 2002; Mies et al. 2007). However, these methods require exposure to toxic concentrations of geneticin, an aminoglycoside antibiotic that is closely related to gentamicin. Because aminoglycoside antibiotics are notably nephrotoxic (Lopez-Novoa et al. 2011), the methods necessary for stable transformation may affect the properties of the A6 kidney cell line.
Research presented here was motivated by a need to develop methods for genetic analysis of the Xenopus inner ear outside of the whole animal model. Our results highlight the need for improved gene delivery methods as evidenced by the array of new reagents continuously being released by manufacturers. For example, the recently developed BacMam 2.0 reagents have been designed to improve transduction efficiency through the addition of a pseudotyped capsid protein for more efficient cell entry and genetic elements (enhanced CMV promoter and Woodchuck Post-transcriptional Regulatory Element) that boost expression levels (Invitrogen 2011). Although results were statistically significant only in the CHO cell line, viral-mediated gene delivery has a tendency to be more effective than chemical or physical gene delivery methods; however the cost of viral transduction is also much higher. The successful transfection of amphibian (A6) cells offers promise for our heterologous expression studies in a cell line that shares species origin with our candidate Xenopus inner ear genes. However, we are intrigued by the observation that transfection efficiencies in the amphibian kidney (A6) cell line are significantly lower than in the mammalian ovary (CHO) cell line, which is notably difficult to transfect. We believe this result underscores the need for greater understanding of amphibian cell biology and cell culture, especially in light of the worldwide decline of amphibian populations and the increased prevalence of contaminants in fresh water habitats (Boyer and Grue 1995).
Acknowledgments
We would like to acknowledge Karla Almaraz for assistance with cell culture. Funding for this research was provided by Graduate Fellowships from NSF to DRG (New Mexico AMP, HRD-0331446; IGERT, DGE0504304) and NIH grants to EES (R01DC03292 and P50GM068762).
References
- Altmann F, Staudacher E, Wilson IB, März L. Insect cells as hosts for the expression of recombinant glycoproteins. Glycoconj J. 1999;16(2):109–123. doi: 10.1023/a:1026488408951. [DOI] [PubMed] [Google Scholar]
- Balls M, Worley RS. Amphibian cells in vitro: II. Effects of variations in medium osmolarity on a permanent cell line isolated from Xenopus. Exp Cell Res. 1973;76(2):333–336. doi: 10.1016/0014-4827(73)90384-4. [DOI] [PubMed] [Google Scholar]
- Banerjee P, Berry-Kravis E, Bonafede-Chhabra D, Dawson G. Heterologous expression of the serotonin 5-HT1A receptor in neural and non-neural cell lines. Biochem. Biophys. Res. Commun. 1993;192:104–110. doi: 10.1006/bbrc.1993.1387. [DOI] [PubMed] [Google Scholar]
- Barry PA. Efficient electroporation of mammalian cells in culture. In: Heiser W. C. (ed) Gene delivery to mammalian cells. Humana Press Inc., Totowa. 2004:207–214. doi: 10.1385/1-59259-649-5:207. [DOI] [PubMed] [Google Scholar]
- Boyer R, Grue CE. The need for water quality criteria for frogs. Environ Health Perspect. 1995;103(4):352–357. doi: 10.1289/ehp.95103352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butel JS, Talas M, Ugur J, Melnick JL. Demonstration of infectious DNA in transformed cells. III. Correlation of detection of infectious DNA-protein complexes with persistence of virus in simian adenovirus SA7-induced tumor cells. Intervirology. 1975;5(1–2):43–56. doi: 10.1159/000149879. [DOI] [PubMed] [Google Scholar]
- Chesneau A, Sachs LM, Chai N, Chen Y, Du Pasquier L, Loeber J, Pollet N, Reilly M, Weeks DL, Bronchain OJ. Transgenesis procedures in Xenopus. Biol Cell. 2008;100(9):503–521. doi: 10.1042/BC20070148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condreay JP, Witherspoon SM, Clay WC, Kost TA. Transient and stable gene expression in mammalian cells transduced with a recombinant baculovirus vector. PNAS. 1999;96:127–132. doi: 10.1073/pnas.96.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai PN, Shrivastava N, Padh H. Production of heterologous proteins in plants: strategies for optimal expression. Biotechnol. Adv. 2010;28(4):427–435. doi: 10.1016/j.biotechadv.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Díaz ME, Varela-Ramírez A, Serrano EE. Quantity, bundle types, and distribution of hair cells in the sacculus of Xenopus laevis during development. Hear. Res. 1995;91(1–2):33–42. doi: 10.1016/0378-5955(95)00159-x. [DOI] [PubMed] [Google Scholar]
- Duncan RK, Fuchs PA. Variation in large-conductance, calcium-activated potassium channels from hair cells along the chicken basilar papilla. J. Physiol. 2003;547:357–371. doi: 10.1113/jphysiol.2002.029785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faletti CJ, Perrotti N, Taylor SI, Blazer-Yost BL. sgk: an essential convergence point for peptide and steroid hormone regulation of ENaC-mediated Na+ transport. Am J Physiol Cell Physiol. 2002;282(3):C494–C500. doi: 10.1152/ajpcell.00408.2001. [DOI] [PubMed] [Google Scholar]
- Fettiplace R, Fuchs PA. Mechanisms of Hair Cell Tuning. Annu. Rev. Physiol. 1999;61:809–834. doi: 10.1146/annurev.physiol.61.1.809. [DOI] [PubMed] [Google Scholar]
- Gabriel K. R. A simple method of multiple comparison of means. J. Amer. Stat. Assoc. 1978;73:724–729. [Google Scholar]
- Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, Blitz IL, Blumberg B, Dichmann DS, Dubchak I, Amaya E, Detter JC, Fletcher R, Gerhard DS, Goodstein D, Graves T, Grigoriev IV, Grimwood J, Kawashima T, Lindquist E, Lucas SM, Mead PE, Mitros T, Ogino H, Ohta Y, Poliakov AV, Pollet N, Robert J, Salamov A, Sater AK, Schmutz J, Terry A, Vize PD, Warren WC, Wells D, Wills A, Wilson RK, Zimmerman LB, Zorn AM, Grainger R, Grammer T, Khokha MK, Richardson PM, Rokhsar DS. The genome of the Western clawed frog Xenopus tropicalis. Science. 2010;328(5978):633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibey BL, Xiao S, Schoenbach KH, Murphy MR, Pakhomov AG. Plasma membrane permeabilization by 60 and 600-ns electric pulses is determined by the absorbed dose. Bioelectromagnetics. 2009;30:92–99. doi: 10.1002/bem.20451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichigi J, Asashima M. Dome formation and tubule morphogenesis by Xenopus kidney A6 cell cultures exposed to microgravity simulated with a 3D clinostat and to hypergravity. In Vitro Cell Dev Biol- Animal. 2001;37:31–44. doi: 10.1290/1071-2690(2001)037<0031:dfatmb>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Invitrogen . CellLight® Reagents *BacMam 2.0* 2011. [1 March 2011]. http://probes.invitrogen.com/media/pis/mp10582.pdf. [Google Scholar]
- Joint Genome Institute [20 January 2009];The Regents of the University of California. 1997 http://genome.jgipsf.org/Xentr4/Xentr4.home.html.
- Kay B, Peng HG. Academic. San Diego: 1991. Methods in cell biology, 36, X. laevis: practical uses in cell and molecular biology. [PubMed] [Google Scholar]
- Kim Y, Klutz AM, Jacobson KA. Systematic investigation of polyamidoamine dendrimers surface- modified with poly(ethylene glycol) for drug delivery applications: synthesis, characterization, and evaluation of cytotoxicity. Bioconjug Chem. 2008;19:1660–1672. doi: 10.1021/bc700483s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight VB, Serrano EE. Tissue and species differences in the application of quantum dots as probes for biomolecular targets in the inner ear and kidney. IEEE Transactions in Nanobioscience. 2006;5:251–262. doi: 10.1109/tnb.2006.886551. [DOI] [PubMed] [Google Scholar]
- Liu MY, Lin SC, Liu H, Candal F, Vafai A. Identification and authentication of animal cell culture by polymerase chain reaction amplification and DNA sequencing. In Vitro Cell Dev Biology-Animal. 2004;39:424–427. doi: 10.1290/1543-706X(2003)039<0424:IAAOAC>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Long G, Pan X, Kormelink R, Vlak JM. Functional entry of baculovirus into insect and mammalian cells is dependent on clathrin-mediated endocytosis. J. Virol. 2006;80(17):8830–8833. doi: 10.1128/JVI.00880-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Novoa JM, Quiros Y, Vicente L, Morales AI, Lopez-Hernandez FJ. New insights into the mechanism of amino—glycoside nephrotoxicity: an integrative point of view. Kidney Int. 2011;79(1):33–45. doi: 10.1038/ki.2010.337. [DOI] [PubMed] [Google Scholar]
- Matsuki N, Ishikawa T, Imai Y, Yamaguchi T. Low voltage pulses can induce apoptosis. Cancer Lett. 2008;269(1):93–100. doi: 10.1016/j.canlet.2008.04.019. [DOI] [PubMed] [Google Scholar]
- Mies F, Spriet C, Héliot L, Sariban-Sohraby S. Epithelial Na+ channel stimulation by n-3 fatty acids requires proximity to a membrane-bound A-kinase-anchoring protein complexed with protein kinase A and phosphodiesterase. J. Biol. Chem. 2007;282(25):18339–18347. doi: 10.1074/jbc.M611160200. [DOI] [PubMed] [Google Scholar]
- Nakahara T, Yaguchi H, Yoshida M, Junji M. Effects of exposure of CHO-K1 cells to a 10-T Static Magnetic Field. Radiology. 2002;224:817–822. doi: 10.1148/radiol.2243011300. [DOI] [PubMed] [Google Scholar]
- Oberholtzer CJ. Frequency tuning cochlear hair cells by differential splicing of BK channel transcripts. Journal Physiol. 1999;518:653–665. doi: 10.1111/j.1469-7793.1999.0629p.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orio P, Rojas P, Ferreira G, Latorre R. New disguises for an old channel: MaxiK channel beta-subunits. News Physiol Sci. 2002;17:156–161. doi: 10.1152/nips.01387.2002. [DOI] [PubMed] [Google Scholar]
- Powers T, Trujillo-Provencio C, Whittaker C, Serrano EE. Gene expression profiling of Xenopus organs yields insight into the Xenopus inner ear transcriptome. ARO. Abstr. 2007;30:741. [Google Scholar]
- Pyott SJ, Meredith AL, Fodor AA, Vazquez AE, Yamoah EN, Aldrich RW. Cochlear function in mice lacking the BK channel α, β1, or β4 subunits. J. Biol. Chem. 2007;282:3312–3324. doi: 10.1074/jbc.M608726200. [DOI] [PubMed] [Google Scholar]
- Ramanathan K, Michael TH, Jiang GJ, Hiel H, Fuchs PA. A molecular mechanism for electrical tuning of cochlear hair cells. Science. 1999;283:215–217. doi: 10.1126/science.283.5399.215. [DOI] [PubMed] [Google Scholar]
- Scolnick EM, Bumgarner SJ. Isolation of infectious xenotropic mouse type C virus by transfection of a heterologous cell with DNA from a transformed mouse cell. J. Virol. 1975;15(5):1293–1296. doi: 10.1128/jvi.15.5.1293-1296.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano EE, Trujillo-Provencio C, Sultemeier DR, Bullock WM, Quick QA. Identification of genes expressed in the Xenopus inner ear. Cell Mol Biol (Noisy-le-grand) 2001;47(7):1229–1239. [PMC free article] [PubMed] [Google Scholar]
- Shin JB, Adams D, Paukert M, Siba M, Sidi S, Levin M, Gillespie PG, Gründer S. Xenopus TRPN1 (NOMPC) localizes to microtubule-based cilia in epithelial cells, including inner-ear hair cells. Proc Natl Acad Sci. 2005;102(35):12572–12577. doi: 10.1073/pnas.0502403102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GE, Summers MD, Fraser MJ. Production of human beta interferon in insect cells infected with a baculovirus expression vector. Mol. Cell. Biol. 1983;3(12):2156–2165. doi: 10.1128/mcb.3.12.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultemeier DR, Knight VB, Manuelito SJ, Hopkins M, Serrano EE. Heterologous and homologous expression systems for functional analysis of Xenopus inner ear genes. Assoc. Res Otolaryngol Abs. 2005:821. [Google Scholar]
- Tanaka M, Asashima M, Atomi Y. Proliferation and differentiation of Xenopus A6 cells under hypergravity as revealed by time-lapse imaging. Vitro Cell Dev Biol Anim. 2003;39(1–2):71–79. doi: 10.1290/1543-706X(2003)039<0071:PADOXA>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Terhag J, Cavara NA, Hollmann M. Cave Canalem: how endogenous ion channels may interfere with heterologous expression in Xenopus oocytes. Methods. 2010;51(1):66–74. doi: 10.1016/j.ymeth.2010.01.034. [DOI] [PubMed] [Google Scholar]
- Trujillo-Provencio C, Powers TR, Sultemeier DR, Serrano EE. RNA isolation from Xenopus inner ear sensory endorgans for transcriptional profiling and molecular cloning. Methods Mol Biol. 2009;493:3–20. doi: 10.1007/978-1-59745-523-7_1. [DOI] [PubMed] [Google Scholar]
- Varela-Ramírez A, Trujillo-Provencio C, Serrano EE. Hear. Res. Vol. 119. 1–2: 1998. Detection of transcripts for delayed rectifier potassium channels in the X. laevis inner ear. pp. 125–134. [DOI] [PubMed] [Google Scholar]
- Wyatt SK, Giorgio TD. DNA delivery to cells in culture using cationic liposomes. In: Heiser WC, editor. Gene delivery to mammalian cells. Humana Press Inc.; Totowa: 2004. pp. 83–86. [DOI] [PubMed] [Google Scholar]