Abstract
Recent studies have demonstrated a relationship between the expression of stem cell-associated genes and relapses in glioblastoma (GBM), suggesting a key role for tumor stem cells in this process. Although there is increasing interest in this field, glioma stem cells (GSCs) are still poorly characterized, their ‘stemness' state and factors maintaining these properties remain largely unknown. We performed an expression profiling analysis of pluripotency in gliomaspheres derived from 11 patients. Comparative analysis between GSCs and H1 and H9 human embryonic stem cells as well as H9-derived neural stem cells indicates major variations in gene expression of pluripotency factors Nanog and OCT4, but a stable pattern for SOX2 suggesting its important function in maintaining pluripotency in GSCs. Our results also showed that all GSC lines have the capacity to commit to neural differentiation and express mesenchymal or endothelial differentiation markers. In addition, hierarchical clustering analysis revealed two groups of GSCs reflecting their heterogeneity and identified COL1A1 and IFITM1 as the most discriminating genes. Similar patterns have been observed in tumors from which gliomaspheres have been established. To determine whether this heterogeneity could be clinically relevant, the expression of both genes was further analyzed in an independent cohort of 30 patients with GBM and revealed strong correlation with overall survival. In vitro silencing of COL1A1 and IFTM1 confirmed the effect of these mesenchymal-associated genes on cell invasion and gliomasphere initiation. Our results indicate that COL1A1 and IFITM1 genes could be considered for use in stratifying patients with GBM into subgroups for risk of recurrence at diagnosis, as well as for prognostic and therapeutic evolution.
Keywords: glioblastoma stem cells, mesenchymal profile, COL1A1, IFITM1, survival
Introduction
Glioblastoma (GBM), the highest grade of glioma tumor (WHO grade IV), is the most frequent and aggressive primary brain tumor in adults. The current standard of care for patients with GBM includes tumor resection followed by adjuvant radiation therapy and chemotherapy. A significant survival benefit was reported for GBM patients treated with temozolomide combined with radiation therapy.1 However, despite advances in surgical and medical neuro-oncology, the prognosis for GBM patients remains poor and median survival is <1 year. As a result of its infiltrating characteristics, complete resection of GBM is not possible and recurrence occurs almost invariably at the primary location of the tumor. Recent data suggest that tumor relapses may be linked to the presence within the tumor of a cell sub-population with stem cell characteristics, called ‘tumor-initiating stem cells' or ‘cancer stem cells' (CSCs). Such cells, designated as glioma stem cells (GSCs) have been isolated from human glioma tissues.2, 3 First reports suggested that tumorigenic cells in GBM were restricted to the CD133+ population.2, 3 However, recent studies have revealed that CD133-negative cells isolated from GBM could also be tumorigenic2, 3, 4, 5, 6, 7, 8, 9 and may in fact represent inter-convertible phenotypic states of the same cell population giving rise to a heterogeneous GSC population.5 These cells are highly resistant to conventional chemotherapeutic drugs, including temozolomide10, 11 and mediate tumor recurrence following radiation therapy.12 Therefore, complete eradication of GSCs may be a prerequisite for successful therapeutic strategies. In order to arrive at such an approach, in vivo and in vitro models that faithfully recapitulate the stem cell compartment of gliomas have been developed. Among these models, gliomaspheres, also termed neurospheres, are cultured in vitro and enriched in GSCs. This in vitro model reflects biological and pathological characteristics of the stem cell compartment. Recently, using a comparative genomic analysis between GSCs and human neural stem cells (NSCs), Sandberg et al.13 identified, alterations of Wnt and Hedgehog signaling pathways as important events conferring self-renewal potential and tumorigenic properties to GSCs. However, the characterization of molecular events underlying GSCs tumorigenesis remains challenging as GBM tumors comprise heterogeneous populations of GSCs with distinct molecular alterations associated with contrasting phenotypes in tumor initiation and differential response to therapy.14, 15 Human embryonic stem cells (hESCs) represent archetypal stem cells that maintain their self-renewal and pluripotency by blocking differentiation through a well-coordinated transcriptional network.16, 17, 18, 19, 20 These biological features are shared by both normal and CSCs.21, 22, 23, 24 The ESC-like transcriptional program was shown to be activated in poorly differentiated tumors including GBMs and appeared to be predictive of metastasis and death.18, 25 However, unlike ESCs, the stemness abilities of different types of CSCs may not be reflected in a single-gene expression program and there are likely to be many routes to stemness as suggested by Müller et al.26 Stem cell traits normally present in human NSCs are shown to be aberrantly co-expressed with pluripotent markers of hESCs in gliomas. This general deregulated expression profile correlates with increased malignancy and probably reflects its cell of origin.27 To date, pluripotency/stemness characteristics, and differentiation potential of GSCs remain largely unexplored. A better understanding of these aspects would be crucial for further improvements in GBM therapy. To address these questions, we took advantage of the unique pluripotential characteristic of human embryonic and NSC lines, and compared the gene expression profiles of 11 GSC lines isolated from GBM patients with those of H1 and H9-ESCs, and with H9-derived NSCs.
Results
TaqMan stem cell pluripotency assays in GSCs
To highlight the pluripotency/stemness status and the differentiation ability of gliomaspheres, which are considered to be enriched in GSCs and represent tumoral stem cells, the expression levels of selected genes were compared with those of hESCs (H1 and H9-ESCs) and human NSCs (H9-HNSCs). The pluripotency and stemness status of H9-HNSCs, which serves as a reference in our strategy, was first compared with the well- characterized H1 and H9-ESCs. We observed a lower expression of Nanog and POU5F1 in H9-HNSCs, but a moderately elevated expression of SOX2 (Supplementary Figure 1A). As expected, NSC and neural morphogenesis genes (Nestin, NeuroD1 and PAX6) as well as GCM1 (glial cells missing gene) having a neurogenic role, were expressed at a higher level in H9-HNSCs. On the other hand, glial fibrillary acidic protein (GFAP) (highly specific for cells of astroglial lineage), oligodendrocyte and neuron development transcription factors (OLIG2, HLXB9 and ISL1) were found to be equally expressed or downregulated (Supplementary Figure 1B). In addition, our results confirmed that H9-HNSCs are multipotent because the genes involved in mesoderm (for example, COL1A1, RUNX2, COL2A1 and CDH5) or in endoderm development (FN1, LAMA1, LAMB1 and LAMC1) have a relative high constitutive expression (Supplementary Figure 1C).
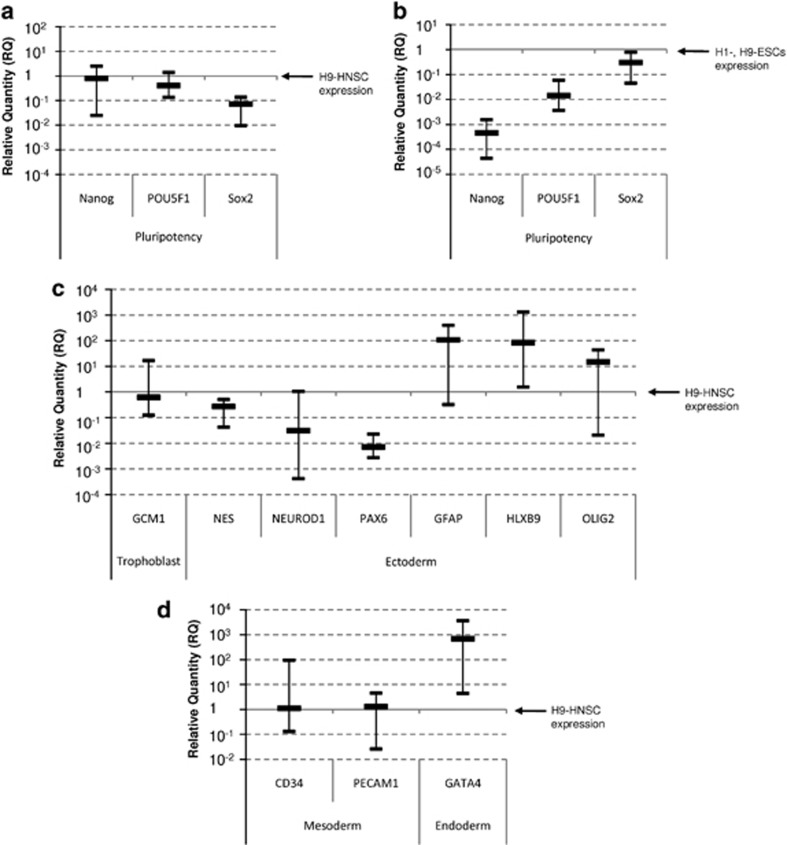
The pluripotency and stemness status of GSCs was next compared with that of H9-HNSCs cultured under the same conditions. Nanog and POU5F1 were found to be equally expressed in H9-HNSCs and GSC lines, whereas the expression of SOX2 appeared to be lower in GSCs (Figure 1a). Interestingly, the expression of SOX2 in all GSCs was quite similar to H1 and H9-ESCs (<1 log; Figure 1b). Nestin and GCM1 were also found to be equally expressed in H9-HNSCs and GSCs lines while NeuroD1 and PAX6 expression were lower in GSCs. In contrast, genes involved in astrocyte (GFAP), neuron (HLXB9) and oligodendrocyte (OLIG2) differentiation were found to have a higher expression in GSCs indicating that GSCs are more committed to neural differentiation than H9-HNSCs (Figure 1c). However, GSC population showed multipotent potential because genes involved in endothelial differentiation such as CD34 or PECAM1 had expression patterns similar to H9-HNSCs and the endoderm-related transcription factors GATA4 were found to be overexpressed in GSCs compared with NSCs (Figure 1d).
Figure 1.
TaqMan low-density array analysis. (a) mRNA expression of stemness and pluripotency-related genes in GSCs as compared with the expression of H9-HNSC. (b) mRNA expression of stemness and pluripotency-related genes in GSCs as compared with the expression of H1, H9-ESC lines. (c,d) Expression analysis of neural (GFAP, NEUROD1 and OLIG2) endothelial (CD34 and PECAM1) and trophoblastic (GCM1) differentiation markers in GSCs as compared with H9-HNSCs. The chart shows median values with range from 11 GSC lines determined by 2−ΔΔCt relative quantification method. Relative expression of target genes determined by the using GAPDH as endogenous control and normalized to H9-HNSC or H1, H9-ESCs calibrator.
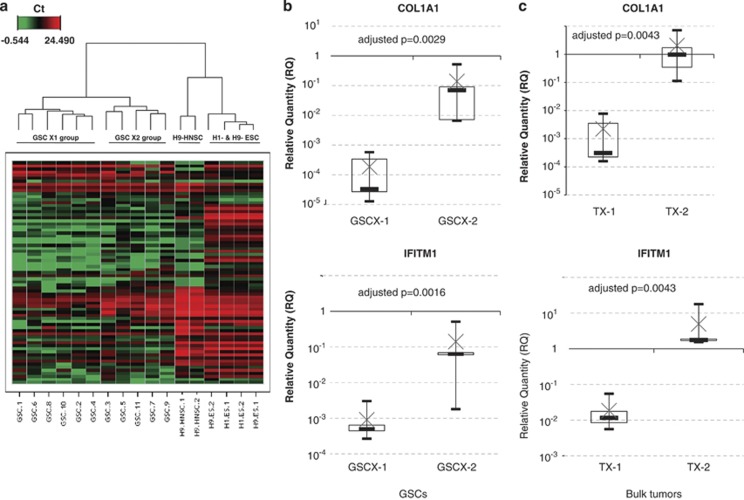
A hierarchical clustering analysis was performed using Ward's linkage method and Euclidean distance. The unsupervised method segregated the 11 GSCs samples from H9-HNSCs, H1- and H9-ESCs, and clearly separated GSCs into two distinct groups (Figure 2a). Group GSCX-1 included GSCs 1, 2, 4, 6, 8, 10 and group GSCX-2, GSCs 3, 5, 7, 9, 11. To identify genes differentially expressed between the two groups, individual mRNA expression levels were compared (Supplementary Table 3). Seven genes (COL1A1, IFITM1, SFRP2, FN1, SST, FOXA2 and GAL) presented a significant higher expression in group GSCX-2 as compared with group GSCX-1using a t-test with Benjamini–Hochberg correction (Supplementary Figure 2). It is noticeable that COL1A1 and IFITM1 expression harbored the most significant difference between the two groups of patients (Figure 2b). The expression level of these two genes was also performed in bulk tumors from which GSCs were established. Interestingly, the expression of both genes was shown to be higher in bulk tumors from group TX-2 than in bulk tumors of group TX-1 (Figure 2c). However, no significant difference in overall survival was observed between both groups of patients (Table 1).
Figure 2.
Hierarchical clustering analysis of GSCs. (a) The unsupervised clustering using Ward's linkage method and Euclidean distance individualized H9 HNSCs, H1-H9-ESCs and two groups of GSCs (GSCX-1 and GSCX-2). Note that culture conditions of H9 HNSC do not influence the expression of pluripotency gene samples H9 HNSC-1 and H9 HNSC-2 samples. (b) Box plot representation of COL1A1 and IFITM1 expression levels between GSC groups GSCX-1 and GSCX-2. The charts show the log10 expression of relative quantification (RQ) values normalized to the expression of H9 HNSC cell line. (c) Box plot representation of COL1A1 and IFITM1 expression levels between bulk tumor groups TX-1 and TX-2. The top edge of the boxes represents the 75th percentile, the bottom edge, the 25th percentile and × the mean. The range is shown as vertical edge.
Table 1. Characteristics of GBM patients from whom neurospheres were derived.
| Groups | Patient | Sex | Age (years) | Therapy | Overall survival (months) | Status |
|---|---|---|---|---|---|---|
| X-1 | #1 (GSC-1) | M | 69 | RX+TMZ | 15 | Dead |
| #2 (GSC-2) | M | 57 | RX+TMZ | 9 | Dead | |
| #4 (GSC-4) | M | 50 | RX+TMZ | >18 | Alive | |
| #6 (GSC-6) | M | 53 | RX+TMZ | 10 | Dead | |
| #8 (GSC-8) | F | 54 | none | 2 | Dead | |
| #10 (GSC-10) | M | 69 | RX+TMZ | >18 | Alive | |
| X-2 | #3 (GSC-3) | M | 56 | RX+TMZ | 9 | Dead |
| #5 (GSC-5) | F | 65 | RX+TMZ | 13 | Dead | |
| #7 (GSC-7) | M | 51 | RX+TMZ | 13 | Dead | |
| #9 (GSC-9) | M | 56 | RX+TMZ | 6 | Dead | |
| #11 (GSC-11) | M | 61 | RX+TMZ | >18 | Alive |
Abbreviations: GBM, glioblastoma; GSC, glioma stem cell; RX, radiotherapy 60 Gy; TMZ, temozolomide.
Effects of COL1A1 and IFITM1 inhibition on cell invasion, proliferation and neurosphere initiation
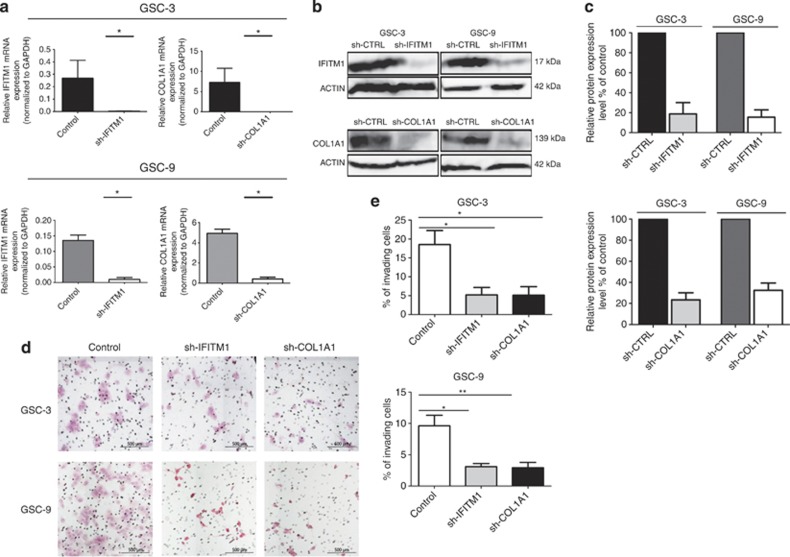
In order to determine the biological significance of differential expression levels of COL1A1 and IFITM1 in GSCs, we inhibited both gene expression and studied their subsequent effect on invasion, proliferation and neurosphere initiation. Lentiviral vectors encoding short hairpin RNA (shRNA) directed against COL1A1 and IFITM1 were used to transduce GSC-3 and GSC-9, which displayed a high basal expression of the two genes. Inhibition of COL1A1 and IFITM1 expression were monitored by quantitative reverse transcriptase–PCR and western blot. For the two genes, a decrease of >90% in mRNA expression was observed in GSC-3 and GSC-9 as compared with control cells (P<0.05; Figure 3a). Similarly, western blot analysis revealed an 80% reduction for COL1A1 and IFITM1 proteins levels in GSC-3 and GSC-9 cells as compared with control cells (Figures 3b and c).
Figure 3.
Knockdown efficiency of COL1A1 and IFITM1 and effect on cell invasion. (a) COL1A1 and IFITM1 mRNA expression was monitored by quantitative reverse transcriptase–PCR in GSC-3 and GSC-9 (n=2). Significant inhibition of 90% of mRNA expression was observed. GADPH was used as internal control. *P<0.05. Each value is expressed as mean±s.e.m. (b) Western blot analysis showed diminution of IFITM1 and COL1A1 proteins expression, β-actin was used as loading control. (c) Densitometry of protein level. The bar graph shows relative IFITM1 and COL1A1 protein expression levels expressed as percentage of IFITM1 and COL1A1 expression in control cells (n=3). We observed >80% inhibition for both genes in GSC-3 and GSC-9 cells, as compared with control cells. (d) Representative fields of Boyden cell invasion assay. After 3 days of invasion, cells that migrated across the membrane were fixed and stained with hematoxylin/eosin and were counted in one hundred different fields. The invaded cells were photographed under the microscope at × 100 magnification. (e) A significant reduction of invasion is observed in both GSC-3 and GSC-9 cells lines with sh-IFITM1 as well as for sh-COL1A1. The results represent a percentage of invaded cells. Data are presented as mean±s.e.m. from three independent experiments. *P<0.05, **P <0.01.
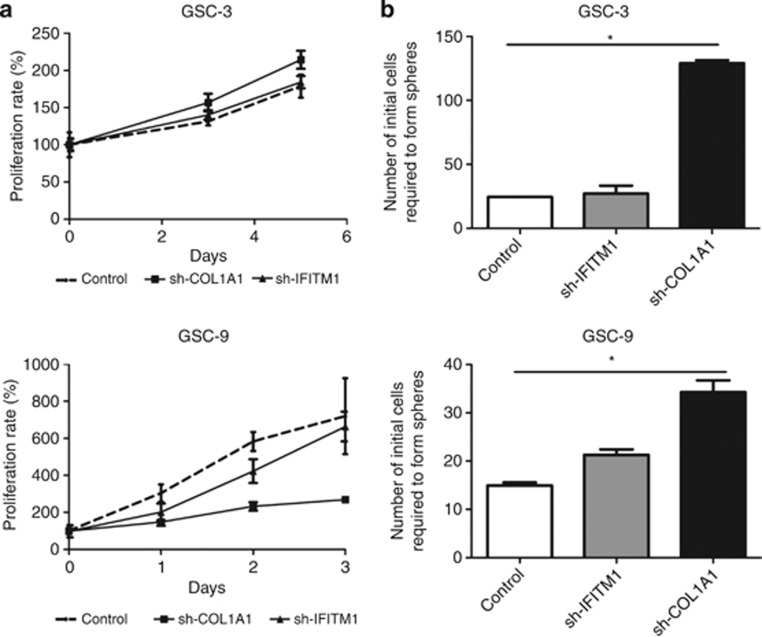
The effect of shRNA-COL1A1 and shRNA-IFITM1 on cell invasion was first studied using Boyden cell invasion assay. After 3 days, we observed a significant reduction in the invasive potency of sh-IFITM1 GSC-3 (72%) and sh-IFITM1 GSC-9 (67%) cells as compared with control cells (P<0.05). An equivalent reduction of invasion was also shown for sh-COL1A1 GSC-3 (72%) and sh-COL1A1 GSC-9 (70%) cells (Figures 3d and e). The effect of inhibiting COL1A and IFITM1 expression on cell proliferation was then analyzed using a MTS assay. GSC-3 and GSC-9 cells have a 6-day and 1-day doubling time, respectively. Inhibition of IFITM1 or COL1A1 expression had no effect on doubling time for either GSC lines (5.7 days for sh-IFITM1 GSC-3, 2 days for sh-IFITM1 GSC-9 and 4.6 days for sh-COL1A1 GSC-3). Nevertheless, we observed a slight increase of doubling time for sh-COL1A1 GSC-9 (2 days; Figure 4a). Finally, the effects of inhibiting COL1A1 and IFITM1 expression on neurosphere-forming capacity were evaluated. The frequency of neurosphere-initiating cells corresponding to the number of cells required to generate at least one tumor sphere per well was 1/25 for GSC-3 and 1/15 for GSC-9. Inhibition of IFITM1 expression only slightly modified the neurosphere-initiating capacity of GSC-3 (1/27) and GSC-9 (1/21). However, inhibition of COL1A1 resulted in a decrease of neurosphere-initiating cell frequency, as the number of cells required to form neurospheres was 1/128 for sh-COL1A1 GSC-3 and 1/35 for sh-COL1A1 GSC-9 (Figure 4b).
Figure 4.
Effects of COL1A1 and IFITM1 inhibition on cell proliferation and on neurosphere-initiating capacity. (a) Cell proliferation was measured by using a MTS assay. Each experiment was carried out in replicates of six and repeated twice and expressed as mean±s.d. (b) Inhibition of COL1A1 resulted in a significant (*P<0.05) decrease of neurosphere-initiating cell (NS-IC) frequency whereas inhibition of IFITM1 expression only slightly modified the neurosphere-initiating capacity. Final cell dilutions yielding 37% of negative wells correspond to the dilution at which there is one NS-ICs per well. Each experiment was performed in duplicate and repeated twice and expressed as mean±s.d.
Altogether, these observations indicate that COL1A1 and IFITM1 knockdown markedly decrease the invasive capacity of GSCs from primary human GBMs. Furthermore, suppressing COL1A1 expression strongly affects the neurosphere-initiating capacity of GSCs.
Expression of COL1A1 and IFITM1 in tumors and relation to survival
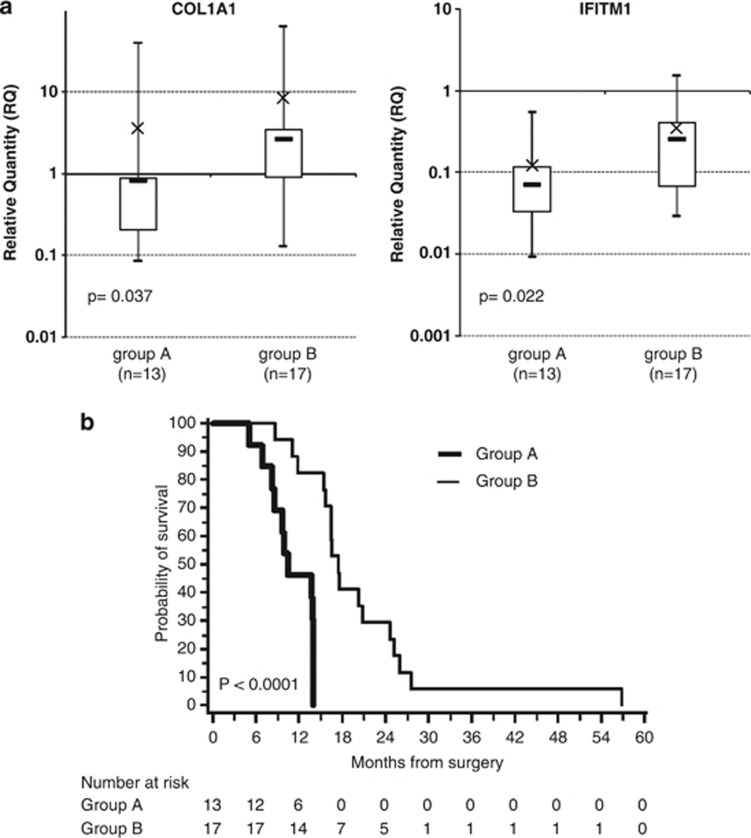
The absence of significant difference in overall survival betweenTX-1 and TX-2 groups may be due to the small number of cases. To test this hypothesis and to determine whether the expression level of COL1A1 and IFITM1 in tumors could be of clinical significance, we analyzed the mRNA expression of COL1A1 and IFITM1 in primary tumor samples of an independent cohort of 30 GBM patients (Table 2). A hierarchical clustering separates patient samples into two statistically distinct subgroups (called A and B). Patients from subgroup A (n=13) showed a significant reduction in the expression of COL1A1 and IFITM1 as compared with patients from subgroup B (P=0.037 and P=0.022, respectively; Figure 5a and Supplementary Table 4). Kaplan–Meier survival analysis demonstrated that patients from subgroup A had a median overall survival of 10.5 months (95% confidence interval: 8.2–14.0) and patients from subgroup B had an overall survival of 17.5 months (95% confidence interval: 15.5–24.7; P<10−4; Figure 5b).
Table 2. Characteristics of 30 GBM patients from an independent cohort study.
| Patient | Sex | Age (years) | Extent of resection | Initial treatment | ECOG grade | IDH1 status | IDH2 status |
MGMT methylation |
PFS (months) | OS (months) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| UM | M | ||||||||||
| Group A (n=13) | |||||||||||
| 1 | F | 68 | Complete | S+RX+TMZ | 3 | Wt | Wt | + | − | 11.4 | 13.9 |
| 2 | F | 65 | Biopsy | S+RX+TMZ | 2 | + | − | 4.4 | 5.1 | ||
| 3 | F | 41 | Complete | S+RX+TMZ | 1 | + | − | 8.1 | 9.7 | ||
| 4 | F | 64 | Complete | S+RX+TMZ | 2 | + | + | 0.4 | 14.0 | ||
| 5 | M | 75 | Complete | S+RX | 2 | + | + | 8.3 | 9.9 | ||
| 6 | M | 68 | Partial | S+RX+TMZ | 2 | + | − | 10.3 | 14.0 | ||
| 7 | M | 53 | Complete | S+RX+TMZ | 2 | + | + | 4.0 | 6.9 | ||
| 8 | M | 65 | Complete | S+RX+TMZ | 1 | + | + | 7.2 | 10.5 | ||
| 9 | M | 66 | Partial | S+RX+TMZ | 2 | + | + | 5.0 | 14.0 | ||
| 10 | M | 63 | Complete | S+RX+TMZ | 2 | + | + | 4.2 | 8.6 | ||
| 11 | F | 61 | Complete | S+RX+TMZ | 2 | + | + | 11.0 | 13.7 | ||
| 12 | F | 66 | Complete | S+RX+TMZ | 2 | + | + | 6.9 | 8.2 | ||
| 13 | M | 46 | Partial | S+RX+TMZ | 2 | + | − | 10.0 | 14.0 | ||
| Group B (n=17) | |||||||||||
| 1 | M | 56 | Complete | S+RX+TMZ | 2 | Wt | Wt | + | + | 40.0 | 56.8 |
| 2 | F | 58 | Complete | S+RX+TMZ | 2 | + | + | 16.6 | 17.6 | ||
| 3 | M | 63 | Partial | S+RX+TMZ | 2 | + | − | 20.3 | 25.2 | ||
| 4 | M | 56 | Complete | S+RX+TMZ | 1 | + | − | 11.8 | 16.5 | ||
| 5 | M | 57 | Complete | S+RX | 2 | + | + | 6.0 | 15.5 | ||
| 6 | F | 72 | Complete | S+RX+TMZ | 3 | + | − | 18.7 | 20.8 | ||
| 7 | F | 53 | Complete | S+RX+TMZ | 2 | + | + | 7.2 | 11.0 | ||
| 8 | M | 73 | Complete | S+RX+TMZ | 2 | + | + | 15.3 | 16.5 | ||
| 9 | F | 60 | Partial | S+RX+TMZ | 2 | + | + | 15.8 | 26.1 | ||
| 10 | M | 54 | Complete | S+RX+TMZ | 2 | + | + | 22.0 | 24.7 | ||
| 11 | M | 68 | Biopsy | S+RX+TMZ | 2 | + | + | 9.7 | 20.3 | ||
| 12 | M | 67 | Complete | S+RX+TMZ | 2 | + | + | 14.2 | 15.7 | ||
| 13 | M | 52 | Biopsy | S+RX+TMZ | 2 | + | − | 7.0 | 8.7 | ||
| 14 | M | 63 | Complete | S+RX+TMZ | 2 | + | − | 12.6 | 17.5 | ||
| 15 | M | 69 | Biopsy | S+RX+TMZ | 2 | + | − | 10.6 | 11.9 | ||
| 16 | M | 72 | Complete | S+RX | 1 | + | + | 4.1 | 27.6 | ||
| 17 | F | 69 | Complete | S+RX+TMZ | 2 | + | + | 15.9 | 16.5 | ||
Abbreviations: GBM, glioblastoma; M, methylated; OS, overall survival; PFS, progression-free survival; RX, radiotherapy 60 Gy; S, surgery; TMZ, temozolomide; UM, unmethylated; wt, wild type.
Figure 5.
COL1A1 and IFITM1 expression levels in an independent cohort of 30 patients' tumors. (a) Box plot representation of the mRNA expression levels. The charts show the log10 expression of relative quantification (RQ) values normalized to the expression of H9 NSC cell line. The top edge of the boxes represents the 75th percentile, the bottom edge, the 25th percentile and × the mean. The range is shown as vertical edge. (b) Kaplan–Meier analysis for overall survival of an independent cohort of 30 GBM patients. Survival distribution of patients with low (group A; n=13) versus high (group B; n=17) expression of COL1A1 and IFITM1 (P<10−4).
Discussion
Recent data suggest that GBMs belong to a tumor class in which the CSC hypothesis may be valid with the presence of a small population of cells displaying self-renewal and differentiation abilities. These progenitors have been detected by the use of in vitro and in vivo transplantation assays2, 3, 4, 5, 6, 7, 8, 9 with a hierarchy similar to that found in hematopoietic stem cells.28 Several malignant features of GBMs such as recurrence, radio- and chemo-resistance may be due to the presence of GSC population, which could represent a therapeutic target. The purpose of the present work was to investigate the pluripotency, stemness characteristics and the differentiation potential of GSCs using a Taqman Low Density Array (TLDA)-based comparative strategy. Our results have highlighted a sizable decrease in Nanog and POU5F1 mRNA expression in hNSCs as well as in GSCs compared with pluripotent hESCs. These data are in line with those of a previous study from Choi et al.29 showing, through cell reprogramming experiments, that these two genes had a key role in controlling the pluripotency and the differentiation status of hNSCs. Among genes involved in the maintenance of pluripotency or expressed in undifferentiated cells, only SOX2 showed a similar expression in GSCs and H9-ESCs. This observation suggests that SOX2 largely contributes to the maintenance of a pluripotent and undifferentiated state in GSCs. SOX2 is known to be involved in NSC homeostasis and its expression is essential for the maintenance of induced pluripotent stem cells as well as long-term in vitro neurosphere cultures.29, 30
On the basis of TLDA results, the differentiation potential of GSCs was consistent with their cellular origin. Genes involved in astrocytic (GFAP), neuronal (NES and NEUROD1) and oligodendrocytic (OLIG2) differentiation were clearly overexpressed as compared with hNSC. Similarly, the GCM1 gene, which is thought to act as a binary switch between neuronal and glial cell determination, displayed higher expression in GSCs. In the Drosophila nervous system, the ortholog of GCM1 is transiently expressed in glial precursors to switch their fate from the neuronal default state to glia.31 Interestingly, the genes involved in bone (RUNX2) and endothelial (CDH5, CD34 and PECAM) differentiations are also overexpressed in hNSCs and GSCs as compared with H1 and H9-ES cells. Plasticity is a characteristic of NSCs and differentiation of NSCs and GSCs into unanticipated cell types such as endothelial cells has already been shown.31, 32, 33, 34
Hierarchical clustering based on TLDA analysis clearly identified two GSC groups. Seven genes appeared significantly overexpressed in group GSCX-2. Among them, COL1A1, FN1 and LAMB1 known to be mesenchymal markers are involved in the extracellular matrix. The role of extracellular matrix components such as fibronectin (FN1) in glioma invasion is now well known. In this regard, microarray studies have revealed an overexpression of FN1 in human gliomas as compared with normal brain tissue.35 A recent study performed by Kim et al.,36 which aimed at identifying predictive survival models, classified GBM into three distinctive subgroups according to the differential expression of mesenchymal-related genes. Interestingly, the mesenchymal-enriched subgroup of GBM was found to significantly overexpress COL1A1 and FN1 genes, as observed in our study in group GSCX-2. Laminin subunit β1 (LAMB1) was also found overexpressed in human gliomas. Among genes differentially expressed in the two GSC groups, COL1A1 and IFITM1 were found to be the most discriminating. COL1A1 is considered as a mesenchymal osteoblast marker37 and was also defined as a glioma endothelial marker selectively expressed in the microvasculature.38 In vitro silencing of COL1A1 in two different GSC lines revealed its prominent role in promoting invasion and neurosphere-initiating capacity. IFITM1 is a transmembrane protein whose expression is induced by interferon γ. It is expressed on the cell surface of primordial germ cells in a developmentally regulated manner and is supposed to modulate cell adhesion and differentiation.39 This protein is also involved in the invasion process and progression of gastric cancer cells,40 colorectal tumors41 and head and neck squamous cell carcinomas42 through the activation of matrix metalloproteases -12 and -13. Recently, gene invalidation studies using shRNA unveiled the role of IFITM1 in proliferation, migration and invasion of GBM cell line U87MG.43 Our results obtained in vitro using two IFITM1 knockdown GSC lines are corroborate previous observations because IFITM1 silencing led to a significant reduction of invasive capacity. Taken together, our results highlight two distinct types of glioma-initiating stem cells according to the expression of mesenchymal markers.
The expression level of COL1A1 and IFITM1 in bulk tumors from which the GSCs were established probably reflects their expression level in GSCs. The lack of difference in overall survival between the two groups is probably due to the small number of cases (n=11). However, analysis of an independent cohort of 30 primary GBM patients clearly identified two groups of patients according to the expression levels of COL1A1 and IFITM1 in their tumors with a statistically significant difference in overall survival, which increased from 10 to 17.5 months. Importantly, the groups of patients were not different in terms of age, ECOG score or treatment regimens consisting in concomitant temozolomide and RT after surgery. Further, no alteration in factors known to influence patients prognosis (IDH1 and IDH2 mutations or MGMT methylation status)29, 30, 44, 45 was observed in either group. In addition, class comparison between very long-term survivors (>24 months) and very short-term survivors (<12 months) in the Rembrandt database revealed a significant clinical value for COL1A1 (P=0.04), in agreement with our results (https://caintegrator.nci.nih.gov/rembrandt/).
A recently established notion postulates that neoplastic transformation in the central nervous system converts neural cells into cell types manifesting a mesenchymal phenotype state associated with uncontrolled ability to invade and stimulate angiogenesis.46, 47 Thus, a mesenchymal phenotype would be the hallmark of tumor aggressiveness in human malignant glioma in contrast to proneural phenotype associated with longer survival.46, 47 Consistent with this concept, recent reports have suggested an epithelial-to-mesenchymal-like transition phenomenon in GBM, similar to epithelial tumors but lacking cadherin reorganization.48
In a recent in vitro study, Mao et al.15 have characterized mesenchymal GSCs using a 5796-gene signature and reported an increase in glycolytic pathways in these cells. These authors also showed promising therapeutic opportunities through inhibition of aldehyde dehydrogenase 1 A3 in these mesenchymal-enriched GSCs. In our study, overexpression of IFITM1 and COL1A1 appears predictive of a longer survival. This result may seem paradoxical as it is known that GBMs with mesenchymal phenotype are aggressive tumors. However, recent data have shown a beneficial effect with better prognostic in patients with the mesenchymal subtype,46 supporting the difference of survival observed in our study for patients with high levels of COL1A1 and IFITM1 expression. In conclusion, the expression of these two genes could represent useful molecular markers, not only to characterize GSCs, but also to stratify GBM patients at diagnosis since both genes appears to correlate with a longer survival.
Materials and methods
Patients
Gliomaspheres were derived from 11 adult patients with high-grade gliomas (Table 1). Tissues were obtained during surgery (Department of Neurological Surgery, Poitiers University Hospital, France) after informed consent of all patients and in accordance with the Declaration of Helsinki. Patients were free from any past medical history, especially with regard to cancer. Tumor diagnosis and grading were established according to the WHO criteria49 and were systematically confirmed by two expert neuropathologists. With the approval of the ethics committee, validation of COL1A1 and IFITM1 expression was performed in a panel of primary tumors from an independent cohort of 30 patients (Table 2).
Determination of IDH1, IDH2 and MGMT promoter status in GBM patients
The genomic region spanning wild-type R132 of IDH1 and R172 of IDH2 was analyzed by direct sequencing as previously described.44 Methylation status of the MGMT promoter was determined by bisulfite modification and subsequent nested methylation specific-polymerase chain reaction.45
In vitro and in vivo GSCs characterization
Tumor samples were obtained within 30 min after surgical resection from 11 adult GBM patients (GSC-1–GSC-11) and tumor sphere cultures were performed and characterized as we described previously.50, 51 Isolation and characterization of GSCs were based upon criteria described by Singh et al.2 and Galli et al.3 All GSC lines were assessed for in vitro clonogenicity by limiting dilution assays. The number of cells required to form one neurosphere, which reflects the frequency of CSCs in the entire population, was determined as previously described50 (Supplementary Table 1).
hESC culture and H9-derived human NSC culture
After authorization from the French Biomedicine Agency (permission to conduct research on human embryonic stem cells, SANB0721164S and SANB0721165S, 9 February 2007), hESC lines H1 and H9 (WiCell Research Institute, Madison, WI, USA) were cultured on mitomycin-treated mouse embryonic fibroblast cells in KnockOut Dulbecco's modified Eagle medium supplemented with 20% KnockOut serum replacement (Invitrogen, Carlsbad, CA, USA), and 5 ng/ml basic fibroblast growth factor. GIBCO Human Neural Stem Cells (H9-HNSCs) are derived from NIH-approved H9 (WA09) hESCs (Life Technologies, Grand Island, NY, USA). Cells were cultured following the manufacturer's instructions (H9-HNSC-1) as well as in neurobasal glioma stem cells medium (H9-HNSC-2). Culture conditions of H9-HNSCs did not influence the expression of pluripotency genes (Figure 2a).
Analysis of gliomaspheres mRNA by human stem cell pluripotency low-density array
TaqMan Human Stem Cell Pluripotency Array (Applied Biosystems, Foster City, CA, USA) consisted in 4 identical 96 gene sets (90 target genes and 6 endogenous controls). The list of target genes was based on a previous work from the International Stem Cell Initiative52 and is detailed in Supplementary Table 2. These genes were selected with the following criteria: (i) expression in undifferentiated cells (embryonic carcinoma cells, primordial germ cells or hESCs), (ii) maintenance of pluripotency, (iii) expression level correlated with stem cell state (stemness) and (iv) determination of the differentiation capability into all three embryonic germ layers (ectoderm, endoderm and mesoderm). Briefly, 2 μg of total RNA obtained from cells at passage 6 were reverse transcribed using the High Capacity RNA-to-cDNA Kit according to the manufacturer's instructions (Applied Biosystems). The genetic stability of early passage GSCs in serum-free conditions has previously been demonstrated.53, 54 Each line of the microfluidic card was loaded with 100 μl of a mixture containing the complementary DNA template and 50 μl of 2X TaqMan Universal PCR Master Mix. Quantitative PCR was then carried out with the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). Each experiment was conducted in triplicate and target genes that showed no amplification, abnormal amplification curves or bad replicates were excluded (Supplementary Table 2). Amplification data were analyzed using the StatMiner 3.0 software (Integromics, Madrid, Spain). Relative quantification of target gene expression was determined by the 2−ΔΔCt method using glyceraldehyde 3-phosphate dehydrogenase (GAPDH, most stable reference gene) as an endogenous control and normalized to the expression in H1-H9-ESCs or H9-HNSCs.
Vector design and preparation of lentiviral supernatant
shRNA constucts against human COL1A1 and IFITM1 were cloned downstream to human H1 promoter into a modified pTRIP lentiviral backbone carrying the selectable cassette GFP-2A-puroR under the control of the human phosphoglycerate kinase promoter. Small interfering RNA sequences and details about the constructs are available on request. Lentiviral particles were produced using standard methods. Briefly, viral plasmid was introduced together with packaging plasmids into 293T cells using calcium phosphate transfection procedure. 293T cells are grown in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum and 100 IU/ml of penicillin–streptomycin (Life Technologies). Medium was replaced after 24 h of transfection, and viral particles were collected after 48 h by ultracentrifugation at 25 000 r.p.m. for 2 h. Viral particles were re-suspended in serum-free medium, aliquoted and stored at −80 °C until use. GSC-3 or GSC-9 cell lines (1 × 105 cells) were incubated with the indicated lentiviral particles at a multiplicity of infection of 1 for 48 h. Control cells were infected with an empty vector. Two days after transduction, cells were selected with 1 μM of puromycin (Sigma, Lyon, France).
Cell proliferation
Doubling times of gliomaspheres as well as the inhibitory effect of shRNA-COL1A1 and shRNA-IFITM1 on their proliferation were assessed by CellTiter 96 Aqueous Non-Radioactive Cell Proliferation Assay (Promega, Lyon, France). Cells were plated in a 96-well plate at a density of 5 × 104 cells per well in 100 μl medium and quantification of viable cells was performed at 492 nm with a micro-plate reader (Dynex Technologies, Chantilly, France).
Cell invasion assay
Cell invasion assay was performed using Boyden chamber with 8 μm pore size (Becton Dickinson Biosciences, Le pont de Claix, France), coated with matrigel (Becton Dickinson Biosciences). Control cells, shRNA-silenced cells were seeded in the upper chamber of each well and NBE medium was added to the lower chamber. Cells were allowed to invade for 3 days and the upper side of the membrane containing non-invasive cells was cleaned with a cotton swab. Invading cells were fixed with cold methanol followed by hematoxylin/eosin staining. The membrane was cut-off and the number of migrating cells was counted under a microscope (Nikon Eclipse 80I, TribVN ICS framework, Châtillon, France).
Limiting dilutions and neurosphere-initiating cell assay
To determine the effect of shRNA-COL1A1 and shRNA-IFITM1 on the frequency of ‘neurosphere-initiating cell', we performed limiting dilution assays using single-cell populations immediately after dissociation of neurospheres. Final cell dilutions ranged from 1 cell per well to 80 cells per well. At day 21, the fraction of wells free of neurosphere for each cell plating density was determined. These results were plotted against the number of cells plated per well. The number of cells required to form one neurosphere, which reflects the frequency of CSCs in the entire population, was determined as described previously.50
Western blot analysis
Cells were lysed in Laemmli buffer and protein concentration was determined using a Bradford assay (Bio-Rad, Marnes-La-Coquette, France). Equal amounts of protein samples were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred onto a nitrocellulose membrane (Amersham Biosciences, Velizy-Villacoublay, France). Membranes were blocked with 5% non-fat milk in phosphate-buffered saline 0.1% Tween for 2 h and incubated overnight at 4 °C with the primary antibody. Antibodies used in this study were goat anti-IFITM1 antibody (R&D Systems Europe, Lille, France), rabbit anti-COL1A1 antibody (Abcam, Paris, France) and mouse β-actin antibody (Abcam). After incubation with the appropriate secondary antibodies (Santa Cruz, Dallas, TX, USA), blots were revealed by chemiluminescence (ECL Plus reagent, Amersham Biosciences). Band intensity was quantified with the use of ImageJ software (Bethesda, MD, USA).
Determination of COL1A1 and IFITM1 mRNA expression by reverse transcriptase–quantitative PCR
Total RNA was extracted from tumor specimens using Qiagen RNeasy Mini Kit according to the manufacturer's instructions (Qiagen, Courtaboeuf, France). In all, 3 μg of total RNA was transcribed into complementary DNA using Superscript II (Life Technologies). Expression of COL1A1, IFITM1, and GAPDH was determined using TaqMan Gene Expression Assays Hs00164004_m1, Hs00705137_s1, Hs Hs99999905_m1 (Applied Biosystems) respectively. Relative quantification was established by the 2−ΔΔCt method with GAPDH as an endogenous control and normalized to the expression of the H9-HNSCs. Analysis of the results was done using the StatMiner software.
Statistical analysis
Clustering gene expression patterns were determined using hierarchical algorithms of StatMiner software. In this study, Euclidean distance and Ward's linkage method were applied although clustering results were identical whatever the linkage method and distance measure used. In order to identify target genes with significant differences in their expression, a parametric paired t-test was used adjusting the P-value with the Benjamini–Hochberg false discovery rate-controlling procedure to obtain reliable significance levels with the high number of tests performed (StatMiner).
An independent cohort of patients (n=30) was analyzed using SAS v.9 software (SAS, Cary, NC, USA). Fisher's exact test was used for testing the association of groups from cluster analysis with those given by patient's characteristics (MGMT). Correlations between gene expression data were analyzed using Spearman's rank test. Overall survival estimates with 95% confidence intervals were provided by the Kaplan–Meier method and compared with the log-rank test.
Acknowledgments
We thank Ms C Marquant, Mr P Rivet and Mr S Martin for technical assistance with cell culture, DNA sequencing and methylation specific-polymerase chain reaction (MS-PCR). We also thank Dr Denis Biard for the small interfering RNA construct and Jeffrey Arsham, an American medical translator, for having reread and reviewed the original English-language text. Anaïs Balbous is recipient of a doctoral fellowship from ‘En avant la vie' and ‘Sport et collections' foundations. Funding for this work was provided by Ligue contre le Cancer de la Vienne et des Deux-Sèvres, Région Poitou-Charentes, Cancéropôle Grand Ouest (Réseau Gliomes), and the ‘Sport et Collection' and ‘Rotary Club de Civray' foundations.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogenesis website (http://www.nature.com/oncsis)
Supplementary Material
References
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJB, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. New Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- Ogden AT, Waziri AE, Lochhead RA, Fusco D, Lopez K, Ellis JA, et al. Identification of A2B5+CD133- tumor-initiating cells in adult human gliomas Neurosurgery 200862505–514.discussion 514–515. [DOI] [PubMed] [Google Scholar]
- Chen R, Nishimura MC, Bumbaca SM, Kharbanda S, Forrest WF, Kasman IM, et al. A hierarchy of self-renewing tumor-initiating cell types in glioblastoma. Cancer Cell. 2010;17:362–375. doi: 10.1016/j.ccr.2009.12.049. [DOI] [PubMed] [Google Scholar]
- Beier D, Hau P, Proescholdt M, Lohmeier A, Wischhusen J, Oefner PJ, et al. CD133(+) and CD133(-) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67:4010–4015. doi: 10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- Wang J, Sakariassen PØ, Tsinkalovsky O, Immervoll H, Bøe SO, Svendsen A, et al. CD133 negative glioma cells form tumors in nude rats and give rise to CD133 positive cells. Int J Cancer J Int Cancer. 2008;122:761–768. doi: 10.1002/ijc.23130. [DOI] [PubMed] [Google Scholar]
- Son MJ, Woolard K, Nam D-H, Lee J, Fine HA. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4:440–452. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo KM, Kim SY, Jin X, Song SY, Kong D-S, Lee J-I, et al. Clinical and biological implications of CD133-positive and CD133-negative cells in glioblastomas. Lab Investig J Tech Methods Pathol. 2008;88:808–815. doi: 10.1038/labinvest.2008.57. [DOI] [PubMed] [Google Scholar]
- Liu G, Yuan X, Zeng Z, Tunici P, Ng H, Abdulkadir IR, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M-K, Kang S-K. Tumorigenesis of chemotherapeutic drug-resistant cancer stem-like cells in brain glioma. Stem Cells Dev. 2007;16:837–847. doi: 10.1089/scd.2007.0006. [DOI] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Sandberg CJ, Altschuler G, Jeong J, Strømme KK, Stangeland B, Murrell W, et al. Comparison of glioma stem cells to neural stem cells from the adult human brain identifies dysregulated Wnt- signaling and a fingerprint associated with clinical outcome. Exp Cell Res. 2013;319:2230–2243. doi: 10.1016/j.yexcr.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Denysenko T, Gennero L, Roos MA, Melcarne A, Juenemann C, Faccani G, et al. Glioblastoma cancer stem cells: heterogeneity, microenvironment and related therapeutic strategies. Cell Biochem Funct. 2010;28:343–351. doi: 10.1002/cbf.1666. [DOI] [PubMed] [Google Scholar]
- Mao P, Joshi K, Li J, Kim S-H, Li P, Santana-Santos L, et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci USA. 2013;110:8644–8649. doi: 10.1073/pnas.1221478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CL, Miura T, Robson P, Lim S-K, Xu X-Q, MY-C Lee, et al. Transcriptome profiling of human and murine ESCs identifies divergent paths required to maintain the stem cell state. Stem Cells Dayt Ohio. 2005;23:166–185. doi: 10.1634/stemcells.2004-0162. [DOI] [PubMed] [Google Scholar]
- Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom DT, Dowell RD, Jacobsen ES, Gordon W, Danford TW, MacIsaac KD, et al. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet. 2007;39:730–732. doi: 10.1038/ng2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assou S, Le Carrour T, Tondeur S, Ström S, Gabelle A, Marty S, et al. A meta-analysis of human embryonic stem cells transcriptome integrated into a web-based expression atlas. Stem Cells Dayt Ohio. 2007;25:961–973. doi: 10.1634/stemcells.2006-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke MF, Fuller M. Stem cells and cancer: two faces of eve. Cell. 2006;124:1111–1115. doi: 10.1016/j.cell.2006.03.011. [DOI] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Kang J, Shakya A, Tantin D. Stem cells, stress, metabolism and cancer: a drama in two Octs. Trends Biochem Sci. 2009;34:491–499. doi: 10.1016/j.tibs.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Schoenhals M, Kassambara A, De Vos J, Hose D, Moreaux J, Klein B. Embryonic stem cell markers expression in cancers. Biochem Biophys Res Commun. 2009;383:157–162. doi: 10.1016/j.bbrc.2009.02.156. [DOI] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller F-J, Laurent LC, Kostka D, Ulitsky I, Williams R, Lu C, et al. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455:401–405. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg J, He X, Peredo I, Orrego A, Hesselager G, Ericsson C, et al. Activation of neural and pluripotent stem cell signatures correlates with increased malignancy in human glioma. PLoS One. 2011;6:e18454. doi: 10.1371/journal.pone.0018454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112:4793–4807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- Choi HW, Kim JS, Choi S, Jang HJ, Kim MJ, Choi Y, et al. Neural stem cells achieve and maintain pluripotency without feeder cells. PLoS One. 2011;6:e21367. doi: 10.1371/journal.pone.0021367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon T, Smith A, Savatier P. Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell Biol. 2002;12:432–438. doi: 10.1016/s0962-8924(02)02352-8. [DOI] [PubMed] [Google Scholar]
- Akiyama Y, Hosoya T, Poole AM, Hotta Y. The gcm-motif: a novel DNA-binding motif conserved in Drosophila and mammals. Proc Natl Acad Sci USA. 1996;93:14912–14916. doi: 10.1073/pnas.93.25.14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berx G, van Roy F. Involvement of members of the cadherin superfamily in cancer. Cold Spring Harb Perspect Biol. 2009;1:a003129. doi: 10.1101/cshperspect.a003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajithlal GB, McGuire TF, Lu J, Beer-Stolz D, Prochownik EV. Endothelial-like cells derived directly from human tumor xenografts. Int J Cancer J Int Cancer. 2010;127:2268–2278. doi: 10.1002/ijc.25251. [DOI] [PubMed] [Google Scholar]
- Dong J, Zhao Y, Huang Q, Fei X, Diao Y, Shen Y, et al. Glioma stem/progenitor cells contribute to neovascularization via transdifferentiation. Stem Cell Rev. 2011;7:141–152. doi: 10.1007/s12015-010-9169-7. [DOI] [PubMed] [Google Scholar]
- Sallinen SL, Sallinen PK, Haapasalo HK, Helin HJ, Helén PT, Schraml P, et al. Identification of differentially expressed genes in human gliomas by DNA microarray and tissue chip techniques. Cancer Res. 2000;60:6617–6622. [PubMed] [Google Scholar]
- Kim Y-W, Koul D, Kim SH, Lucio-Eterovic AK, Freire PR, Yao J, et al. Identification of prognostic gene signatures of glioblastoma: a study based on TCGA data analysis. Neuro-Oncol. 2013;15:829–839. doi: 10.1093/neuonc/not024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori-Akiyama Y, Akiyama H, Rowitch DH, de Crombrugghe B. Sox9 is required for determination of the chondrogenic cell lineage in the cranial neural crest. Proc Natl Acad Sci USA. 2003;100:9360–9365. doi: 10.1073/pnas.1631288100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Carson-Walter EB, Cooper A, Winans BN, Johnson MD, Walter KA. Vascular gene expression patterns are conserved in primary and metastatic brain tumors. J Neurooncol. 2010;99:13–24. doi: 10.1007/s11060-009-0105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka SS, Yamaguchi YL, Tsoi B, Lickert H, Tam PPL. IFITM/Mil/fragilis family proteins IFITM1 and IFITM3 play distinct roles in mouse primordial germ cell homing and repulsion. Dev Cell. 2005;9:745–756. doi: 10.1016/j.devcel.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Yang G, Xu Y, Chen X, Hu G. IFITM1 plays an essential role in the antiproliferative action of interferon-gamma. Oncogene. 2007;26:594–603. doi: 10.1038/sj.onc.1209807. [DOI] [PubMed] [Google Scholar]
- Andreu P, Colnot S, Godard C, Laurent-Puig P, Lamarque D, Kahn A, et al. Identification of the IFITM family as a new molecular marker in human colorectal tumors. Cancer Res. 2006;66:1949–1955. doi: 10.1158/0008-5472.CAN-05-2731. [DOI] [PubMed] [Google Scholar]
- Hatano H, Kudo Y, Ogawa I, Tsunematsu T, Kikuchi A, Abiko Y, et al. IFN-induced transmembrane protein 1 promotes invasion at early stage of head and neck cancer progression. Clin Cancer Res Off J Am Assoc Cancer Res. 2008;14:6097–6105. doi: 10.1158/1078-0432.CCR-07-4761. [DOI] [PubMed] [Google Scholar]
- Yu F, Ng SSM, Chow BKC, Sze J, Lu G, Poon WS, et al. Knockdown of interferon-induced transmembrane protein 1 (IFITM1) inhibits proliferation, migration, and invasion of glioma cells. J Neurooncol. 2011;103:187–195. doi: 10.1007/s11060-010-0377-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, JC-H Lin, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegi ME, Diserens A-C, Gorlia T, Hamou M-F, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. New Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- Verhaak RGW, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Kahlert UD, Nikkhah G, Maciaczyk J. Epithelial-to-mesenchymal(-like) transition as a relevant molecular event in malignant gliomas. Cancer Lett. 2013;331:131–138. doi: 10.1016/j.canlet.2012.12.010. [DOI] [PubMed] [Google Scholar]
- Kleihues P, Louis DN, Scheithauer BW, Rorke LB, Reifenberger G, Burger PC, et al. The WHO classification of tumors of the nervous system J Neuropathol Exp Neurol 200261215–225.discussion 226–229. [DOI] [PubMed] [Google Scholar]
- Villalva C, Martin-Lannerée S, Cortes U, Dkhissi F, Wager M, Le Corf A, et al. STAT3 is essential for the maintenance of neurosphere-initiating tumor cells in patients with glioblastomas: a potential for targeted therapy. Int J Cancer J Int Cancer. 2011;128:826–838. doi: 10.1002/ijc.25416. [DOI] [PubMed] [Google Scholar]
- Villalva C, Cortes U, Wager M, Tourani J-M, Rivet P, Marquant C, et al. O6-Methylguanine-methyltransferase (MGMT) promoter methylation status in glioma stem-like cells is correlated to temozolomide sensitivity under differentiation-promoting conditions. Int J Mol Sci. 2012;13:6983–6994. doi: 10.3390/ijms13066983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adewumi O, Aflatoonian B, Ahrlund-Richter L, Amit M, Andrews PW, Beighton G, et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Ernst A, Hofmann S, Ahmadi R, Becker N, Korshunov A, Engel F, et al. Genomic and expression profiling of glioblastoma stem cell-like spheroid cultures identifies novel tumor-relevant genes associated with survival. Clin Cancer Res Off J Am Assoc Cancer Res. 2009;15:6541–6550. doi: 10.1158/1078-0432.CCR-09-0695. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.