Abstract
Light-induced proton pumping in bacteriorhodospin is carried out through five proton transfer steps. We propose that the proton transfer to Asp85 from the Schiff base in the L-to-M transition is accompanied by the relocation of a water cluster on the cytoplasmic side of the Schiff base from a site close to the Schiff base in L to the Phe219-Thr46 region in M. The water cluster present in L, formed at 170 K, is more rigid than that at room temperature. This may be responsible for blocking the conversion of L to M at 170 K. In the photocycle at room temperature, this water cluster returns to the site close to the Schiff base in N, with a rigid structure similar to that of L at 170 K. The increase in the proton affinity of Asp85, which is a prerequisite for the one-way proton transfer in the M-to-N transition, is suggested to be facilitated by a structural change which disrupts interactions between Asp212 and the Schiff base, and between Asp212 and Arg82. We propose that this liberation of Asp212 is accompanied by a rearrangement of the structure of water molecules between Asp85 and Asp212, stabilizing the protonated Asp85 in M.
INTRODUCTION
Bacteriorhodopsin is an excellent enzyme system in which to investigate the underlying chemistry of protein function. The reaction can be started by light, which is a noninvasive trigger of the protein function, and allows precise comparison of the intermediates to the initial unphotolyzed state having the all-trans retinal chromophore (BR). Fourier transform infrared spectroscopy (FTIR) studies are especially powerful for investigating changes in the polar groups around active sites when combined with structural information obtained by X-ray diffraction studies.
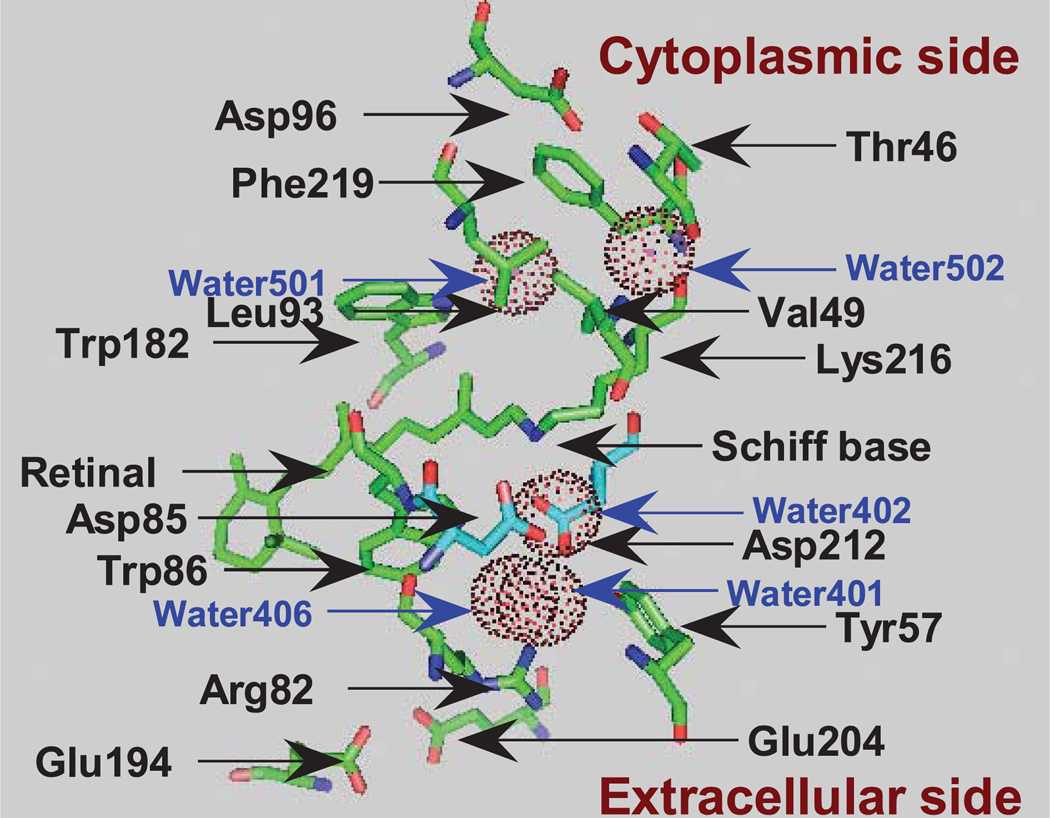
Light causes the trans-to-cis isomerization of the C13 = C14 bond of the retinal chromophore, leading to a sequence of photointermediates that unfold over the course of femtoseconds to milliseconds, resulting in conservation of a fraction of the photon’s energy by means of the electrogenic transfer of a proton across the bacterial membrane. Unidirectional proton pumping by bacteriorhodopsin is the result of the sum of intramolecular proton transfer processes, each of which is closely synchronized with the transitions between the photo-intermediates, L, M, N and O (concisely summarized with references in Morgan et al. [1]). Residues and water molecules relevant to the current studies are shown in Fig. 1 on the basis of the crystallographic structure of protein databank entry 1c3w (2). The first proton transfer from the Schiff base to Asp85, on the extracellular side of the Schiff base, occurs in the L-to-M transition. Concomitant with this, a proton is released (3) from the proton release group (PRG), which is composed of Glu204, Glu194, Arg82, Tyr57 and water molecules surrounded by these residues (4). The deprotonated Schiff base of M then accepts a proton from Asp96, on the opposite side of the Schiff base, resulting in the formation of N. A proton is taken up by Asp96 from the cytoplasm in the N-to-0 transition, and this is also accompanied by the cis-to-trans reisomerization of the C13 = C14 bond. The final step is the proton transfer from Asp85 to the PRG, resetting the structure to BR. In the whole cycle, one proton moves from the cytoplasm to the extracellular medium driven by one photon. These proton transfer reactions are, in principle, driven by successive proton affinity changes (conventionally expressed by pKa) of the Schiff base, Asp85, the PRG and Asp96. The pathways of proton transfer between the residues have been discussed from the point of view of accessibility changes in the Schiff base (5,6), and have also recently been investigated by using QM/MM calculations for the L-to-M transition (7–9) and for the M-to-N transition (10). Here, on the basis of our FTIR studies and other data in the literature, we discuss our view of what regulates the affinity changes of the Schiff base and Asp85.
Figure 1.
Structure of bacteriorhodopsin showing the relative locations of the residues and water molecules (dot circles) that are discussed in the text, depicted on the basis of protein databank entry 1c3w (1) using PyMol (80).
A WATER CLUSTER ON THE CYTOPLASMIC SIDE STABILIZES L RELATIVE TO M
In pH titrations of BR, the pKa of the Schiff base in bacteriorhodopsin has been estimated to be ~13 (11), much higher than the value of ~7 for the retinal–butylamine complex in water-methanol solution (12), a model Schiff base. A solid state 15N-NMR study on bacteriorhodopsin, in which the Schiff base was labeled with 15N, has indicated that this elevated pKa is caused by a directly interacting water molecule (Water402) and the negative charge of the counterion (Asp85) delocalized through a hydrogen bonding complex composed of the polar residues and additional water molecules (13) (Water401, Water406, Arg82 and Asp212) (Fig. 1). Lowering the pH of BR below 3 changes its color from purple to blue. This change results from the protonation of Asp85, indicating that the pKa of Asp85 in BR is 3 (14).
The effect of light on bacteriorhodopsin is exerted primarily through the trans-to-cis isomerization of the C13 = C14 bond of the chromophore retinal. Resonance Raman studies have shown that the L, M and N intermediates at ambient temperature have the chromophore in the 13-cis, 14-trans, 15-trans conformation (15,16), in which the N–H bond of the Schiff base should be oriented opposite to that in BR, facing the cytoplasmic side. The Schiff base, however, is protonated in the L and N states but deprotonated in M. Proton transfer from the Schiff base to Asp85 in the L → M transition occurs even at pH 3. The presence of the unprotonated state of the Schiff base, even in the pH range lower than the pKa of the model Schiff base, may be the result of the absence of water molecules interacting directly with the Schiff base in M. A crystallographic model of BR at 1.55 Å resolution (2) shows two water molecules (Water501 and Water502 in Fig. 1) on the cytoplasmic side, but they are not near the Schiff base. In contrast, the protonation of the Schiff base in L and N may be the result of interactions with water molecules that stabilize the protonated form of the Schiff base.
L is the state immediately before the first proton transfer. The L-minus-BR spectrum can be recorded at 170 K as a steady state under continuous illumination. The spectrum exhibits a broad feature between 3550 and 3450 cm−1, which is composed of at least three water O-H vibration bands (17,18). In this spectrum, the positive bands (due to L) are more intense than the negative bands (BR) reflecting the fact that water molecules become polarized upon L formation. No vibrational bands of this nature appear in either the K intermediate at 80 K or KL at 135 K (19). The intensity of these bands is affected by mutants of residues located on the cytoplasmic side of the Schiff base: Thr46, Asp96, Leu93 and Val49.
The details of these mutant studies have been reviewed previously (20,21), but will be repeated concisely below to emphasize the effect of the water cluster on the protonated state of the Schiff base. The intensity of the water bands in the L-minus-BR spectrum becomes smaller in T46V, but is restored in T46V/D96N in parallel with a shift of the L-to-M equilibrium to M, which is also reversed in the double mutant (22). These mutations did not affect the vibrational bands of water at 3643 cm−1 or of Asp85 at 1761 cm−1, both of which are on the extracellular side of the Schiff base. The shift of equilibrium toward L was also observed in V49A and L93M, mutants in which there is an increase in intensity of the water vibrational bands (18,23) apparently related to removal of steric hindrance, which then allows accommodation of the water cluster. This effect of the side chain of Leu93 has also been suggested by molecular dynamics calculations (24) and crystallographic analysis (25). A water cluster forms in the cavity surrounded by the protein stretching from the Schiff base to the region around Asp96 and Gly220 (26). This structure has sufficient structural integrity to be almost completely preserved at 80 K, even without the direct support of the Schiff base. This can be observed in the L′ state, in which the Schiff base N–H is oriented toward Asp85, probably through Water402. However, this structure cannot be maintained if the sample is warmed to 170 K, indicating that the hydrogen bonding interaction of the Schiff base N–H that orients toward the cytoplasmic side is of primary importance for the L structure, including the water cluster (27). These results suggest that the water cluster, together with the surrounding protein environment on the cytoplasmic side, works to regulate the L↔M equilibrium under the influence of the protonated Schiff base N–H.
The structure proposed on the basis of our FTIR results can be depicted on the basis of the X-ray structural model in protein databank entry 1ucq (25), which contains two water molecules with high occupancy and a space to accommodate two additional water molecules extending from the Schiff base to the region around Thr46 on the cytoplasmic side. However, the FTIR data are difficult to reconcile with other models: 1r3p (28) and 2ntw (29), which do not show such a water cluster. Also, the Schiff base N–H in 2ntw (29) is connected to Asp85 through Water402 on the extracellular side. This is hard to reconcile with our proposed structure of L.
Both FTIR and X-ray studies were performed at low temperatures. The FTIR studies were performed using purple membrane, whereas the latter two X-ray models were obtained with different crystals from the former (1ucq). Relatively small doses of X-ray cause change in the visible spectrum (25), which suggests generation of a different structure around the Schiff base.
WATER CLUSTER AT ROOM TEMPERATURE
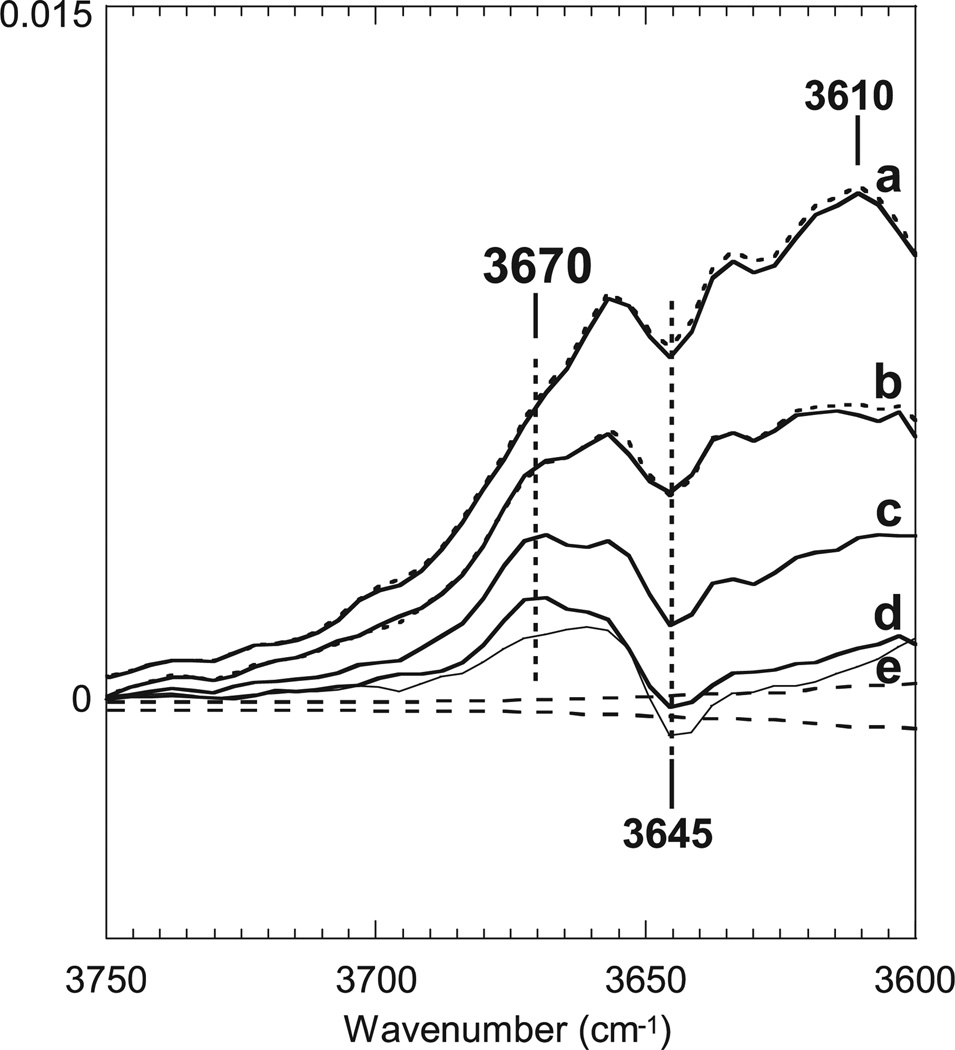
The FTIR data on L described above were obtained at cryogenic temperature. When L is formed at 170 K, it does not convert to M even upon warming to 230 K (a temperature where M can be produced by illumination of BR). Instead, upon warming, the L produced at 170 K returns to BR (30,31). It is, thus, important to study the structure of L at physiologic temperature. To this end, time-resolved step-scan FTIR spectroscopy was conducted at 25°C, and the data set in the 1800–900 cm−1 region from the microsecond to millisecond time range was subjected to a global fit using a function of six exponential components with different time constants (1). The rate constants from this analysis were used to calculate component spectra for the full acquired spectral region, and the output of this fit was then used to calculate the spectral components for a linear series of intermediates using the method of Chizhov et al. (32). It is important to note that these are intermediates in the kinetic model and do not have a one-to-one relationship with the canonical mechanistic intermediates of the bacteriorhodopsin photocycle. The results are presented in Fig. 2 as a series of difference spectra vs BR for this series of kinetic intermediates (1). The earliest component, which decays with a time constant of 43 µs, is mainly composed of L. Contributions from M and then N appear in subsequent spectra. The corresponding spectra in the 1800–900 cm−1 region for the chromophore and the protein are basically coincident with previous results (33–36).
Figure 2.
Spectra obtained by a six-component kinetic fit to the data set in the 1800–900 cm−1 region due to the vibrations of the chromophore and the protein in the light-induced reaction of bacteriorhodopsin (see text for details). Superimposed dotted lines show the corresponding spectra from a five-exponential fit for the same data set in the 3750–3630 cm−1 region. Averaged residuals are shown as broken lines. Spectra are reproduced from our data files for the published spectra in Morgan et al. (1) (© 2007 American Chemical Society).
All bands in the 3700–3600 cm−1 region in Fig. 2 result from water vibrations, confirmed by the fact that they undergo a 12 cm−1 frequency shift in 18O water. The first spectrum (a), mainly due to L (75%), exhibits a broad intense feature with a peak around 3610 cm−1. This feature becomes smaller in the second spectrum (30% of L; b), and disappears in the third spectrum (c), which is due to pure M, and the fourth (d) and fifth (e, thin line) spectra, which have the appearance of N and O (1). In order to know whether the spectral changes in these water vibrations of L take place in concert with the changes in the chromophore, the data set for the water vibrational region (3750–3630 cm−1) was separately fit as described above (1) and the results overlaid on corresponding spectra in Fig. 2, shown as dotted lines. The first and second spectra are almost completely coincident in shape and are similar in associated rate constant with the corresponding spectra obtained by fitting the vibrations due to the chromophore and the protein. The results strongly suggest that at least a large part of the broad water feature is associated with the intrinsic structure of L. These water features in the FTIR difference spectra are similar to those observed with L at 170 K, which have been ascribed to a water cluster on the cytoplasmic side of the Schiff base. The water bands observed at 25°C, however, appear at higher frequencies, indicating weaker H-bonding.
It is possible that water vibrational features of this kind could be caused by heating produced by energy dissipated from the chromophore, as has been reported in measurements on a dye–water mixture (37), in which light absorbed by the dye molecules changes to heat and induces spectral changes in the region of 1750–1600 cm−1 and above 3000 cm−1 where water O–H bending and stretching vibration bands are located. However, the signals observed in solvent water disappear within 20 µs, which is faster than the first and second components in the L-to-M transitions (described above). In bacteriorhodopsin the heat or free energy from the chromophore is transferred, during its relaxation process, into the protein moiety and internal water molecules during each exergonic transition, such as KL to L. Water molecules that acquire free energy from the chromophore could contribute to properties of L, which has a less rigid structure at 25°C than that at 170 K. The water structure observed in L disappears in the ensuing, irreversible L-to-M transition.
WATER RELOCATION IN THE L-TO-M TRANSITION
Several structured vibrational bands resulting from water vibrations are superimposed on the broad FTIR features (Fig. 2). Among them is a positive band at 3670 cm−1 that can be ascribed to M (1). The same band was previously observed in M at 230 K, and shown to be depleted in the F219L mutant (38). In the BR state, F219L has three water molecules in this region (protein databank entry 1p8i [39]). The FTIR results suggest that at least one of these water molecules retains a similar structure in M. We propose that M has a water-containing cavity surrounded by Trp182, Leu93, Thr46 and Phe219. The 3670 cm−1 band is not affected by V49A (38,40), which is a mutant of the residue, Val49, close to the Schiff base. Hence, M has water molecules close to Phe219, but not close to the Schiff base. These results can be reconciled with published X-ray structure models of M, from protein databank entries 1f4z (41), 1cwq (42), 1p8h (39) and 1iw9 (43).
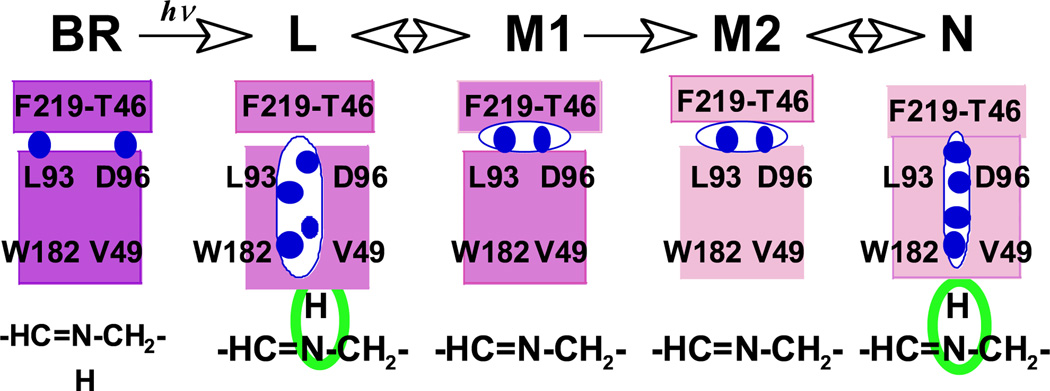
On the basis of the results, obtained at cryogenic temperature, we have proposed that in L there is a string of water molecules, which fills a cavity surrounded by Trp182, Leu93, Thr46 and Val49, and which stabilizes L with the protonated Schiff base (21). A similar water cluster is present in L at room temperature but with a less rigid structure, as evidenced by the weaker hydrogen-bonding water. Relocation of this water-filled cavity to the Phe219-Thr46 region corresponds to a decrease in the proton affinity to the Schiff base and thus stabilizes the conformation of M (Fig. 3).
Figure 3.
Schematic representation of the water cluster on the cytoplasmic side. The string of blue circles represents the water cluster. The larger cavity shown in L represents a less rigid water cluster than in N.
STRUCTURAL FEATURES OF THE SCHIFF BASE IN L
The Schiff base in L at room temperature is similar to the Schiff base in N (1), which is possibly undistorted. In contrast, L at 170 K shows unusual vibrational features due to the perturbation of the Schiff base, which manifest themselves as hydrogen out-of-plane bending vibrations (HOOP) at 1073, 1064 and 1056 cm−1 (44), and in-plane bending vibrations at 1312 and 1300 cm−1 (45), as compared to the Schiff base vibrations at ambient temperature: a HOOP band at 984 cm−1 in L and at 1005 cm−1 in N, and in-plane vibrations of the Schiff base at 1300 cm−1 in L and at 1302 cm−1 in N. It is probable that the HOOP vibrations in L at 170 K become higher in frequency because of hydrogen bonding interactions with the water cluster on the cytoplasmic side.
An FTIR study of L at low temperature has shown that the stretching vibration of the C14–C15-bond is located at 1155 cm−1 (46), a frequency lower than the corresponding band observed at 1172 cm−1 in a resonance Raman study at room temperature (47). However, the observed frequency of 1155 cm−1 is far above that of the conformationally locked, authentic, 14-s-cis Schiff base of 12-N-ethano retinal (48), at 1096 cm−1, indicating that the C14–C15-bond of L has a conformation close to trans. Distortion of the Schiff base that was suggested by other studies (49,50) was not detected in the models of 1cuq (25) and 1r3p (28), which show full rotation of the C13 = C14 bond without other changes. This 1155 cm−1 band, due to the C14–C15 stretching vibration, is located at the same frequency in D20 as in H2O (46), indicating that the C = N configuration is trans on the basis of the proposal in Smith et al. (51). In contrast, a substantial increase in the frequency might have been expected for the half-way rotation of the C=N bond shown in 2ntw, the structure on which the proposed orientation of the Schiff base toward the extracellular side was largely based (29). As judged from the 13-cis, 14-s-trans, 15-trans configuration deduced from these vibrational studies, even with distortion around C15, the N–H bond of L may be oriented to the cytoplasmic side. However, these arguments on the vibrational results should be further examined by measuring the distances between relevant atoms in solid state NMR studies like that applied to the C14–C15 bond in M (52).
On the other hand, L at room temperature seems to have an undistorted conformation around the Schiff base. Resonance Raman spectra of L at room temperature show that the chromophore has the configuration 13-cis, 14-s-trans, 15-trans (47,51). L in the time-resolved FTIR study did not exhibit the 1155 cm−1 band (see fig. 5 in Morgan et al. [1]), suggesting that there is no distortion in the C14–C15 bond.
The complex structure of the Schiff base interacting with the rigid protein, as suggested by neutron diffraction measurements (53), does not allow large amplitude motions. In the L intermediate, produced at 170 K, this structural rigidity may block a structural change, which would involve the relocation of the water cluster toward the site it characteristically occupies in M. Time-resolved visible spectroscopy has revealed two consecutive L states, L1 and L2 (54). The visible spectrum of L1, which arises in the sub-microsecond time domain before the appearance of normal L (presumably L2 from the time constant), shows a more substantial tail toward longer wavelengths, like L at low temperature (30), probably due to some distortion around the Schiff base. The FTIR spectrum in a time range similar to that for L1 shows changes in the bands due to the chromophore and Asp96/Asp115 similar to those in which occur in advance of the Schiff base changes observed in normal L (55). However, this precursor of L did not show changes in vibrational bands characteristic of those of L at low temperatures: the Schiff base bands at 1310 and 1075/1064/1056 cm−1.
THE SCHIFF BASE–WATER CLUSTER IN N COMPARED TO L
The vibrational features of the Schiff base resulting from L at room temperature are rather similar to those of N (1), suggesting that the orientation and environment on the cytoplasmic side of the Schiff base in L is similar to that in N. However, the fourth and fifth time-resolved spectra (d and e in Fig. 2), which include considerable contributions from N, do not exhibit the broad feature with a peak at 3610 cm−1 which is observed for L. The Schiff base vibrations of N, recorded at 80 K after trapping it at 260 K, are similar to those at room temperature (56). A water cluster in N, deduced from this spectrum, is also similar to that of L at 170 K, having a more rigid structure than the corresponding structure of L at room temperature.
A perturbed Val49-Pro50 backbone linkage is observed upon formation of N in the wild type but not in the V49A mutant (38). This could be related to the shift of the M↔N equilibrium toward N in V49A. The X-ray structure model of N in the V49A mutant (39) has a water cluster extending from the Schiff base to Asp96. This water cluster between the Schiff base and Asp96 is similar to the structure of the water cluster in L that we are proposing. The restoration of the proton affinity of the Schiff base in N might be accomplished by relocation of water molecules to the Schiff base.
As a whole, the isomerization of the retinal to the 13-cis form may induce the reorganization of the protein on the cytoplasmic side of the Schiff base to accommodate the water cluster. The movement of the side chains of Val49 and Leu93 is suggested as above. The perturbation of Asp96 is accompanied by changes in hydrogen bonding with Thr46. This structure, in turn, stabilizes the protonated Schiff base in L relative to M. The conversion of the protein into the form characteristic of M, with the unprotonated Schiff base, occurs concurrently with removal of water from the Schiff base. Fluctuation between these two states would amount to the equilibrium between L and M. The other equilibrium, between N and M, may be established in a similar way, by returning the water cluster to the Schiff base in N with an accompanying protonation of the Schiff base. These changes, which are depicted in a cartoon in Fig. 3, are composed of two similar equilibria, one between L and M and one between M and N.
ASP85 IN THE ONE-WAY PROTON TRANSFER
A crucial aspect of the mechanism for producing one-way proton transfer consists of enforcing the tendency of the Schiff base to only accept a proton from Asp96 in the M → N transition, and not from Asp85 by reverse flow, which would be the reverse reaction of the L → M transition. This selective proton transfer should be assured by increasing the proton affinity of Asp85 during the lifetime of M. Váró and Lanyi (57,58) have proposed from their kinetics study that the early M intermediate, M1, which is in equilibrium with L, proceeds irreversibly, with a large decrease in free energy, to the late M intermediate, M2, which is observed after L has disappeared. This transition comprises a key step for one-way proton transport, and may be brought about by the stabilization of the protonated Asp85 of M.
Titration studies on the BR state show that deprotonation of the PRG causes the pKa of Asp85 to increase to around 7 (5,59). In contrast, other observations of the properties of M show that Asp85 remains protonated even at pH 10 (49,60,61). The pKa of Asp85 may be much higher than that of the Schiff base of N, 8.2 (62), preventing the reverse flow of the proton of Asp85 to the Schiff base. Below we will discuss several factors that may stabilize the protonated Asp85.
THE ROLE OF ASP212 IN INCREASING THE PKA OF ASP85 IN M
Asp212 and Arg82 are the nearest charged residues to Asp85. The spatial relationship of these residues in BR is shown in Fig. 1. D212N is inactive in proton pumping even though it undergoes the L to N transition in the photocycle (63,64). Proton pumping in D212N can be partially restored by the addition of chloride to the sample, though only below pH 7 (65,66), and also by a second mutation that restores the charge balance in D212N/R82Q (67). The photocycle of D212N/R82Q proceeds from L to N without apparent accumulation of M, but replacement of the retinal by 14-fluoro retinal, to decrease the pKa of the Schiff base, restores the M formation. A lowered proton affinity of Asp85 in this mutant may be responsible for the apparent absence of M. Thus, the involvement of Asp212 in increasing the pKa of Asp85 in the photocycle is suggested.
Spectroscopic data showing more rapid formation of M in the mutants of Arg82 (68,69), and the absence of M in E194Q (70) both suggest that the increased pKa of Asp85 in M is brought about by disruption of the interaction between Asp212 and Arg82 under the influence of Glu194, which attracts Arg82. Crystallographic models of M of wild type bacteriorhodopsin (see protein databank entries 1cwq [42], 1kg8 [71], 1m0m [6] and 1iw9 [43]) as well as the M intermediate structure for the E204Q mutant in 1f4z (41) show that the guanidium group of Arg82 moves away from Asp212 toward Glu194 in M, whereas a structure that has been claimed to be M1 (obtained at acidic pH where the PRG remains protonated in M), 1p8h (39), retains contact between Arg82 and Asp212. Thus, Asp212, the Schiff base and the PRG are all expected to regulate the proton affinity of Asp85 in M. We propose that Asp212 breaks its interaction with Arg82 and the Schiff base in M. Removal of the positive charges of the Schiff base and Arg82 pushes the pKa of the carboxylic acids higher. Asp212 forms a di-carboxylic acid pair together with Asp85, in which one of the pKa values is elevated under influence of the other, as observed in malonic acid (2.8 and 5.8) compared to acetic acid (4.75) (72). These experimental results, however, are not consistent with an effect by which Asp212 would destabilize the protonated Asp85, as was concluded by QM/MM calculations based on the simple model including Asp85, Asp212, Thr89 and the Schiff base (7,8). The reason is unclear, but an active role of mobile water molecules is anticipated (see below).
WATER AROUND ASP85 AND ASP212
Besides exerting an electrostatic effect of this kind, Asp212 may form a different structure with Asp85 in M through a rearrangement of the water molecules between Asp85 and Asp212. Though the protonated Asp85 in the M intermediate has been supposed to be present in an environment free from water because of the high C=O stretching frequency of the 1761 cm−1 band, it is not appropriate to apply such a macroscopic view to the C=O bond in an inhomogeneous environment. It should be noted that dehydrated bacteriorhodopsin produces M, but remains at M1 (73).
In BR, Asp212 interacts with Asp85 through two water molecules (Water401 and Water406 in Fig. 1), and interacts with the Schiff base through a different water molecule (Wat402), and also with Arg82 through one of these water molecules (Water406) which also hydrates the carboxylic oxygen of Asp212 (2). The FTIR spectra in Fig. 2 show a band resulting from BR at 3645 cm−1. This band is the O–H bond of water near Asp85 in the BR state (1,74,75), and moves to 3566 cm−1 in M (40). This 3566 cm−1 band is depleted in W86F (76). These data are consistent with Water401 forming a single hydrogen bond with Asp85 in the BR state with a dangling O–H (75), which then forms a hydrogen bond to Trp86 in M.
Crystallographic structural models of the M intermediate, around Asp85, reported by various groups, though not completely coincident with each other, invariably show at least one water molecule (Water401) (1c8s [77], 1kg8 [71], 1p8h [39], 1cwq [42] and 1iw9 [43]); the latter two of these models show one additional diffuse water molecule. The positions of water molecules between Asp85 and Asp212 and nearby Trp86, in the crystallographic structure model of BR (Fig. 1), seem to be largely retained in M. Water molecules may bridge a gap between the carboxylic oxygen atoms of Asp85 and Asp212. The QM/MM calculations as stated above (7,9) have shown the stabilized pair of protonated Asp85/unprotonated Schiff base to be present, rather than the zwitterion pair, in the presence of Water401. Crystallographic results have shown that Water 402, a water molecule that had been interacting with Asp85, in BR, via the same oxygen atom that was interacting with Water401, has been removed in M. Theoretical studies have shown that the removal of Water402 is crucial in stabilizing the protonated Asp85.
The presence of Water401 and Trp86 between Asp212 and Asp85 may promote the stabilization of the protonated form of Asp85 in M, if water molecules can stabilize the protonated form of the carboxylic acid moiety (HO–C=O) of Asp85 by forming hydrogen bonding to Asp85 with the appropriate orientation and bond length. This could be facilitated when Asp212 and intervening water molecules become free in M from the constraints that had been imposed in BR by the interactions with Arg82 and the Schiff base. When one arm of the carboxylic acid moiety is in the canonical C–OH state, this forces the other arm toward the C=O double bond state, which is reflected in a high frequency C=O stretching vibration at 1761 cm−1. This part of Asp85 would be in a nonpolar environment in M (78). The frequency shift of the 1761 cm−1 band of Asp85 to 1755 cm−1 in N (33,49) could be the result of loosening of the interaction between Asp85 and Asp212, rendering the C–OH form less rigid. The eventual decrease in pKa of Asp85 in the O intermediate leads to the transient proton transfer to Asp212 in the O′ state (79).
PERSPECTIVE
The protonated form of the Schiff base in L and N is stabilized relative to M by a water cluster in a cavity produced on the cytoplasmic side of the Schiff base. The protonated form of Asp85 in M is stabilized by water molecules in a cavity created on the extracellular side of the Schiff base in the region surrounded by Asp212, Trp86 and Tyr57. These transient, water-mediated structures determine the direction of proton transfer between the Schiff base and Asp85, and may be produced by water molecules which act both as spacers, to stabilize the conformation, and as cofactors, to stabilize the dipole orientation.
Acknowledgements
The authors are thankful to T. Ebrey, J.K. Lanyi, S.P. Balashov, J. Herzfeld, J. Lugtenburg, F.L. Tomson and Y. Shichida for their support.
Footnotes
This invited paper is part of the Symposium-in-Print: Photoreceptors and Signal Transduction.
Note Added in Proof: After the submission of the revised text, we noticed an article by Lórenz-Fonfria et al. (81), showing that the average intensity of water vibrations between 3750–2700 cm−1 undergoes monoexponential decay and that the time-constant of this process is dependent on the sample thickness, while the decay of L is unaffected. On this basis, the authors argued that the cytoplasmic side in L at room temperature does not have internal water molecules. Studies in (1) were carried out with purple membranes under hydration sufficient to attain the same time constants and occupancies of the intermediates in the photocycle as those in solution.
However, the data in (81) were obtained under lower hydration which disturbs the photocycle, making it slower, and resulting in decreased occupancies of N and O. Even the spectrum of L in (81) differs from that in (1): The intensities of water vibrations are about half of those in (1). Though not shown, in the data used in (1) the decay process of the average intensity of water vibrations between 3750–3630 cm−1 is not monoexponential, and the earliest component, mainly due to L, decays at a rate independent of the sample thickness. Our proposal for the presence of internal water molecules in L is not undermined by the results in (81).
REFERENCES
- 1.Morgan JE, Vakkasoglu AS, Gennis RB, Maeda A. Water structural changes in the L and M intermediates of bacteriorhodopsin as revealed by time-resolved step-scan FTIR spectroscopy. Biochemistry. 2007;46:2787–2796. doi: 10.1021/bi0616596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luecke H, Schobert B, Richter H-T, Cartailler J-P, Lanyi JK. Structure of bacteriorhodopsin at 1.55 Å resolution. J. MoL Biol. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- 3.Heberle J, Dencher NA. Surface-bound optical probes monitor proton translocation and surface potential changes during the bacteriorhodopsin photocyle. Proc. Natl Acad. Sci. USA. 1992;89:5996–6000. doi: 10.1073/pnas.89.13.5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garczarek F, Brown LS, Lanyi JK, Gerwert K. Proton binding within a membrane protein by a protonated water cluster. Proc. Natl Acad. Sci. USA. 2005;102:3633–3638. doi: 10.1073/pnas.0500421102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richter H-T, Brown LS, Needleman R, Lanyi JK. A linkage of the pKa’s of asp-85 and glu-204 forms part of the reprotonation switch of bacteriorhodopsin. Biochemistry. 1996;35:4054–4062. doi: 10.1021/bi952883q. [DOI] [PubMed] [Google Scholar]
- 6.Lanyi JK, Schobert B. Crystallographic structure of the retinal and the protein after deprotonation of the Schiff base: The switch in the bacteriorhodopsin photocycle. J. Mod. Biol. 2002;321:727–737. doi: 10.1016/s0022-2836(02)00682-4. [DOI] [PubMed] [Google Scholar]
- 7.Hayashi S, Ohmine I. Proton transfer in bacteriorhodopsin: Structure, excitation, IR spectra, and potential energy surface analyses by an ab initio QM/MM method. J. Phys. Chem. B. 2000;104:10678–10691. [Google Scholar]
- 8.Bondar A-N, Fischer S, Smith JC, Elstner M, Suhai S. Key role of electrostatic interactions in bacteriorhodopsin proton transfer. J. Am. Chem. Soc. 2004;126:14668–14677. doi: 10.1021/ja047982i. [DOI] [PubMed] [Google Scholar]
- 9.Bondar A-N, Suhai S, Fischer S, Smith JC, Elstner M. Suppression of the back proton-transfer from Asp85 to the retinal Schiff base in bacteriorhodopsin: A theoretical analysis of structural elements. Struct. Biol. 2007;57:454–469. doi: 10.1016/j.jsb.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 10.Lee Y-S, Krauss M. Dynamics of proton transfer in bacteriorhodopsin. J. Am. Chem. Soc. 2004;126:2225–2230. doi: 10.1021/ja036115v. [DOI] [PubMed] [Google Scholar]
- 11.Sheves M, Albeck A, Friedman N, Ottolenghi M. Controlling the pKa of the bacteriorhodopsin Schiff base by use of artificial retinal analogues. Proc. Natl Acad. Sci. USA. 1986;83:3262–3266. doi: 10.1073/pnas.83.10.3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Govindjee R, Balashov S, Ebrey T, Oesterhelt D, Steinberg G, Sheves M. Lowering the intrinsic pKa of the chromophore’s Schiff base can restore its light-induced deprotonation in the inactive Try57 → Asn mutant of bateriorhodopsin. J. Biol. Chem. 1994;269:14353–14354. [PubMed] [Google Scholar]
- 13.Hezfeld J, Lansing JC. Magnetic resonance studies on the bacteriorhodopsin pump cycle. Annu. Rev. Biophys. Biomol. Struct. 2002;31:73–95. doi: 10.1146/annurev.biophys.31.082901.134233. [DOI] [PubMed] [Google Scholar]
- 14.Subramaniam S, Marti T, Khorana HG. Protonation state of Asp (Glu)-85 regulates the purple-to-blue transition in bacteriorhodopsin mutants Arg-82 → Ala and Asp-85 → Glu: The blue form is inactive in proton translocation. Proc. Natl Acad. Sci. USA. 1990;87:1013–1017. doi: 10.1073/pnas.87.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fodor SPA, Ames JB, Gebhard R, van den Berg EMM, Stoeckenius W, Lugtenburg J, Mathies RA. Chromophore structure in bacteriorhodpsin’s N intermediate: Implications for the proton pumping mechanism. Biochemistry. 1988;27:7097–7101. doi: 10.1021/bi00418a064. [DOI] [PubMed] [Google Scholar]
- 16.Ames AB, Fodor SPA, Gebhard R, Raap J, van den Berg EMM, Lugtenburg J, Mathies RA. Bacteriorhodopsin’s M412 intermediate contains a 13-cis, 14-s-trans, 15-anti-retinal Schiff base chromophore. Biochemistry. 1989;28:3681–3687. doi: 10.1021/bi00435a009. [DOI] [PubMed] [Google Scholar]
- 17.Maeda A, Sasaki J, Shichida Y, Yoshizawa T. Water structural changes in the bacteriorhodopsin photocycle: Analysis by Fourier-transform infrared spectroscopy. Biochemistry. 1992;31:462–467. doi: 10.1021/bi00117a023. [DOI] [PubMed] [Google Scholar]
- 18.Maeda A, Tomson FL, Gennis RB, Balashov SP, Ebrey TG. Water molecule rearrangements around Leu93 and Trp182 in the formation of the L intermediate in bacteriorhodopsin’s photocycle. Biochemistry. 2003;42:2535–2541. doi: 10.1021/bi020532n. [DOI] [PubMed] [Google Scholar]
- 19.Maeda A, Verhoeven MA, Lugtenburg J, Gennis RB, Balashov SP, Ebrey TG. Water rearrangement around the Schiff base in the late K (KL) intermediate of the bacteriorhodopsin photocycle. J. Phys. Chem. B. 2004;108:1096–1101. [Google Scholar]
- 20.Maeda A, Balashov SP, Ebrey TG. Participation of internal water molecules and clusters in the unidirectional light-induced proton transfer in bacteriorhodopsin. In. In: Wada M, Shimazaki K, lino M, editors. Light Sensing in Plants. Tokyo: Springer Verlag; 2005. pp. 213–221. [Google Scholar]
- 21.Maeda A, Morgan JE, Gennis RB, Ebrey TG. Water as a cofactor in the unidirectional light-driven proton transfer steps in bacteriorhodopsin. Photochem. Photobiol. 2006;82:1396–1405. doi: 10.1562/2006-01-16-IR-779. [DOI] [PubMed] [Google Scholar]
- 22.Yamazaki Y, Hatanaka M, Kandori H, Sasaki J, Karstens WFJ, Raap J, Lugtenburg J, Bizounok M, Herzfeld J, Needleman R, Lanyi JK, Maeda A. Water structural changes at the proton uptake site (the Thr46-Asp96 domain) in the L intermediate of bacteriorhodopsin. Biochemistry. 1995;34:7088–7093. doi: 10.1021/bi00021a021. [DOI] [PubMed] [Google Scholar]
- 23.Brown LS, Gat Y, Sheves M, Yamazaki Y, Maeda A, Needleman R, Lanyi JK. The retinal Schiff base–counterion complex of bacteriorhodopsin: Changed geometry during the photocycle is a cause of proton transfer to aspartate 85. Biochemistry. 1994;33:12001–12011. doi: 10.1021/bi00206a001. [DOI] [PubMed] [Google Scholar]
- 24.Roux B, Nina M, Pomes R, Smith JC. Thermodynamic stability of water molecules in the bacteriorhodopsin proton channel: A molecular dynamics free energy perturbation study. Biophys. J. 1996;71:670–681. doi: 10.1016/S0006-3495(96)79267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kouyama T, Nishikawa T, Tokuhisa T, Okumura H. Crystal structure of the L intermediate of bacteriorhodopsin: Evidence for vertical translocation of a water molecule during the proton pumping cycle. J. Mol. Biol. 2004;335:531–546. doi: 10.1016/j.jmb.2003.10.068. [DOI] [PubMed] [Google Scholar]
- 26.Maeda A, Herzfeld J, Belenky M, Needleman R, Gennis RB, Balashov SP, Ebrey TG. Water-mediated hydrogen-bonded network on the cytoplasmic side of the Schiff base of the L photointermediate of bacteriorhodopsin. Biochemistry. 2003;42:14122–14129. doi: 10.1021/bi0301542. [DOI] [PubMed] [Google Scholar]
- 27.Maeda A, Tomson FL, Gennis RB, Ebrey TG, Balashov SP. Chromophore-protein-water interactions in the L intermediate of bacteriorhodopsin: FTIR study of the photoreaction of L at 80 K. Biochemistry. 1999;38:8800–8807. doi: 10.1021/bi9907072. [DOI] [PubMed] [Google Scholar]
- 28.Edman K, Royant A, Larsson G, Jacobson F, Taylor T, van der Spoel D, Landau EM, Pebay-Peyroula E, Neutze R. Deformation of helix C in the low temperature L-intermediate of bacteriorhodopsin. J. Biol. Chem. 2004;279:2147–2158. doi: 10.1074/jbc.M300709200. [DOI] [PubMed] [Google Scholar]
- 29.Lanyi JK, Schobert B. Structural changes in the L photointermediate of bacteriorhodopsin. J. Mol. Biol. 2007;365:1379–1392. doi: 10.1016/j.jmb.2006.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwasa T, Tokunaga F, Yoshizawa T. A new pathway in the photoreaction cycle of trans-bacteriorhodopsin and the absorption spectra of its intermediates. Biophys. Struct. Mech. 1980;6:253–270. [Google Scholar]
- 31.Kalisky O, Ottolenghi M, Honig B, Korenstein R. Environmental effects on formation and photoreaction of the M412 photoproduct of bacteriorhodopsin: Implications for the mechanism of proton pumping. Biochemistry. 1981;20:649–655. doi: 10.1021/bi00506a031. [DOI] [PubMed] [Google Scholar]
- 32.Chizhov I, Chernavskii DS, Engelhard M, Mueller K-H, Zubov BV, Hess B. Spectrally silent transitions in the bacteriorhodopsin photocycle. Biophys. J. 1996;71:2329–2345. doi: 10.1016/S0006-3495(96)79475-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braiman MS, Bousche O, Rothschild KJ. Protein dynamics in the bacteriorhodopsin photocycle: Submillisecond Fourier transform infrared spectra of the L, M, and N photointermediates. Proc. Natl Acad. Sci. USA. 1991;88:2388–2392. doi: 10.1073/pnas.88.6.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heßling B, Souvignir G, Gerwert K. A model-independent approach to assigning bacteriorhodopsin’s intramolecular reactions to photocycle intermediates. Biophys. J. 1993;65:1929–1941. doi: 10.1016/S0006-3495(93)81264-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zscherp C, Heberle J. Infrared difference spectra of the intermediates L, M, N, and O of the bacteriorhodopsin photoreaction obtained by time-resolved attenuated total reflection spectroscopy. J. Phys. Chem. B. 1997;101:10542–10547. [Google Scholar]
- 36.Rödig C, Chizov I, Weidlich O, Siebert F. Time-resolved step-scan Fourier transform infrared spectroscopy reveals differences between early and late M intermediates of bacteriorhodopsin. Biophys. J. 1999;76:2687–2701. doi: 10.1016/S0006-3495(99)77421-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garczarek F, Wang J, El-Sayed M, Gerwert K. The assignment of the different infrared continuum absorbance changes observed in the 3000–1800 cm−1 region during the bacteriorhodopsin photocycle. Biophys. J. 2004;87:2676–2682. doi: 10.1529/biophysj.104.046433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamazaki Y, Kandori H, Needleman R, Lanyi JK, Maeda A. Interaction of the protonated Schiff base with the peptide backbone of valine 49 and the intervening water molecule in the N photointermediate of bacteriorhodopsin. Biochemistry. 1998;37:1559–1564. doi: 10.1021/bi972044z. [DOI] [PubMed] [Google Scholar]
- 39.Schobert B, Brown LS, Lanyi JK. Crystallographic structures of the M and N intermediates of bacteriorhodopsin: Assembly of a hydrogen-bonded chain of water molecules between Asp-96 and the retinal Schiff base. J. Mol. Biol. 2003;330:553–570. doi: 10.1016/s0022-2836(03)00576-x. [DOI] [PubMed] [Google Scholar]
- 40.Maeda A, Tomson FL, Gennis RB, Kandori H, Ebrey TG, Balashov SP. Relocation of internal bound water in bacteriorhodopsin during the photoreaction of M at low temperature: An FTIR study. Biochemistry. 2000;39:10154–10162. doi: 10.1021/bi000190q. [DOI] [PubMed] [Google Scholar]
- 41.Luecke H, Schobert B, Cartailler J-P, Richter H-T, Rosengarth A, Needleman R, Lanyi JK. Coupling photoisomerization of retinal to direction transport in bacteriorhodopsin. J. Mol. Biol. 2000;300:1237–1255. doi: 10.1006/jmbi.2000.3884. [DOI] [PubMed] [Google Scholar]
- 42.Sass HJ, Büldt G, Gessenich R, Hehn D, Neff D, Schlessinger R, Berendzen J, Ormos P. Structural alterations for proton translocation in the M state of wild-type bacteriorhodopsin. Nature. 2000;406:549–653. doi: 10.1038/35020607. [DOI] [PubMed] [Google Scholar]
- 43.Takeda K, Matsui Y, Kamiya N, Adachi S, Okumura H, Kouyama T. Crystal structure of the M intermediate of bacteriorhodopsin: Allosteric structural changes mediated by sliding movement of a transmembrane helix. J. Mol. Biol. 2004;341:1023–1037. doi: 10.1016/j.jmb.2004.06.080. [DOI] [PubMed] [Google Scholar]
- 44.Maeda A, Balashov SP, Lugtenburg J, Verhoeven MA, Herzfeld J, Belenky M, Gennis RB, Tomson FL, Ebrey TG. Interaction of internal water molecules with the Schiff base in the L intermediate of the bacteriorhodopsin photocycle. Biochemistry. 2002;41:3803–3809. doi: 10.1021/bi011923p. [DOI] [PubMed] [Google Scholar]
- 45.Maeda A, Sasaki J, Pfefferlé J-M, Shichida Y, Yoshizawa T. Fourier transform infrared spectral studies on the Schiff base mode of all-trans bacteriorhodopsin and its photointermediates, K and L. Photochem. Photobiol. 1991;54:911–921. [Google Scholar]
- 46.Gerwert K, Siebert F. Evidence for light-induced 13-cis, 14-s-cis isomerization in bacteriorhodopsin obtained by FTIR difference spectroscopy using isotopically labeled retinal. EMBO J. 1986;5:805–811. doi: 10.1002/j.1460-2075.1986.tb04285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith SO, Hornung I, van der Steen R, Pardoen JA, Braiman MS, Lugtenburg J, Mathies RA. Are C14–C15 single bond isomerization of the retinal chromophore involved in the proton-pumping mechanism of bacteriorhodopsin? Proc. Nall Acad. Sci. USA. 1986;83:967–971. doi: 10.1073/pnas.83.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cromwell MEM, Gebhard R, Li X-Y, Batenburg ES, Hopman JCP, Lugtenburg J, Mathies RA. Synthesis and vibrational analysis of a locked 14-s-cis conformer of retinal. J. Am. Chem. Soc. 1992;114:10860–10869. [Google Scholar]
- 49.Pfefferlé J-M, Maeda A, Sasaki J, Yoshizawa T. Fourier transform infrared study of the N intermediate of bacteriorhodopsin. Biochemistry. 1991;30:6548–6556. doi: 10.1021/bi00240a027. [DOI] [PubMed] [Google Scholar]
- 50.Hu JG, Sun BQ, Petkova AT, Griffin RG, Herzfeld J. The predischarge chromophore in bacteriorhodopsin: A 15N solid-state NMR study of the L photointermediate. Biochemistry. 1997;36:9316–9322. doi: 10.1021/bi970416y. [DOI] [PubMed] [Google Scholar]
- 51.Smith SO, Meyers AB, Pardoen JA, Winkel C, Mulder PPJ, Lugtenburg J, Mathies RA. Determination of retinal Schiff base configuration in bacteriorhodopsin. Proc. Nall Acad. Sci. USA. 1984;81:2055–2059. doi: 10.1073/pnas.81.7.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lansing JC, Hohwy M, Jaroniec CP, Creemers AFL, Lugtenburg J, Herzfeld J, Griffin RG. Chromophore distortions in the bacteriorhodopsin photocycle: Evolution of the H–C14–C15–H dihedral angle measured by solid state NMR. Biochemistry. 2002;41:431–438. doi: 10.1021/bi011529r. [DOI] [PubMed] [Google Scholar]
- 53.Ferrand M, Dianoux AJ, Petry W, Zaccai G. Thermal motions and function of bacteriorhodopsin in purple membranes: Effects of temperature and hydration studied by neutron scattering. Proc. Natl Acad. Sci. USA. 1993;90:9668–9672. doi: 10.1073/pnas.90.20.9668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zimanyi L, Saltiel J, Brown LS, Lanyi JK. A priori resolution of the intermediate spectra in the bacteriorhodopsin photocycle: The time evolution of the L spectrum revealed. J. Phys. Chem. A. 2006;110:2318–2321. doi: 10.1021/jp056874v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hage W, Kim M, Frei H, Mathies RA. Protein dynamics in the bacteriorhodopsin photocycle: A nanosecond step-scan FTIR investigation of the KL to L transition. J. Phys. Chem. 1996;100:16026–16033. [Google Scholar]
- 56.Maeda A, Gennis RB, Balashov SP, Ebrey TG. Relocation of water molecules between the Schiff base and the Thr46-Asp96 region during light-driven unidirectional proton transport by bacteriorhodopsin: An FTIR study of the N intermediate. Biochemistry. 2005;44:5960–5968. doi: 10.1021/bi047469h. [DOI] [PubMed] [Google Scholar]
- 57.Váró G, Lanyi JK. Kinetics and spectroscopic evidence for an irreversible step between deprotonation and reprotonation of the Schiff base in the bacteriorhodopsin photocycle. Biochemistry. 1991;30:5008–5015. doi: 10.1021/bi00234a024. [DOI] [PubMed] [Google Scholar]
- 58.Váró G, Lanyi JK. Thermodynamics and energy coupling in the bacteriorhodopsin photocycle. Biochemistry. 1991;30:5016–5022. doi: 10.1021/bi00234a025. [DOI] [PubMed] [Google Scholar]
- 59.Balashov SP, Imasheva ES, Govindjee R, Ebrey TG. Titration of aspartate-85 in bacteriorhodopsin: What it says about chromophore isomerization and proton release. Biophys. J. 1996;70:473–481. doi: 10.1016/S0006-3495(96)79591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kouyama T, Nasuda-Kouyama A, Ikegami A, Mathew MK, Stoeckenius W. Bacteriorhodopsin photoreaction: Identification of a long-lived intermediate N (P, R350) at high pH and its M-like photoproduct. Biochemistry. 1988;27:5855–5863. doi: 10.1021/bi00416a006. [DOI] [PubMed] [Google Scholar]
- 61.Braiman MS, Dioumaev A, Lewis JR. A large photolysis-induced pKa increase of the chromophore counterion in bacteriorhodopsin: Implications for ion transport mechanisms of retinal proteins. Biophys. J. 1996;70:939–946. doi: 10.1016/S0006-3495(96)79637-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brown LS, Lanyi JK. Determination of the transiently lowered pKa of the retinal Schiff base during the photocycle of bacteriorhodopsin. Proc. Natl Acad. Sci. USA. 1996;93:1731–1734. doi: 10.1073/pnas.93.4.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Needleman R, Chang M, Ni B, Váró G, Lanyi JK. Properties of Asp212 → Asn bacteriorhodopsin suggest that Asp212 and Asp85 both participate in a counterion and proton acceptor complex near the Schiff base. J. Biol. Chem. 1991;266:11478–11484. [PubMed] [Google Scholar]
- 64.Cao Y, Váró G, Klinger AL, Czajkowsky DM, Braiman MS, Needleman R, Lanyi JK. Proton transfer from Asp-96 to the bacteriorhodopsin Schiff base is caused by a decrease of the pKa of Asp-96 which follows a protein backbone conformational change. Biochemistry. 1993;32:1981–1990. doi: 10.1021/bi00059a015. [DOI] [PubMed] [Google Scholar]
- 65.Moltke S, Krebs MP, Mollaaghababa R, Khorana HG, Heyn MP. Intramolecular charge transfer in the bacteriorhodopsin mutants Asp85 → Asn and Asp212 → Asn: Effects of pH and anions. Biophys. J. 1995;69:2074–2083. doi: 10.1016/S0006-3495(95)80078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shibata M, Yoshitugu M, Mizuide N, Ihara K, Kandori H. Halide binding by the D212N mutant of bacteriorhodopsin affects hydrogen bonding of water in the active site. Biochemistry. 2007;46:7525–7535. doi: 10.1021/bi7004224. [DOI] [PubMed] [Google Scholar]
- 67.Brown LS, Váró G, Hatanaka M, Sasaki J, Kandori H, Maeda A, Friedman N, Sheves M, Needleman R, Lanyi JK. The complex extracellular domain regulates the deprotonation and reprotonation of the retinal Schiff base during the bacteriorhodopsin photocycle. Biochemistry. 1995;34:12903–12911. doi: 10.1021/bi00039a053. [DOI] [PubMed] [Google Scholar]
- 68.Balashov SP, Govindjee R, Kono M, Imasheva ES, Lukashev E, Ebrey TG, Crouch RK, Menick DR, Feng Y. Effect of the arginine-82 to alanine mutation in bacteriorhodopsin on dark adaptation, proton release, and the photochemical cycle. Biochemistry. 1993;32:10331–10343. doi: 10.1021/bi00090a008. [DOI] [PubMed] [Google Scholar]
- 69.Brown LS, Sasaki J, Kandori H, Maeda A, Needleman R, Lanyi JK. Glutamic acid 204 in the terminal proton release group at the extracellular surface of bacteriorhodopsin. J. Biol. Chem. 1995;270:27122–27126. doi: 10.1074/jbc.270.45.27122. [DOI] [PubMed] [Google Scholar]
- 70.Lazarova T, Sanz C, Querol E, Padrós E. Fourier transform infrared evidence for early deprotonation of Asp85 at alkaline pH in the photocycle of bacteriorhodopsin mutants containing E194Q. Biophys. J. 2000;78:2022–2030. doi: 10.1016/S0006-3495(00)76749-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Facciotti MT, Rouhani S, Burkard FT, Betancourt FM, Downing KH, Rose RB, McDermott G, Glaeser RM. Structure of an early intermediate in the M state phase of the bacteriorhodopsin photocycle. Biophys. J. 2001;81:3442–3455. doi: 10.1016/S0006-3495(01)75976-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Edsall JT, Wyman J. Biophysical Chemistry. Vol. 1. New York: Academic Press; 1958. [Google Scholar]
- 73.Ganea C, Gergely C, Ludmann K, Váró G. The role of water in the extracellular half channel of bacteriorhodopsin. Biophys. J. 1997;73:2718–2726. doi: 10.1016/S0006-3495(97)78300-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maeda A, Sasaki J, Yamazaki Y, Needleman R, Lanyi JK. Interaction of aspartate-85 with a water molecule and the protonated Schiff base in the L intermediate of bacteriorhodopsin: A Fourier transform infrared spectroscopic study. Biochemistry. 1994;33:1713–1717. doi: 10.1021/bi00173a013. [DOI] [PubMed] [Google Scholar]
- 75.Garczarek F, Gerwert K. Functional waters in intra-protein proton transfer monitored by FTIR difference spectroscopy. Nature. 2006;439:109–112. doi: 10.1038/nature04231. [DOI] [PubMed] [Google Scholar]
- 76.Hatanaka M, Kashima R, Kandori H, Friedman N, Sheves M, Needleman R, Lanyi JK, Maeda A. Trp86 → Phe replacement in bacteriorhodopsin affects a water molecule near Asp85 and light adaptation. Biochemistry. 1997;36:5493–5498. doi: 10.1021/bi970081k. [DOI] [PubMed] [Google Scholar]
- 77.Luecke H, Schobert B, Richter H-T, Cartailler J-P, Lanyi JK. Structural changes in bacteriorhodopsin during ion transport at 2 angstrom resolution. Science. 1999;286:255–260. doi: 10.1126/science.286.5438.255. [DOI] [PubMed] [Google Scholar]
- 78.Dioumaev AK, Braiman M. Modeling vibrational spectra of amino acid side chains in proteins: The carbonyl stretch frequency of buried carboxylic residues. J. Am. Chem. Soc. 1995;117:10572–10574. [Google Scholar]
- 79.Dioumaev AK, Richter H-T, Brown LS, Tanio M, Tuzi S, Saitô H, Kimura Y, Needleman R, Lany JK. Existence of a proton transfer chain in bacteriorhodopsin: Participation of Glu-194 in the release of protons to the extracellular surface. Biochemistry. 1998;37:2496–2506. doi: 10.1021/bi971842m. [DOI] [PubMed] [Google Scholar]
- 80.DeLano W. PYMOL. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 81.Lórenz-Fonfria VA, Furutani Y, Kandori H. Active internal waters in the bacteriorhodopsin photocycle. A comparative study of the L and M intermediates at room and cryogenic temperatures by infrared spectroscopy. Biochemistry. 2008;47:4071–4081. doi: 10.1021/bi7024063. [DOI] [PubMed] [Google Scholar]