Abstract
Rotational-Echo DOuble-Resonance (REDOR) NMR is a powerful and versatile solid-state NMR measurement that has been recruited to elucidate drug modes of action and to drive the design of new therapeutics. REDOR has been implemented to examine composition, structure, and dynamics in diverse macromolecular and whole-cell systems, including taxol-bound microtubules, enzyme-cofactor-inhibitor ternary complexes, and antibiotic-whole-cell complexes. The REDOR approach involves the integrated design of specific isotopic labeling strategies and the selection of appropriate REDOR experiments. By way of example, this digest illustrates the versatility of the REDOR approach, with an emphasis on the practical considerations of experimental design and data interpretation.
Keywords: REDOR, solid-state NMR, whole-cell NMR, drug discovery, taxol, vancomycin
The translation of discoveries at the bench into therapies to prevent and treat human disease and to improve overall health is a grand challenge and ultimate goal of many basic science research programs and of much larger laboratories and companies that ultimately develop drugs and bring them to the clinic. Drug design and discovery efforts are enormously interdisciplinary endeavors, involving small and large molecules, biological targets, and the complexities of human physiology and drug pharmacology. The notion of “the magic bullet,” popularized by Dr. Ehrlich, illustratively emphasizes the ultimate goal of identifying a highly selective therapeutic strategy to exert its action with no side effects or toxicity. While there are tremendous opportunities and active research in the areas of protein therapy, immunotherapy, and gene therapy, most current therapies employ small-molecule drugs to influence biological and biochemical phenomena in the host. Determining the bio-active bound conformations of drugs and mapping their interactions with their targets with high resolution are crucial to understanding the molecular and chemical basis for drug modes of action.
Solid-state NMR has emerged as a powerful tool to examine biologically relevant drugbound complexes of systems that pose a challenge to analysis by conventional methods. Rotational-Echo DOuble-Resonance (REDOR) NMR1, in particular, has played an often unique role in elucidating drug modes of action and in driving the design of new therapeutic candidates2–4. Since its introduction in 1989, REDOR has been implemented to examine composition, structure, and dynamics in diverse biological macromolecular and whole-cell systems including enzyme-cofactor-inhibitor ternary complexes, protein-protein complexes, lipid-embedded membrane proteins, bacteria-antibiotic complexes, and intact leaves2–5. REDOR measurements can be implemented to determine the strength of dipolar couplings, and hence precise distances, between heteronuclei and can also be used as a spectroscopic filter.
The REDOR approach is powerful, versatile, and applicable to many systems, yet there is no set protocol and is best appreciated by way of example. This digest will provide an introduction to the REDOR measurement and will highlight three examples that implement REDOR in distinct ways to deliver insights in drug design and discovery. Emphasis will be placed on the practical considerations of the examples presented, rather than discussing the in depth biological implications of the discoveries that resulted from the REDOR studies. The hope is to describe and illustrate key parameters and considerations in designing and performing REDOR measurements for drug design and discovery problem-solving efforts. The specific examples have been selected from the author’s current and former laboratories, although REDOR is a robust experiment and has been performed and utilized in creative and important ways in laboratories around the world. This digest is dedicated to Professor Jacob Schaefer in celebration of his 75th Birthday and nearly 25 years of REDOR.
REDOR access to dipolar couplings
Rotational-echo double-resonance (REDOR) provides a direct measure of short and long-range dipolar couplings, typically between isolated pairs of heteronuclei1. The dipolar coupling between two spins harbors contributions from the internuclear distance and the orientation of the internuclear vector with respect to the applied magnetic field,6allowing REDOR to function as a spectroscopic ruler and protractor. A brief primer on the REDOR experiment follows. More complete descriptions of the evolution of the dipolar coupling are beyond the scope of this digest and are available in the references cited here3, 7, 8.
The dipole-dipole coupling between heteronuclei within a magnetic field is dependent upon both spatial and spin coordinates, as described by the truncated dipolar Hamiltonian below (1).
| (1) |
The angle, θ, is the angle between the I-S internuclear vector and the applied magnetic field; γ1 and γ2 are the gyromagnetic ratios of the I and S nuclei; ħ is the reduced Planck constant; and r is the internuclear vector between the two spins. See Figure 2 for values of constants. Magic-angle spinning, which is performed to yield high-resolution NMR spectra, averages over the spatial coordinates and suppresses the dipolar interaction in a coherent manner9. In solution-state NMR, molecular tumbling averages the dipolar coupling in a noncoherent manner. Thus, in solution NMR, unless strategies are used to prevent this averaging (such as using aligned media or viscous solvents)10, the dipolar couplings are averaged and cannot be accessed. Magic-angle spinning averages the dipolar couplings in a coherent manner, at a precise angle and frequency. Thus, in solid-state NMR, it is possible to defeat the averaging of the spatial coordinates (by magic-angle spinning) through the manipulation of the spin coordinates (by the application of radiofrequency pulses). The REDOR measurement utilizes the application of rotor-synchronized radiofrequency pulses to operate exclusively on the spin coordinates and interfere with the complete suppression of the dipolar coupling by magic-angle spinning1. This recoupling, or reintroduction of the dipolar coupling, is the basic principle for other homonuclear recoupling techniques that are also used to measure dipolar couplings and obtain distance parameters11–13.
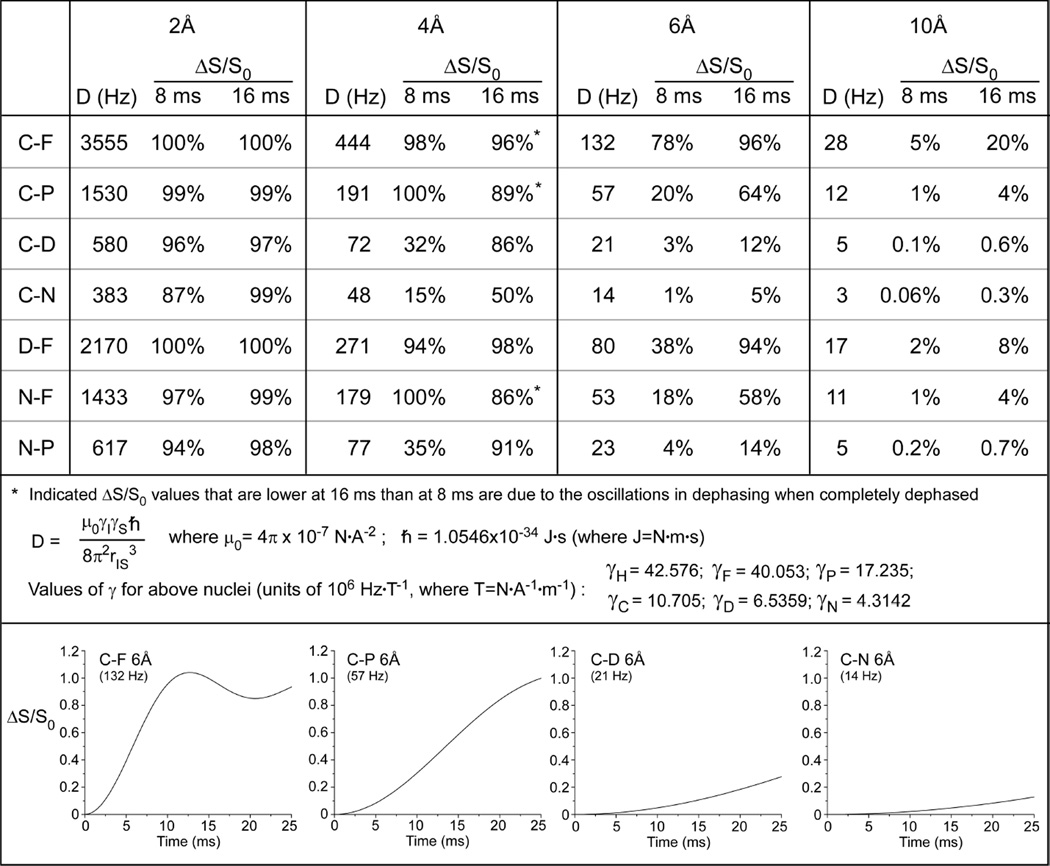
Figure 2. Dipolar couplings to distances.
Strengths of the dipolar couplings (D) between pairs of nuclei at different distances and the percent dephasing (ΔS/S0) that would be expected in REDOR experiments as a function of evolution time to for 2, 4, 6, and 10 Å distances between isolated spin pairs (top). Values of constants used in calculations of D (middle). Calculated plots of ΔS/S0 for C–F, C–P, C–D, and C–N pairs at 6Å for 25 ms of dipolar evolution time (bottom).
The REDOR measurement
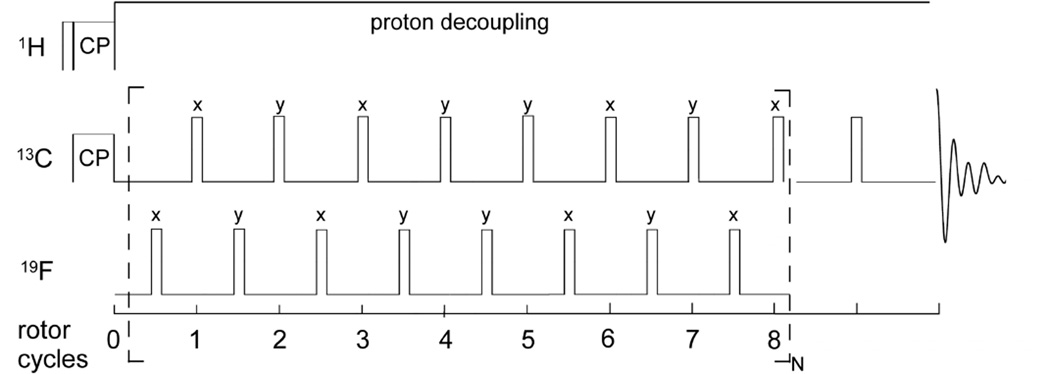
In practice, the REDOR measurement is performed in two parts (Figure 1, left), once without and once with rotor-synchronized dephasing pulses, yielding the full echo (S0) spectrum and the dephased (S) spectrum, respectively. Maximum intensity rotational echoes are formed at the end of each rotor period when no dephasing pulses are applied (S0spectrum). During the second half of the measurement, rotor-synchronized π pulses are applied to the dephasing spin to flip the sign of the dipolar coupling, yielding an average precessional frequency, ωD (α, β; t) for each coupled spin in the powder1. The pulses coincident with the rotor period serve to add the dephasing of subsequent rotor periods.
Figure 1. The REDOR measurement.
REDOR is performed in two parts, once with dephasing pulses (S spectrum) and once without (full echo, S0 spectrum). REDOR spectra are typically collected with standard xy-8 phase cycling55, on both observed and dephasing channels.
The time for which the dipolar coupling operates in the REDOR measurement is termed the dipolar evolution time (Nr * tr). Each spin accumulates a net phase due to dipolar transitions during this time, as defined in expression 2:
| (2) |
where Δφ is the accumulation of phase; tr is the time of one rotor period; Nr is the number of rotor cycles; ωD is the average precessional frequency for each coupled spin; α and β are the azimuthal and polar angles, respectively, in a coordinate system with the z-axis parallel to the rotor axis; t1 is the time of application of the π pulse from the start of the rotor period. ωD is in cycles per second, thus multiplying ωD by trNr yields a phase.
Thus, dipolar couplings result in signal attenuation at the end of every rotor period when dephasing pulses are applied. The difference in signal intensity (ΔS = S0 − S) for the observed spin in the two parts of the REDOR experiment is compared to the full-echo reference spectrum, S0, for each REDOR evolution time. Weak couplings can be amplified by increasing the number of rotor cycles (Nr) over which the dephasing occurs or by decreasing the spinning speed (tr). Figure 2 provides the strength of the dipolar coupling between pairs of nuclei at different distances and indicates the percent dephasing (ΔS/S0) that would be expected in REDOR experiments as a function of evolution time for 2, 4, 6, and 10 Å distances between isolated spin pairs. Full plots of the dipolar evolution for C–F, C–P, C–D, and C–N pairs at 6Å are also illustrated. Numerical simulations can be used to generate dephasing curves that correspond to distributions of distances14, or for dipolar couplings in the presence of motion15, that can be compared to the experimental data. In terms of nomenclature, we employ the Schaefer convention of identifying the observe and dephase spins for a specific measurement as 13C{19F}REDOR, for example, in a carbon-observed fluorine-dephased REDOR experiment.
Naturally, an understanding of the REDOR pulse sequence and measurement is crucial to designing and implementing REDOR experiments. Yet, equally important is the selection and implementation of the appropriate isotopic labeling strategy to introduce pairs or clusters of labeled nuclei into the system of interest. Many creative and highly selective isotopic labeling strategies have been reported across all realms of NMR, both in solution and solids, and only a few will be described here in the context of the examples below. Some universal considerations include (1) the gyromagnetic ratio of the nuclei (Figure 1) since the dipolar coupling to be measured scales with the γ of each nucleus. As seen in Figure 2, larger couplings exhibit larger dephasing (ΔS/S0) in REDOR spectra and, thus, are easier to determine accurately. In addition, the γ of the observe nucleus is important, as (2) the sensitivity of the NMR measurement is a function of γ. One must also consider (3) the natural abundance background of the target isotope label, whether as an observe spin or a dephasing spin, and the extent to which isotope labels are resolved by chemical shift from other labels or the natural abundance background or whether NMR strategies to achieve selectivity are desired, such as selecting only carbons that are bonded to a nitrogen (as in protein carbonyls and alpha carbons) or in proximity to phosphorous (as in lipid phosphate headgroups). 19F is often a desired nucleus to include in a REDOR approach to access long-range distances due to its large γ, close to that of 1H. However, it is generally used as a dephaser in REDOR experiments. From experience with fluorinated analogs of taxol and oritavancin, as described further in this digest, the chemical shift anisotropy of 19F is large and yields numerous spinning sidebands in 19F observe spectra. In addition, the 19F isotropic shift is very sensitive to external shielding effects16, 17, including solvent molecules and local conformations, which results in substantial broadening. 19F is often generally included individually at only one or very few places in a sample in REDOR approaches and, thus, serves well as a specific dephaser. Although possible and potentially appropriate for a certain system, one could instead observe 19F, as in a 19F{13C} REDOR experiment. However, this would require frequency-selective dephasing pulses to select one carbon type of interest, for example, and would still involve multiple carbon dephasers in a typical biological application. Overall, the three examples selected to highlight the REDOR approach in this digest include examples which involved challenging synthetic chemistry in order to introduce isotope labels where they would provide the most valuable information as probes of structure as well as new biosynthetic strategies to achieve selective labeling of whole-cell systems, together with distinct spectroscopic challenges and considerations.
Bio-active bound ligand conformations: Microtubule-bound taxol
The determination of target-bound drug conformations is crucial to understanding drug modes of action and to driving the design of new molecules with altered properties, including but not limited to: (i) increased potency, (ii) enhanced selectivity, and (iii) reduced toxicity. In systems inaccessible to X-ray crystallography and solution-state NMR techniques, or in systems where such information is available but new information in the biologically relevant context of a large multi-protein assembly or membrane or the whole cell is desired, REDOR provides an avenue to determine drug-bound conformations as well to examine proximities between the drug and its target.
The determination of the bio-active bound conformation of the microtubule-bound anticancer drug taxol (also known as paclitaxel) was sought after in order to understand its structure-based ability to bind to tubulin and to guide the development of improved and perhaps simplified analogs18. Taxol has a relatively rigid central tetracyclic ring system, but has four flexible side chains that could be accommodated in numerous conformations. Although the structure of the αβ-tubulin dimer, prepared as taxolstabilized Zn-induced polymerized sheets, had been solved by electron crystallography in 2001 and the binding site of taxol could be identified, the 3.7-Å resolution was too low to permit the determination of the taxol conformation. Thus, several REDOR measurements were designed and employed to determine the conformation of taxol bound to intact microtubules. The latter of two major studies18, 19determined distances between synthetically incorporated 2H and 19F labels in taxol using 2H{19F} REDOR18. This selection of labels is attractive in having no natural abundance background contributions from the microtubules in the sample, whereas 13C and 15N are present at natural abundance as 1.11% and 0.37%, respectively. In the former REDOR study, using a 13C-carbonyl labeled taxol, installation of an 15N adjacent to the 13C in taxol was required in order to select the taxol 13C from among the much larger 13C contributions from the natural abundance carbonyls in the microtubules, as there is only one taxol bound per tubulin dimer (100kD)19. Deuterium, however, is quadrupolar and has a large chemical shift anisotropy. Thus, by way of detail, processing of 2H-observe REDOR experiments is aided by synchronous sampling where the spinning sideband intensities are folded into the centerband20, 21. The choice of 19F as one of the labels is attractive since 19F has a γ that is almost as high as 1H and, thus, enables longer distance measurements than is possible when selecting nuclei with lower values of γ. This REDOR example also represents one of the experimentally toughest set of distance measurements reported due to the very long accumulation time necessary to obtain high-quality spectra to permit the quantification of 5% dephasing while observing the low-sensitivity 2H nucleus (2Hγ ≈ 1Hγ/6.5).
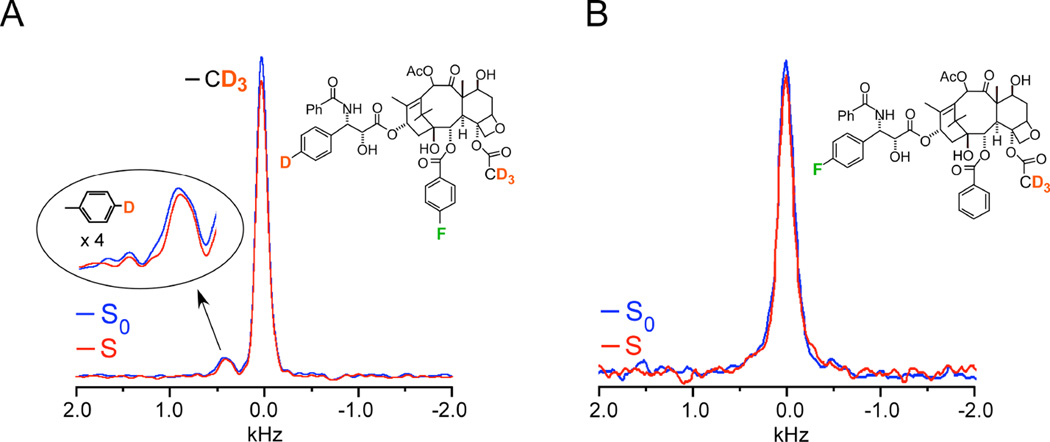
Strategically targeted 2H-19F distances were measured on two labeled analogues, named here according to their original designation as analogues 4 (Figure 3A) and 5 (Figure 3B)18. Analogue 4 had a single 19F to be used as the dephaser and two types of deuterons that would be observed and would be shift-resolved from one another, thus allowing the REDOR experiment to report on two sets of distances in the same experiment (Figure 3A). The 2H 3-19F distance in analogue 4 was determined to be 7.8 ± 0.5 Å, based on the observed dephasing of 8%, i.e. a difference between the full-echo (S0) and dephased spectrum (S) of 8% (Figure 3). The single aromatic deuterium did not exhibit significant dephasing by the 19F and thus a distance was not obtained for that 2H -19F pair (Figure 3A). However, this provided key information as one of the possible taxol models under consideration would have resulted in a 2H -19F distance of 4.5 Å. This close proximity would have been accompanied by 100% dephasing after 8 ms of dipolar evolution time--64 rotor cycles×0.125 ms, where 0.125 ms is the rotor period corresponding to 8 kHz MAS. Thus, this REDOR result demonstrated that the 2H -19F distance must be greater than 8 Å, an important parameter in the final modeling. The 2H 3-19F distance in analogue 5 was determined to be 6.3 ± 0.5 Å, corresponding to approximately 6% dephasing (Figure 3B). The error bars in the distances resulted from the consideration of the integrals of the peak heights compared to the integrals of an equivalent frequency width of noise18. 2H{19F} REDOR NMR provided these key distances that were able to rule out certain conformational models that had been proposed and support a T-shaped conformation when it is bound to tubulin (Figure 3)18. It was based on these collective distances that Ojima and coworkers designed and synthesized a structurally restrained analogue to enforce the “REDOR-taxol” conformation that was as potent as taxol, emphasizing the power in the structure-based drug design approach22, 23. The generation of potent analogues, for cancer therapy as well as other indications, is invaluable towards the identification of compounds with improved efficacy, reduced toxicity, and the ability to evade resistance mechanisms.
Figure 3. A 7.8 Å 2H-19F distance in taxol.
(A) 2H{19F} 64-Tr REDOR spectra of tubulin-bound taxol analogue 4, corresponding to 8 ms dipolar evolution time with MAS at 8 kHz, resulted in 8 % dephasing and a calculated distance of 7.8 (± 0.5)Å. The full-echo spectrum is shown in blue, and the dephased spectrum in red. The time domain signals were detected synchronously with the rotor so that all sidebands have been folded into the centerbands. The CD3 peak is assigned zero frequency and the aromatic-D peak appears about 5 ppm to low field. Each spectrum resulted from the accumulation of 640,000 scans with a 1.5 sec recycle delay (approximately 11 days each and, thus, days for the ΔS spectrum). (B) 2H{19F}32-Tr REDOR spectra of tubulin-bound taxol analogue 5, corresponding to 4 ms dipolar evolution time with MAS at 8 kHz, resulted in 6 % dephasing and a calculated distance of 6.3 (± 0.5)Å. Each spectrum resulted from the accumulation of 1,056,000 scans (18 days each and, thus, 36 days for the ΔS spectrum). Spectra were obtained on a 500 MHz spectrometer. Figure adapted from Paik et al.18
In terms of sensitivity, for typical REDOR experiments performed at moderate field strengths (300–600 MHz) in complex biosolids, one typically aims to prepare samples which have at least one micromole of labeled pairs. For samples such as these noncrystalline taxol-bound microtubules as well as other assemblies and whole cell samples with heterogeneity, typical carbon linewidths range from 1–2 ppm up to 5 ppm. This is in contrast to solid-state NMR studies on microcrystalline proteins, where much less sample is required due to narrower linewidths, on the order of 0.1 ppm. Microtubules, in particular, are comprised of the 100 kD αβ-tubulin dimers, with one taxol binding to each dimer. Thus, with one site of interest among the 100 kD dimer, 1µmol corresponds to 100 mg microtubules complexed to taxol in a one-to-one ratio. Although some complexes contained nearly this much material, the experiments with taxol analogue 5 contained only 0.1 micromoles of microtubule-bound drug. The one million scan experiment in Figure 3 required 36 days of spectrometer time (one million scans (18 days) for the full-echo S0 spectrum and one million scans for the dephased S spectrum)18. In this study, a control experiment was also performed on a sample without 19F to additionally demonstrate that the spectrometer and the probe, which is designed with superb isolation of the 1H and 19F channels and to permit stable long-term signal averaging, would yield perfectly equal S0 and S spectra in the absence of the 19F dephaser in a one million scan experiment. The reader should note that with just twice as much sample, the million-scan experiment would have taken four times less time. Ideally, one wants to measure more than one point on a REDOR curve, but as demonstrated in the taxol example, even one point can provide crucial information in a precious sample. Concentrated effort is often made to prepare large samples, balancing the amount of protein or complex with the amount of lyophilization buffer to protect the complex in the lyophilized solid state. While some studies have involved REDOR experiments on hydrated samples, most of the complex biological systems we have worked with, including whole cells and antimicrobial peptide-lipid vesicle complexes, have not yielded enhancements in resolution due to hydration and, rather, exhibited reduced signal intensity, making lyophilized samples more amenable to extensive REDOR measurements24–26. Lyophilization conditions for each sample must be determined or optimized towards maintaining native state conformations. In the case of taxol-bound microtubules, electron microscopy was performed to assess microtubules. Without adequate stabilization with lyoprotectants, microtubules were of disparate lengths; thus, buffer components were varied to achieve preparations that were identical pre- and post-lyophilization and which minimized buffer volume in the final sample (unpublished data). For other proteins and enzymes, drug-binding or activity assays can be implemented on samples pre- and post-lyophilization, whereas whole-cell samples can be assayed for viability.
Targeting bacterial cell-wall assembly
REDOR has emerged as a powerful tool to determine chemical composition, including the quantification of cross-links and various bond densities in complex systems, including insect cuticle, mussel byssus, plant cells, and bacterial cell walls27–31. This approach has found tremendous value in the determination of the modes of action of antibiotics that target cell-wall assembly. Such quantitative determinations pose a challenge to analysis by conventional methods due to the insolubility of the bacterial cell wall. The major component of the S. aureus cell wall is peptidoglycan, consisting of polymers of a repeating motif consisting of a disaccharide with a pentapeptide stem with a pentaglycine bridge attached to the sidechain of lysine, the third residue in the stem (Figure 4A). Different types of linkages exist in other Gram-positive and Gram-negative organisms. In S. aureus, the intact peptidoglycan precursor is synthesized inside the cell and transported to the membrane exoface where it is then polymerized through the action of transglycosylation (to link the glycans) and transpeptidation (to crosslink the glycine of one stem with the penultimate D-Ala of a neighboring stem). Major solid-state NMR efforts to dissect S. aureus cell-wall composition and structure and to understand the structure-based influence of antibiotics (with an emphasis on vancomycin and potent vancomycin analogues) began in the Schaefer laboratory and are now continuing pursuits in our laboratories and also in others. Selected results from these studies will be described to articulate how REDOR approaches, relying on unique isotopic labeling strategies and NMR selection, can yield key atomic-level distances and quantitative parameters of chemical composition in both isolated cell walls and intact whole cells.
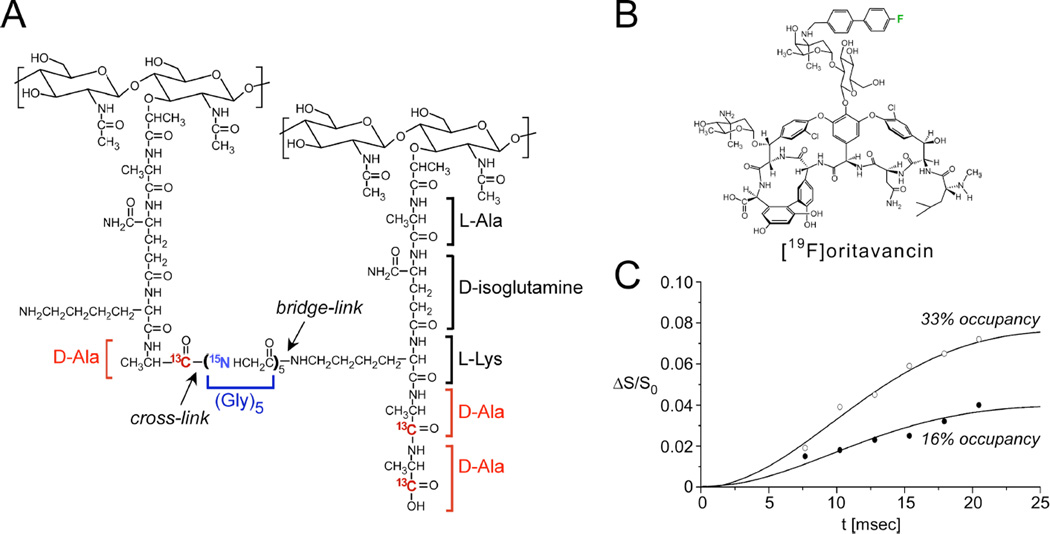
Figure 4. Distances between an antibiotic and the bacterial cell wall.
(A) Chemical schematic of the S. aureus peptidoglycan, highlighting the unique bridge-link and crosslink sites that are often targeted in REDOR experiments through the use of selective D-[1-13C]Ala, [15N]Gly, and L-[ε-15N]Lys labeling. (B) Chemical structure of [19F]oritavancin. (C) 13C{19F} REDOR dephasing (ΔS/S0) as a function of the dipolar evolution time, t, for complexes of [19F]oritavancin with peptidoglycan isolated from S. aureus whole cells grown on media containing D-[1-13C]alanine. The binding site occupancy of [19F]oritavancin complexed to cell walls was either 33% (open circles) or 16% (closed circles), wherein 2 µmol antibiotic was bound to 20 or 40 mg, respectively, of isolated peptidoglycan (dry mass). The calculated cell wall dephasing (solid lines) assumed a Gaussian distribution of distances for isolated 13C-19F pairs centered at 7.6 Å with a width of 1.5 Å. The dashed line also provides a reasonable fit to the data. Spectra were obtained on a 500 MHz spectrometer. Figure adapted from Kim et al.35
The earliest structural studies described the structural interactions of the peptidoglycan with oritavancin, a vancomycin analogue that has been in clinical development (Figure 4B)32, 33. REDOR measurements were performed using the fluorinated version of the drug, [19F]oritavancin (where replacement of the drug’s Cl with F has no influence on activity), also known as LY329332. Figure 4C provides REDOR data points and simulated dephasing plots corresponding to the measurements of the dipolar coupling between the drug and D-Ala in isolated S. aureus cell walls33. Quantification of this distance resulted from two parameters: knowledge of the amount of cell wall material and the percent occupancy of the drug determined in binding assays. In particular, 2 µmol LY329332 was bound to 20 mg of cell walls (about 12 µmol of peptidoglycan stems) and corresponds to 33% binding site occupancy. The REDOR experiments were performed on two samples in which the binding occupancy was either 33% or 16%. An accounting of the contributions to the S0 spectra and an analysis of how many 13C-19F pairs are present are central aspects of quantification in REDOR experiments such as this. In this regard, the dephasing plateau harbors important information. For isolated 13C-19F pairs, with 33% occupancy, one would start the analysis by expecting a dephasing plateau of 33% if all carbons were coupled to a 19F. However, the dephasing reached a plateau of 8%. In this case, a factor of about 2 was attributed to the determination that half of the peptidoglycan stems represented actual binding sites (harboring D-Ala-D-Ala stem termini rather than the shorter D-Ala termini) and another factor of 2 to the coupling of each 19F to two carbons (the L-Ala and D-Ala of a neighboring stem), yielding the reduced plateau of 8%. In these early studies, both L-Ala and D-Ala were labeled due to alanine racemization, whereas a racemase inhibitor was used in all subsequent studies which confirm and extend this early work34. This early example emphasizes the need to dissect and account for labeling patterns and efficiencies due to in vivo biosynthesis in REDOR studies in complex biosystems. The second sample with 16% binding occupancy accordingly reached a plateau of 4%, one-half that of the first sample (Figure 4C). Thus, by scaling the dephasing based on the plateau (rather than 100%), the data were fit to a distance centered at 7.6 Å with a width of 1.5 Å. More recent work in more selectively labeled samples has demonstrated that oritavancin binding in S. aureus is homogenous and exhibits a single comparable distance34. Other REDOR measurements indicated that the fluorine on [19F]oritavancin was placed co-linearly along the glycine bridge35. That is, it was closest to one glycine with each other glycine subsequently farther away, rather than possible placement near the middle of the bridge35. In the latter case, the REDOR measurements would have revealed one close distance and sets of nearest distances rather than the clear arrangement of five carbons each successively more distant than the previous one. Collectively, these REDOR measurements determined that [19F]oritavancin binds to a secondary peptidoglycan binding site, hugging the pentaglycine bridge, and explains the ability of analogues with abrogated primary binding sites (binding the D-Ala-D-Ala terminus) to still bind to the cell wall and prevent cell-wall assembly36, as well as its activity against S. aureus with altered D-Ala-D-Lac termini. It is crucial that these types of measurements regarding antibiotic-cell wall complexes are made within the context of native cell walls rather than synthetic cell-wall mimics as the intact cell wall contains compact glycine bridges, for example, with a certain length and orientation that are not recapitulated in soluble mimics amenable to solution-state NMR and other methods. A comprehensive recent publication by Kim, Schaefer, and coworkers has examined the locations of the hydrophobic sidechain of several important lipoglycopeptide analogues bound to the cell wall to correlate structure with function34.
This work also employed 31P{19F}REDOR to demonstrate, with other supporting data, that the hydrophobic sidechain is not embedded in the membrane indicating that at therapeutic-dosage levels, the sidechain does not act as a membrane anchor34.
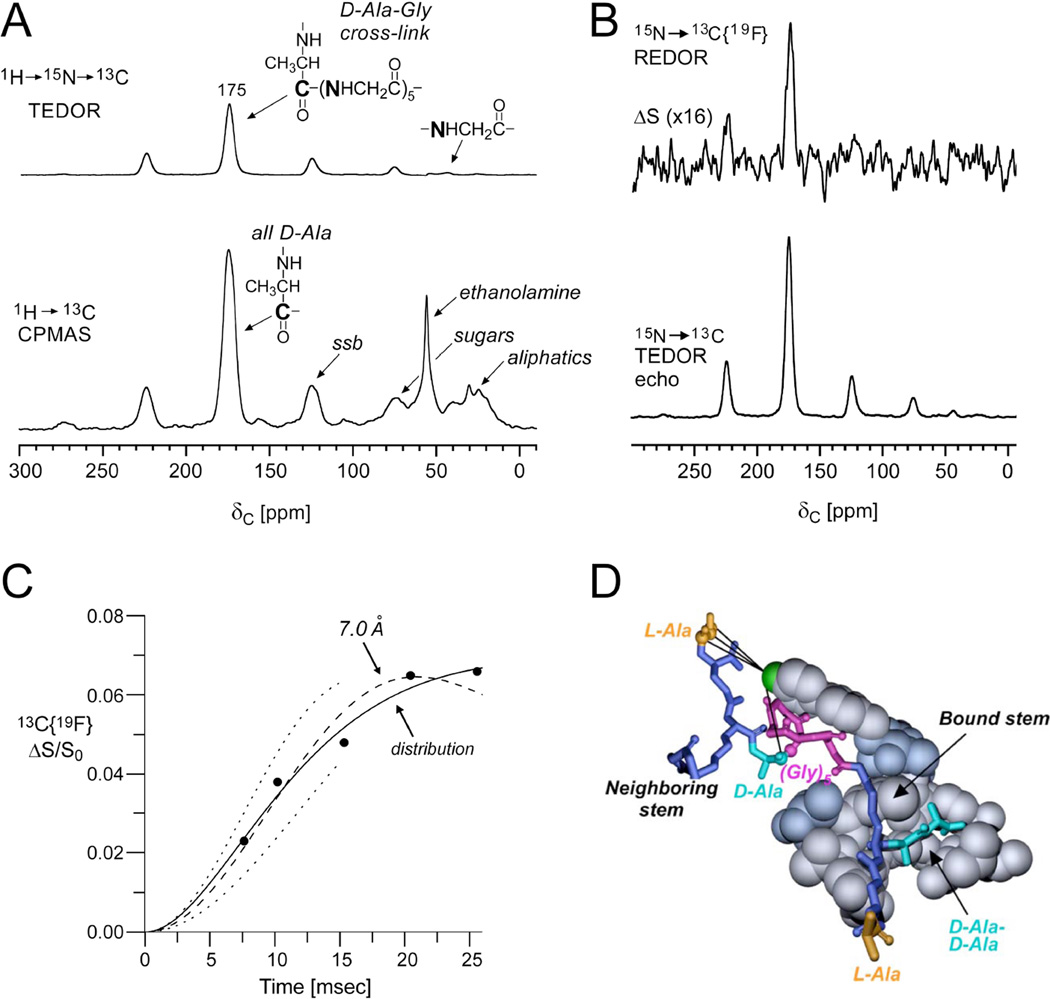
A whole-cell strategy was also developed to characterize [19F]oritavancin binding directly in intact whole cells37. Prior to these experiments, REDOR analysis from whole-cell samples required using a cell-wall S0 spectrum in order to normalize the whole-cell S0 spectrum to distinguish the cell-wall contributions from those of the cytoplasmic constituents35. In the study described here, the first use of TEDOR-REDOR in whole cells enabled the site-specific selection of the cross-linked D-Ala carbonyl carbon (proposed to be near to the 19F of the drug as described above) and subsequent distance measurement to the fluorine37. Figure 5 illustrates the ability to use a TEDOR (transferred-echo double-resonance)38transfer to first select the 13C nuclei that are directly bonded to 15N in cells that were biosynthetically labeled with D-[1-13C]Ala and [15N]Gly. In the TEDOR-REDOR experiment, the TEDOR-selected carbons (Figure 5A) were then dephased by 19F (Figure 5B) to measure the selected 13C-19F distance. One set of REDOR spectra, corresponding to 15 ms evolution time, is shown in Figure 5B and resulted from 120,000 scans, thus requiring about 4 days of acquisition. The data from five REDOR evolution times and simulated plots demonstrated close agreement with the earlier measurements, indicating that the C–F pairs could be fit best by a distribution of distances centered at 7.4 Å and a width of 1.6 Å (Figure 5C). In the case of whole cells, it was determined that there is heterogeneity in the binding sites in the cell wall, perhaps with slightly different conformations among layers close to the cell membrane and those towards the outer edges of the cell wall. The resulting model consistent with several distance measurements between [19F]oritavancin and labeled peptidoglycan in S. aureus whole-cell and cell walls is provided in Figure 5D.
Figure 5. Site-specific antibiotic-cell wall distances in whole cells.
(A) Full echo 13C CPMAS and TEDOR selected spectrum of lyophilized whole cells of S. aureus grown on media containing D-[1-13C]Ala and [15N]Gly in the presence of an alanine racemase inhibitor to ensure selective alanine labeling. The 13C CPMAS spectrum (bottom) was the result of 256 scans. The TEDOR-selected 13C spectrum (top) was the result of 120,0scans and harbors contributions only from labeled and natural abundance carbons that are bonded to 15N. (B) 15N→ 13C{19F} TEDOR-selected REDOR spectra of the whole-cell sample complexed to [19F]oritavancin after 15 ms dipolar evolution time (96 rotor cycles with MAS at 6250 Hz). S0 and S spectra resulted from the accumulation of 120,000scans. (C) 15N→13C{19F} TEDOR-selected REDOR dephasing (ΔS/S0) as a function of dipolar evolution time. The calculated dephasing using a Gaussian distribution of distances centered at 7.4 Å (solid line) provided a better match to the experimental dephasing than a single 7.0 Å distance (dashed line). The dotted lines show the calculated dephasing for single distances of 6.5 Å (upper) and 7.5 Å (lower). Spectra were obtained on a 500 MHz spectrometer. (D) Molecular model of [19F]oritavancin bound to the peptidoglycan of S. aureus generated from several distance measurements including the featured TEDOR-REDOR distance. Figure adapted from Cegelski et al.37
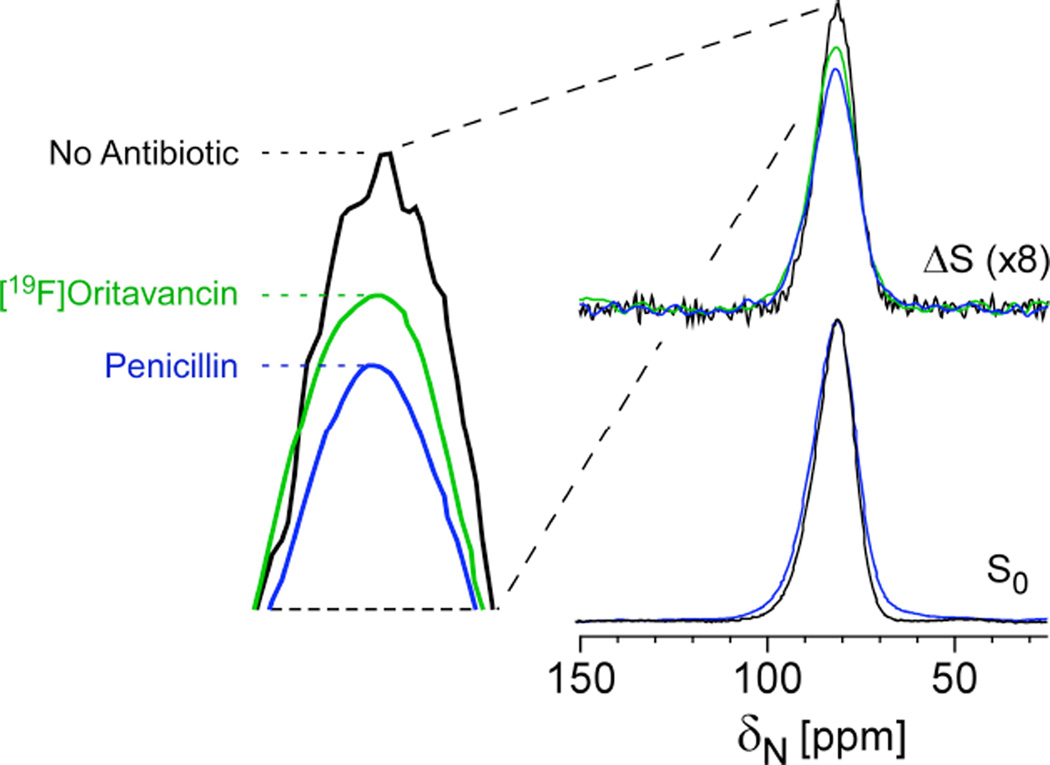
In the spirit of the above strategy, one could also observe 15N and select just the [15N]glycines that are directly bonded to the D-[113C]Ala. In fact, this approach was employed as a whole-cell NMR assay to determine whether an antibiotic can block crosslink formation without the need to isolate cell walls or to perform perturbative hydrolysis measures associated with HPLC and mass spectrometry-based profiling. In this example, the full-echo 15N spectrum harbors contributions from all glycines in the cell wall and cytoplasm and any metabolic products that resulted from transformations of labeled glycine (Figure 6). The REDOR difference spectrum, ΔS, reveals only nitrogens that are dephased by D-[13C]Ala, i.e. cross-link sites in the cell wall. Penicillin is known to prevent crosslink formation and, as anticipated, a clear decrease in 13C-15N bond density was observed for cells after treatment with penicillin39. Thus, again, bond selectivity can be achieved in the context of intact cells. This result demonstrated that oritavancin can prevent crosslink formation and supported the full analysis that oritavancin exhibits a dual mode of action in S. aureus, preventing both transglycosylation and transpeptidation39.
Figure 6. Cross-link density and inhibition of crosslinking in whole cells.
REDOR selection and quantification of peptidoglycan crosslinks in S. aureus whole cells labeled with [15N]glycine and D-[1-13C]alanine and a racemase inhibitor in the absence (black) and presence of 0.15 µg/mL penicillin (blue) and 20 µg/mL oritavancin (green). The REDOR difference measures the relative number of cross-links per pentagycyl bridging segment. Penicillin has a large effect, as a known crosslinking inhibitor and oritavancin also inhibited transpeptidation. Spectra were obtained on a 300 MHz spectrometer. Figure adapted from Kim et al.39
Membrane-associated proximities
REDOR has also been implemented to examine the structures of membrane proteins and the interactions of proteins, peptides, and other molecules, in and within membrane systems25, 40–51. Solid-state NMR, in general, has been invaluable in examining the structures of membrane proteins in native membrane environments. This final section highlights the ability to employ 13C{31P} REDOR and 13C{19F} REDOR to examine the relative proximities of guest molecules to lipid headgroups and lipid tails within a membrane system, particularly utilizing the comparative power of internal lipid rulers of reference.
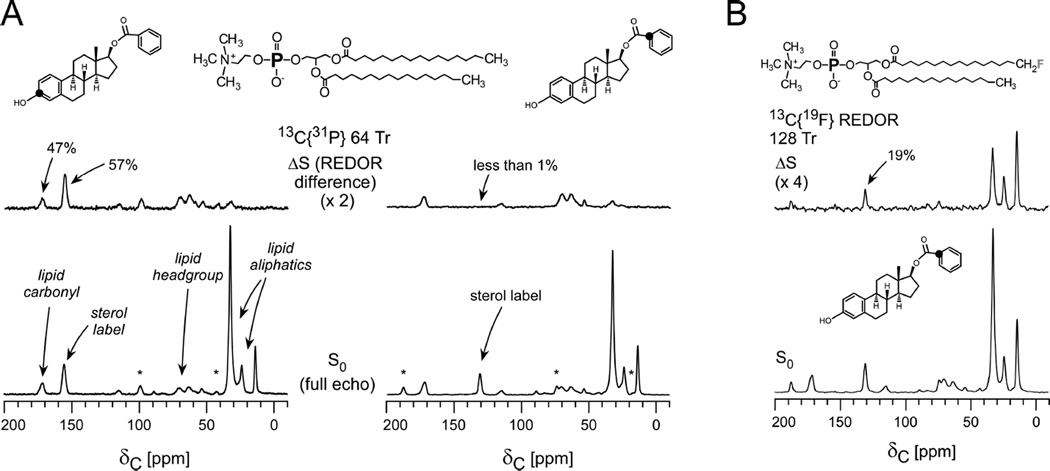
In the selected example, a series of comparative REDOR measurements were performed in order to map the orientation and depth of a panel of estradiol analogues in DPPC lipid vesicles41. The placement of the analogues correlated with their neuroprotective potency and provided support for the possibility that the more potent analogues confer enhanced protection against oxidation of unsaturated fatty acids in membrane lipids. The 13C{31P} REDOR full-echo spectra in Figure 7 include all carbon contributions in the system. Each sterol 13C label is readily resolved from the lipid carbons. The observed dephasing (ΔS/S0) reveals the relative proximity of each carbon type to phosphorous. As expected, the carbonyls of the lipid headgroups exhibit strong dephasing, whereas no 13C{31P} dephasing is observed for the lipid tail methyl groups. If, however, the membrane was perturbed, and kinks were formed, then some lipid tails would exhibit dephasing due to their increased proximity to the phosphate headgroups. This can occur, for example, when a high percentage of lipids with fluorinated tails are employed in the sample preparation52. For this reason, preparations of this type restrict the percentage of fluorinated lipids to less than or equal to 5% of the total lipid content.
Figure 7. Lipid cartography and guest-lipid proximities.
(A) 13C{31P} REDOR spectra of multilamellar vesicles containing 5% fluorinated DPPC and 95% DPPC with [3-13C]estradiol benzoate (left) and estradiol [113C]benzoate (right) after 9 ms of of dipolar evolution (64 Tr with MAS at 7143 Hz). (B) 13C{19F} 128-Tr REDOR spectra of the estradiol [1-13C]benzoate-containing vesicles. The substantial dephasing for the label in the benzoate ring placed that carbon proximate to the lipid tails. Spectra were obtained on a 500 MHz spectrometer. Figure adapted from Cegelski et al.41
The dephasing of two different 13C-labeled versions of the same sterol, [3-13C]estradiol benzoate and [1-13C]estradiol benzoate, were compared to map the molecular orientation of the compound in the bilayer. As seen in Figure 7A, it could immediately be determined that the sterol was situated with its phenolic group near to the headgroups and the benzoate group within the bilayer. In particular, the [3-13C]estradiol benzoate label exhibited stronger 31P dephasing than the lipid headgroup carbonyls and, thus, was determined to be relatively closer to the phosphate headgroup than the lipid carbonyls. Additional 13C{19F} REDOR confirmed the proximity of the benzoate group on the other side of the molecule to fluorinated lipid tails (Figure 7B). In contrast to the 13C{31P} REDOR spectra to assess headgroup proximity, the lipid methyl carbons and methylenes near to the tails exhibited significant dephasing to 19F, whereas dephasing of the headgroup carbonyls was not observed due to their long distance from the fluorinated tails. In the case of potential kinking (or interdigitation and shrinking of bilayer thickness), as mentioned above, one would observe 19F dephasing of the carbonyls (Figure 7A)52. Overall, one should always show the full width of the REDOR spectra in such studies so that the dephasing of the guest molecule and its proximity to certain lipid moeities can be evaluated with respect to proximities of the internal lipid landmarks and the lipid-centric dephasing41. This analysis could similarly be performed to evaluate the relative proximity of labels in membrane proteins or other membrane-associated components with lipid atoms. Furthermore, the incorporation of guest molecules or proteins may even alter lipid packing or thickness and the lipid-centric REDOR dephasing can be compared among samples with and without additional components. In the above study, a lipid-to-compound ratio of 20:1 was used as employed previously in studies using antimicrobial peptides25, 26. This ratio would need to be evaluated and optimized in cases of membrane proteins or ion channels, where protein activity can be assayed as a function of composition. REDOR measurements designed to access proximities to the phosphate headgroups could also be performed in intact cells or membrane extracts if there was sufficient resolution of the desired carbon resonance(s) from the natural abundance background in the cellular system, or if the guest molecule was labeled with 19F or 2H. However, in the context of whole cells, one would not immediately benefit from the use of the internal lipid landmarks since carbonyl contributions in the S0 spectrum, for example, would harbor all proteins and carboxyl-group containing molecules. Nevertheless, as shown in work with the antibiotic, oritavancin, lack of REDOR dephasing between 19F (incorporated in the antibiotic) and 31P in whole cell preparations of S. aureus with the antibiotic added at therapeutically relevant concentrations, revealed that the hydrophobic substituent on the antibiotic was not near to lipid headgroups, i.e. membrane anchors were not detected34. Thus, the lack of dephasing yielded important insight in the context of a relevant, whole-cell system.
Conclusions and Outlook
REDOR is a versatile tool in the drug discovery toolbox. Looking towards the future, challenges exist in examining large membrane proteins, such as G-protein coupled receptors (GPCRs), yet nearly half of all clinically available drugs target GPCRs. While pioneering crystallography and NMR efforts have delivered tremendous structural insights into the β-adrenergic receptor53and other important ion transporters and membrane proteins, the structures open up many additional and important avenues of study needed to further understand structure-function relationships, particularly in membrane environments, and the modes of action of molecular inhibitors. A challenge for REDOR in these systems is the need for adequate amounts of sample, as emphasized in the examples above. These examples were not microcrystalline proteins or assemblies that yield extremely narrow lines, thus more signal averaging and/or sample was required to obtain high-resolution spectra than on proteins which exhibit extremely high order or crystallinity. Yet, the taxol example illustrates the extent to which one can push the envelope on achieving a balance between sensitivity and the selection of the REDOR measurement to deliver key distances needed to solve crucial problems in drug design. Indeed, the ability to implement REDOR and report directly on the conformation of particularly large natural product molecules, such as taxol, is imperative to driving efforts to re-design and re-engineer the architecture of complex molecules, potentially simplifying synthetic efforts and enabling the discovery and generation of new analogues. In addition, advancements in dynamic nuclear polarization (DNP) should also enable the extension of REDOR to sample-limited studies.
Enormous efforts are also underway to map the human kinome and to identify inhibitors of kinases that are also integral to signal transduction cascades. Even when crystal structures of proteins and protein-inhibitor complexes exist, there are sometimes questions as to whether crystallization alters the conformation or reduces inherent heterogeneity to a single population. Dynamics may also play a role in drug binding and, in addition to other NMR dynamics measurements, REDOR can be integrated with experiments like CODEX54to map selected structural changes with dynamics. Similarly, as also discussed, there is an ever-increasing need for new strategies to prevent and treat infectious diseases due to the dwindling number of candidate antibiotics in the drug development pipeline and the continued and inevitable emergence of bacterial strains resistant to current antibiotics. For antibiotic candidates that interact with the cell wall and influence cell-wall assembly, REDOR is uniquely suited to examine drug modes of action in intact and insoluble samples, whereas most other methods rely on degradation of the cell-wall or the use of cell-wall surrogates that do not typically capture the structure and architecture of the intact cell wall. In summary, REDOR has emerged as a key problem-solving tool in the structural biology and biochemical toolbox and is needed to address outstanding problems in biology and medicine. The future is extremely bright for new REDOR approaches to deliver exciting discoveries to drive the design and development of new therapeutics.
Acknowledgements
L.C. gratefully acknowledges her PhD mentor, Professor Jacob Schaefer, for tremendously insightful discussions over the years and dedicates this digest to him in celebration of his 75th Birthday. L.C. holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund and gratefully acknowledges funding from the NIH Director’s New Innovator Award (DP2OD007488), the Stanford Terman Fellowship, the Hellman Faculty Scholar Award, and Stanford’s Center for Molecular Analysis and Design.
References
- 1.Gullion T, Schaefer J. Journal of Magnetic Resonance. 1989;81:196. doi: 10.1016/j.jmr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Toke O, Cegelski L. eMagRes. John Wiley & Sons, Ltd; 2007. [Google Scholar]
- 3.Matsuoka S, Inoue M. Chemical Communications. 2009:5664. doi: 10.1039/b910230b. [DOI] [PubMed] [Google Scholar]
- 4.Gullion T, Graham AW. Annual Reports on NMR Spectroscopy. Vol. Volume 65. Academic Press; 2009. p. 111. [Google Scholar]
- 5.McDowell LM, Poliks B, Studelska DR, O'Connor RD, Beusen DD, Schaefer J. Journal of Biomolecular NMR. 2004;28:11. doi: 10.1023/B:JNMR.0000012864.70184.48. [DOI] [PubMed] [Google Scholar]
- 6.O'Connor RD, Schaefer J. Journal of Magnetic Resonance. 2002;154:46. doi: 10.1006/jmre.2001.2461. [DOI] [PubMed] [Google Scholar]
- 7.Mueller KT, Jarvie TP, Aurentz DJ, Roberts BW. Chemical Physics Letters. 1995;242:535. [Google Scholar]
- 8.Grage SL, Watts A. Annual Reports on NMR Spectroscopy, Vol 60. 2007;60:191. [Google Scholar]
- 9.Andrew ER. Philosophical Transactions of the Royal Society a-Mathematical Physical and Engineering Sciences. 1981;299:505. [Google Scholar]
- 10.Prestegard JH, Kishore AI. Current Opinion in Chemical Biology. 2001;5:584. doi: 10.1016/s1367-5931(00)00247-7. [DOI] [PubMed] [Google Scholar]
- 11.Ishii Y, Balbach JJ, Tycko R. Chemical Physics. 2001;266:231. [Google Scholar]
- 12.Rienstra CM, Hatcher ME, Mueller LJ, Sun BQ, Fesik SW, Griffin RG. Journal of the American Chemical Society. 1998;120:10602. [Google Scholar]
- 13.Gullion T. Concepts in Magnetic Resonance. 1998;10:277. [Google Scholar]
- 14.Stueber D, Yu T-Y, Hess B, Kremer K, O'Connor RD, Schaefer J. The Journal of Chemical Physics. 2010;132:104901. doi: 10.1063/1.3330412. [DOI] [PubMed] [Google Scholar]
- 15.Goetz JM, Schaefer J. Journal of Magnetic Resonance. 1997;127:147. doi: 10.1006/jmre.1997.1198. [DOI] [PubMed] [Google Scholar]
- 16.Feeney J, McCormick JE, Bauer CJ, Birdsall B, Moody CM, Starkmann BA, Young DW, Francis P, Havlin RH, Arnold WD, Oldfield E. Journal of the American Chemical Society. 1996;118:8700. [Google Scholar]
- 17.Pearson JG, Montez B, Le HB, Oldfield E, Chien EYT, Sligar SG. Biochemistry. 1997;36:3590. doi: 10.1021/bi961664h. [DOI] [PubMed] [Google Scholar]
- 18.Paik Y, Yang C, Metaferia B, Tang SB, Bane S, Ravindra R, Shanker N, Alcaraz AA, Johnson SA, Schaefer J, O'Connor RD, Cegelski L, Snyder JP, Kingston DGI. Journal of the American Chemical Society. 2007;129:361. doi: 10.1021/ja0656604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li YK, Poliks B, Cegelski L, Poliks M, Gryczynski Z, Piszczek G, Jagtap PG, Studelska DR, Kingston DGI, Schaefer J, Bane S. Biochemistry. 2000;39:281. doi: 10.1021/bi991936r. [DOI] [PubMed] [Google Scholar]
- 20.Holl SM, Marshall GR, Beusen DD, Kociolek K, Redlinski AS, Leplawy MT, Mckay RA, Vega S, Schaefer J. Journal of the American Chemical Society. 1992;114:4830. [Google Scholar]
- 21.Mehta AK, Shayo Y, Vankayalapati H, Hurley LH, Schaefer J. Biochemistry. 2004;43:11953. doi: 10.1021/bi049697h. [DOI] [PubMed] [Google Scholar]
- 22.Geney R, Sun L, Pera P, Bernacki RJ, Xia SJ, Horwitz SB, Simmerling CL, Ojima I. Chemistry & Biology. 2005;12:339. doi: 10.1016/j.chembiol.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Sun L, Geng XD, Geney R, Li Y, Simmerling C, Li Z, Lauher JW, Xia SJ, Horwitz SB, Veith JM, Pera P, Bernacki RJ, Ojima I. Journal of Organic Chemistry. 2008;73:9584. doi: 10.1021/jo801713q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tong G, Pan Y, Dong H, Pryor R, Wilson GE, Schaefer J. Biochemistry. 1997;36:9859. doi: 10.1021/bi970495d. [DOI] [PubMed] [Google Scholar]
- 25.Toke O, Maloy WL, Kim SJ, Blazyk J, Schaefer J. Biophysical Journal. 2004;87:662. doi: 10.1529/biophysj.103.032706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toke O, O'Connor RD, Weldeghiorghis TK, Maloy WL, Glaser RW, Ulrich AS, Schaefer J. Biophysical Journal. 2004;87:675. doi: 10.1529/biophysj.103.032714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cegelski L, O'Connor RD, Stueber D, Singh M, Poliks B, Schaefer J. Journal of the American Chemical Society. 2010;132:16052. doi: 10.1021/ja104827k. [DOI] [PubMed] [Google Scholar]
- 28.Cegelski L, Kim SJ, Hing AW, Studelska DR, O'Connor RD, Mehta AK, Schaefer J. Biochemistry. 2002;41:13053. doi: 10.1021/bi0202326. [DOI] [PubMed] [Google Scholar]
- 29.Zhou XX, Cegelski L. Biochemistry. 2012;51:8143. doi: 10.1021/bi3012115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDowell LM, Burzio LA, Waite JH, Schaefer J. Journal of Biological Chemistry. 1999;274:20293. doi: 10.1074/jbc.274.29.20293. [DOI] [PubMed] [Google Scholar]
- 31.Kramer KJ, Hopkins TL, Schaefer J. Insect Biochemistry and Molecular Biology. 1995;25:1067. [Google Scholar]
- 32.Cegelski L, Kim SJ, Hing AW, Studelska D, O'Connor R, Mehta A, Schaefer J. Biophysical Journal. 2002;82:468a. [Google Scholar]
- 33.Kim SJ, Cegelski L, Studelska D, O'Connor R, Mehta A, Schaefer J. Biophysical Journal. 2002;82:469a. [Google Scholar]
- 34.Kim SJ, Tanaka KSE, Dietrich E, Rafai Far A, Schaefer J. Biochemistry. 2013;52:3405. doi: 10.1021/bi400054p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim SJ, Cegelski L, Studelska DR, O'Connor RD, Mehta AK, Schaefer J. Biochemistry. 2002;41:6967. doi: 10.1021/bi0121407. [DOI] [PubMed] [Google Scholar]
- 36.Kim SJ, Matsuoka S, Patti GJ, Schaefer J. Biochemistry. 2008;47:3822. doi: 10.1021/bi702232a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cegelski L, Steuber D, Mehta AK, Kulp DW, Axelsen PH, Schaefer J. Journal of Molecular Biology. 2006;357:1253. doi: 10.1016/j.jmb.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 38.Hing AW, Vega S, Schaefer J. Journal of Magnetic Resonance. 1992;96:205. [Google Scholar]
- 39.Kim SJ, Cegelski L, Stueber D, Singh M, Dietrich E, Tanaka KSE, Parr TR, Far AR, Schaefer J. Journal of Molecular Biology. 2008;377:281. doi: 10.1016/j.jmb.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuoka S, Ikeuchi H, Umegawa Y, Matsumori N, Murata M. Bioorganic & Medicinal Chemistry. 2006;14:6608. doi: 10.1016/j.bmc.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 41.Cegelski L, Rice CV, O'Connor RD, Caruano AL, Tochtrop GP, Cai ZY, Covey DF, Schaefer J. Drug Development Research. 2005;66:93. [Google Scholar]
- 42.Mani R, Tang M, Wu X, Buffy JJ, Waring AJ, Sherman MA, Hong M. Biochemistry. 2006;45:8341. doi: 10.1021/bi060305b. [DOI] [PubMed] [Google Scholar]
- 43.Kandasamy SK, Lee DK, Nanga RPR, Xu J, Santos JS, Larson RG, Ramamoorthy A. Biochimica Et Biophysica Acta-Biomembranes. 2009;1788:686. doi: 10.1016/j.bbamem.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 44.Ghosh U, Xie L, Weliky DP. Journal of Biomolecular NMR. 2013;55:139. doi: 10.1007/s10858-013-9709-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saito H, Tuzi S, Yamaguchi S, Kimura S, Tanio M, Kamihira M, Nishimura K, Naito A. Journal of Molecular Structure. 1998;441:137. [Google Scholar]
- 46.Watts JA, Watts A, Middleton DA. Journal of Biological Chemistry. 2001;276:43197. doi: 10.1074/jbc.M104808200. [DOI] [PubMed] [Google Scholar]
- 47.Smith SO, Smith C, Shekar S, Peersen O, Ziliox M, Aimoto S. Biochemistry. 2002;41:9321. doi: 10.1021/bi012117l. [DOI] [PubMed] [Google Scholar]
- 48.Wang JX, Balazs YS, Thompson LK. Biochemistry. 1997;36:1699. doi: 10.1021/bi962578k. [DOI] [PubMed] [Google Scholar]
- 49.Porcelli F, Buck B, Lee DK, Hallock KJ, Ramamoorthy A, Veglia G. Journal of Biological Chemistry. 2004;279:45815. doi: 10.1074/jbc.M405454200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sackett K, Nethercott MJ, Epand RF, Epand RM, Kindra DR, Shai Y, Weliky DP. Journal of Molecular Biology. 2010;397:301. doi: 10.1016/j.jmb.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murphy OJ, Kovacs FA, Sicard EL, Thompson LK. Biochemistry. 2001;40:1358. doi: 10.1021/bi0015109. [DOI] [PubMed] [Google Scholar]
- 52.Hirsh DJ, Lazaro N, Wright LR, Boggs JM, McIntosh TJ, Schaefer J, Blazyk J. Biophysical Journal. 1998;75:1858. doi: 10.1016/S0006-3495(98)77626-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Granier S, Kobilka B. Nature Chemical Biology. 2012;8:670. doi: 10.1038/nchembio.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.deAzevedo ER, Hu WG, Bonagamba TJ, Schmidt-Rohr K. Journal of the American Chemical Society. 1999;121:8411. [Google Scholar]
- 55.Gullion T, Baker DB, Conradi MS. Journal of Magnetic Resonance. 1990;89:479. [Google Scholar]