Abstract
During ribosome recycling, post-termination complexes are dissociated by ABCE1 and eRF1 into 60S and tRNA/mRNA-associated 40S subunits, after which tRNA/mRNA are released by eIF1/eIF1A, Ligatin or MCT1/DENR. In some instances, 40S subunits remain associated with mRNA and reinitiate at nearby AUGs. Here, we recapitulated reinitiation in vitro using a reconstituted mammalian translation system. The presence of eIFs 2/3/1/1A and Met-tRNAiMet was sufficient for recycled 40S subunits to remain on mRNA, scan bidirectionally and reinitiate both at upstream and downstream AUGs if mRNA regions flanking the stop codon were unstructured. Imposition of 3’-directionality additionally required eIF4F. Strikingly, post-termination ribosomes were not stably anchored on mRNA, and migrated bidirectionally to codons cognate to the P-site tRNA. Migration depended on the mode of peptide release (puromycin>eRF1>eRF1/eRF3), the nature of tRNA, and was enhanced by eEF2. The mobility of post-termination ribosomes suggests that some reinitiation events could involve 80S ribosomes rather than 40S subunits.
Keywords: reinitiation, recycling, ribosome, ABCE1, eRF1, eRF3, eIF3, eEF2
INTRODUCTION
Translation is a cyclical process, consisting of initiation, elongation, termination and ribosome recycling. Eukaryotic initiation begins with assembly of a 43S preinitiation complex, containing a 40S subunit, an eIF2-GTP/Met-tRNAiMet ternary complex (eIF2-TC), and eIFs 3, 1 and 1A (Jackson et al., 2010). 43S complexes attach to the 5’-end of mRNA and scan to the initiation codon, where they form 48S initiation complexes with established codon-anticodon base-pairing. Attachment is mediated by eIF4A, eIF4B, and eIF4F, which consists of three subunits: eIF4E (a cap-binding protein), eIF4A (an RNA helicase) and eIF4G (a scaffold for eIF4E and eIF4A, which also binds to eIF3). eIFs 4A/4B/4F unwind the cap-proximal region of mRNA and stimulate attachment of 43S via the eIF3/eIF4G interaction. eIFs 4A/4B/4G also assist 43S complexes during scanning, which on structured mRNAs additionally requires the DExH-box protein DHX29. After start codon recognition, eIF5 and eIF5B induce hydrolysis of eIF2-bound GTP, release of eIFs and joining of a 60S subunit, resulting in formation of an 80S ribosome. Subsequent translation is mediated by elongation factors eEF1A and eEF2, which deliver aa-tRNA to the ribosomal A site and catalyze ribosomal translocation after formation of the peptide bond, respectively.
Termination involves the concerted action of release factors eRF1 and eRF3 (Alkalaeva et al., 2006). eRF1 is responsible for stop codon recognition and inducing hydrolysis of peptidyl-tRNA, whereas eRF3 strongly stimulates peptide release by eRF1 in a GTP-dependent manner. After peptide release, eRF1 remains bound to post-termination complexes (post-TCs) and together with the ATP-binding cassette protein ABCE1 splits them into 60S and tRNA/mRNA-associated 40S subunits (Pisarev et al., 2010). Subsequent dissociation of tRNA can be promoted by eIFs 3, 1 and 1A (Pisarev et al., 2010), Ligatin, or MCT1/DENR (Skabkin et al, 2010). tRNA release is followed by dissociation of mRNA, but if release of tRNA is induced by eIFs 3/1/1A, it also requires eIF3’s loosely associated j subunit. Thus, there is considerable redundancy in factors promoting tRNA/mRNA release from recycled 40S subunits. Importantly, at low [Mg2+], eIFs 3, 1 and 1A can mediate the entire recycling process, with eIF3 responsible for splitting post-termination ribosomes (Pisarev et al., 2007a).
In some cases post-TCs do not undergo complete recycling: 40S subunits remain bound to mRNA, and termination is followed by reinitiation, usually downstream of the stop codon (Jackson et al., 2012). It has become apparent from genome-wide bioinformatic analyses that >45% of mammalian mRNAs contain at least one upstream ORF (uORF) (Calvo et al., 2009), and ribosome profiling revealed that these uORFs are frequently translated (Ingolia et al., 2011). Analogous experiments indicate that only 13% of yeast mRNAs contain uORFs, but that many of them are also translated (Ingolia et al., 2009; Lawless et al., 2009). Reinitiation can be modulated in response to environmental changes (Jackson et al., 2012), and is thus emerging as an important regulatory event.
Efficient reinitiation commonly occurs only after translation of short open reading frames (ORFs), and depends on the duration of elongation, being inhibited by pseudoknot-induced ribosomal pausing (Kozak, 2001). These results are generally consistent with the initial suggestion that some eIFs remain transiently associated with ribosomes through elongation and termination, and those ribosomes that retain eIFs are able to resume scanning and reinitiate after post-termination dissociation of the 60S subunit (Kozak, 1987). More recently, investigation of reinitiation after translation of short ORFs that was driven by different IRES-dependent mechanisms showed that efficient reinitiation occurred only if the original initiation event involved eIF4F, or at least eIF4A and eIF4G’s middle domain (Pöyry et al., 2004). This led to the suggestion that resumption of scanning, eventual eIF2-TC acquisition and reinitiation would depend on ribosomal retention of eIF4F, presumably via the eIF4G-eIF3-40S chain of interactions.
Although the majority of mammalian short ORFs are permissive for reinitiation, efficient reinitiation does not occur after randomly generated short ORFs in yeast (Yun et al., 1996), implying that in contrast to mammals, reinitiation in yeast depends on specific mRNA sequences. The best characterized example of reinitiation in yeast is translation of GCN4 mRNA: it has four short uORFs, the first of which is permissive for reinitiation (Hinnebusch, 2005). Its permissiveness depends on upstream and downstream mRNA sequences. The downstream sequence includes ~10 AU-rich nucleotides following the stop codon and the last sense codon (Grant and Hinnebusch, 1994), whereas the upstream sequence extends for up to ~180 nts and interacts with eIF3a (e.g. Szamecz et al., 2008). The differences between reinitiation in yeast and mammals might therefore be determined by differences in eIF3’s composition and/or its interaction with eIF4G (Hinnebusch, 2006), leading to a requirement for specific sequences in yeast mRNAs so that they can interact with these factors to ensure their prolonged retention.
Reinitiation after long ORFs is rare and mainly occurs on viral mRNAs. The best-characterized examples are bicistronic subgenomic calicivirus mRNAs, in which the second cistron, encoding a minor capsid protein, is translated by reinitiation. The process depends on a short segment upstream of the restart AUG called TURBS (“termination codon upstream ribosome binding site”), which contains a conserved sequence that is complementary to 18S rRNA (e.g. Lütterman and Meyers, 2007), and also specifically interacts with eIF3 (Pöyry et al., 2007).
To investigate the molecular mechanism of eukaryotic reinitiation, we recapitulated this and other unconventional post-termination events, using a mammalian in vitro reconstituted translation system.
RESULTS
Instability of mammalian post-TCs and migration of post-termination ribosomes
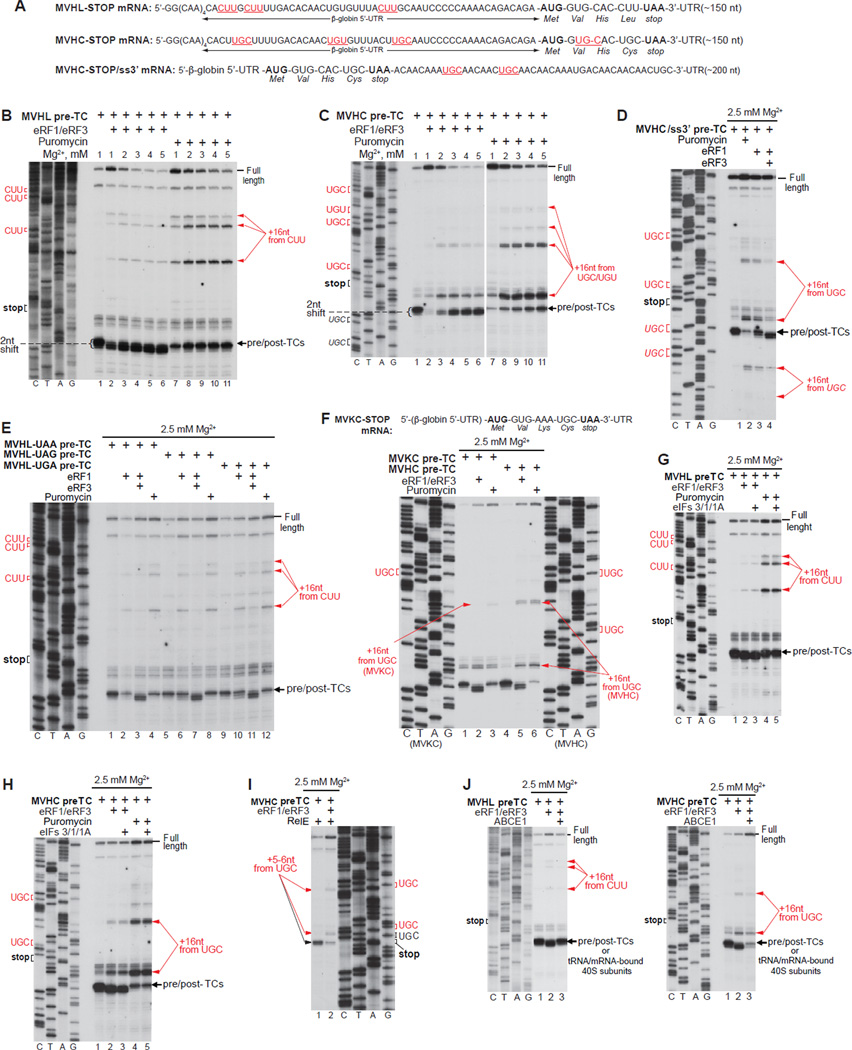
Although it is widely assumed that reinitiation involves recycled 40S subunits, we nevertheless first examined post-termination 80S ribosomes. For this, pre-termination complexes (pre-TCs) were formed on MVHL-STOP and MVHC-STOP mRNAs comprising 12 unstructured 5’-terminal nucleotides (four CAA triplets) followed by the β-globin 5’-UTR, MVHL or MVCH ORFs, a UAA stop codon and an ~150-nt 3’-UTR (Alkalaeva et al., 2006; Fig. 1A), using 40S and 60S subunits, eIFs 2, 3, 1, 1A, 4A, 4B, 4F, 5 and 5B, eEF1H, eEF2 and aa-tRNAs. Pre-TCs were then purified by sucrose density gradient (SDG) centrifugation and incubated over a range of [Mg2+] with eRFs or puromycin to induce peptide release. The position on mRNA of post-termination ribosomes was determined by toe-printing (Figs. 1B–C).
Figure 1. Mammalian post-termination ribosomes can migrate to nearby codons that are cognate to the P-site deacylated tRNA.
(A; F, upper panel) Structure of (A) MVHL-STOP, MVHC-STOP, MVHC-STOP/ss3’ and (F) MVKC-STOP mRNAs. Upstream and downstream codons that are identical to the last codon of the encoded tetrapeptide are in red and underlined (A). (B-H, J) Toe-printing analysis of ribosomal complexes obtained by treating pre-TCs formed on MVHL-STOP, MVHC-STOP, MVHC-STOP/ss3’ or MVKC-STOP mRNAs with combinations of eRF1, eRF3, puromycin, eIFs 3/1/1A and ABCE1 at the indicated free [Mg2+]. Black arrows show the positions of pre-TCs, post-TCs and recycled 40S subunits. Red arrows indicate the positions of migrated post-termination ribosomes. Upstream and downstream codons that are identical to the last codon of the encoded tetrapeptide are in red. (I) Primer-extension analysis of A-site mRNA cleavage induced by RelE in pre- and post-TCs assembled on MVHC-STOP mRNA. The positions of cleavages and of corresponding P-site codons are indicated.
Incubation of pre-TCs with eRF1/eRF3 led to the appearance not only of toe-prints corresponding to eRF1-associated post-TCs (which are characterized by a 2nt shift forward), but also of additional stops +16 nts from upstream CUU or UGC/UGU codons on MVHL-STOP and MVHC-STOP mRNA, respectively (Figs. 1B–C, lanes 1–6). Their intensity was inversely proportional to the distance from the stop codon. The CUU and UGC/UGU codons are cognate to the P-site tRNALeu and tRNACys of the corresponding pre-TCs, which indicates that post-termination ribosomes are not stably anchored on mRNA and can migrate to nearby codons that are cognate to the P-site tRNA. Migration was most efficient at 2–3 mM free Mg2+ (Figs. 1B–C, lanes 3, 4). Elevation of [Mg2+] increased the stability of post-TCs and reduced migration (Figs. 1B–C, lanes 5, 6), whereas lowering it to 1 mM caused substantial dissociation of deacylated tRNA (manifested by the appearance of prominent full-length cDNA), which in turn decreased the amount of migrated complexes (Figs. 1B–C, lanes 2). Post-TCs assembled on MVHC-STOP mRNA were less stable than on MVHL-STOP mRNA, and more full-length cDNA (at lower [Mg2+]) and stronger toe-prints corresponding to migrated ribosomal complexes (at higher [Mg2+]) were observed in this case (Figs. 1B–C, lanes 2–6). Triggering peptide release with puromycin resulted in enhancement of toe-prints corresponding to migrated ribosomes (Figs. 1B–C, lanes 7–11; Fig. 1E), and toe-prints of intermediate intensity were observed when peptide release was induced by eRF1 alone (Fig. 1E). Thus, continued association of eRFs with ribosomal complexes after peptide release increased their stability.
Even though MVHL- and MVHC-STOP mRNAs also contain cognate downstream Leu and Cys codons, toe-prints corresponding to migrated post-termination ribosomes occurred only upstream of the stop codon. To investigate whether 3’-directional migration was restricted by potential secondary structure, pre-TCs were formed on MVHC/ss3’ mRNA with an unstructured downstream region (Fig. 1A). On this mRNA, post-termination ribosomes were able to move downstream, but migration upstream was nevertheless preferred (Fig. 1D).
The proportion of migrated ribosomes did not depend on the stop codon sequence, irrespective of the mode of peptide release (Fig. 1E). However, it was influenced by the nucleotides in the E-site. Thus, ribosomes migrated less efficiently on mRNA encoding the MVKC than the MVHC tetrapeptide (Fig. 1F, compare lanes 2, 3 with lanes 5, 6). Since SDG-purified pre-TCs lack E-site tRNA (Taylor et al., 2012), the only difference between these post-TCs was the triplet in the E-site: AAA on MVKC-STOP mRNA, but CAC on MVHC-STOP mRNA.
To confirm that the observed toe-prints corresponded to 80S ribosomes rather than to individual 40S subunits, post-TCs were further incubated with eIFs 3/1/1A, which at higher [Mg2+] induce tRNA/mRNA release from 40S subunits but do not affect post-termination 80S complexes (Pisarev et al., 2007a, 2010). eIFs 3/1/1A did not influence these toe-prints (Figs. 1G, H), indicating that they were caused by 80S ribosomes. To verify that migration of post-termination ribosomes was not induced during toe-printing, ribosomal complexes were instead treated with the bacterial toxin RelE, which cleaves mRNA in the A-site (e.g. Neubauer et al, 2009). In pre-TCs, RelE induced cleavage after the first and second nucleotides following the UGC codon of the MVHC ORF (Fig. 1I, lane 1), whereas in post-TCs, RelE induced additional cleavages that corresponded to ribosomal complexes arrested at upstream UGC triplets (Fig. 1I, lane 2). Thus, ribosome migration occurred before toe-printing and was not induced by reverse transcriptase.
Next, we compared the association of tRNALeu and tRNACys with 40S subunits after ABCE1-mediated splitting of post-TCs. At 2.5 mM free Mg2+, tRNALeu remained bound to 40S subunits, which was manifested as a quantitative reversal of the 2 nt toe-print shift indicative of dissociation of post-TCs into 60S and tRNA/mRNA-associated 40S subunits that were now responsible for the toe-print (Fig. 1J, left panel). In contrast, tRNACys dissociated spontaneously, yielding a weak toe-print corresponding to tRNA/mRNA-associated 40S subunits and prominent full-length cDNA (Fig. 1J, right panel). Thus, the weaker association of tRNACys with 40S subunits, which in the presence of 60S subunits resulted in efficient migration of post- termination 80S ribosomes, now led to its spontaneous dissociation from 40S subunits following ABCE1-mediated splitting of post-TCs.
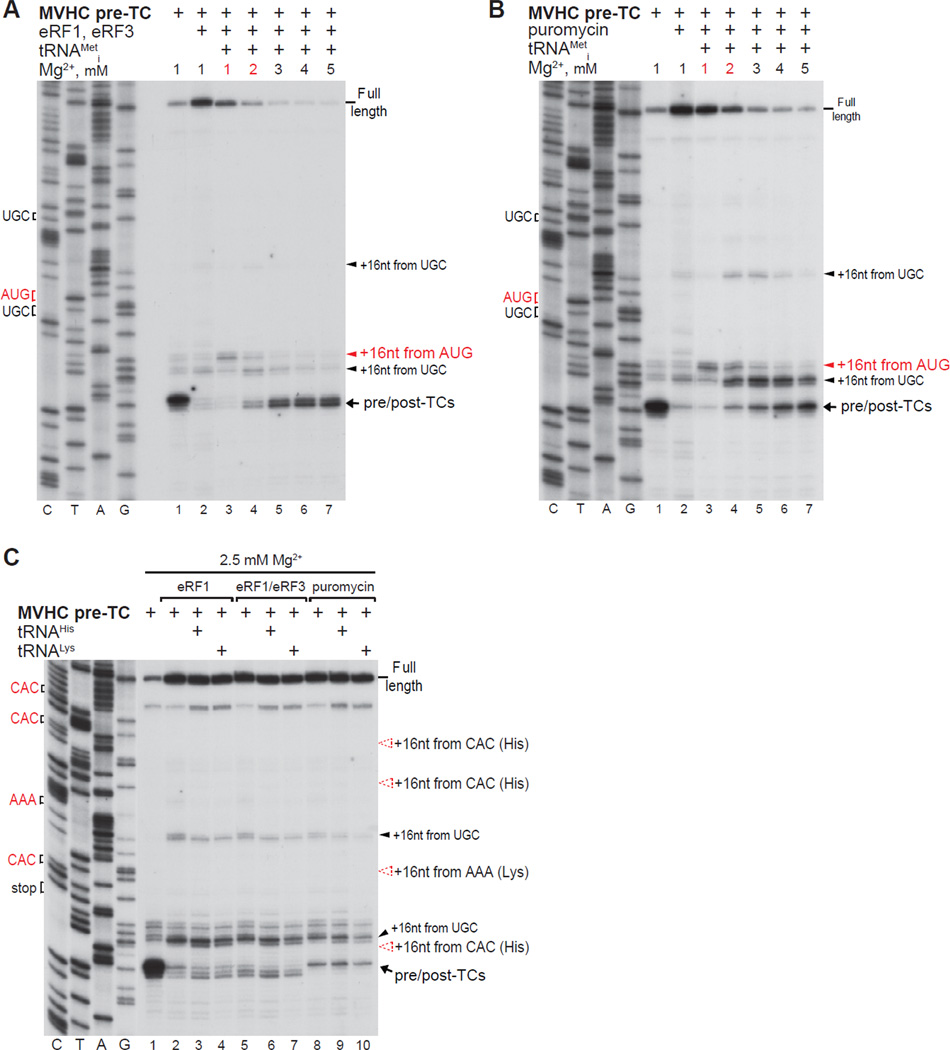
Because deacylated tRNA dissociated readily from post-TCs at low [Mg2+] (Fig. 1C, lane 2), we investigated whether post-termination ribosomes with a vacant P-site but still potentially bound to mRNA, could accept other tRNAs and stop at their cognate codons. At 1 mM Mg2+ (and less so at 2 mM) a small proportion of post-termination ribosomes could rebind tRNAiMet and migrate to the upstream AUG (Figs. 2A, B, lanes 3, 4). None of the tested elongator tRNAs showed similar activity: ribosomal complexes were not arrested at CAC or AAA triplets in the presence of cognate tRNAHis or tRNALys, respectively (Fig. 2C).
Figure 2. The ability of post-termination ribosomes with a vacant P-site to rebind tRNAs.
(A–C) Toe-printing analysis of ribosomal complexes obtained by treating pre-TCs formed on MVHC-STOP mRNA with eRF1, eRF1/eRF3 or puromycin in the presence/absence of (A, B) tRNAiMet and (C) tRNAHis or tRNALys at the indicated free [Mg2+]. Red arrows show the positions of ribosomal complexes arrested at AUG (Met), CAC (His) or AAA (Lys) codons.
Influence of eEF2 on the stability of post-TCs
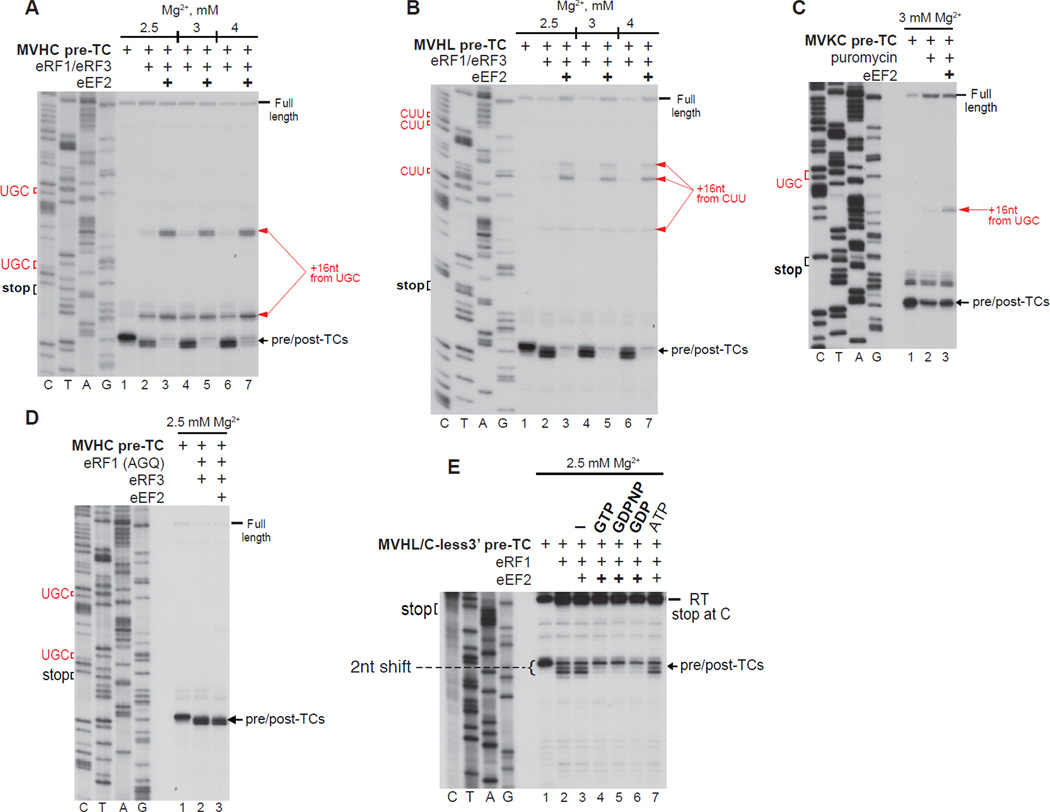
Strikingly, eEF2 destabilized post-TCs obtained with eRF1/eRF3 and strongly enhanced ribosome migration (Figs. 3A, B), altering the proportion of complexes arrested at different cognate codons by inducing bypassing of the closest codon and promoting arrest at the next one upstream. eEF2 also eliminated the +2nt toe-print shift (Figs. 3A, B), indicating that it induced dissociation of eRFs. Consistently, in SDG centrifugation experiments, eEF2 strongly reduced association of 32P-labeled eRF1 with post-TCs (not shown). Although destabilization of the eRF1/post-TC interaction was likely one underlying reason for stimulation of ribosome migration, eEF2 also enhanced migration when peptide release was mediated by puromycin (Fig. 3C). eEF2 did not influence ribosomal association of eRF1(AGQ) (a mutant that can bind pre-TCs but is inactive in peptide release; Frolova et al., 1999) (Fig. 3D), indicating that peptide release is a prerequisite for eEF2’s effect on ribosomal association of eRF1. To investigate whether eEF2 depends on GTP hydrolysis to influence ribosomal association of eRF1, pre-TCs were formed on MVHL-STOP/C-less mRNA, which lacks cytidines between the stop codon and the primer so that dGTP could be omitted from primer extension (Alkalaeva et al., 2006). This allowed the possibility that eEF2 acted during toe-printing by utilizing dGTP to be excluded. To avoid the need for inclusion of GTP for eRF3, eEF2’s activity was monitored by reversal of the toe-print shift of post-TCs obtained with eRF1 alone. eEF2 required guanine nucleotides, but was similarly active with GTP, GDPNP and GDP (Fig. 3E).
Figure 3. eEF2 destabilizes post-TCs and promotes ribosome migration.
(A–E) Toe-printing analysis of ribosomal complexes obtained by treating pre-TCs formed on MVHL-STOP, MVHC-STOP, MVKC-STOP or MVHL-STOP/C-less mRNAs with combinations of eRF1, eRF3, puromycin, eRF1(AGQ) and eEF2 at the indicated free [Mg2+]. Black arrows show the positions of pre- and post-TCs. Red arrows indicate the positions of migrated post-termination ribosomes. Upstream codons that are identical to the last codon of the encoded tetrapeptide are shown in red.
Influence of the E-site tRNA on the stability of post-TCs
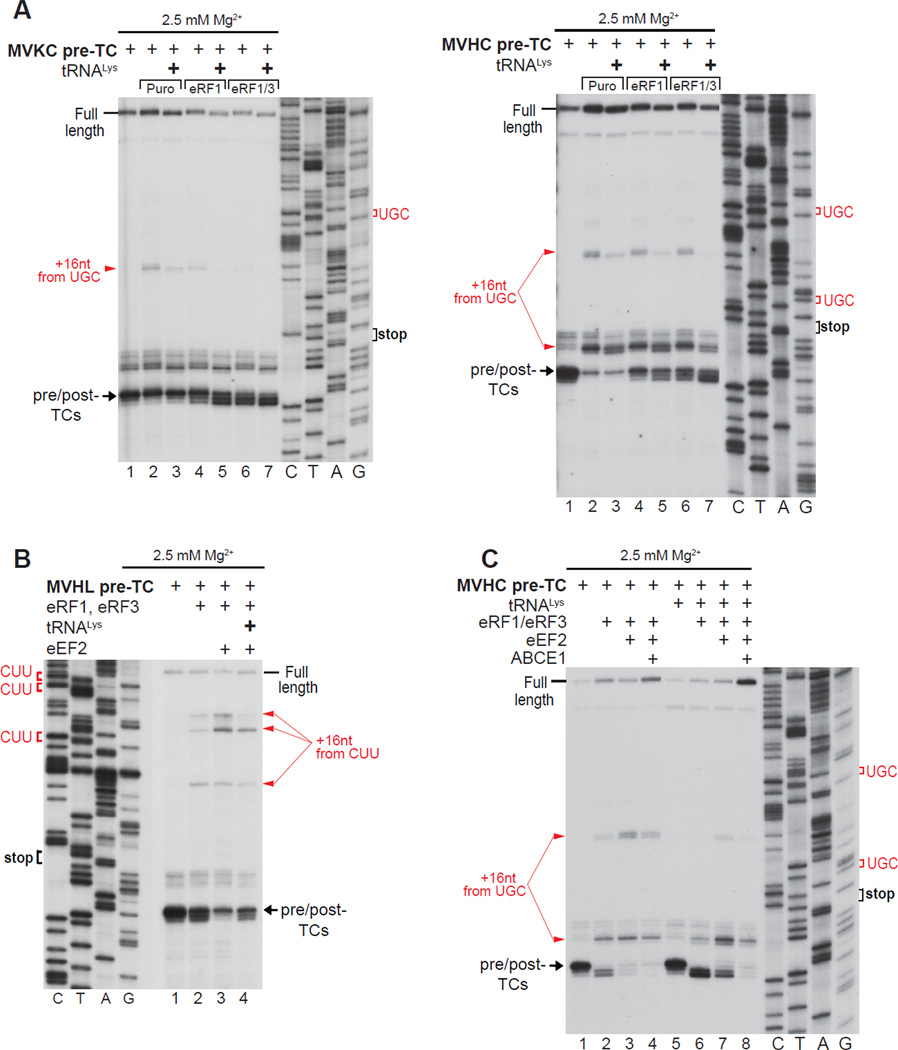
Because the instability of post-TCs would interfere with ABCE1-mediated recycling, we attempted to identify ligands that could stabilize post-TCs and their association with eRF1. SDG-purified pre-TCs lack E-site tRNA (Taylor et al., 2012) and we therefore investigated whether pre-incubation of pre-TCs with deacylated tRNA could stabilize ribosomal complexes after peptide release. tRNALys was preincubated with pre-TCs assembled on MVKC-STOP mRNA and thus containing a cognate Lys codon in the E-site, or on MVHC-STOP mRNA and therefore containing a non-cognate His codon. In both cases, preincubation reduced ribosome migration irrespective of the mode of peptide release, and stabilized association of post-TCs with eRFs, evidenced by enhancement of the toe-print shift (Fig. 4A). It also reduced the destabilizing effect of eEF2 (Fig. 4B). Other tRNAs, including tRNAiMet, had a similar effect (not shown). Notably, although pre-incubation with deacylated tRNA stabilized post-TCs, some migration still occurred when equimolar amounts of eRFs, eEF2 and ABCE1 were added simultaneously to such complexes (Fig. 4C).
Figure 4. The E-site tRNA increases the stability of post-TCs.
(A–C) Toe-printing analysis of ribosomal complexes obtained by treating pre-TCs formed on MVHL-STOP, MVHC-STOP or MVKC-STOP mRNAs with combinations of eRF1, eRF3, puromycin, eEF2 and ABCE1 in the presence/absence of tRNALys at the indicated free [Mg2+]. Black arrows show the positions of pre- and post-TCs. Red arrows indicate the positions migrated post-termination ribosomes. Upstream codons that are identical to the last codon of the encoded tetrapeptide are shown in red.
Reinitiation after ABCE1-mediated recycling
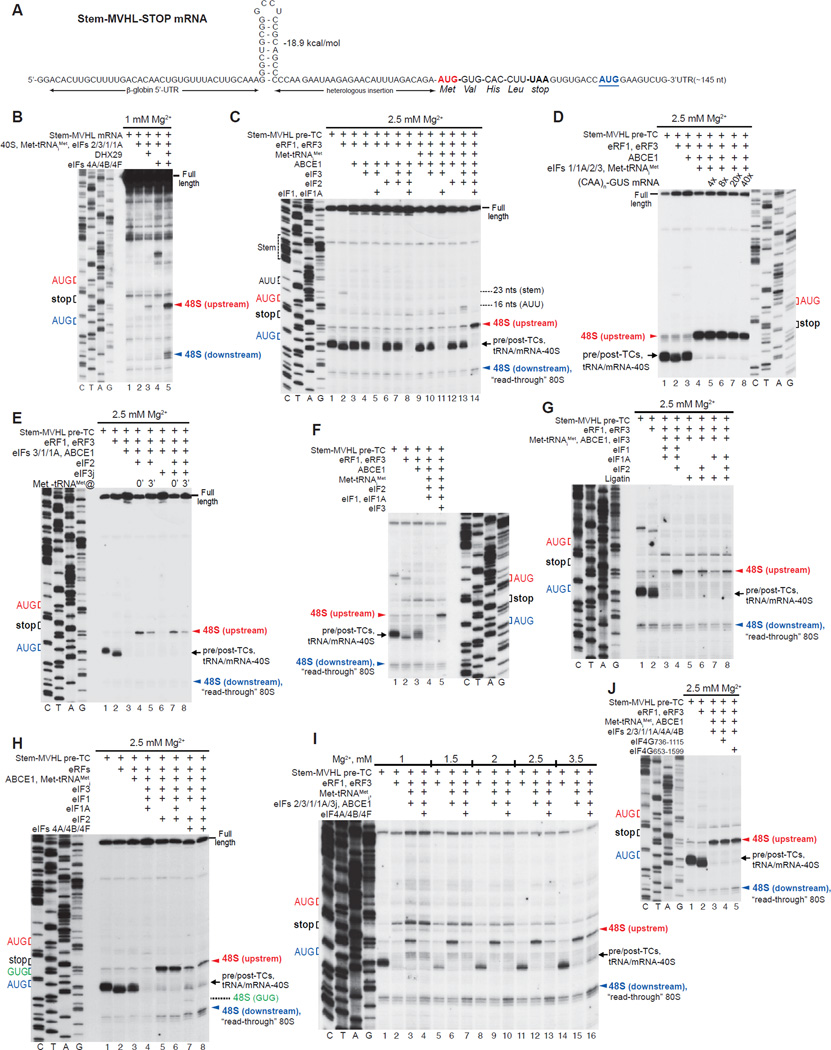
The data described above suggest that after peptide release, post-termination ribosomes should be capable of reinitiating at upstream or downstream codons that are cognate to the P-site deacylated tRNA. However, recycled 40S subunits rather than post-termination 80S ribosomes have been so far implicated in reinitiation, and we therefore investigated whether 40S subunits can remain bound to mRNA and reinitiate following ABCE1-mediated recycling. For this, we used mRNA comprising a relatively unstructured 5’-UTR interrupted by a stable stem, the MVHL ORF, a UAA stop codon, and a downstream region with a random sequence and containing an additional AUG in good context (Stem-MVHL-STOP mRNA, Fig. 5A). The stable stem imposed a strict requirement for DHX29 for initiation on this mRNA (Fig. 5B). Consequently, 48S complexes formed in recycling reactions containing eIFs but lacking DHX29 would be unambiguously attributable to reinitiation rather than to a new round of 5’-end dependent initiation.
Figure 5. Reinitiation following ABCE1-mediated recycling.
(A) Structure of Stem-MVHL-STOP mRNA. (B) Toe-printing analysis of 48S complex formation on Stem-MVHL-STOP mRNA in the presence of combinations of eIFs. (C -J) Toe-printing analysis of ribosomal complexes obtained by treating pre-TCs formed on Stem-MVHL-STOP mRNA with combinations of eRF1, eRF3, ABCE1, Met-tRNAiMet, eIFs 2/3/1/1A/3j/4A/4B/4F/4G, Ligatin and excess competitor (CAA)n-GUS mRNA at the indicated free [Mg2+]. The positions of pre-TCs, post-TCs and tRNA/mRNA-associated recycled 40S subunits are indicated by black arrows; positions of 48S complexes formed on AUG triplets upstream and downstream of the stop codon are shown in red and blue, respectively.
Pre-TCs were assembled using 40S and 60S subunits, eIFs 2/3/1/1A/4A/4B/4F/5/5B, DHX29, eEFs and aa-tRNAs. Although 48S complexes formed mostly at the AUG of the MVHL ORF, some also assembled at the downstream AUG (Fig. 5B, lane 5), and purified pre-TCs therefore contained “read-through” 80S initiation complexes formed at this codon (Fig. 5C, lane 1). Pre-TCs were then incubated with eRFs, ABCE1, Met-tRNAiMet and combinations of eIFs at 2.5 mM free Mg2+ to minimize ABCE1-independent recycling by eIFs. Incubation of pre-TCs with eRFs caused a 2nt toe-print shift and also yielded a low-intensity toe-print 23 nts downstream of the stem, likely caused by its arrest of migrating ribosomes (Fig. 5C, lane 2). ABCE1 partially eliminated the toe-print shift due to reversible dissociation of post-TCs into 60S and tRNA/mRNA-associated 40S subunits, whereas additional inclusion of eIFs 3/1/1A yielded a prominent full-length cDNA due to dissociation of tRNA and mRNA from recycled 40S subunits (Fig. 5C, lanes 3, 5). However, in the presence of eRFs, ABCE1, eIFs 3/1/1A, eIF2 and Met-tRNAiMet, very efficient 48S complex formation occurred at the upstream AUG of the MVHL ORF (Fig. 5C, lane 14). Since the reaction mixture did not contain DHX29, 48S complexes could not have formed by 5’-end dependent initiation. Moreover, their formation was not affected by a large excess of competitor (CAA)n-GUS mRNA, comprising the GUS coding region and an unstructured 5’-UTR consisting of 19 CAA repeats (Fig. 5D), and no 48S complexes assembled on the competitor itself (not shown). Thus, 48S complexes assembled on the upstream AUG in the presence of eIFs 2/3/1/1A and Met-tRNAiMet were formed by reinitiation when recycled 40S subunits remained associated with mRNA following eIF3/1/1A-mediated dissociation of deacylated tRNA and could scan in the 5’-direction and acquire eIF2-TC. Delayed addition of Met-tRNAiMet strongly reduced reinitiation (Fig. 5E, lanes 4, 5), either due to rapid dissociation of mRNA from 40S subunits or to their migration past the AUG. Reinitiation was reduced (by ~35%) but not abrogated by eIF3j (Fig. 5E, compare lanes 4, 5 with lanes 7, 8).
In the absence of eIF1/eIF1A, tRNALeu remained bound to 40S subunits, so that only the small proportion, from which tRNALeu had spontaneously dissociated, could participate in 48S complex formation (Fig. 5C, lane 13). As a result, only low-level 48S complex assembly occurred at the upstream AUG of the MVHL ORF. Interestingly, some complexes also formed at the further upstream AUU, possibly due to slower binding of eIF2-TC to migrating 40S subunits with a vacant P-site, or to delayed dissociation of tRNALeu from migrating 40S subunits, if like post-termination 80S ribosomes, recycled 40S subunits containing tRNALeu can migrate along mRNA. In either case, a proportion of recycled 40S subunits would acquire eIF2-TC only after scanning past the proximal AUG. Whereas low-level reinitiation could occur in the absence of eIF1/eIF1A, eIF3 was essential (Fig. 5C, lane 12). Importantly, although at 2.5 mM free Mg2+, eIF1 and eIF1A did not efficiently dissociate tRNA from recycled 40S subunits in the absence of eIF3 (Fig. 5C, lane 12, Pisarev et al., 2010), they could do so at 1 mM free [Mg2+] (Fig. 5F, lane 4). However, even at 1 mM Mg2+, eIF3 was required for reinitiation (Fig. 5F, lanes 4, 5). Ligatin could replace eIF1/eIF1A in promoting dissociation of tRNALeu from 40S subunits, and medium-level eIF2-mediated 48S complex formation at the upstream AUG occurred in its presence (Fig. 5G, lanes 6, 8). However, it could not substitute for eIF2 in promoting reinitiation (Fig. 5G, lanes 5, 7).
Although recycled 40S subunits were capable of efficient reinitiation upstream, only trace amounts of 48S complex formed at the downstream AUG in the presence of eIFs 2/3/1/1A and Met-tRNAiMet (Fig. 5C, lane 14). eIFs 4A/4B/4F strongly reduced upstream reinitiation and promoted 48S complex formation at the downstream AUG over a wide range of [Mg2+] (Figs. 5H, lane 8; 5I). In eIF1A’s absence, 48S complexes also formed at the GUG preceding the downstream AUG (Fig. 5H, lane 7), providing further support for reinitiation resulting from linear scanning from the stop codon. Surprisingly, eIFs 4A, 4B and eIF4G736–1115 were almost inactive in imposing 3’ directionality on reinitiation, and the activity of eIF4A, eIF4B and eIF4G653–1599 was low (Fig. 5J).
Reinitiation after ribosome recycling mediated by eIFs
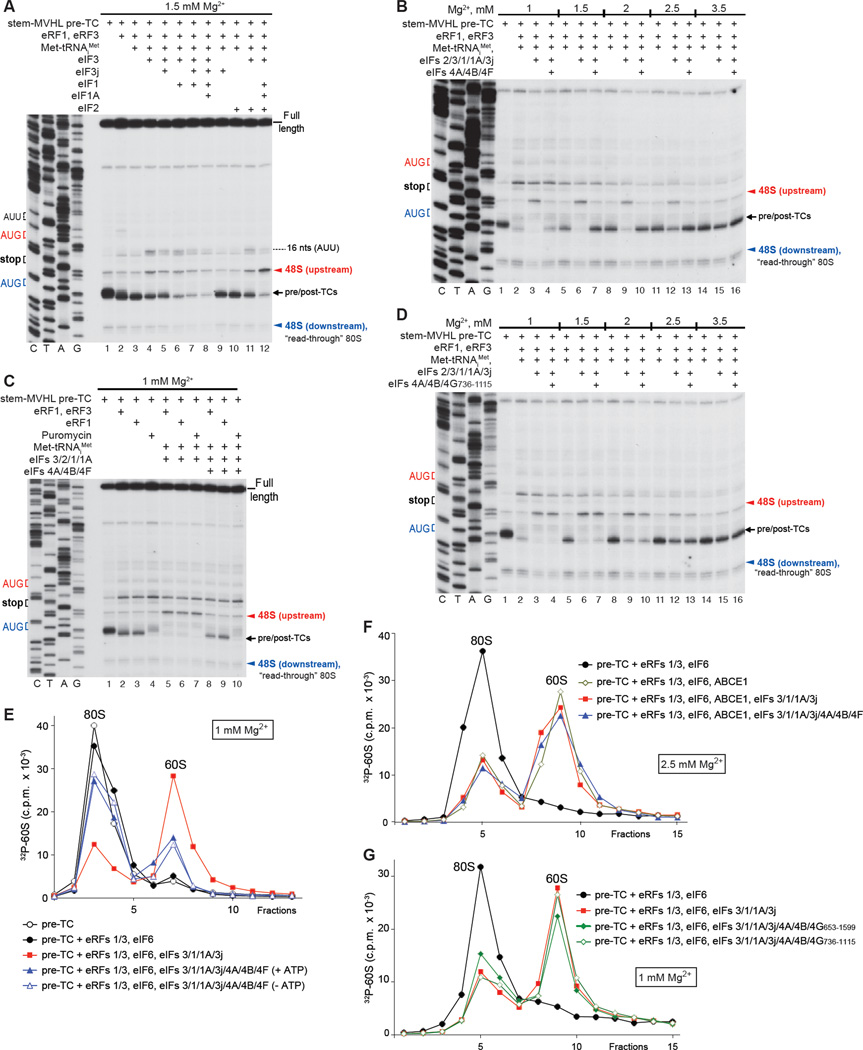
We next investigated reinitiation after ABCE1-independent recycling by eIFs at lower [Mg2+]. For this, pre-TCs assembled on Stem-MVHL-STOP mRNA were incubated with eRFs, Met-tRNAiMet and combinations of eIFs at 1.5 mM free Mg2+. Surprisingly, eIF3 alone not only induced dissociation of post-TCs, but also promoted low-level reinitiation at upstream AUG and AUU triplets (Fig. 6A, lane 4). The inability of eIF3 alone to promote reinitiation during ABCE1-mediated recycling (Fig. 5C) could be due to inefficient spontaneous dissociation of tRNALeu from recycled 40S subunits at higher [Mg2+]. Consistent with a previous report (Pisarev et al., 2007a), combinations of eIF3j, eIF1 and eIF1A increased recycling efficiency, but concomitantly also diminished eIF3-mediated reinitiation, in the ordereIF3j<eIF1<eIF1/3j<eIF1/eIF1A/eIF3j (Fig. 6A, lanes 5–8). The negative influence of eIF3j could be due to its activity in dissociating mRNA in the absence of P-site tRNA (e.g. Unbehaun et al., 2004), which would reduce the amount of mRNA-associated 40S subunits that are capable of accepting Met-tRNAiMet into the vacant P-site, whereas eIF1/eIF1A could promote dissociation of the P site Met-tRNAiMet when it is not stabilized by eIF2 (Lomakin et al., 2006). Inclusion of eIF2 with eIFs 3/1/1A led to both efficient recycling and reinitiation at the AUG of the MVHL ORF (Fig. 6A, lane 12).
Figure 6. Reinitiation following eIFs-mediated recycling.
(A–D) Toe-printing analysis of ribosomal complexes obtained by treating pre-TCs formed on Stem-MVHL-STOP mRNA with combinations of eRF1, eRF3, puromycin, ABCE1, Met-tRNAiMet and eIFs 2/3/1/1A/3j/4A/4B/4F/4G at the indicated free [Mg2+]. The positions of pre- and post-TCs are indicated by black arrows; positions of 48S complexes formed at the AUG triplets upstream and downstream of the stop codon are shown in red and blue, respectively. (E-G) Dissociation of pre-TCs, assembled on Stem-MVHL-STOP mRNA with [32P]60S subunits after incubation with combinations of eRFs, ABCE1 and eIFs at the indicated [Mg2+], assayed by SDG centrifugation. Upper fractions were omitted for clarity.
However, when reaction mixtures were supplemented with eIFs 4A/4B/4F, instead of imposing 3’ directionality, as in the case of ABCE1-mediated recycling (Fig. 5I), these factors abrogated recycling at all tested [Mg2+] (Fig. 6B). This inhibitory effect did not depend on the mode of peptide release (Fig. 6C), and did not require eIF4B (not shown). eIFs 4A/4B/4G736–1115 did not inhibit recycling, but also did not impose 3’ directionality on reinitiation (Fig. 6D).
To validate toe-printing data, the influence of eIFs 4A/4B/4F on recycling was assayed by SDG centrifugation using pre-TCs assembled with [32P]60S subunits. eIFs 4A/4B/4F strongly inhibited splitting of post-TCs by eIFs 3/1/1A, and their activity did not require ATP (Fig. 6E). In a control reaction, eIFs 4A/4B/4F did not affect ribosomal splitting by ABCE1 (Fig. 6F). eIFs 4A/4B/4G736–1115 did not influence splitting of post-TCs by eIFs 3/1/1A, and inhibition by eIFs 4A/4B/4G653–1599 was weak (Fig. 6G).
Reinitiation on mRNA with a single-stranded region downstream of the stop codon
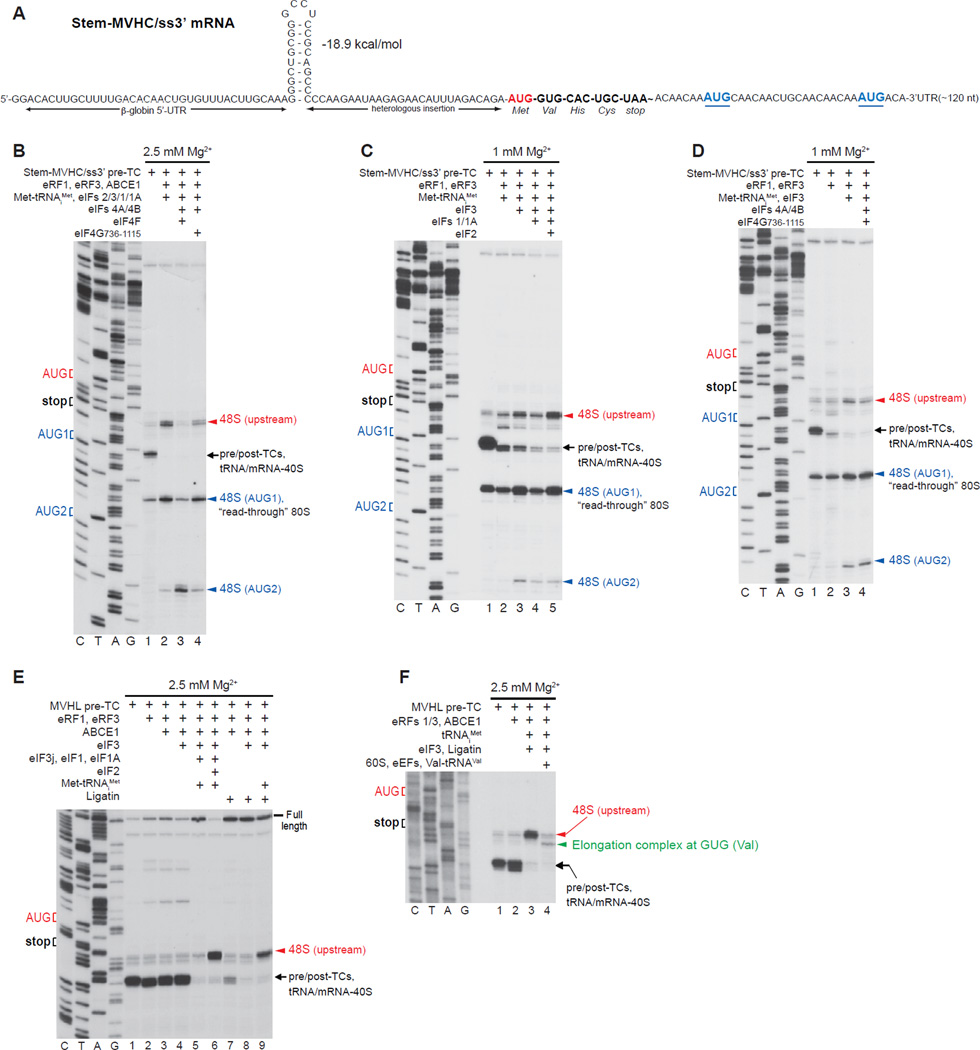
To investigate whether recycled 40S subunits are capable of 3’-directional reinitiation independently of eIFs 4A/4B/4F if the region downstream of the stop codon is unstructured, we used pre-TCs assembled on Stem-MVHC/ss3’ mRNA comprising a 5’-UTR identical to that of Stem-MVHL-STOP mRNA, the MVHC ORF, a UAA stop codon, and an unstructured downstream region with two additional AUGs (Fig. 7A). As on Stem-MVHL-STOP mRNA, some 48S complexes also assembled at the first downstream AUG, and purified pre-TCs therefore contained “read-through” 80S complexes formed on this triplet (Fig. 7B, lane 1).
Figure 7. Reinitiation on mRNA with an unstructured region downstream of the stop codon.
(A) Structure of Stem-MVHC/ss3’ mRNA. (B-D) Toe-printing analysis of ribosomal complexes obtained by treating pre-TCs formed on Stem-MVHC/ss3’ mRNA with combinations of eRF1, eRF3, ABCE1, Met-tRNAiMet and eIFs 2/3/1/1A/4A/4B/4F/4G at the indicated free [Mg2+]. The positions of pre-TCs, post-TCs and tRNA/mRNA-associated recycled 40S subunits are indicated by black arrows; positions of 48S complexes formed at the AUG triplets upstream and downstream of the stop codon are shown in red and blue, respectively. (E, F) Toe-printing analysis of ribosomal complexes obtained by treating pre-TCs formed on MVHL-STOP mRNA with different combinations of eRF1, eRF3, ABCE1, Met-tRNAiMet, tRNAiMet, eIFs 2/3/1/1A, Ligatin, eEFs, 60S subunits and Val-tRNAVal at the indicated free [Mg2+]. The positions of pre-TCs, post-TCs and tRNA/mRNA-associated recycled 40S subunits are indicated by black arrows; positions of 48S complexes formed at the upstream AUG triplet and to elongation complexes formed at the GUG codon are shown in red and green, respectively.
When ABCE1-mediated splitting of post-TCs assembled on Stem-MVHC/ss3’ mRNA was done in the presence of Met-tRNAiMet and eIFs 2/3/1/1A, 40S subunits were able to scan bidirectionally and form 48S complexes at the first and even second downstream AUGs (Fig. 7B, lane 2). eIFs 4A/4B/4F strongly reduced reinitiation at the upstream and first downstream AUGs, leading to 48S complex formation predominantly at the second downstream AUG (Fig. 7B, lane 3), which could be due to rapid migration of 40S subunits past the first downstream AUG before acquiring eIF2-TC. As on Stem-MVHL-STOP mRNA (Fig. 5J), eIFs 4A/4B/4G736–1115 did not affect reinitiation (Fig. 7B, lane 4).
When ribosome recycling was induced by eIFs at 1 mM Mg2+, efficient downstream reinitiation occurred even in the presence of eIF3 alone (Fig. 7C, lane 3). eIF3-mediated reinitiation on Stem-MVHC/ss3’mRNA was more efficient than on Stem-MHVL-STOP mRNA (compare Figs. 7C, 6A), likely due to stronger spontaneous dissociation of tRNACys. Reinitiation mediated by eIF3 alone was reduced by eIFs 1/1A (Fig. 7C, lane 4) but was not influenced by eIFs 4A/4B/4G736–1115 (Fig. 7D). Efficient downstream reinitiation also occurred in the presence of eIFs 2/3/1/1A (Fig. 7C, lane 5).
Ligatin-mediated reinitiation
Although Ligatin could not replace eIF2 during reinitiation on Stem-MVHL-STOP mRNA (Fig. 5G), eIF3 and Ligatin together promoted reinitiation at the upstream AUG after ABCE1-mediated splitting of post-TCs formed on MVHL-STOP mRNA (Fig. 7E, lane 9). This might be due to potential differences in the stability of association of MVHL-STOP and Stem-MVHL-STOP mRNAs with 40S subunits after Ligatin-induced release of tRNALeu. 48S complexes assembled with eIF3 and Ligatin joined 60S subunits directly to form 80S ribosomes that in the presence of eEFs and Val-tRNAVal underwent one cycle of elongation (Fig. 7F).
DISCUSSION
Instability of mammalian post-TCs and the possibility of reinitiation by post-termination 80S ribosomes
We found that post-termination ribosomes are not stably anchored on mRNA, and can readily migrate to nearby upstream and downstream codons that are cognate to the P-site deacylated tRNA if the mRNA regions flanking the stop codon are unstructured (Fig. 1). The efficiency of migration depends on the mode of peptide release (puromycin>eRF1>eRF1/eRF3) and the nature of the P-site tRNA (e.g. tRNACys >tRNALeu). eEF2 strongly promotes migration and also destabilizes association of post-termination ribosomes with eRF1 (Fig. 3A–C), whereas E-site tRNA and elevated [Mg2+] both reduce migration and stabilize ribosomal binding of eRF1 (Fig. 4).
Similar mRNA slippage and re-pairing of the P-site tRNA with a nearby upstream codon were observed in bacterial ribosomal complexes with deacylated tRNA in the P site and a vacant A site, and were attributed to destabilization of P-site codon-anticodon base-pairing due to adoption by the P-site tRNA of the P/E hybrid state (McGarry et al., 2005), in which its CCA-end and acceptor stem move to the E site of the large subunit (e.g. Dunkle et al., 2011). The transition of tRNA to the P/E state in bacterial ribosomal complexes also involves a 6Å lateral movement of its anticodon stem loop (ASL) and mRNA relative to the large subunit, coupled with movement of the platform of the small subunit, loss of the ASL’s contact with H69, and conformational changes in the tRNA that include kinking between the ASL and the D stem (Dunkle et al., 2011). Deacylated tRNA can fluctuate between P/P and P/E states, and formation of the hybrid state is loosely coupled with spontaneous intersubunit rotation that switches the ribosome to the ‘unlocked’ conformation, which is essential for translocation (e.g. Valle et al., 2003; Cornish et al, 2008). Eukaryotic ribosomal complexes are even more likely to spontaneously adopt the hybrid state than equivalent bacterial complexes (Munro et al., 2010; Budkevich et al., 2011), and individual eukaryotic 80S ribosomes also favor a rotated conformation (Ben-Shem et al., 2010).
During elongation, the decreased stability of the codon-anticodon interaction in the P-site of pretranslocation complexes is likely compensated by base-pairing of the A site codon with the anticodon of peptidyl-tRNA, which allows the open reading frame to be maintained. eRF1 appears to play a similar stabilizing role in post-TCs (Figs. 1B–D), where it directly interacts with the stop codon and likely promotes the nonrotated conformation of the ribosome, which is also required for binding of ABCE1 (Becker et al., 2012). Stabilization of post-TCs by E site tRNA (Fig. 4) is consistent with its inhibition of adoption of the P/E hybrid state (e.g. Budkevich et al., 2011). The observation that mRNA slippage occurred less frequently on MVKC-STOP than MVHC-STOP mRNA (Fig. 1F) suggests that the sequence of mRNA in the mRNA-binding channel, at least in the E-site, also affects the stability of post-TCs, possibly due to specific interactions with the 40S subunit, which may impair ribosome migration or antagonize conformational changes that are required for adoption of the hybrid state. The observed dependence of the stability of post- TCs on the nature of the P site deacylated tRNA (Figs. 1B–C) is consistent with the reported variability in affinities of different tRNAs to the P/E hybrid state (Shoji et al., 2009).
Binding of eEF2 or its prokaryotic analogue EF-G to ribosomes with a vacant A site and deacylated tRNA in the P site strongly promotes the P/E hybrid state and the rotated conformation independently of GTP hydrolysis (e.g. Valle et al., 2003; Taylor et al., 2007). Facilitation by eEF2 of migration of post-termination ribosomes (Fig. 3A–C) is thus consistent with mRNA slippage being induced by the P/E hybrid state. Dissociation of eRF1 by eEF2 in the presence of GTP, GDPNP or GDP could be explained by intersubunit rotation, which is incompatible with binding of eRF1. This activity of eEF2 parallels the release of RF1 from bacterial ribosomes by RF3, which also promotes intersubunit rotation that is not compatible with binding of RF1 (Gao et al., 2007).
Given that ribosomes containing P-site deacylated tRNA can accept aa-tRNA in the A-site and then undergo translocation, albeit at a substantially reduced rate (Semenkov et al., 2000), migrated post-termination ribosomes should be capable of reinitiating translation from upstream or downstream codons that are cognate to P-site deacylated tRNA, or from nearby AUG and possibly other codons if a corresponding exchange of deacylated P site tRNA had occurred (Fig. 2). Post-termination migration and binding of aa-tRNA to the A site followed by translocation could thus plausibly account for the resumption of translation from multiple locations on the same mRNA following puromycin treatment of polysomes in rabbit reticulocyte lysate (RRL) (Williamson and Schweet, 1964). But can migration of post-termination ribosomes occur in vivo? In prokaryotes, the E site tRNA can dissociate during elongation spontaneously and independently of binding of aa-tRNA to the A site (Uemura et al., 2010). Thus, it is reasonable to assume that at least in some ribosomal complexes, dissociation of the E site tRNA precedes binding of eRFs, yielding post-termination ribosomes with a vacant E site, which are prone to migration. On the other hand, the observation that ABCE1 stimulates peptide release by eRF1 (Shoemaker and Green, 2011) suggests that it can bind to ribosomal complexes before this step so that recycling would follow termination immediately, eliminating the possibility of migration. However, as eRF1 is twice as abundant as ABCE1 in yeast (Ghaemmaghami et al., 2003), some termination events would likely occur without ABCE1, leading to a delay between peptide release and recycling. Moreover, conditions such as oxidative stress that reduce the level of active ABCE1 (Alhebshi et al., 2012) would impair recycling, potentially favoring ribosome migration, and in turn, reinitiation. Notably, bidirectional post-termination migration of bacterial ribosomes can lead to reinitiation (Adhin and van Duin, 1990) and is enhanced by mutations in the ribosome recycling factor that impair dissociation of post-TCs (Janosi et al., 1998).
The ability of post-termination ribosomes to reinitiate translation from non-AUG codons could account for some aspects of termination-reinitiation on dicistronic viral mRNAs with overlapping stop/start codons, such as efficient initiation at nearby non-AUG codons when the native start codon is mutated (e.g. Luttermann and Meyers, 2007). This mechanism might also promote some aberrant initiation events, such as those that lead to translation of cryptic ORFs lacking AUG codons in the 3’-UTR to yield peptides that are presented by MHC class I molecules for immune surveillance (Schwab et al., 2003). More speculatively, it could permit ribosomal access to the 3’UTR or to alternative reading frames overlapping the stop codon, leading to translation of coding sequences that could ultimately evolve into novel genes (e.g. Carvunis et al., 2012).
Except in the few instances when reinitiation is programmed, post-termination ribosome migration would generally reduce translational fidelity by promoting aberrant initiation events, and mRNA sequences have likely evolved to minimize its frequency. Reinitiation would be reduced if there were biases for the last codon to correspond to tRNAs that have a higher affinity to P/P over P/E states, for mRNA regions flanking the initiation codon to establish stabilizing contacts with the 40S subunit, and for increased secondary structure around the stop codon to minimize ribosome migration. Consistently, there is an increase in GC content downstream of stop codons in mammalian mRNAs that could function to suppress migration of post-termination ribosomes (Shabalina et al, 2004). However, whether sequence bias immediately upstream of the stop codon (e.g. Cridge et al., 2006) also reflects the requirement for stabilization of post-TCs remains to be determined.
Reinitiation following recycling of post-TCs
We found that when ABCE1-dependent or independent splitting of post-TCs proceeds in the presence of Met-tRNAiMet and eIFs 2/3/1/1A, 40S subunits remain bound to mRNA and efficiently reinitiate at nearby upstream and downstream AUGs, if the region flanking the stop codon is unstructured (Figs. 5C–F, 6A, 7B–C). Although eIF1/eIF1A are required for dissociation of tRNA from recycled 40S subunits (Pisarev et al., 2007a), low level reinitiation can occur in their absence (Figs. 5C, 6A), particularly when spontaneous dissociation of deacylated tRNA is strong, e.g. at low [Mg2+] or on MVHC-STOP mRNA. eIF3, on the other hand, is essential and likely plays a key role in ensuring retention of mRNA on recycled 40S subunits (Kolupaeva et al. 2005; Unbehaun et al., 2004) and promoting scanning (e.g. Szamecz et al., 2008). Interestingly, eIF3-associated recycled 40S complexes can acquire initiator tRNA and form 48S complexes at nearby AUGs independently of eIF2, albeit with low efficiency (Fig. 6A). Such complexes are, however, susceptible to dissociation by eIF1/eIF1A (Lomakin et al., 2006).
Although eIFs 2/3/1/1A can promote reinitiation, they cannot impose 3’-directionality on scanning recycled 40S subunits, and formation of 48S complexes at either upstream or downstream AUGs is determined by the lack of secondary structure in the corresponding region between the AUG and the stop codon. 3’-directionality of reinitiation is promoted by the eIF4 group of factors. However, in contrast to eIF4F, eIF4A and the central domain of eIF4G (eIF4G736–1115) did not have this activity (Figs. 5H–J), even though eIF4G736–1115 is active during 5’-dependent initiation (Pestova and Kolupaeva, 2002). eIF4G736–1115 contains the eIF3-binding site and one of eIF4G’s two eIF4A-binding sites, but lacks an upstream RNA-binding region (~ a.a. 681–720) that is required for eIF4G’s full activity in 5’-end-dependent scanning (Prévôt et al., 2003). The analogous RNA-binding domain in yeast eIF4G is required to ensure the 5’-3’ directionality of RNA unwinding by eIF4F (Rajagopal et al., 2012). Thus, the lack of activity of eIF4G736–1115 during reinitiation could be due to its inability to establish a functional interaction with mRNA that is already accommodated in the mRNA-binding cleft. Notably, the N-terminally extended eIF4G682–1115, containing the upstream RNA-binding region, was able to support reinitiation after a short ORF in RRL (Pöyry et al., 2004).
Another unexpected effect of the eIF4 group of factors was that eIF4F (but not eIF4G736–1115) strongly inhibited eIF3’s activity in splitting post-TCs in the absence of ABCE1 (Figs. 6B, C, E). The location of eIF4F on eIF3-bound 40S subunits is not known, and thus whether it causes steric hindrance that impairs binding of eIF3 to 40S subunits that are part of post-TCs, or acts indirectly by inducing conformational changes in eIF3 is not yet apparent. We were not able to generate recombinant full-length eIF4G, and it therefore also remains to be determined whether eIF4E is required for eIF4F’s inhibitory activity.
A striking result is that reinitiation in the in vitro reconstituted system was very efficient, so that almost all recycled 40S subunits retained mRNA, acquired eIF2-TC, scanned and formed 48S complexes at nearby AUGs. Although pre-TCs used in our studies were formed after translation of short ORFs, they had been purified by SDG centrifugation that strips off eIFs, which might have remained on 40S subunits during initial cycles of elongation, and therefore resemble pre-TCs formed in vivo after translation of long ORFs, which do not support efficient reinitiation. A potential difference in reinitiation in vivo after short and long ORFs is that the former would proceed with retained eIF3 that is no longer associated with eIF3j (Unbehaun et al., 2004), whereas the latter could be promoted by eIF3 that has been able to rebind eIF3j. However, inclusion of eIF3j only moderately decreased reinitiation in the in vitro reconstituted system (Fig. 5E), which could not account for the significant difference in reinitiation after short and long ORFs observed in vivo.
It has been suggested that efficient reinitiation in vivo requires the continuous presence of eIF3/eIF4G on the 40S subunit throughout elongation, termination and recycling (Pöyry et al., 2004). In the reconstituted system, eIF3 and eIF4F were also critical for efficient reinitiation at downstream AUGs, but could obviously be acquired de novo. This raises the question of why this does not occur in vivo after translation of long ORFs. Although it cannot be strictly excluded that cells contain an as-yet unidentified factor specifically dedicated to promoting tRNA/mRNA release from recycled 40S subunits if the latter are not associated with eIF3 (i.e. after translation of long ORFs), this would imply an additional mechanism to dissociate it from 40S subunits to prepare them for the next round of initiation. The existence of such a factor would strongly complicate recycling, and it would therefore be more logical to assume that the difference in reinitiation after long ORFs in vivo and reinitiation in the in vitro reconstituted system is determined by differences in the relative levels of free individual eIFs that are available for reinitiation. Thus, if the in vivo concentrations of ribosome/mRNA-unbound eIF3/eIF4F are low compared to those of eIF1/eIF1A, Ligatin and MCT1/DENR, tRNA would be released from recycled 40S subunits that are not yet bound to eIF3, leading to prompt dissociation of mRNA due to the absence of eIF3’s stabilizing influence. Moreover, some deacylated tRNAs that are particularly prone to spontaneous dissociation (e.g. tRNACys) would not even require these factors, and splitting of post-TCs will be followed by quick tRNA/mRNA release.
Our data suggest that continued association of eIF3 with 40S subunits in vivo following translation of short ORFs should be sufficient to promote a reasonable level of downstream reinitiation if mRNA flanking the stop codon is unstructured. The suggested requirement for continued association of eIF4F or eIF4G with 40S subunits (Pöyry et al., 2004) could be accounted for by the necessity to unwind downstream mRNA secondary structure or by eIF4G’s stabilizing effect on ribosomal association of eIF3 to ensure its retention during translation of short ORFs.
EXPERIMENTAL PROCEDURES
Plasmid construction, purification of factors and ribosomal subunits, phosphorylation of 60S subunits, and aminoacylation of tRNA are described in Supplemental Information, which also contains detailed protocols for all experimental procedures.
Assembly of ribosomal complexes
Pre-TCs were assembled and purified by SDG centrifugation (Alkalaeva et al., 2006).
Toe-printing analysis of ribosomal complexes
To investigate the stability of post-termination 80S ribosomes, 3.75 nM SDG-purified pre-TCs were incubated for 10 min at 37°C with 1.25 µM eRF1 alone, 100 nM eRF1 together with 100 nM eRF3, 100 nM eRF1(AGQ) together with 100 nM eRF3, or 1 mM puromycin in the presence/absence of 75 nM eIF3, 250 nM eIF1, 250 nM eIF1A, 100 nM eEF2, 100 nM ABCE1, 75 nM tRNAiMet, 75 nM tRNAHis or 75 nM tRNALys in 40 µl buffer A (20 mM Tris, pH 7.5, 100 mM KCl, 0.25 mM spermidine, 2 mM DTT) supplemented with 0.5 mM GTP and 0.5 mM ATP, or with 0.4 mM GTP, GDP, GDPNP or ATP and corresponding amounts of MgCl2 to achieve the free [Mg2+] indicated on each panel. The resulting ribosomal complexes were analyzed by primer extension (Pisarev et al., 2010). For analysis by RelE, ribosomal complexes were incubated with 40 pmol RelE for 10 min at 37°C. mRNA was then phenol-extracted and subjected to primer extension.
To investigate reinitiation by recycled 40S subunits, 3.75 nM SDG-purified pre-TCs were incubated for 10 min at 37°C with 100 nM eRF1 together with 100 nM eRF3, 1.25 µM eRF1 alone, or 1 mM puromycin in the presence of different combinations of 150 nM ABCE1, 100 nM eIF2, 75 nM eIF3, 250 nM eIF1, 250 nM eIF1A, 375 nM eIF4A, 100 nM eIF4B, 250 nM eIF4F, 250 nM eIF4G736-1115, 250 nM eIF4G653-1599, 250 nM eIF3j, 125 nM Ligatin, 125 nM in vitro transcribed Met-tRNAiMet, 15–150 nM (CAA)n-GUS mRNA, 50 nM 60S subunits, 100 nM eEF1H, 250 nM eEF2 and 125 nM Val-tRNAVal in 40 µl buffer A supplemented with 0.2 mM GTP and 0.2 mM ATP and corresponding amounts of MgCl2 to achieve the free [Mg2+] indicated on each panel. Resulting ribosomal complexes were analyzed by primer extension.
Dissociation of post-termination 80S ribosomes into subunits analyzed by SDG centrifugation
4.2 nM SDG-purified pre-TCs assembled on Stem-MVHL-STOP mRNA with [32P]60S subunits were incubated for 10 minutes at 37°C with 125 nM eRF1, 125 nM eRF3, 83 nM eIF6 and indicated combinations of 117 nM ABCE1, 58 nM eIF3, 208 nM eIF1, 208 nM eIF1A, 167 nM eIF3j, 167 nM eIF4A, 83 nM eIF4B, 125 nM eIF4F, 125 nM eIF4G736-1115 and 125 nM eIF4G653-1599 in 120 µl buffer A supplemented with 0.4 mM ATP, 0.4 mM GTP and corresponding amounts of MgCl2 to achieve the free [Mg2+] indicated on each panel. Resulting ribosomal complexes were analysed by centrifugation through 10–30% SDGs followed by Cerenkov counting (Pisarev et al., 2007a).
Supplementary Material
Highlights.
eIFs 2/3/1/1A promote bidirectional reinitiation by recycled 40S subunits
Imposition of 3’ -directionality on reinitiation additionally requires eIF4F
Post-termination 80S ribosomes are mobile and can migrate to cognate codons
eEF2 induces dissociation of eRF1 and promotes 80S ribosomal migration
ACKNOWLEDGMENTS
We thank C. Neubauer and V. Ramakrishnan for their gift of RelE, L. Yu. Frolova for release factor expression vectors, and R. Jackson for many insightful discussions during the course of this study. This work was supported by NIH Grants GM59660 and GM80623 and HFSP Grant RGP0062/2012 to TVP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adhin MR, van Duin J. Scanning model for translational reinitiation in eubacteria. J. Mol. Biol. 1990;213:811–818. doi: 10.1016/S0022-2836(05)80265-7. [DOI] [PubMed] [Google Scholar]
- Alhebshi A, Sideri TC, Holland SL, Avery SV. The essential iron-sulfur protein Rli1 is an important target accounting for inhibition of cell growth by reactive oxygen species. Mol. Biol. Cell. 2012;23:3582–3590. doi: 10.1091/mbc.E12-05-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- Becker T, Franckenberg S, Wickles S, Shoemaker CJ, Anger AM, Armache JP, Sieber H, Ungewickell C, Berninghausen O, Daberkow I, et al. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature. 2012;482:501–506. doi: 10.1038/nature10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- Budkevich T, Giesebrecht J, Altman RB, Munro JB, Mielke T, Nierhaus KH, Blanchard SC, Spahn CM. Structure and dynamics of the mammalian ribosomal pretranslocation complex. Mol. Cell. 2011;44:214–224. doi: 10.1016/j.molcel.2011.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl. Acad. Sci. USA. 2009;106:7507–7512. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvunis AR, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, Simonis N, Charloteaux B, Hidalgo CA, Barbette J, Santhanam B, et al. Proto-genes and de novo gene birth. Nature. 2012;487:370–374. doi: 10.1038/nature11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol. Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cridge AG, Major LL, Mahagaonkar AA, Poole ES, Isaksson LA, Tate WP. Comparison of characteristics and function of translation termination signals between and within prokaryotic and eukaryotic organisms. Nucleic Acids Res. 2006;34:1959–1973. doi: 10.1093/nar/gkl074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JH. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332:981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova LY, Tsivkovskii RY, Sivolobova GF, Oparina NY, Serpinsky OI, Blinov VM, Tatkov SI, Kisselev LL. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Zhou Z, Rawat U, Huang C, Bouakaz L, Wang C, Cheng Z, Liu Y, Zavialov A, Gursky R, et al. RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors. Cell. 2007;129:929–941. doi: 10.1016/j.cell.2007.03.050. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Grant CM, Hinnebusch AG. Effect of sequence context at stop codons on efficiency of reinitiation in GCN4 translational control. Mol. Cell. Biol. 1994;14:606–618. doi: 10.1128/mcb.14.1.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem. Sci. 2006;31:553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell. Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. Termination and post-termination events in eukaryotic translation. Adv. Protein Chem. Struct. Biol. 2012;86:45–93. doi: 10.1016/B978-0-12-386497-0.00002-5. [DOI] [PubMed] [Google Scholar]
- Janosi L, Mottagui-Tabar S, Isaksson LA, Sekine Y, Ohtsubo E, Zhang S, Goon S, Nelken S, Shuda M, Kaji A. Evidence for in vivo ribosome recycling, the fourth step in protein biosynthesis. EMBO J. 1998;17:1141–1151. doi: 10.1093/emboj/17.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva VG, Unbehaun A, Lomakin IB, Hellen CU, Pestova TV. Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA. 2005;11:470–486. doi: 10.1261/rna.7215305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol. Cell. Biol. 1987;7:3438–3445. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Constraints on reinitiation of translation in mammals. Nucleic Acids Res. 2001;29:5226–5232. doi: 10.1093/nar/29.24.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawless C, Pearson RD, Selley JN, Smirnova JB, Grant CM, Ashe MP, Pavitt GD, Hubbard SJ. Upstream sequence elements direct post-transcriptional regulation of gene expression under stress conditions in yeast. BMC Genomics. 2009;10:7. doi: 10.1186/1471-2164-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Shirokikh NE, Yusupov MM, Hellen CU, Pestova TV. The fidelity of translation initiation: reciprocal activities of eIF1, IF3 and YciH. EMBO J. 2006;25:196–210. doi: 10.1038/sj.emboj.7600904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttermann C, Meyers G. A bipartite sequence motif induces translation reinitiation in feline calicivirus RNA. J. Biol. Chem. 2007;282:7056–7065. doi: 10.1074/jbc.M608948200. [DOI] [PubMed] [Google Scholar]
- McGarry KG, Walker SE, Wang H, Fredrick K. Destabilization of the P site codon-anticodon helix results from movement of tRNA into the P/E hybrid state within the ribosome. Mol. Cell. 2005;20:613–622. doi: 10.1016/j.molcel.2005.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Altman RB, Tung CS, Sanbonmatsu KY, Blanchard SC. A fast dynamic mode of the EF-G-bound ribosome. EMBO J. 2010;29:770–781. doi: 10.1038/emboj.2009.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer C, Gao YG, Andersen KR, Dunham CM, Kelley AC, Hentschel J, Gerdes K, Ramakrishnan V, Brodersen DE. The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell. 2009;139:1084–1095. doi: 10.1016/j.cell.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Hellen CU, Pestova TV. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007a;131:286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CU, Pestova TV. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol. Cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöyry TA, Kaminski A, Jackson RJ. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes Dev. 2004;18:62–75. doi: 10.1101/gad.276504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöyry TA, Kaminski A, Connell EJ, Fraser CS, Jackson RJ. The mechanism of an exceptional case of reinitiation after translation of a long ORF reveals why such events do not generally occur in mammalian mRNA translation. Genes Dev. 2007;21:3149–3162. doi: 10.1101/gad.439507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévôt D, Décimo D, Herbreteau CH, Roux F, Garin J, Darlix JL, Ohlmann T. Characterization of a novel RNA-binding region of eIF4GI critical for ribosomal scanning. EMBO J. 2003;22:1909–1921. doi: 10.1093/emboj/cdg175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal V, Park EH, Hinnebusch AG, Lorsch JR. Specific domains in yeast translation initiation factor eIF4G strongly bias RNA unwinding activity of the eIF4F complex toward duplexes with 5'-overhangs. J. Biol. Chem. 2012;287:20301–20312. doi: 10.1074/jbc.M112.347278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab SR, Li KC, Kang C, Shastri N. Constitutive display of cryptic translation products by MHC class I molecules. Science. 2003;301:1367–1371. doi: 10.1126/science.1085650. [DOI] [PubMed] [Google Scholar]
- Semenkov YP, Rodnina MV, Wintermeyer W. Energetic contribution of tRNA hybrid state formation to translocation catalysis on the ribosome. Nat. Struct. Biol. 2000;7:1027–1031. doi: 10.1038/80938. [DOI] [PubMed] [Google Scholar]
- Shabalina SA, Ogurtsov AY, Rogozin IB, Koonin EV, Lipman DJ. Comparative analysis of orthologous eukaryotic mRNAs: potential hidden functional signals. Nucleic Acids Res. 2004;32:1774–1782. doi: 10.1093/nar/gkh313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc. Natl. Acad. Sci. USA. 2011;108:E1392–E1398. doi: 10.1073/pnas.1113956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji S, Abdi NM, Bundschuh R, Fredrick K. Contribution of ribosomal residues to P-site tRNA binding. Nucleic Acids Res. 2009;37:4033–4042. doi: 10.1093/nar/gkp296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skabkin MA, Skabkina OV, Dhote V, Komar AA, Hellen CU, Pestova TV. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 2010;24:1787–1801. doi: 10.1101/gad.1957510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szamecz B, Rutkai E, Cuchalová L, Munzarová V, Herrmannová A, Nielsen KH, Burela L, Hinnebusch AG, Valásek L. eIF3a cooperates with sequences 5' of uORF1 to promote resumption of scanning by post-termination ribosomes for reinitiation on GCN4 mRNA. Genes Dev. 2008;22:2414–2425. doi: 10.1101/gad.480508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DJ, Nilsson J, Merrill AR, Andersen GR, Nissen P, Frank J. Structures of modified eEF2 80S ribosome complexes reveal the role of GTP hydrolysis in translocation. EMBO J. 2007;26:2421–2431. doi: 10.1038/sj.emboj.7601677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D, Unbehaun A, Li W, Das S, Lei J, Liao HY, Grassucci RA, Pestova TV, Frank J. Cryo-EM structure of the mammalian eukaryotic release factor eRF1-eRF3-associated termination complex. Proc. Natl. Acad. Sci. USA. 2012;109:18413–18418. doi: 10.1073/pnas.1216730109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura S, Aitken CE, Korlach J, Flusberg BA, Turner SW, Puglisi JD. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature. 2010;464:1012–1017. doi: 10.1038/nature08925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unbehaun A, Borukhov SI, Hellen CU, Pestova TV. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev. 2004;18:3078–3093. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- Williamson AR, Schweet R. Role of the genetic message in initiation and release of the polypeptide chain. Nature. 1964;202:435–437. doi: 10.1038/202435a0. [DOI] [PubMed] [Google Scholar]
- Yun DF, Laz TM, Clements JM, Sherman F. mRNA sequences influencing translation and the selection of AUG initiator codons in the yeast Saccharomyces cerevisiae . Mol. Microbiol. 1996;19:1225–1239. doi: 10.1111/j.1365-2958.1996.tb02468.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.