Abstract
We have performed a detailed population study of patients with genetic muscle disease in the northern region of England. Our current clinic population comprises over 1100 patients in whom we have molecularly characterized 31 separate muscle disease entities. Diagnostic clarity achieved through careful delineation of clinical features supported by histological, immunological and genetic analysis has allowed us to reach a definitive diagnosis in 75.7% of our patients. We have compared our case profile with that from Walton and Nattrass’ seminal study from 1954, also of the northern region, together with data from other more recent studies from around the world. Point prevalence figures for each of the five major disease categories are comparable with those from other recent studies. Myotonic dystrophies are the most common, comprising 28.6% of our clinic population with a point prevalence of 10.6/100 000. Next most frequent are the dystrophinopathies and facioscapulohumeral muscular dystrophy making up 22.9% (8.46/100 000) and 10.7% (3.95/100 000) of the clinic population, respectively. Spinal muscular atrophy patients account for 5.1% or 1.87/100 000 patients. Limb girdle muscular dystrophy, which was described for the first time in the paper by Walton and Nattrass (1954) and comprised 17% of their clinic population, comprises 6.2% of our clinic population at a combined prevalence of 2.27/100 000. The clinic population included patients with 12 other muscle disorders. These disorders ranged from a point prevalence of 0.89/100 000 for the group of congenital muscular dystrophies to conditions with only two affected individuals in a population of three million. For the first time our study provides epidemiological information for X-linked Emery–Dreifuss muscular dystrophy and the collagen VI disorders. Each of the X-linked form of Emery–Dreifuss muscular dystrophy and Ullrich muscular dystrophy has a prevalence of 0.13/100 000, making both very rare. Bethlem myopathy was relatively more common with a prevalence of 0.77/100 000. Overall our study provides comprehensive epidemiological information on individually rare inherited neuromuscular conditions in Northern England. Despite the deliberate exclusion of relatively common groups such as hereditary motor and sensory neuropathy (40/100 000) and mitochondrial disorders (9.2/100 000), the combined prevalence is 37.0/100 000, demonstrating that these disorders, taken as a group, encompass a significant proportion of patients with chronic disease. The study also illustrates the immense diagnostic progress since the first regional survey over 50 years ago by Walton and Nattrass.
Keywords: population study, prevalence, Northern England, inherited neuromuscular diseases, muscular dystrophy
Introduction
In 1954, Walton and Nattrass published their classic paper (Walton and Nattrass, 1954) in which they described 105 cases of muscle disease from Northumberland and Durham in the North of England. They proposed a new classification of muscle disorders, basing this on their detailed clinical observations. An interval of ~50 years provided an interesting time point at which to review and compare our current case profile with that described in 1954. We therefore undertook a comprehensive survey of all known cases of inherited muscle disease in children and adults in the population of the northern region of England. This region comprises the counties of Northumberland, Durham, Cumbria and parts of Yorkshire and Lancashire (Fig. 1), thus including but much larger than the region encompassed by Walton and Nattrass (1954). The geographical area for the new study is extensive with most of the population clustered in and around the large cities of Newcastle and Middlesbrough. The estimated total population according to the last census is 2.99 million (Supplementary Table S1).
Figure 1. The northern region of England showing catchment area for this study and location of Newcastle muscle clinic within the region.
The catchment area includes the counties of Northumberland, Durham, Cumbria and parts of Yorkshire and Lancashire with an estimated total population of 2.99 million.
Relatively low levels of population migration compared with regions such as London and the South East of England reflect the many established communities with multi-generation families in which members maintain close family ties. Those with inherited muscle disease have received specialist care via a ‘hub and spoke’ (regional centre with outreach) model for over 50 years facilitating ascertainment of patients and in many instances their relatives and descendants. The current model of care encompasses both adult and child patients providing continuity throughout life.
A major strength of the 1954 study was the detailed clinical assessment made of each patient. This remains an important cornerstone of current clinical practice, with the additional ability to confirm clinical impressions with an array of laboratory-based diagnostic techniques at the protein and molecular genetic level. It is no longer sufficient to classify a patient as, for example, ‘limb girdle muscular dystrophy (LGMD)’ without making strenuous attempts to confirm the subtype. Techniques used in this study to confirm the molecular diagnosis included muscle biopsy assessment with detailed immunohistochemical and western blot analysis of primary and secondary changes with all relevant diagnostic antibodies (Anderson and Davison, 1999) and genetic investigations to discover the pathogenic sequence variants. This rigorous approach has led to a confirmed molecular diagnosis in 75.7% of the patients with a total number of 31 different genetic entities now identifiable in this patient group.
Methods
The precise geographical boundaries of the northern region are defined at present by government-determined healthcare organizations termed primary care trusts (PCTs; Supplementary Table and Fig. 1). The northern region is the catchment area for the Institute of Human Genetics at Newcastle University.
Our inclusion criteria were all registered patients with inherited muscle diseases diagnosed and currently seen by the neuromuscular team at the Institute of Human Genetics. Child and adult cases are seen by the same team and so all age groups were included. Cases were ascertained predominantly from the database compiled from attendance at muscle clinics held either at the Institute of Human Genetics in Newcastle or at one of the outreach clinics throughout the northern region. The outreach clinics are a service of the Institute of Human Genetics and patients are seen by the same clinicians. The service has a high profile and patients have been referred from primary, secondary and tertiary care throughout the region for decades and as such are unlikely to be referred elsewhere in the country. Other sources available for this study included disease-specific databases such as the Duchenne (DMD) and Becker muscular dystrophy (BMD) databases, a database maintained by the Muscular Dystrophy Campaign Regional Care Advisor for the northern region and the regional myotonic dystrophy database, which was established after an audit conducted by our genetic nurse found that almost half of the probands with a genetically confirmed diagnosis of myotonic dystrophy type 1 had no cascade screening of their at-risk relatives. This was addressed systematically throughout the region with a detailed review of the pedigree in each family. Every at-risk relative was then contacted and invited to a myotonic dystrophy nurse-led clinic. We have not yet audited how many of the at-risk relatives refused the offer of genetic testing. All of these databases were part of the neuromuscular service structure at the Newcastle Institute of Human Genetics. Data were cross-checked across databases to eliminate duplication. The prevalence day was August 1, 2007.
All patients considered to have a potential genetic cause for their muscle disease are seen by the muscle team at the Institute of Human Genetics and were included in the study, regardless of family history. The clinic population is drawn mainly from tertiary referral sources and so the majority of the patients with conditions such as isolated hyperCKaemia or with myopathic features on neurophysiological testing would not be included automatically unless there was additional evidence suggestive of a primary muscle disorder. To avoid incomplete data collection, a number of diagnostic categories were excluded from the study due to other specialist provision for these patient groups within the region. Thus, our exclusion criteria were patients with inherited disorders such as mitochondrial and metabolic myopathies, ion channel disorders, congenital myasthenic syndromes and hereditary motor and sensory neuropathies as well as those with acquired neuromuscular disease including inflammatory myopathies, myasthenia gravis and motor neuron diseases.
Disease groups and diagnostic standards are defined as listed in Table 1. Current classifications and diagnostic criteria are drawn from those established by the European Neuromuscular Centre (Emery, 1998) and from more recent key references for individual conditions as proposed in the gene table of monogenic neuromuscular disorders (Kaplan, 2009). The approach was driven by clinical assessment with diagnostic tests employed in logical sequence. The approach for individual conditions varied from the relatively straightforward confirmation of a gene expansion in a condition such as myotonic dystrophy type 1 to the more complex approach required to define, for example, the subtype of LGMD (Bushby et al., 2007; Norwood et al., 2007). In addition, all unclassified LGMD patients have been screened for muscular dystrophy (DM)2 and presently known genetic causes of myofibrillar myopathies (desmin, myotilin, Z-band alternatively spliced PDZ-motif protein (ZASP), filamin C and B-crystalline mutations). Table 1 presents the key features of the diagnostic tests required for each condition to be considered confirmed to our diagnostic standards. The most difficult area is those patients with a phenotype of spinal muscular atrophy (SMA) and consistent muscle biopsy features in whom no genetic confirmation could be achieved. For some conditions, the diagnostic criteria are still evolving as further genetic and protein information becomes available.
Table 1. The table lists the diagnostic tests that we used for the diagnostic workup of our patient cohort beside standard clinical assessment.
| Disease group | Diagnostic standard |
|
|---|---|---|
| Muscle biopsy analysis: key changes | Genetic analysis | |
| Muscular dystrophies | ||
| Dystrophinopathies | Dystrophic. Reduced or absent dystrophin immunolabelling. | Deletion, duplication or point mutation in the DMD gene. |
| Facioscapulohumeral muscular dystrophy | Biopsy not indicated unless genetic analysis negative. | Deletion in subtelomeric region of 4q35 |
| LGMD (1A–1G) | LGMD1A: unspecific myopathic or dystrophic changes and/or accumulation of myofibrillar proteins; | MYOT mutation |
| LGMD1B/ADEDMD: unspecific myopathic or dystrophic changes; | LNA mutation; | |
| LGMD1C: myopathic or dystrophic, reduction in caveolin-3 | CAV3 mutation; | |
| LGMD (2A–2O) | LGMD2A: dystrophic, variable change in calpain-3 levels on immunoblotting; | CAPN3 mutation; |
| LGMD2B: dystrophic, may be inflammatory features, reduction in dysferlin immunolabelling; | DYSF mutation; | |
| LGMD2C-F: dystrophic, primary and secondary reduction in sarcoglycans and dystrophin immunolabelling; | SGCG, SGCA, SGCB or SGCD mutation; | |
| LGMD2G: dystrophic, abnormal telethonin expression on sections or WB | TCAP mutation | |
| LGMD2l: dystrophic, secondary reduction in laminin α2 on WB and α-dystroglycan | FKRP mutation; | |
| LGMD2K, M-O: abnormal α-dystrogylcan and laminin α2 expression | POMT1/2, FCMD or POMGNT1 mutation | |
| X-linked Emery–Dreifuss muscular dystrophy (EDMD-X) | Dystrophic, emerin immunolabelling reduced. | EMD mutation |
| (CMD) | ||
| Oculopharyngeal muscular dystrophy | Biopsy not indicated. | PABPN1 expansion |
| Congenital muscular dystophy (CMD) | ||
| MDC1A (laminin α2 chain deficient CMD) | Dystrophic, primary alteration in laminin α2 and secondary reduction in α-dystroglycan immunolabelling. | LAMA2 mutation. |
| Walker–Warburg syndrome | Abnormal α-dystroglycan and laminin α2 immunolabelling. | FKRP, POMT1/2, FCMD, POMGNT1 or LARGE mutation. |
| Rigid spine muscular dystrophy | Myopathic, fibre type disproportion, maybe multi-minicores. | SEPN1 mutation. |
| Congenital myopathies | ||
| Nemaline myopathy | Rod-like structures seen on light and electron microscopy. | More common: NEB or ACTA1 mutations |
| Central core disease | Central cores showing lack of oxidative enzyme activity, predominance of type1 fibres. Cores seen on electron microscopy. | RYR1 mutation. |
| Collagen Vl-related disorders | ||
| Bethlem myopathy | Unspecific biopsy findings, abnormal immunolabelling for collagen Vl in cultivated skin fibroblasts | COL6A1, COL6A2 or COL6A3 mutation |
| Ullrich congenital muscular dystrophy (UCMD) | Dystrophic; abnormal collagen Vl expression in muscle and cultivated skin fibroblasts | COL6A1, COL6A2 or COL6A3 mutation |
| Myofibrillar myopathies | Aggregates of desmin, myotilin and other myofibrillar proteins; EM. | DES, MYOT, CRYAB, ZASP, VCP or FLNC mutation. |
| Myotonic dystrophies | Biopsy unnecessary if genetic analysis confirmed. | DMPK or ZNF9 mutation. |
| SMA types I–III | Biopsy not required. If performed shows neurogenic changes. | SMN1 gene deletion. |
Muscle biopsies underwent standard histochemical analysis and if indicated were subsequently processed for immunoanalysis including immunoblotting. Genetic testing was provided by the molecular laboratory of the Institute of Human Genetics at Newcastle University, the National Commissioning Group (NCG) Referral Centre for Congenital Muscular Dystrophies and Congenital Myopathies at Great Ormond Street Hospital in London and the NCG Referral Centre for LGMD at the Institute of Human Genetics at Newcastle University. Key references for the listed diseases and their diagnostic workup can be found at: http://www.musclegenetable.org/.
WB = Western blot;
Results
Overall central muscle clinic database profile
One thousand one hundred and five cases were identified as eligible for inclusion in the study, representing 31 different disease entities. These were compiled into the central muscle clinic database, hereafter termed ‘the database’. The combined prevalence was 37.0/100 000 total population. Of these 1105 cases, 836 were confirmed to the diagnostic standards listed in Table 1, representing 75.7% of the patients in the database. In 145 patients, the diagnosis is uncertain at present or tests are in progress. A detailed breakdown of the data is given in Table 2.
Table 2. Analysis of cases by disease group.
| Disease group | No. of cases based on clinical diagnosis | No.of cases confirmed according to diagnostic standard (Table 1) | Proportion (%) of clinic population affected (with 95% confidence intervals) | Estimated point prevalence per 100 000 (with 95% confidence intervals) |
|---|---|---|---|---|
| Muscular dystrophies | ||||
| Duchenne (DMD) | 124 | 124 | 11.2 (9.4–13.1) | 8.29a (6.8–9.8) |
| Intermediate (IMD) | 7 | 7 | 0.6 (0.2–1.1) | 0.47a (0.1–0.8) |
| Becker (BMD) | 109 | 109 | 9.9 (8.1–11.6) | 7.29a (5.9–8.7) |
| Manifesting carriers | 13 | 13 | 1.2 (0.5–1.8) | 0.43 (0.2–0.7) |
| Total | 253 | 253 | 22.9 (20.4–25.4) | 8.46 (7.4–9.5) |
| Facioscapulohumeral muscular dystrophy (FSHD) | 118 | 116 | 10.7 (8.9–12.5) | 3.95 (3.2–4.7) |
| LGMD1B (AD EDMD) | 6 | 6 | 8.8b (2.1–15.6) | 0.20 (0–0.4) |
| LGMD2A | 18 | 15 | 26.5b (16.0–37.0) | 0.60 (0.3–0.9) |
| LGMD2B | 4 | 2 | 5.9b (0.3–11.5) | 0.13 (0–0.3) |
| LGMD2C | 4 | 4 | 5.9b (0.3–11.5) | 0.13 (0–0.3) |
| LGMD2D | 2 | 2 | 2.9b (0–7.0) | 0.07 (0–0.2) |
| LGMD2E | 2 | 2 | 2.9b (0–7.0) | 0.07 (0–0.2) |
| LGMD2I | 13 | 12 | 19.1b (9.8–28.5) | 0.43 (0.2–0.7) |
| LGMD unconfirmed | 19 | NA | 27.9b (17.3–38.6) | 0.64 (0.4–0.9) |
| Total LGMD | 68 | 43 | 6.15 (4.7–7.6) | 2.27 (1.7–2.8) |
| Emery–Dreifuss muscular dystrophy X-linked (EDMD-X) | 4 | 4 | 0.36 (0–0.7) | 0.13 (0–0.3) |
| Oculopharyngeal muscular dystrophy | 4 | 4 | 0.36 (0–0.7) | 0.13 (0–0.3) |
| CMD | ||||
| MDC1A | 18 | 18 | 1.62 (0.9–2.4) | 0.60 (0.3–0.9) |
| Walker–Warburg syndrome | 1 | 1 (POMGnT1) | 0.09 (0–0.3) | 0.03 (0.01) |
| UCMD | 4 | 3 | 0.36 (0–0.7) | 0.13 (0–0.3) |
| RSMD | 4 | 1 | 0.36 (0–0.7) | 0.13 (0–0.3) |
| Congenital myopathies | ||||
| Nemaline myopathy | 6 | 5 | 0.54 (0.1–1.0) | 0.20 (0–0.4) |
| Bethlem myopathy | 23 | 18 | 2.08 (1.2–2.9) | 0.77 (0.5–1.1) |
| Central core disease | 12 | 10 | 1.09 (0.5–1.7) | 0.40 (0.2–0.6) |
| Distal myopathy | 10 | 0 | 0.90 (0.4–1.5) | 0.33 (0.1–0.5) |
| Myofibrillar myopathy | 7 | 2 (MYOT) | 0.18 (0–0.4) | 0.07 (0–0.2) |
| 5 (DES) | 0.45 (0.1–0.9) | 0.17 (0–0.3) | ||
| Myotonic dystrophies | ||||
| DM1 | 311 | 311 | 28.1 (25.5–30.9) | 10.4 (9.3–11.6) |
| DM2 | 5 | 5 | 0.45 (0.1–0.9) | 0.17 |
| Spinal muscular atrophies | ||||
| MA type I | 3 | 3 | 0.27 (0–0.6) | 0.10 (0–0.2) |
| SMA type II | 17 | 17 | 1.54 (0.8–2.3) | 0.57 (0.3–0.8) |
| SMA type III | 36 | 19 | 1.72 (1.0–2.5) | 0.64 (0.4–0.9) |
| 17 inconclusive | 1.54 (0.8–2.3) | 0.57 (0.3–0.8) | ||
| Total | 56 | 39 | 5.07 (3.8–6.4) | 1.87 (1.4–2.4) |
| Others | ||||
| Excluded categories (e.g. HMSN etc.) | 5 | NA | 5.25 (3.9–6.6) | 1.94 (1.4–2.4) |
| Uncertain diagnosis/tests in progress | 145 | NA | 13.1 (11.1–15.1) | NA |
| Patients with confirmed diagnosis | NA | 836 | 75.7 (73.1–78.2) | NA |
| Total number of patients in database | 1105 | NA | NA | 37.0 |
The table lists the number of cases in each disease group category and those confirmed to the diagnostic standard listed in Table 1. It shows the proportion of our clinic population affected by each condition and the estimated population prevalence for each condition.
Males only.
Proportion of total LGMD patients.
RSMD = CMD with rigidity of the spine; HMSN = hereditary motor and sensory neuropathies.
Major diagnostic categories
Approximately two-thirds of the patients fell into five major diagnostic groups: myotonic dystrophy type 1 (DM1), DMD/BMD, facioscapulohumeral muscular dystrophy (FSHD), SMA and LGMD. The largest group was the one with DM1. These patients comprised just over a quarter of the total sample (28.1%) with a prevalence of 10.4/100 000 (Table 2). Although all undiagnosed patients with proximal muscle weakness and muscle pain were screened for the CCTG repeat expansion in intron 1 of the zinc finger protein 9 (ZNF9) gene, myotonic dystrophy type 2 (DM2) was very uncommon in the northern region, with only five patients confirmed with this diagnosis. All DM1 and DM2 patients had a proven expansion mutation in the respective gene.
FSHD patients accounted for 10.7% (prevalence 3.95/10.0000) of the total patient population. Of the 118 cases, 116 were confirmed to the diagnostic standard (Tables 1 and 2) with the presence of a contraction of the polymorphic microsatellite repeat D4Z4 on chromosome 4qter.
DMD and BMD comprised ~10% each giving a combined prevalence for the dystrophinopathies of 8.46/100 000 total population (Table 2). For males only, the prevalences for DMD and BMD were 8.29 and 7.29, respectively. The dystrophinopathy category included those with intermediate muscular dystrophy (IMD, 0.47/100 000 males) and manifesting carriers of either DMD or BMD (0.43/100 000 total population). All cases were confirmed to the diagnostic standard of having a deletion, duplication or point mutation within the dystrophin gene.
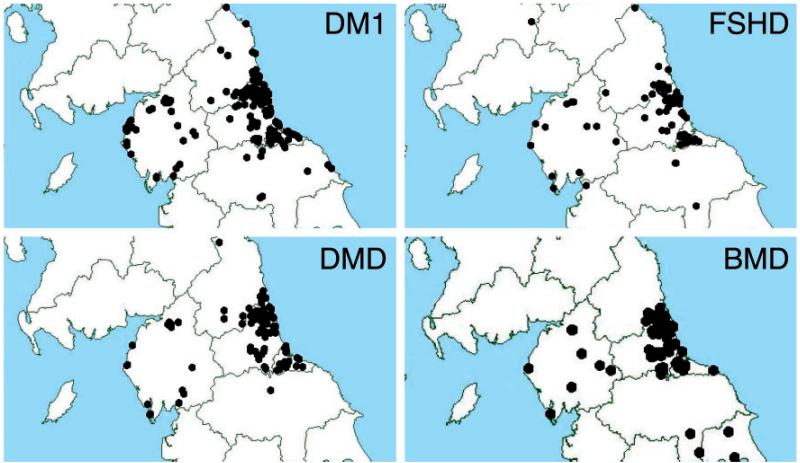
The geographical distribution of patients with DM1, FSHD, DMD and BMD is shown in Fig. 2, with data points illustrated by postcode. As may be expected, the majority of patients are clustered around the major population centres. These cities have relatively easy access to services whereas much of the northern region consists of open countryside which patients with restricted mobility may find more challenging.
Figure 2. Geographical distribution of cases within northern region of England.
Myotonic dystrophy type 1 (DM1), facioscapulohumeral muscular dystrophy (FSHD), Duchenne muscular dystrophy (DMD) and Becker muscular dystrophy (BMD).
The combined prevalence of SMA was 5.1%. Seventy per cent of the patients had a deletion of the SMN1 gene and were either classified as SMA I, II or III. The remainder of cases were assigned a diagnosis of SMA III on the basis of clinical and neurophysiological features and exclusion of other diagnoses (Table 2).
Limb girdle muscular dystrophy
The fifth major category was those with LGMD. These patients comprised 6.15% of the clinic population with a combined prevalence of 2.27/100 000 for the northern region (Table 2). We have attained a definite diagnosis in 49 patients or 72% of the LGMD group. Nineteen patients, who all showed limb girdle weakness and a dystrophic muscle biopsy pattern, remain unclassified at present despite extensive testing. It should be noted that our study is of the northern region population as distinct from our national LGMD referral clinic, which accepts referrals from throughout the UK in a government-funded initiative for the management of rare diseases.
By subtype, LGMD2A (calpainopathy) was the most common with a prevalence of 0.60/100 000 or 26.5% of the total LGMD group. Fifteen out of 18 patients were confirmed to the diagnostic standard of detection of homozygous or compound heterozygous mutations in the calpain-3 gene. The other three patients showed a clinical phenotype consistent with LGMD2A, calpain-3 deficiency on western blots without other protein abnormalities by immunoanalysis, and no mutation in any of the other investigated genes. All three showed one missense mutation in the calpain 3 gene.
The next largest category was LGMD2I, which made up 19.1% of the total LGMD group with a prevalence of 0.43/100 000. Twelve of the 13 patients had a confirmed mutation in the fukutin-related protein (FKRP) gene, meeting our diagnostic standard. One patient was diagnosed based on family history.
The other categories were individually much smaller. The combined sarcoglycanopathy (LGMD2C-E) patients together comprised 11.7% giving a prevalence of 0.27/100 000. Laminopathy cases, combining LGMD1B and autosomal dominant Emery–Dreifuss muscular dystrophy (ADEDMD) (but listed together in Table 2 as LGMD1B), were more uncommon, comprising only 8.8% of the LGMD patients. Dysferlinopathy (LGMD2B) was marginally less common still at 5.9% with a prevalence of 0.13/100 000. To date, all but two of these cases have been confirmed through detection of a mutation in the relevant gene. Two LGMD 2B cases without a mutation showed a clinical picture consistent with dysferlin deficiency and loss of dysferlin expression in biopsy samples.
Of the remaining patients in the LGMD group, 27.9% are at present unclassified despite strenuous efforts to encompass all reasonable diagnoses. As this is a dynamic database, diagnostic work on patients continues and it is possible that a definite diagnosis will be reached in future years.
Finally, from our total clinic population, no patient has been found to have the conditions LGMD1A, 1C or 2G, H, J or K.
Minor diagnostic categories
Of the remaining patients, 91 were classified within 1 of 12 disorders (Tables 1 and 2).
Collagen VI-related disorders
Bethlem myopathy was the most common disorder overall within the 12 minor categories. Out of 100 000, 0.77 (2.08%) were affected. All patients showed a clinical phenotype of Bethlem myopathy and an abnormal collagen VI expression on skin fibroblast cultures. In 14 of the patients, we have already identified mutations in one of the three genes encoding collagen VI with further work in progress on the remainder. Prevalence of Ullrich congenital muscular dystrophy (UCMD), allelic to Bethlem myopathy, was much less common at 0.13/100 000. All UCMD patients showed either complete or partial loss of collagen VI on muscle biopsy sections or abnormal collagen VI expression in skin fibroblast cultures. Mutations in the genes encoding the alpha chains of collagen VI were subsequently identified in all UCMD patients.
Congenital myopathies
Of the congenital myopathies, central core disease was the most common form with 1.09% or 0.40/100 000 prevalence. Of the 12 patients, 10 had a mutation confirmed within the RYR1 gene. Nemaline myopathy was approximately half as common with 0.54% or 0.20/100 000.
Congenital muscular dystrophies
Congenital muscular dystrophies (CMD) had a combined prevalence of 0.76/100 000 (2.08% total) with the majority being CMD with laminin α2 chain deficiency (MDC1A) at 1.62% or 0.60/100 000. All of our MDC1A patients showed the characteristic clinical pattern of MDC1A, loss of laminin α2 expression on muscle biopsy sections and white matter changes on magnetic resonance images of the brain. In a subset of the patients genetic confirmation of the diagnosis was achieved by mutation analysis of the LAMA2 gene, whereas in the remaining patients the genetic result is still pending. In our population, CMD with rigidity of the spine (RSMD) due to mutations in the selenoprotein 1 gene showed a prevalence of 0.13/100 000, similar to UCMD.
Of the remaining groups, X-linked Emery–Dreifuss muscular dystrophy (EDMD-X) was also very uncommon with a prevalence of 0.13/100 000, the same as UCMD and RSMD. All of the EDMD-X patients had confirmed mutations in the emerin gene. There were initially only four patients with oculopharyngeal muscular dystrophy (OPMD) included in our cohort of patients, but we assumed that OPMD patients also attended other clinics in the region. The national referral genetics laboratory for OPMD informed us that there have been only five genetically confirmed diagnoses of OPMD from our region within the last five years, suggesting that our clinic number does not reflect full ascertainment.
The final three categories are the myofibrillar and distal myopathies. Within these groups, myotilin and desmin mutations were found in 0.18% (0.07/100 000) and 0.45% (0.17/100 000) of the clinic population, respectively. Presently, 0.9% of patients in the database are categorized as having a pure distal myopathy based on clinical symptoms, serum creatine kinase activity, neurophysiological examinations and/or magnetic resonance imaging. We have excluded patients with Miyoshi myopathy and desmin or myotilin mutations in this group of patients. Diagnostic work on this latter group is ongoing and may result in reclassification of some patients in due course.
Discussion
Overall summary of northern region population data and comparison with other studies
The combined population prevalence figure for all inherited muscle disease categories in our clinic database was 37.0/100 000. Thus, although many of the conditions represented are individually rare, cumulatively they are an important proportion of those with chronic disease. Prevalence studies are likely to underestimate those diseases that lead to early death, such as DMD, as prevalence reflects both incidence and duration. Our combined prevalence figure compares well with previous population studies. The figure from Hughes’ study was 34.5/100 000 (Hughes et al., 1996), but this included patients with the relatively common hereditary motor and sensory neuropathies, metabolic myopathies, mitochondrial myopathy and X-linked bulbospinal neuronopathy, all of which were excluded from our study. Darin’s study of the epidemiology of neuromuscular disorders in childhood in Western Sweden (Darin and Tulinius, 2000) produced an inherited neuromuscular disorders point prevalence figure of 53.1/100 000. This group also included hereditary motor and sensory neuropathies, mitochondrial disease and metabolic myopathies. Emery estimated a total prevalence of inherited neuromuscular disease of ~1 in 3500 (28.6/100 000) (Emery, 1991). Given that this group included hereditary motor and sensory neuropathies, familial motor neurone diseases and familial myasthenia gravis, our data suggest that we have better ascertainment of inherited muscle disease cases. Emery estimates the inherited neuromuscular disease prevalence as exceeding 1 in 3000 (33.3/100 000) if disorders such as congenital myopathies, mitochondrial myopathies, rarer forms of muscular dystrophy and SMA are included.
Thus, rigorous clinical assessment and laboratory diagnostic work has allowed us to ascertain 1105 patients with inherited muscle disease within the Northern region of England. To date, 836 patients have had their clinical diagnosis confirmed to the respective diagnostic standard, equating to 75.7% of the total number of patients. One would expect this figure to increase even further in the coming years with the increasing ease of large-throughput genetic screening technology, together with the availability of new genetic tests applicable to categories such as the myofibrillar myopathies. Our rates of diagnostic confirmation were highest for the dystrophinopathies and myotonic dystrophies, probably reflecting that these conditions are reasonably straightforward to diagnose clinically and molecular testing relatively quick to accomplish. In previous population studies such as those by Hughes and Darin (Hughes et al., 1996; Darin and Tulinius, 2000), the number of cases definitely confirmed by genetic analysis is not stated explicitly.
Some conditions such as mitochondrial disease and ion channel disorders were deliberately excluded from our database due to other provision for these patient groups within the northern region, although in other geographical areas these groups of patients are seen in the same clinics. Thus, as a speculative exercise, adding the published prevalence figures for mitochondrial disease (9.2/100 000) (Schaefer et al., 2008), metabolic myopathies such as McArdle’s (1/100 000) (Haller, 2000) and late-onset Pompe disease (1/60 000) (Ausems et al., 1999) and myotonia congenita (1/100 000) (Emery, 1991) to our prevalence data, the combined total for inherited muscle disease is 49.9/100 000, despite still excluding hereditary motor and sensory neuropathies. Further addition of the prevalence figures for acquired myasthenia gravis (15/100 000) (Robertson et al., 1998) and inflammatory myopathies including inclusion body myositis (0.93/100 000) (Phillips et al., 2000) and polymyositis and dermatomyositis (21.5/100 000) (Bernatsky et al., 2009) brings the combined prevalence for inherited and acquired muscle disease to an estimated figure of 87.3/100 000. For context, the prevalence figure quoted for multiple sclerosis in North America and Northern Europe is between 60 and 200/100 000 (Lunemann and Martin,, 2007), showing that patients with neuromuscular disease are, despite popular perception, a relatively common group of chronic disease patients with a combined prevalence similar to those conditions often considered to be far more prevalent. This has implications for long-term disease management and attendant health economic implications.
Comparison of major diagnostic groups with other population prevalence studies
The most common condition in our database was myotonic dystrophy type 1 (DM1) with a prevalence figure of 10.4/100 000. The combined child and adult population study from Northern Ireland (Hughes et al., 1996) produced a similar figure of 8.4/100 000. Darin’s study (Darin and Tulinius, 2000) found five per 100 000 cases in children; one would expect fewer in this younger population. Other estimates of the prevalence of DM1 vary widely according to geography, ranging from one per 100 000 in Japan (Davies et al., 1992) to 10/100 000 in Iceland (Leifsdottir et al., 2005). In some regions such as Quebec, the prevalence is much higher due to founder effects (Yotova et al., 2005). Precise data for the overall worldwide prevalence are not available but one estimate is 1:8 000 (Harper 1989). Our relatively high prevalence figure reflects systematic family tracing, extrapolating from the proband in an attempt to ascertain all family members potentially at risk of having inherited the condition. One aim was to find cases at risk of cardiac arrhythmia and to institute medical intervention when required. A second aim was to offer timely genetic counselling, particularly to mothers at risk of having a child with congenital myotonic dystrophy. The prevalence number of DM1 in our region might well be higher than 10.4/100 000 as not all asymptomatic at-risk family members have accepted the offer for genetic testing.
Prevalence of myotonic dystrophy type 2 also varies geographically. The condition is not described in comparable population study papers, but separate studies show a much higher prevalence in Southern Germany, again possibly due to a founder effect (Bachinski et al., 2003). The estimate discussed in the report of the 115th European Neuromuscular Centre workshop of 1/100 000 in Germany was considered to be the minimum prevalence figure (Udd et al., 2003).
FSHD prevalence was 3.95/100 000 in our study. Figures allowing direct comparison are listed in Emery’s review from 1991 and range from 0.22 to 6.69/100 000. The two most widely differing figures are from two individual states in the USA although this apparent divergence is based on only two cases in each study. The figure quoted from the Netherlands of 1.87/100 000 had been re-assessed and was estimated to be ~5/100 000 (Padberg et al., 1995) with an assumed complete ascertainment. A recent study from Italy, although of a smaller population, reported a prevalence of 4.4/100 000 (Mostacciuolo et al., 2009). Thus, our figure of 3.95/100 000, based on 118 cases, may well be a fairly accurate one.
Dystrophinopathies comprised 22.9% of our clinic population with a combined prevalence of 8.46/100 000. Of these, DMD prevalence was 8.29, IMD 0.47 and BMD 7.29/100 000 males. Hughes remarked that his ascertainment of DMD and other severe cases was probably complete and his figure of 8.2/100 000 (Hughes et al., 1996) males aligns very well with ours. For BMD, Hughes’ prevalence was 3.26/100 000 males, which is under half of our figure, presumably reflecting the difference in severity affecting complete ascertainment. Darin’s figures were 16.8 for DMD and 1.6 for BMD per 100 000 males under 16 years (Darin and Tulinius, 2000). The higher figure for DMD in the childhood population is perhaps a more accurate representation for this group in the past, although increasing life expectancy will have increased the point prevalence in our current clinic population. Eagle noted that the mean survival of DMD patients had improved in each successive decade since the 1960s, increasing from 14 years in the 1960s to 19 years for those in 1990 (not ventilated), presumably reflecting better overall care (Eagle et al., 2002), with the most striking change in mean survival to 25.3 years for those using nocturnal non-invasive ventilation.
The next major group was those with SMA. Hughes produced a figure of 1.4 (Hughes et al., 1996), Darin 2.8 (Darin and Tulinius, 2000) and our study 1.87/100 000. All three studies included patients under 16 years and so would be expected to capture all forms of SMA although the proportion of the total clinic population will be relatively higher in the childhood-only study. Approximately a third of our SMA III patients did not show a mutation in the SMN1 gene, which is in accordance with the literature (Wirth, 2000). It is also possible that genetic advances have re-assigned patients from this group to other categories, thus lowering the prevalence in our study compared with Darin’s (Darin and Tulinius, 2000).
Limb girdle muscular dystrophy patients from the northern region and comparison with other studies
Our study population of 1105 patients contains 68 classified as having a form of LGMD, representing a population prevalence of 2.27/100 000. Several recent papers have examined the relative frequencies of the LGMD subtypes in their patient populations whereas others have focused on a particular subtype.
Using a methodical approach of immunohistochemical analysis and then DNA sequencing successive genes, van der Kooi and colleagues updated a previous study of 105 LGMD patients from the Netherlands and were able to classify 51% of them into a definite subtype (van der Kooi et al., 2007). Calpainopathy was the most frequent diagnosis, similar to our study, affecting 21% of the families, with sarcoglycanopathy more common than LGMD2I. This is in marked contrast to the relatively low prevalence of LGMD2A in Denmark (Duno et al., 2008), which the authors estimate to be 5- to 6-fold lower in ethnic Danes compared with other European countries. Conversely LGMD2I appears to be relatively more common in Northern Europe compared with North America. A study from Denmark showed a high proportion of LGMD2I (38% of LGMD2-classified patients) (Sveen et al., 2006). Although there were phenotypic differences between the patients homozygous or heterozygous for the 826C>A mutation, testing for this mutation in this population detected all the cases. Our population had a prevalence of LGMD2I closest to the Danish study, perhaps reflecting previous migration from Denmark to the Northern region of England.
A study from Italy examined 181 LGMD patients (155 families) and was able to confirm a diagnosis in 72.9% (Guglieri et al., 2008). They found the most common groups to be calpainopathy with 28.4%, which has also been shown in other Southern European epidemiological studies (Angelini, 2004; Fanin et al., 2005; Saenz et al., 2005; Balci et al., 2006). The authors emphasize the importance of confirming the suspected diagnosis through molecular genetic testing, especially given the fairly poor predictive value of calpain-3 protein analysis, for example, with CAPN3 gene mutations detected in only 61% of those in whom a reduction in the level of calpain-3 was found on western blot. The next most common LGMD group in this study (Guglieri et al., 2008) was dysferlinopathy at 18.7% and the combined sarcogly-canopathies at 18.1% (all percentages are of families). The percentage of combined sarcoglycanopathy (LGMD2C-E) patients in our study was 11.7% (0.27/100 000), slightly lower than in the Italian studies. A Dutch study detected a sarcoglycanopathy in 23% of their LGMD2 patients, in whom α- and β-sarcoglycan deficiency was equally common (Ginjaar et al., 2000). In our patients, γ-sarcoglycan mutations were twice as frequent as the other subtypes.
The frequency of dysferlinopathy varied among the study populations. In our study, these patients were relatively few (5.9% or 0.13/100 000) compared with other areas, including 18.7% in the Italian population (Guglieri et al., 2008), but in keeping with the low frequency in the Dutch LGMD population where only one patient was identified (van der Kooi et al., 2007). In contrast, there seem to be founder effects in certain regions such as Israel, markedly raising the local prevalence (Leshinsky-Silver et al., 2007). With 18% LGMD2B was also the most common diagnosis in a study that examined paediatric and adult LGMD subtypes from six centres in the USA (Moore et al., 2006). The second most common LGMD forms were sarcoglycanopathy and dystroglycanopathy (LGMD2I), both at 15%, with calpainopathy fourth most frequent at 12%. The authors comment that their study population was ethnically diverse but are unable to provide prevalence data as their patients were from a referral rather than population base.
Thus, these studies concur on certain points. First, the studies emphasize the comprehensive diagnostic effort required to classify LGMD patients, noting that careful attention to the combination of clinical, immunohistochemical and molecular genetic data is required and that reliance on just one aspect may be misleading. Nevertheless, all studies had a significant proportion of patients classified as LGMD but without a definite diagnosis, mostly in a similar range to our figure of 27.9% (van der Kooi et al., 2007, 41.3%; Guglieri et al., 2008, 29%). Second, all re-affirm that autosomal recessive LGMD is much more common than autosomal dominant LGMD. In our study, the proportion of patients affected with dominant subtypes was 8.8% and 13.5% in the Dutch study, all of whom were affected by laminopathy. There was a striking lack of LMNA mutations detected in the Italian study with all of their 10 patients with dominant inheritance affected by mutations in caveolin-3 (in contrast to our study where we found none). Moore et al. (2006) found <1% with LGMD1A and 4% with LGMD1B but none with LGMD1C. Finally, the studies show the relative frequencies of confirmed LGMD subtypes in different populations. Calpainopathy is the most frequent diagnosis in those European populations studied but only fourth most frequent in the referral pool in the USA, which might be explained by a less comprehensive immunoanalysis of muscle biopsies and/or genetic testing. Dysferlinopathy seems less frequent in the North of UK and caveolinopathy is very rare in our catchment area.
Comparison of minor diagnostic groups with other population prevalence studies
Even when taken as a group of conditions, epidemiological data for many of the rarer inherited muscle diseases from other studies are limited. Emery (1991) does not mention the congenital myopathies at all. Hughes et al. (1996) estimated their prevalence to be 3.5/100 000 and Darin and Tulinius (2000) 5.0/100 000. Central core myopathy prevalence is unknown but it is probably more common than other congenital myopathies (Jungbluth, 2007).
Here, for the first time, we are able to present data on the prevalence of three myopathies. The most common group in our study was Bethlem myopathy with a prevalence of 0.77/100 000, the second most common central core disease with a prevalence of 0.40/100 000 and lastly nemaline myopathy at 0.20/100 000. The combined prevalence of 1.37/100 000 is still rather lower than the figures from Hughes et al. (1996) and Darin and Tulinius (2000), but all of our cases have been rigorously assessed according to clear diagnostic criteria. We found no patients with other forms of congenital myopathy such as centronuclear myopathy.
Congenital muscular dystrophies
The CMD are a complex group of conditions whose classification is made through the combination of clinical and laboratory studies (Lisi and Cohn, 2007; Muntoni et al., 2008). A recent study examines the frequency of CMD subtypes in a cohort of 101 patients from Australia (Peat et al., 2008). The authors were able to reach a definitive diagnosis in 24% of their patients and to assign a diagnosis on the basis of immunofluorescence changes in 45%. The most common group were those with defects in glycosylated α-dystroglycan (25%), the second most common were those with collagen VI abnormalities (12%) and 8% with a primary laminin α2 (merosin) abnormality (Peat et al., 2008). The study was not population-based and no prevalence figures were given. The most common category in our study was MDC1A (merosin deficiency) at a prevalence of 0.60/100 000. All patients had a confirmed mutation in the LAMA2 gene. A study from Japan found that primary collagen VI deficiency accounted for the second most common CMD, with Fukuyama CMD being the most frequent (Okada et al., 2007). Although 34 patients were found to have a collagen VI deficiency, only 26 of these had a mutation in one of the collagen VI genes. The percentage of their total CMD cases with collagen VI deficiency was 7.2%. In our study, UCMD and rigid spine muscular dystrophy were equally as common as each other with a prevalence of 0.13/100 000. We had only one patient with a defect in α-dystroglycan. Our combined CMD prevalence figure including UCMD was 0.89/100 000. Figures from previous studies were 2.5 (Darin and Tulinius, 2000) and 0.6/100 000 (Hughes et al., 1996).
X-linked Emery–Derifuss muscular dystrophy (EDMD-X) was very uncommon in our study with a prevalence of 0.13/100 000. Prevalence data for this disease are difficult to find but overall it is far less common than autosomal dominant EDMD.
There have been many advances in myofibrillar and distal myopathies in recent years but prevalence data for this set of diseases are sparse (Udd, 2007, 2009; Goebel et al., 2008; Selcen, 2008). Prevalence of some disorders such as tibial muscular dystrophy vary geographically with a particularly high prevalence of this disorder in Finland (8/100 000) but less certain frequencies elsewhere. Only 0.9% of our patients were found to have a distal myopathy.
Comparison with the 1954 study
Walton and Nattrass (1954) proposed a new classification based on three major groups of disorders (Table 3). Eighty-four patients were classified in the muscular dystrophy group: 48 Duchenne-type, 18 limb girdle and 15 facioscapulohumeral. Those with ‘myotonic syndrome’ comprised 15 with dystrophia myotonica and 6 with myotonia congenita. The group of 18 patients classified as ‘limb-girdle muscular dystrophy’ were noted to have ‘commencement of muscular weakness in either the shoulder or pelvic girdle’ and slower progression than in the Duchenne group but more rapid than in those with facioscapulohumeral type. Inheritance was usually autosomal recessive and they noted that the natural history of disease progression in these patients showed more variation than in the other groups, both observations in keeping with current knowledge.
Table 3. Comparative proportions and estimated point prevalences of affected patients from 1954 (Walton and Nattrass) and current studies.
| Walton and Natrass | Number (%) affected (n = 105) | Current study | Number (%) affected (n = 1105) | Estimated point prevalence |
|---|---|---|---|---|
| ‘Duchenne type’ | 48 (46) | DMD | 124 (11.2) | 8.29a |
| BMD/IMD | 116 (10.5) | |||
| Total | 253 (22.9) | |||
| ‘Facioscapulohumeral’ | 15 (14) | Facioscapulohumeral | 118 (10.7) | 3.95 |
| ‘Limb girdle’ | 18 (17) | All limb girdle including specific subgroups | 68 (6.15) | 2.27 |
| Dystrophia myotonica subgroup of ‘myotonic syndrome’ | 15 (14) | Myotonic dystrophy type 1 | 311 (28.1) | 10.4 |
Males only
Diagnostic categories and proportions of the affected population from our current study are interesting to compare with those from the original paper (Table 3) and demonstrate the enhanced clarity provided by molecular analysis. With time, the number of conditions that we can now definitively identify has increased greatly although, other than spinal muscular atrophy, the major categories remain unchanged in type.
The proportion defined as LGMD in our study (6.15%) is lower than from the 1954 study (17%). BMD patients, a condition not defined until 1955, may have been part of this earlier group due to a relatively late-onset and similar distribution of weakness. SMA patients may also have been included here, as may other categories.
The most marked difference in the proportions of patients between the two studies is for dystrophinopathy patients. ‘Duchenne type’ muscular dystrophy patients comprised almost half of Walton and Nattrass’ patients but only about 11% of our population, with our BMD patients another 10%. It is likely that some of those originally classified as ‘Duchenne type’ had other conditions such as BMD and LGMD2I (three had macroglossia); 11 of the 48 ‘Duchenne type’ patients from 1954 had an age of onset of >8 years. In contrast, some had an unexpectedly early age of onset of under 1 year and may represent other conditions. While some of the patients in the Duchenne type group almost certainly had other diseases it is likely that the Duchenne group as currently recognized has probably always had relatively full ascertainment due to the severity of the condition. Their proportion of the total muscle patient population will therefore fall as patients with milder conditions are now more likely to be included.
Our current population prevalence for BMD cannot be compared directly with 1954 as the condition was not yet described at that point. A study from 1991 (Bushby et al., 1991) attempted to ascertain all BMD cases within the northern region of England. The prevalence rate of BMD was found to be 2.38/100 000 (total population) and that for DMD 2.48/100 000. Review of these cases showed that a number had previously received other diagnoses such as limb girdle dystrophy or spinal muscular atrophy. These figures are lower than ours from the current study, perhaps differences reflecting improved survival, particularly for the DMD patients (Eagle et al., 2002).
As indicated, advances in investigative techniques, particularly genetic testing, during the past 50 years now often enable a precise diagnosis to be established with confidence. In addition, it is often possible to assign patients with relatively mild symptoms and signs or even those seemingly unaffected to a precise diagnostic category at an earlier stage in their clinical course than possible through clinical examination alone. Systematic genetic tracing for conditions such as myotonic dystrophy type 1 frequently reveals that several members of a proband’s family have a trinucleotide expansion in the DMPK gene, even though many have relatively few symptoms or signs. This may account in part for this group of patients comprising over a quarter of our population compared with 14% in the earlier study. It also suggests that our figures are still likely to be an underestimate as factors such as incomplete family pedigree information prevent full ascertainment.
Supplementary Material
Acknowledgements
The authors thank the National Commissioning Group (NCG) for supporting the diagnostic work in the LGMD population. We are grateful to Prof. Carol Bower, Telethon Institute for Child Health Research, University of Western Australia and Prof. Heather Cordell, Institute of Human Genetics at Newcastle University (UK) for helpful comments and discussion. We are thankful to Jane Barnes, Jessica Hoogendijk and Geoff Bell for their support with the databases. P.F.C is a Wellcome Trust Senior Fellow in Clinical Science.
Funding: This work was supported by the Medical Research Council (UK) as part of the MRC Centre for Neuromuscular Diseases and the Muscular Dystrophy Campaign (UK). V.S. is a member of the German Muscular Dystrophy Network (MD-NET 01GM0601) funded by the German Ministry of Education and Research (BMBF, Bonn, Germany); www.md-net.org. MD-NET is a partner of TREAT-NMD (EC, 6th FP, proposal #036825; www.treat-nmd.eu).
Abbreviations
- BMD
Becker muscular dystrophy
- CMD
Congenital muscular dystrophy
- DMD
Duchenne muscular dystrophy
- EDMD
Emery–Dreifuss muscular dystrophy
- FSHD
facioscapulohumeral muscular dystrophy
- IMD
intermediate muscular dystrophy
- LGMD
limb girdle muscular dystrophy
- OPMD
oculopharyngeal muscular dystrophy
- PCTs
primary care trusts
- SMA
spinal muscular atrophy
- UCMD
Ullrich congenital muscular dystrophy
References
- Anderson LV, Davison K. Multiplex Western blotting system for the analysis of muscular dystrophy proteins. Am J Pathol. 1999;154:1017–22. doi: 10.1016/S0002-9440(10)65354-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini C. Limb-girdle muscular dystrophies: heterogeneity of clinical phenotypes and pathogenetic mechanisms. Acta Myol. 2004;23:130–6. [PubMed] [Google Scholar]
- Ausems MG, Verbiest J, Hermans MP, Kroos MA, Beemer FA, Wokke JH, et al. Frequency of glycogen storage disease type II in The Netherlands: implications for diagnosis and genetic counselling. Eur J Hum Genet. 1999;7:713–6. doi: 10.1038/sj.ejhg.5200367. [DOI] [PubMed] [Google Scholar]
- Bachinski LL, Udd B, Meola G, Sansone V, Bassez G, Eymard B, et al. Confirmation of the type 2 myotonic dystrophy (CCTG)n expansion mutation in patients with proximal myotonic myopathy/proximal myotonic dystrophy of different European origins: a single shared haplotype indicates an ancestral founder effect. Am J Hum Genet. 2003;73:835–48. doi: 10.1086/378566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balci B, Aurino S, Haliloglu G, Talim B, Erdem S, Akcoren Z, et al. Calpain-3 mutations in Turkey. Eur J Pediatr. 2006;165:293–8. doi: 10.1007/s00431-005-0046-3. [DOI] [PubMed] [Google Scholar]
- Bernatsky S, Joseph L, Pineau CA, Belisle P, Boivin JF, Banerjee D, et al. Estimating the prevalence of polymyositis and dermatomyositis from administrative data: age, sex, and regional differences. Ann Rheum Dis. 2009;68:1192–6. doi: 10.1136/ard.2008.093161. [DOI] [PubMed] [Google Scholar]
- Bushby K, Norwood F, Straub V. The limb-girdle muscular dystrophies—diagnostic strategies. Biochim Biophys Acta. 2007;1772:238–42. doi: 10.1016/j.bbadis.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Bushby KM, Thambyayah M, Gardner-Medwin D. Prevalence and incidence of Becker muscular dystrophy. Lancet. 1991;337:1022–4. doi: 10.1016/0140-6736(91)92671-n. [DOI] [PubMed] [Google Scholar]
- Darin N, Tulinius M. Neuromuscular disorders in childhood: a descriptive epidemiological study from western Sweden. Neuromuscul Disord. 2000;10:1–9. doi: 10.1016/s0960-8966(99)00055-3. [DOI] [PubMed] [Google Scholar]
- Davies J, Yamagata H, Shelbourne P, Buxton J, Ogihara T, Nokelainen P, et al. Comparison of the myotonic dystrophy associated CTG repeat in European and Japanese populations. J Med Genet. 1992;29:766–9. doi: 10.1136/jmg.29.11.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duno M, Sveen ML, Schwartz M, Vissing J. cDNA analyses of CAPN3 enhance mutation detection and reveal a low prevalence of LGMD2A patients in Denmark. Eur J Hum Genet. 2008;16:935–40. doi: 10.1038/ejhg.2008.47. [DOI] [PubMed] [Google Scholar]
- Eagle M, Baudouin SV, Chandler C, Giddings DR, Bullock R, Bushby K. Survival in Duchenne muscular dystrophy: improvements in life expectancy since 1967 and the impact of home nocturnal ventilation. Neuromuscul Disord. 2002;12:926–9. doi: 10.1016/s0960-8966(02)00140-2. [DOI] [PubMed] [Google Scholar]
- Emery AE. Population frequencies of inherited neuromuscular diseases—a world survey. Neuromuscul Disord. 1991;1:19–29. doi: 10.1016/0960-8966(91)90039-u. [DOI] [PubMed] [Google Scholar]
- Emery AE. Diagnostic criteria for neuromuscular disorders. 2nd edn. Pub. Royal Soc of Med; 1998. [Google Scholar]
- Fanin M, Nascimbeni AC, Fulizio L, Angelini C. The frequency of limb girdle muscular dystrophy 2A in northeastern Italy. Neuromuscul Disord. 2005;15:218–24. doi: 10.1016/j.nmd.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Ginjaar HB, van der Kooi AJ, Ceelie H, Kneppers AL, van Meegen M, Barth PG, et al. Sarcoglycanopathies in Dutch patients with autosomal recessive limb girdle muscular dystrophy. J Neurol. 2000;247:524–9. doi: 10.1007/s004150070151. [DOI] [PubMed] [Google Scholar]
- Goebel HH, Fardeau M, Olive M, Schroder R. 156th ENMC International Workshop: desmin and protein aggregate myopathies, 9-11 November 2007, Naarden, The Netherlands. Neuromuscul Disord. 2008;18:583–92. doi: 10.1016/j.nmd.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Guglieri M, Magri F, D’Angelo MG, Prelle A, Morandi L, Rodolico C, et al. Clinical, molecular, and protein correlations in a large sample of genetically diagnosed Italian limb girdle muscular dystrophy patients. Hum Mutat. 2008;29:258–66. doi: 10.1002/humu.20642. [DOI] [PubMed] [Google Scholar]
- Haller RG. Treatment of McArdle disease. Arch Neurol. 2000;57:923–4. doi: 10.1001/archneur.57.7.923. [DOI] [PubMed] [Google Scholar]
- Harper PS. Myotonic dystrophy. 2nd edn. W. B. Saunders Co.; London: 1989. [Google Scholar]
- Hughes MI, Hicks EM, Nevin NC, Patterson VH. The prevalence of inherited neuromuscular disease in Northern Ireland. Neuromuscul Disord. 1996;6:69–73. doi: 10.1016/0960-8966(94)00017-4. [DOI] [PubMed] [Google Scholar]
- Jungbluth H, Sewry CA, Muntoni F. The congenital myopathies. In: Schapira AHV, et al., editors. Neurology and clinical neuroscience. Elsevier; Amsterdam: 2007. [Google Scholar]
- Kaplan JC. Gene table of monogenic neuromuscular disorders (nuclear genome only) Neuromuscul Disord. 2009;19:77–98. doi: 10.1016/j.nmd.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Leifsdottir G, Benedikz JE, Johannesson G, Jonsson JJ, Sveinbjornsdottir S. Prevalence of myotonic dystrophy in Iceland. Laeknabladid. 2005;91:829–34. [PubMed] [Google Scholar]
- Leshinsky-Silver E, Argov Z, Rozenboim L, Cohen S, Tzofi Z, Cohen Y, et al. Dysferlinopathy in the Jews of the Caucasus: a frequent mutation in the dysferlin gene. Neuromuscul Disord. 2007;17:950–4. doi: 10.1016/j.nmd.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Lisi MT, Cohn RD. Congenital muscular dystrophies: new aspects of an expanding group of disorders. Biochim Biophys Acta. 2007;1772:159–72. doi: 10.1016/j.bbadis.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Lunemann JD, Martin R. Epidemiology and genetics of multiple sclerosis. In: Schapira AHV, Byrne E, DiMauro S, Frackowiak RSJ, Johnson RT, Mizuno Y, et al., editors. Neurology and clinical neuroscience. Elsevier; Amsterdam: 2007. [Google Scholar]
- Moore SA, Shilling CJ, Westra S, Wall C, Wicklund MP, Stolle C, et al. Limb-girdle muscular dystrophy in the United States. J Neuropathol Exp Neurol. 2006;65:995–1003. doi: 10.1097/01.jnen.0000235854.77716.6c. [DOI] [PubMed] [Google Scholar]
- Mostacciuolo ML, Pastorello E, Vazza G, Miorin M, Angelini C, Tomelleri G, et al. Facioscapulohumeral muscular dystrophy: epidemiological and molecular study in a north-east Italian population sample. Clin Genet. 2009;75:550–5. doi: 10.1111/j.1399-0004.2009.01158.x. [DOI] [PubMed] [Google Scholar]
- Muntoni F, Torelli S, Brockington M. Muscular dystrophies due to glycosylation defects. Neurotherapeutics. 2008;5:627–32. doi: 10.1016/j.nurt.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norwood F, de Visser M, Eymard B, Lochmuller H, Bushby K. EFNS guideline on diagnosis and management of limb girdle muscular dystrophies. Eur J Neurol. 2007;14:1305–12. doi: 10.1111/j.1468-1331.2007.01979.x. [DOI] [PubMed] [Google Scholar]
- Okada M, Kawahara G, Noguchi S, Sugie K, Murayama K, Nonaka I, et al. Primary collagen VI deficiency is the second most common congenital muscular dystrophy in Japan. Neurology. 2007;69:1035–42. doi: 10.1212/01.wnl.0000271387.10404.4e. [DOI] [PubMed] [Google Scholar]
- Padberg GW, Frants RR, Brouwer OF, Wijmenga C, Bakker E, Sandkuijl LA. Facioscapulohumeral muscular dystrophy in the Dutch population. Muscle Nerve Suppl. 1995;2:S81–4. [PubMed] [Google Scholar]
- Peat RA, Smith JM, Compton AG, Baker NL, Pace RA, Burkin DJ, et al. Diagnosis and etiology of congenital muscular dystrophy. Neurology. 2008;71:312–21. doi: 10.1212/01.wnl.0000284605.27654.5a. [DOI] [PubMed] [Google Scholar]
- Phillips BA, Zilko PJ, Mastaglia FL. Prevalence of sporadic inclusion body myositis in Western Australia. Muscle Nerve. 2000;23:970–2. doi: 10.1002/(sici)1097-4598(200006)23:6<970::aid-mus20>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Robertson NP, Deans J, Compston DA. Myasthenia gravis: a population based epidemiological study in Cambridgeshire, England. J Neurol Neurosurg Psychiatry. 1998;65:492–6. doi: 10.1136/jnnp.65.4.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz A, Leturcq F, Cobo AM, Poza JJ, Ferrer X, Otaegui D, et al. LGMD2A: genotype-phenotype correlations based on a large mutational survey on the calpain 3 gene. Brain. 2005;128:732–42. doi: 10.1093/brain/awh408. [DOI] [PubMed] [Google Scholar]
- Schaefer AM, McFarland R, Blakely EL, He L, Whittaker RG, Taylor RW, et al. Prevalence of mitochondrial DNA disease in adults. Ann Neurol. 2008;63:35–9. doi: 10.1002/ana.21217. [DOI] [PubMed] [Google Scholar]
- Selcen D. Myofibrillar myopathies. Curr Opin Neurol. 2008;21:585–9. doi: 10.1097/WCO.0b013e32830a752b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sveen ML, Schwartz M, Vissing J. High prevalence and phenotype-genotype correlations of limb girdle muscular dystrophy type 2I in Denmark. Ann Neurol. 2006;59:808–15. doi: 10.1002/ana.20824. [DOI] [PubMed] [Google Scholar]
- Udd B. Molecular biology of distal muscular dystrophies—sarcomeric proteins on top. Biochim Biophys Acta. 2007;1772:145–58. doi: 10.1016/j.bbadis.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Udd B. 165th ENMC International Workshop: Distal myopathies 6-8th February 2009 in Naarden, The Netherlands. Neuromuscul Disord. 2009;19:429–38. doi: 10.1016/j.nmd.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Udd B, Meola G, Krahe R, Thornton C, Ranum L, Day J, et al. Report of the 115th ENMC workshop: DM2/PROMM and other myotonic dystrophies. 3rd Workshop, 14-16 February 2003, Naarden, The Netherlands. Neuromuscul Disord. 2003;13:589–96. doi: 10.1016/s0960-8966(03)00092-0. [DOI] [PubMed] [Google Scholar]
- van der Kooi AJ, Frankhuizen WS, Barth PG, Howeler CJ, Padberg GW, Spaans F, et al. Limb-girdle muscular dystrophy in the Netherlands: gene defect identified in half the families. Neurology. 2007;68:2125–8. doi: 10.1212/01.wnl.0000264853.40735.3b. [DOI] [PubMed] [Google Scholar]
- Walton JN, Nattrass FJ. On the classification, natural history and treatment of the myopathies. Brain. 1954;77:169–231. doi: 10.1093/brain/77.2.169. [DOI] [PubMed] [Google Scholar]
- Wirth B. An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA) Hum Mutat. 2000;15:228–37. doi: 10.1002/(SICI)1098-1004(200003)15:3<228::AID-HUMU3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Yotova V, Labuda D, Zietkiewicz E, Gehl D, Lovell A, Lefebvre JF, et al. Anatomy of a founder effect: myotonic dystrophy in Northeastern Quebec. Hum Genet. 2005;117:177–87. doi: 10.1007/s00439-005-1298-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.