A new in vivo role is defined for the yeast F-BAR protein Hof1 in polarized cell growth. Hof1 dimers bind to the FH1 domains of the formin Bnr1 and inhibit actin polymerization while the formin remains attached to filament ends. This activity is required in vivo for normal actin cable architecture and polarized secretion.
Abstract
Asymmetric cell growth and division rely on polarized actin cytoskeleton remodeling events, the regulation of which is poorly understood. In budding yeast, formins stimulate the assembly of an organized network of actin cables that direct polarized secretion. Here we show that the Fer/Cip4 homology–Bin amphiphysin Rvs protein Hof1, which has known roles in cytokinesis, also functions during polarized growth by directly controlling the activities of the formin Bnr1. A mutant lacking the C-terminal half of Hof1 displays misoriented and architecturally altered cables, along with impaired secretory vesicle traffic. In vitro, Hof1 inhibits the actin nucleation and elongation activities of Bnr1 without displacing the formin from filament ends. These effects depend on the Src homology 3 domain of Hof1, the formin homology 1 (FH1) domain of Bnr1, and Hof1 dimerization, suggesting a mechanism by which Hof1 “restrains” the otherwise flexible FH1-FH2 apparatus. In vivo, loss of inhibition does not alter actin levels in cables but, instead, cable shape and functionality. Thus Hof1 tunes formins to sculpt the actin cable network.
INTRODUCTION
Asymmetric cell division plays an essential role in a variety of processes, including polar body extrusion in oocytes (Li and Albertini, 2013), tissue patterning during development (Gonczy, 2008), and stem cell renewal and differentiation (Fuchs and Chen, 2013). In each of these settings, there is a requirement for cells to maintain an axis of polarity, which directs intracellular traffic of cargoes to one end of the cell to complete division and ensure selective inheritance of materials to the daughter cell. The budding yeast Saccharomyces cerevisiae divides asymmetrically during vegetative (mitotic) growth, and the general mechanisms it uses to accomplish this appear to be widely conserved with other eukaryotes (Bi and Park, 2012).
S. cerevisiae grows asymmetrically by initiating bud formation at one end of the cell at a cortical landmark and then rapidly reorganizing its actin cable network such that it grows from this site and directs secretion to the bud (Pruyne et al., 2004; Moseley and Goode, 2006; Johnson et al., 2011). Actin cables are essential for polarized cell growth and are continuously polymerized and turned over with a life cycle of 5–15 s (Pruyne et al., 1998; Yang and Pon, 2002; Yu et al., 2011). The formation of cables depends on the ongoing actin assembly-promoting activities of formins (Evangelista et al., 2002; Sagot et al., 2002). This family of proteins is highly conserved in eukaryotes and performs essential roles in a wide range of actin-based processes (Firat-Karalar and Welch, 2011; Breitsprecher and Goode, 2013). S. cerevisiae expresses two formins, Bni1 and Bnr1, which localize during polarized growth to the bud tip and bud neck, respectively (Pruyne et al., 1998; Buttery et al., 2007). From these positions, Bni1 and Bnr1 assemble actin cables that extend toward the back of the mother cell (retrograde) at 0.5–1.0 μm/s (Yang and Pon, 2002; Yu et al., 2011), providing linear tracks for myosin V (Myo2)–dependent transport of secretory vesicles and other cargoes in the anterograde direction at a rate of 3.0 μm/s (Schott et al., 2002). Because this system of cable assembly and turnover is so highly dynamic, maintaining proper cable length, shape, and mechanical properties requires continuous and precise control of formin activities, and small changes in formin activity regulated at the molecular level can lead to pronounced changes in cable architecture and function (Chesarone et al., 2009; Chesarone-Cataldo et al., 2011).
Formins catalyze both the nucleation and elongation phases of actin filament assembly using their C-terminal formin homology 1 (FH1) and FH2 domains and adjacent diaphanous autoregulatory domain (DAD). FH2 is an antiparallel homodimer that binds tightly to and processively tracks the growing end of the filament (Higashida et al., 2004; Kovar and Pollard, 2004; Xu et al., 2004; Breitsprecher and Goode, 2013). This activity of FH2 helps to shield growing ends from capping proteins while allowing insertion of new actin subunits (Zigmond et al., 2003; Harris et al., 2004; Moseley et al., 2004). The adjacent FH1 domains contain multiple polyproline stretches that bind profilin–actin monomer complexes and guide monomers onto the filament end to accelerate elongation (Romero et al., 2004; Kovar et al., 2006). Precisely how formins catalyze filament nucleation is less clear, but it involves recruitment of actin monomers by the FH2 and DAD domains, and possibly FH1 domains, and can be strongly stimulated by formin-interacting nucleation promoting factors, such as adenomatous polyposis coli, Spire, and Bud6 (Li and Higgs, 2003; Paul and Pollard, 2008; Okada et al., 2010; Gould et al., 2011; Graziano et al., 2011; Vizcarra et al., 2011). In cells, these potent activities of formins must also be tightly controlled, both spatially and temporally, in order to generate actin networks with precise architectures tailored to their functions (Chesarone et al., 2010).
Fer/Cip4 homology–Bin amphiphysin Rvs (F-BAR) family proteins play prominent roles in endocytosis and cytokinesis and typically have a dimeric N-terminal F-BAR domain that binds lipid membranes and one or two C-terminal Src homology 3 (SH3) domains (Roberts-Galbraith and Gould, 2010). S. cerevisiae expresses three F-BAR proteins: Syp1, Bzz1, and Hof1. Syp1 is an early-arriving component of cortical endocytic patches and directly inhibits Wiskott–Aldrich syndrome protein (WASP)/Las17 to block Arp2/3 complex–mediated actin assembly until the proper stage of endocytosis (Rodal et al., 2003; Sun et al., 2006; Boettner et al., 2009; Feliciano and Di Pietro, 2012). Bzz1 is a later-arriving component of endocytic actin patches and directly binds WASP/Las17 to help trigger Arp2/3 complex–mediated actin assembly (Sun et al., 2006). In contrast, Hof1 localizes exclusively to the bud neck during bud growth and cell division and directly interacts with septins through one of its N-terminal coiled-coil domains (Meitinger et al., 2013; Oh et al., 2013). Hof1 function has only been well studied during cytokinesis, in which it plays an important role in assembling the primary septum and regulating actomyosin ring contraction (Lippincott and Li, 1998; Korinek et al., 2000; Vallen et al., 2000; Meitinger et al., 2011). However, Hof1 also has reported physical interactions with the actin regulatory proteins Las17/WASP, Vrp1/WASP-interacting protein (WIP), and Bnr1 (Kamei et al., 1998; Naqvi et al., 2001; Tong et al., 2002; Rajmohan et al., 2009). Further, hof1Δ cells have enlarged mother cells (Vallen et al., 2000), which leaves open the possibility of Hof1 functioning to promote polarized growth.
Prompted by these earlier observations, we investigate the potential for Hof1 regulation of the actin cytoskeleton during polarized growth. We find that the C-terminal half of Hof1 is required for proper actin cable organization and polarized secretory vesicle trafficking in vivo and that this same region of Hof1 directly inhibits the actin assembly activities of Bnr1 in vitro. Further analysis shows that the SH3 domains of Hof1 interact with the FH1 domains of Bnr1 to “restrain” the formin, inhibiting actin filament nucleation and elongation. In vivo, the loss of these inhibitory effects does not alter the levels of actin in cables, but instead, the shape and geometry of the cable network. Thus formin inhibition by Hof1 at the bud neck is used as a mechanism to sculpt the growing actin network.
RESULTS
The C-terminal half of Hof1 regulates actin cable organization in mother cells
To investigate the potential functions of Hof1 in actin regulation during polarized growth, we compared actin cytoskeleton organization and function in hof1 mutant and wild-type cells. For this, we generated both a hof1Δ strain and a hof1ΔCT strain (Figure 1A). The hof1ΔCT mutant lacks the C-terminal half of the protein, which is predicted to mediate interactions with the actin regulatory proteins mentioned previously (see Introduction). Our hof1Δ strain was impaired for cell growth at elevated temperatures, consistent with previous studies (Lippincott and Li, 1998; Vallen et al., 2000; Oh et al., 2013), and the hof1ΔCT strain showed similar growth defects (Figure 1B).
FIGURE 1:
Cell growth and F-actin organization defects of hof1 mutants. (A) Domain layout of Hof1 and Hof1ΔCT construct. CC2, coiled-coil domain 2. (B) Fivefold serial dilutions of yeast strains grown on YEPD plates at 25, 30, 34, and 37°C. (C) Quantification of F-actin fluorescence intensity in individual actin patches. Each triangle represents a single actin patch. Error bars, SEM. (D) Endocytic patch dynamics in wild-type and hof1Δ cells. Sla1-GFP, an early endocytic coat marker, and Abp1-mRFP, a late actin marker, were coexpressed in hof1Δ cells and an isogenic wild-type strain. Top, single frames of representative cells. Bottom, kymographs of a single endocytic patch in the WT cell. Time is shown on the x-axis and distance on the y-axis, with the interior of the cell on the bottom. (E) Average Sla1-GFP and Abp1-mRFP lifetime in WT and hof1Δ strains. Lifetime was calculated from fluorescence intensity plots as the longest sequence of consecutive positive values. Each symbol represents the fluorescence lifetime of one patch. Error bars, SEM. (F) Cellular F-actin organization. Cells were grown to log phase at 25°C in YEPD, fixed, and stained with Alexa Fluor 488–phalloidin. All images are composites of multiple individual images, separated by white lines. Scale bar, 5 μm. (G) Visual scoring of actin cable phenotypes for strains in F. Parallel, cells for which almost all cables in the mother were aligned along the bud–mother axis; misoriented, cells for which most cables ran across the width of the mother (transverse) or cables appeared to be randomly oriented within the mother (meshworks). n > 150 cells for each strain. Error bars, SD. (H) Live-cell imaging of indicated strains expressing ABP140-3GFP in log phase. Cells are representative selections from images in Supplemental Figure S1B. Scale bar, 5 μm. (I) Actively elongating cables in live cells were visualized using an Abp140-3GFP marker. Cable “extension angles” (area shaded in red) correspond to the angles of cable elongation relative to the mother–bud axis. (J) “Extension angles” of individual cables of indicated strains were quantitatively measured as in I using custom Matlab code. Each triangle denotes a single cable. Trials 1 and 2 denote independent experiments performed on different days. Angles >90° indicate cables making a U-turn and growing back toward the bud neck. n > 25 cables per strain per trial. Error bars, SEM. (K) Cells expressing GFP-Sec4 from a 2-μm plasmid were observed over a period of 60 s. Vesicles were categorized as anterograde (rapid vectoral movement toward the bud) or random/retrograde (rapid movement away from the bud or slow, less-directed movements). Error bars, SD. In all cases in which statistical significant was determined, Student's t test was used.
Although Hof1 has been reported to localize to the bud neck (Lippincott and Li, 1998; Korinek et al., 2000; Vallen et al., 2000), it is a member of the F-BAR protein family, which has conserved roles in endocytosis (Roberts-Galbraith and Gould, 2010). Further, Hof1 has reported physical interactions with endocytic/actin patch proteins Las17/WASP, Vrp1/WIP, and Bbc1 (Naqvi et al., 2001; Tong et al., 2002; Rajmohan et al., 2009). For these reasons, we investigated whether Hof1 influences cortical actin patch function. We first quantitatively compared the F-actin fluorescence intensities of actin patches in fixed wild-type, hof1Δ, and hof1ΔCT cells using Alexa Fluor 488–phalloidin. All three strains had similar average patch intensities, suggesting that Hof1 does not play a significant role in regulating the F-actin levels of these structures (Figure 1C). To assess potential defects in endocytosis, we used live-cell imaging to compare cortical patch lifetimes in wild-type and hof1Δ strains, simultaneously monitoring an early endocytic coat marker (Sla1–green fluorescent protein [GFP]) and a late F-actin marker (Abp1-monomeric red fluorescent protein [mRFP]) in the same cells (Figure 1D and Supplemental Figure S1A). Endocytosis proceeds in discrete stages that are highly stereotyped, with components arriving at and departing from the cortical site with a high degree of temporal precision (Weinberg and Drubin, 2012). Thus, if Hof1 has an important role in regulating this process, hof1Δ cells should exhibit differences in the lifetimes of the markers at endocytic sites. However, we found that the average lifetimes of Sla1-GFP and Abp1-mRFP at endocytic sites were not significantly different between hof1Δ and wild-type cells (Figure 1E). These results suggest that Hof1, unlike the other two yeast F-BAR proteins, Syp1 and Bzz1, does not play a significant role during endocytosis.
We next compared F-actin organization in fixed cells from wild-type, hof1Δ, and hof1ΔCT strains. Consistent with previous studies, we observed no obvious defects in the distribution of actin patches (Lippincott and Li, 1998; Korinek et al., 2000; Vallen et al., 2000). However, in hof1Δ and hof1ΔCT cells, there were visible defects in actin cable organization. Whereas most wild-type cells had cable networks aligned along the mother–bud axis, hof1Δ and hof1ΔCT cells showed a substantial increase in the percentage of cells with “transverse” cables or cable “meshworks” lacking polarity (Figure 1, F and G). As a second independent test of this phenotype, we generated wild-type, hof1Δ, and hof1ΔCT strains expressing an integrated cable marker (Abp140-3GFP) and imaged cable network organization in live cells (Figure 1H and Supplemental Figure S1B). This yielded results similar to the analysis in fixed cells, confirming that both hof1Δ and hof1ΔCT cells have defects in actin cable organization (Supplemental Figure S1C). Further, it revealed that cables in hof1Δ and hof1ΔCT cells are abnormally decorated by Abp140-3GFP, with a more fragmented pattern in mutants than in wild-type cells (Figure 1H and Supplemental Figure S1B). Because Abp140 is an actin-bundling protein (Asakura et al., 1998), the fragmented decoration pattern suggests that hof1Δ and hof1ΔCT cells may have altered cable architecture (see Discussion).
We considered two hypotheses that might account for the increase in misoriented cables in hof1 mutants. First, the cables could become overgrown and buckle upon hitting the rear of the mother cell. Second, the cables could grow with an altered orientation well before they reach the mother cell cortex. To distinguish between these two possibilities, we used live-cell imaging to measure the angle of growth, with respect to the mother–bud axis, for Abp140-3GFP–marked cables while they were elongating in mother cells (Figure 1I). The average extension angles of cables (calculated for two independent experiments) were 43 and 55° for hof1ΔCT cells and 18 and 25° for wild-type cells (Figure 1J). These observations suggest that the misoriented cable phenotype of hof1ΔCT cells (Figure 1G) arises at least in part from cables growing transversely before they are long enough to contact the back of the mother cell.
Because cables serve as tracks for myosin V (Myo2)–dependent trafficking of post-Golgi secretory vesicles from the mother cell to the bud (Govindan et al., 1995), we also used live-cell imaging to compare secretory vesicle movements (marked by GFP-Sec4) in wild-type, hof1Δ, and hof1ΔCT cells (Supplemental Videos S1–S6). Nearly 80% of the vesicles tracked in wild-type cells showed persistent, directed movements toward the bud in the 60-s observation window. In contrast, these bud-directed movements were greatly reduced in hof1Δ and hof1ΔCT cells (Figure 1K). Many of the vesicles in the mutants followed meandering (less-directed) paths, consistent with the misoriented cables in the hof1 mutants. Taken together, these observations show that the C-terminal half of Hof1 is required for proper cable organization in mother cells and in turn for efficient polarized secretion.
The C-terminal half of Hof1 regulates actin cable organization through Bnr1
The actin cable network in S. cerevisiae is assembled by two formins, Bni1 and Bnr1 (Evangelista et al., 2002; Sagot et al., 2002). Therefore we investigated whether Hof1’s effects on cable organization depended on Bni1 and/or Bnr1. To accomplish this, we generated a hof1Δ bnr1Δ strain and compared its cable organization to wild-type and single-mutant strains by the same criteria as in Figure 1G. Deletion of BNR1 suppressed the misoriented cable phenotype caused by hof1Δ (Figure 2, A and B). Because in the absence of BNR1 all cable assembly depends on Bni1, this result suggests that the C-terminal half of Hof1 regulates Bnr1, instead of Bni1-dependent actin cable organization.
FIGURE 2:
Analysis of hof1 phenotypes in the absence of Bni1 or Bnr1 function. (A) F-actin staining of indicated strains, performed as in Figure 1F. Scale bar, 5 μm. (B) Scoring of actin cable phenotypes for strains in A, using same criteria as in Figure 1G. Data for wild-type and hof1Δ strains in Figure 1G are shown to aid in comparison. n > 150 cells for each strain. Error bars, SD. (C) Cellular F-actin organization. Cells were grown to log phase at 25°C in YEPD and either fixed immediately (left) or incubated at 35°C for 5 min before fixation (right). Staining and imaging were performed as in Figure 1F. Scale bar, 5 μm. (D) Scoring of actin cable phenotypes for strains in C, using the same criteria as in Figure 1G. n > 100 cells for each strain. Error bars, SD. (E, F) Quantification of mother cell volumes. Each triangle represents a single cell. Error bars, SEM. (G) Quantification of normalized F-actin levels among the indicated strains. Each triangle indicates a single cell. Error bars, SEM. (H) Schematic for Hof1 functions in polarized growth. The C-terminal half (red boxes) functions upstream of Bnr1 to regulate actin cable organization. The N-terminal half (white boxes) operates by a Bnr1-independent pathway to regulate cell size and also regulates actin cable abundance. In all cases in which statistical significant was determined, Student's t test was used.
Because hof1Δ is synthetic lethal with bni1Δ (Kamei et al., 1998), we could not test whether bni1Δ suppresses the cable defects caused by hof1Δ. Moreover, we attempted to generate a bni1Δ hof1ΔCT double mutant by crossing bni1Δ and hof1ΔCT single mutants; however, all of the predicted double-mutant spores were inviable (Supplemental Figure S2A). As an alternative strategy, we generated bni1-1 hof1Δ and bni1-1 hof1ΔCT strains. The bni1-1 allele is temperature sensitive, such that bni1-1 bnr1Δ cells have cables and are viable at 25°C but at elevated temperatures rapidly lose cables and arrest growth (Sagot et al., 2002). We observed that bni1-1 hof1Δ and bni1-1 hof1ΔCT strains grew normally at 25°C but not at higher temperatures, at which BNI1 function was disrupted (Supplemental Figure S2B). After a 5-min shift to the nonpermissive temperature (35°C), control bni1-1 bnr1Δ cells showed complete loss of cables (Figure 2C, top), demonstrating that the shift abolishes BNI1 function. Because neither of the bni1-1 hof1 double-mutant strains showed a significant increase in cable organization defects compared with bni1-1 after the short temperature shift (Figure 2, C and D), our observations further support the view that the C-terminal half of Hof1 regulates organization of actin cables produced by Bnr1 rather than Bni1. In addition, since hof1Δ cells show similar amounts of Bnr1 at the bud neck as wild-type cells (Supplemental Figure S2, C and D), the cable organization defects we observed in our hof1Δ strain do not appear to arise from aberrant Bnr1 localization.
The N-terminal half of Hof1 governs mother cell size and actin cable levels
Two earlier studies showed that hof1Δ cells are substantially larger than isogenic wild-type cells (Vallen et al., 2000; Jendretzki et al., 2009), suggesting a defect in polarized growth. To test whether this cell size defect depends on Hof1’s role in actin cable organization, we measured the volumes of mother cells during polarized growth in wild-type, hof1Δ, and hof1ΔCT strains (see Materials and Methods). Whereas wild-type cells had an average volume of 27 ± 1 μm3, similar to the value reported using z-series differential interference contrast imaging (Ferrezuelo et al., 2012), hof1Δ cells were about twice as large, with an average volume of 51 ± 3 μm3. hof1ΔCT cells had an average volume of 36 ± 4 μm3 (Figure 2E), suggesting that proper cell size is controlled by activities contained in the N-terminal half of Hof1. Further, whereas the average cell size for the hof1ΔCT strain was slightly larger than that for the wild-type strain, the distribution of cell sizes in the hof1ΔCT strain was similar to that for wild type, with only a small fraction of cells approaching hof1Δ-like sizes. These results suggest that the N-terminal half of Hof1 performs an important function in polarized growth that affects cell size, which is separate from the function of its C-terminus in cable organization. Consistent with this view, we additionally found that deletion of BNR1 failed to suppress the increased cell size of the hof1Δ strain (Figure 2F).
In our initial inspection of the actin phenotypes (Figure 1F), we also noticed that hof1Δ cells had higher levels of actin cables than wild-type and hof1ΔCT cells. To investigate this further, we quantified cable levels in mother compartments by acquiring z-series of mother cells stained for F-actin, measuring total fluorescence intensity for each focal plane and summing the intensities to determine the total amount of F-actin. This analysis showed that hof1Δ mother cells have substantially higher levels of F-actin incorporated into the cable network than do wild-type mother cells (Supplemental Figure S2E). However, as mentioned earlier, mother cells are substantially larger in the hof1Δ than in the wild-type strain, which prompted us to also normalize F-actin levels for cell volume. After normalization, hof1Δ mother cells still had ∼25% higher F-actin cable levels per unit volume than wild-type or hof1ΔCT mother cells (Figure 2G). Our data show that expression of the N-terminal half of Hof1 (in the hof1ΔCT strain) complements the increase in cell size and elevated cable levels (Figure 2, E and G) but not the cable misorientation defects (Figure 1G and Supplemental Figure S1C). Thus the N-terminus of Hof1 is sufficient to govern cell size and cable levels, whereas the C-terminal half is required for proper organization of Bnr1-assembled cables (Figure 2H).
Hof1 directly inhibits Bnr1-mediated actin assembly in vitro
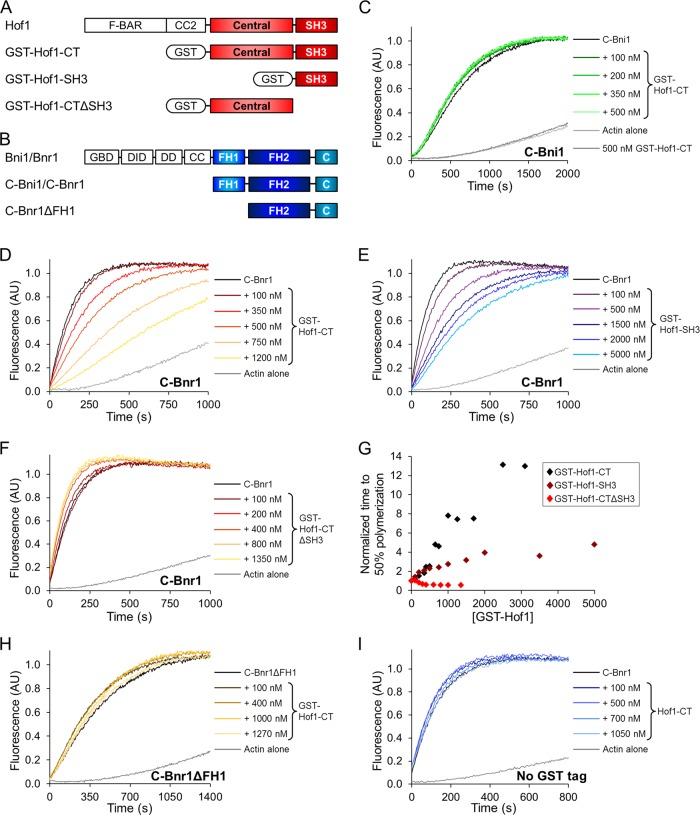
The foregoing genetic analyses indicated that the C-terminal half of Hof1 regulates Bnr1-dependent actin cable organization, and previously it was reported that the SH3 domain of Hof1 interacts with Bnr1 (Kamei et al., 1998). Therefore we investigated whether the C-terminal half of Hof1 directly influences the actin assembly–promoting activities of Bnr1. To accomplish this, we purified glutathione S-transferase (GST)–Hof1-CT (residues 350–669) and tested its effects on C-Bnr1 and C-Bni1 (Figure 3, A and B). We attempted to purify full-length Hof1, but expression levels were extremely low. Because the N-terminal F-BAR domain is predicted to dimerize Hof1 (Roberts-Galbraith and Gould, 2010), we kept the GST tag on our Hof1 constructs to retain this function. In bulk actin assembly assays, GST-Hof1-CT potently inhibited the effects of C-Bnr1 but not C-Bni1 (Figure 3, C and D, and Supplemental Figure S3A). Of importance, GST-Hof1-CT did not affect actin assembly in the absence of C-Bnr1 (Figure 3C, gray curves). Hof1 showed similar inhibitory effects on Bnr1 when yeast actin was used instead of rabbit muscle actin (RMA; Supplemental Figure S3B).
FIGURE 3:
Biochemical effects of purified Hof1 truncations on formin activity in bulk assembly assays performed in the absence of profilin. (A) Domain layouts of purified Hof1 truncations. Unless otherwise noted, all Hof1 truncations contained an N-terminal GST tag. Hof1-CT includes residues 350–669; Hof1-SH3, residues 575–669; Hof1-CTΔSH3, residues 350x574. (B) Domain layouts of purified formins. GBD, GTPase-binding domain; DID, diaphanous inhibitory domain; DD, dimerization domain; CC, coiled coils. C-Bni1 and C-Bnr1 encompass the FH1 domain to the C-terminus (residues 1227–1953 and 757–1375 respectively); C-Bnr1ΔFH1 includes the FH2 domain to the C-terminus (residues 868–1375). (C) Monomeric actin, 2 μM, was polymerized in the presence of 10 nM C-Bni1 and the indicated concentrations of GST-Hof1-CT. (D–F) Monomeric actin, 2 μM, was polymerized in the presence of 2 nM C-Bnr1 and the indicated concentrations of GST-Hof1-CT (D), GST-Hof1-SH3 (E), or GST-Hof1-CTΔSH3 (F). AU, arbitrary units. (G) Concentration-dependent effects of Hof1 truncations on C-Bnr1–mediated actin assembly. Each diamond indicates a single reaction. (H) Monomeric actin, 2 μM, was polymerized in the presence of 2 nM C-Bnr1ΔFH1 and the indicated concentrations of GST-Hof1-CT. (I) Monomeric actin, 2 μM, was polymerized in the presence of 2 nM C-Bnr1 and the indicated concentrations of Hof1-CT lacking a GST tag.
Next we asked which regions of Hof1-CT mediate inhibition of C-Bnr1 by testing two additional constructs, GST-Hof1-SH3 (residues 575–669) and GST-Hof1-CTΔSH3 (residues 350–574, or “central domain”). In bulk actin assembly assays, GST-Hof1-SH3 showed inhibitory effects on C-Bnr1 (Figure 3E), but they were less potent than those of GST-Hof1-CT (Figure 3G, compare black and crimson data points), and GST-Hof1-CTΔSH3 had no effect on C-Bnr1 activity (Figure 3, F and G). Together these results indicate that the SH3 domain of Hof1 is required for the inhibitory effects on C-Bnr1 and that the central domain makes additional contributions.
In a reciprocal set of experiments, we asked which domains of C-Bnr1 (Figure 3B) are required for inhibition by Hof1. GST-Hof1-CT failed to inhibit C-Bnr1ΔFH1, even at high concentrations (Figure 3H), consistent with Hof1 binding to the FH1 domain of Bnr1 (Kamei et al., 1998; Tonikian et al., 2009). To test the importance of dimerization by Hof1 for its inhibitory activities, we proteolytically removed the GST tag, producing Hof1-CT. This construct had no effect on C-Bnr1 activity (Figure 3I), indicating that dimerization of Hof1 is critical for its ability to inhibit Bnr1.
Hof1 inhibitory effects on Bnr1-mediated actin filament nucleation and elongation
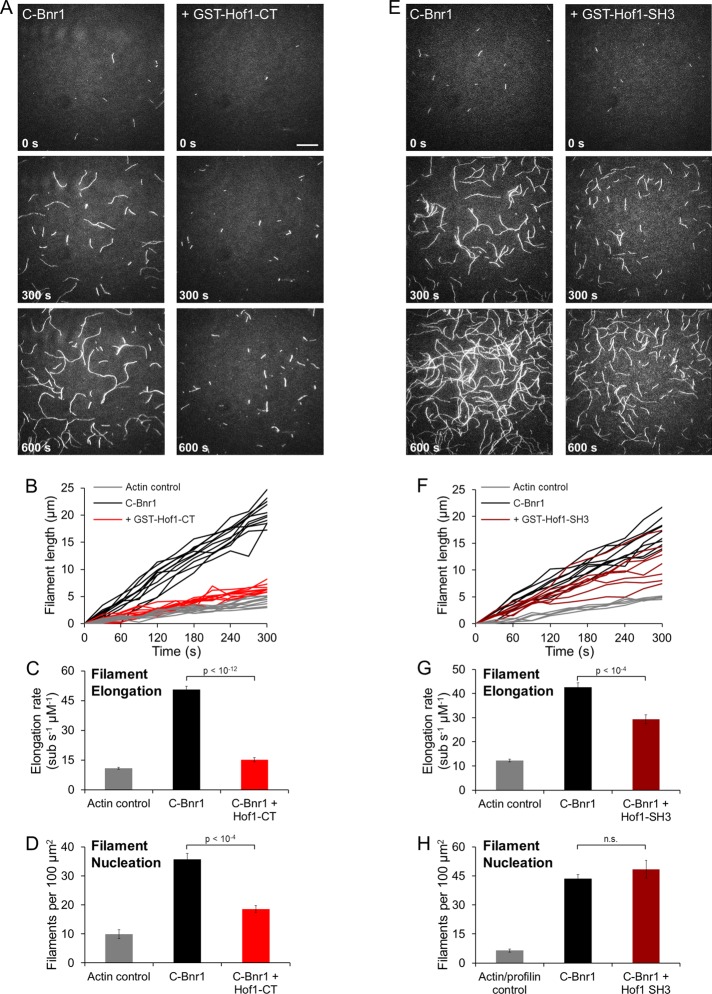
The foregoing results leave open the possibility that Hof1 inhibits the activities of Bnr1 in actin nucleation and/or elongation, since bulk assays do not distinguish between these effects. Therefore we next used total internal reflection fluorescence (TIRF) microscopy to directly visualize the effects of Hof1 on individual filaments. In the presence of profilin, C-Bnr1 increased the average elongation rate of filaments from 11 ± 1 (control) to 51 ± 2 subunits μM−1 s−1. GST-Hof1-CT inhibited C-Bnr1 effects, reducing the elongation rate to 15 ± 1 subunits μM−1 s−1 (Figure 4, A–C). In the same experiments, we measured the average density of filaments per field of view to assess nucleation efficiency. In control reactions (actin and profilin, no C-Bnr1), the average density was 10 ± 2 filaments/100 μm2. Addition of 0.1 nM C-Bnr1 increased the density to 36 ± 2 filaments/100 μm2. GST-Hof1-CT inhibited C-Bnr1, reducing the density to 19 ± 1 filaments/100 μm2 (Figure 4, A and D). Thus GST-Hof1-CT suppresses both the actin nucleation and elongation effects of C-Bnr1. Of importance, the smaller GST-Hof1-SH3 construct failed to inhibit C-Bnr1–mediated actin nucleation and only partially suppressed elongation (Figure 4, E–H), consistent with its relatively weak inhibitory effects in bulk assays.
FIGURE 4:
Effects of purified Hof1 truncations on Bnr1-dependent actin nucleation and elongation performed in the presence of profilin using single-color TIRF microscopy. (A, E) Monomeric actin, 0.5 μM, 10% Oregon green labeled, was polymerized in the presence of 0.1 nM C-Bnr1 with or without 100 nM GST-Hof1-CT (A) or 0.2 nM C-Bnr1 with or without 500 nM GST-Hof1-SH3 (E) and observed for 600 s, with representative fields of view shown for indicated time points. Scale bar, 20 μm. (B, F) Filament length vs. time for individual filaments observed in A (B) or E (F). Filaments were measured every 30 s and normalized to a length of 0 μm at 0 s. (C, G) Mean elongation rates for filaments observed in B (C) or F (G). Elongation rates were determined from the mean slope of individual filament traces (e.g., B and F). n > 10 filaments for each condition. Error bars, SEM. (D, H) Quantification of filament densities for reactions in A (D) or E (H). Each bar represents a mean of at least six fields of view from at least two independent experiments. Error bars, SEM. In all cases in which statistical significant was determined, Student's t test was used.
Together our observations from bulk and TIRF analysis show that the SH3 domain of Hof1 is required for all inhibitory effects on nucleation and elongation but alone is sufficient to only partially inhibit elongation. The central domain of Hof1 is required for any inhibition of nucleation and for full inhibition of elongation but on its own has no effect.
Hof1 decelerates actin filament elongation without displacing Bnr1 from growing ends
A variety of mechanisms could account for the inhibitory effects of GST-Hof1-CT on filament elongation. In particular, Hof1-CT could displace C-Bnr1 from growing ends of filaments or, alternatively, associate with C-Bnr1 and interfere with actin subunit addition while the formin is still on the ends of filaments. To distinguish between these two possibilities, we performed a two-color TIRF assay in which we tethered SNAP-649-PEG-Biotin-C-Bnr1 molecules to the surface and grew untethered actin filaments (10% Oregon green [OG] labeled) from these sites (Figure 5A). Of importance, only the formin is anchored in these experiments, and therefore dissociation of the FH2 domain from the barbed end of the filament results in the filament diffusing away. Thus we could ask whether GST-Hof1-CT can reduce the elongation rate of filaments while the formin is still attached to the barbed end. In control reactions containing actin and profilin, filaments grew from Bnr1-anchored sites at an average rate of 39 ± 1 subunits μM−1 s−1 (Figure 5B), similar to the elongation rates of filaments generated by non–SNAP-tagged, untethered C-Bnr1 (compare Figures 4C and 5C, black bars). Further addition of GST-Hof1-CT reduced the elongation rate to 19 ± 1 subunits μM−1 s−1 (Figure 5C). These results demonstrate that GST-Hof1-CT decreases the rate of elongation while C-Bnr1 remains attached to filament ends.
FIGURE 5:
Two-color TIRF microscopy analysis of GST-Hof1-CT effects on Bnr1-capped filaments performed in the presence of profilin. (A) Cartoon of experimental setup. In all experiments, SNAP649-PEG-Biotin-C-Bnr1 was tethered to coverslips coated with biotin-PEG using streptavidin, allowing simultaneous visualization of the formin molecules and the actin filaments grown from the tethered-formin sites. (B) Monomeric actin, 1.0 μM, 10% Oregon green labeled, was polymerized in the presence of 0.1 nM SNAP-649-PEG-Biotin-C-Bnr1 in the absence (left) or presence (right) of 100 nM GST-Hof1-CT. Formin, red channel; actin, green channel. Formin-capped filaments were identified by the presence of a red dot (insets) at the barbed end (yellow arrowhead). Blue arrowheads indicate pointed ends. Scale bar, 10 μm. (C) Mean elongation rates for filaments observed in B. Elongation rates were determined from the mean slope of individual filament traces. n > 10 filaments for each condition. Error bars, SEM. (D) Percentage of filaments displaced from tethered formin over a 7-min observation window for the indicated conditions. Filaments were scored as displaced if their barbed ends drifted >1 μm from a tethered formin at any point during the time course. Error bars, SEM. (E) A working model for Hof1 inhibitory effects on Bnr1 (see Discussion for details). In all cases in which statistical significant was determined, Student's t test was used.
In addition, we asked whether GST-Hof1-CT changes the fraction of filaments that remain attached to formins over time. We performed these tests on GST-Hof1-CT in the presence and absence of capping protein (CapZ), since mounting evidence suggests capping proteins compete with formins for barbed ends (Kovar et al., 2005; Chesarone et al., 2009; Bartolini et al., 2012). In these experiments, we first polymerized actin filaments (10% OG labeled) for 3 min, again from tethered SNAP-649-PEG-Biotin-C-Bnr1 molecules. Then we flowed in fresh actin monomers and profilin, with or without GST-Hof1-CT and/or CapZ, and monitored filament displacement for 7 min. CapZ alone competitively displaced about ∼25% of the filaments from anchored SNAP-649-PEG-Biotin-C-Bnr1 (Figure 5D). GST-Hof1-CT alone had no effect, nor did it alter the displacement effects of CapZ (Figure 5D). These observations further support the view that GST-Hof1-CT inhibits C-Bnr1-mediated elongation while the formin is processively attached to the filament end.
DISCUSSION
In this study, we showed that the F-BAR protein Hof1 directly regulates formin activity to control actin cable organization and secretory vesicle trafficking during polarized cell growth. This defines a new function for Hof1 in polarized growth, separate from its established role in cytokinesis (Lippincott and Li, 1998; Vallen et al., 2000; Meitinger et al., 2011, 2013). We found that the C-terminal half of Hof1 (Hof1-CT) inhibits, or restricts, the actin assembly activities of the formin Bnr1 but not Bni1. Because Hof1 and Bnr1 both localize to the bud neck throughout polarized growth, these proteins are ideally positioned to interact. Further, Hof1 and Bnr1 each associate with septins (Buttery et al., 2012; Oh et al., 2013), so their spacing and organization may be controlled by interactions with the septin network. Hof1 was identified as a potential binding partner of the Bnr1 FH1 domain in an earlier study (Kamei et al., 1998), as well as in a Bayesian modeling analysis of the yeast SH3 interactome, which predicted a Hof1 binding sequence in the FH1 domain of Bnr1 but not Bni1 (Tonikian et al., 2009). Here we defined the functional effects of this interaction. Given that Bnr1 and Hof1 are expressed at similar levels in cells (Ghaemmaghami et al., 2003), we propose that Hof1 plays an important role in controlling the activity of Bnr1 at the bud neck to influence actin cable architecture and cable network organization. Indeed, we generated a hof1ΔCT allele, which caused defects in the global organization of cable networks, the angles of extension of individual cables growing from the bud neck, in cable decoration by the actin-bundling protein Abp140. Together our in vivo and in vitro observations suggest that proper spatial and/or temporal inhibition of Bnr1 activity at the bud neck by Hof1 plays an important role in shaping the cable network to optimize intracellular transport.
Our data suggest that during polarized growth, Hof1 performs two distinct functions, mediated by different halves of the protein. The C-terminal half of Hof1 directly controls Bnr1 activity to influence the shape and organization of actin cables, keeping them approximately parallel to the mother–bud axis. As such, either complete deletion of HOF1 or a truncation removing the C-terminal half increases the prevalence of cables running orthogonal to this axis, and these defects correlate with wandering (less-directed) movements of secretory vesicles in the mother compartment. We also observed that loss of Hof1-CT in vivo did not result in elevated cable levels. This is likely because there is a finite pool of actin monomers in cells, which means that formins are limited in the total amount of actin cable polymer they can assemble. Under these conditions, loss of a formin inhibitor such as Hof1-CT should result in increase in the number of active Bnr1 molecules attempting to polymerize actin from the same limiting monomer pool, giving rise to disorganized networks of more numerous and shorter or thinner cables. Indeed, this is precisely what we previously observed for a constitutively active bnr1ΔDAD mutant (Chesarone et al., 2009) and what we again observe here for hof1ΔCT cells. However, the high degree of cable disorganization prevented us from being able to reliably count or measure lengths of individual cables.
Although our biochemical and genetic results strongly support a role for Hof1 in tuning the activities of Bnr1 to control the geometry and properties of the actin cable network, Hof1’s role in promoting cable organization might be more complex than our working model portrays. For instance, our live-cell cable imaging (Figure 1H) revealed that the F-actin–binding protein Abp140-3GFP decorates cables more sparsely in hof1Δ or hof1ΔCT cells than in wild-type cells. Hof1 may therefore have an additional role in ensuring that actin-associated proteins are properly loaded onto cables, affecting cable architecture and dynamics. However, as mentioned earlier, constitutively active bnr1ΔDAD mutants have disorganized cable networks (Chesarone et al., 2009), similar to hof1Δ and hof1ΔCT mutants. Thus negative regulation of Bnr1 activity at the bud neck appears to be an important control point in maintaining correct cable shape and function.
Mechanism of formin inhibition by Hof1
Using a combination of bulk actin assembly assays and TIRF microscopy, we defined the mechanism by which Hof1 influences Bnr1 activity. We showed that Hof1 potently inhibits both the actin nucleation and elongation activities of Bnr1 without displacing the formin from growing filament ends. On the basis of all our in vitro observations, we propose the following working model for Hof1 control of Bnr1 activities (Figure 5E). The primary interaction mediating inhibition is between the SH3 domain of Hof1 and the FH1 domain of Bnr1. This interaction is sufficient to reduce the rate of filament elongation by the formin only when the SH3 domain is dimerized, perhaps by reducing the freedom or flexibility of the FH1 domains, thereby interfering with FH1’s ability to “hand off” profilin–actin complexes to FH2 (a “restraint” model). We considered the alternative explanation of competition between the Hof1 SH3 domain and profilin for binding FH1. However, our comparison of dimeric (GST-Hof1-CT) and monomeric (Hof1-CT) constructs showed that even high concentrations of the monomeric Hof1-CT failed to inhibit formin-mediated elongation, which is inconsistent with such a model. The observation that Hof1 dimerization is critical for inhibition favors a “restraint” mechanism of inhibition. In addition to the effects on elongation, we observed that GST-Hof1-CT inhibited actin nucleation by C-Bnr1 and that this required both the SH3 and central domains of Hof1. These results suggest that the central domain plays an important role in either directly interacting with the FH2 domain to inhibit nucleation or indirectly obstructing nucleation by the FH2. Because all inhibitory effects require the SH3 domains, even a very weak interaction between the central domain and FH2 would be sufficient to mediate this block.
The enigmatic F-BAR domain
Although this study defines a role for the C-terminal half of Hof1 in Bnr1 regulation, it raises several new questions with regard to the function of the N-terminal half (F-BAR and coiled-coil [CC] 2 domains) in mediating cell polarization. The chief observations implicating the F-BAR/CC2 in polarity regulation are as follows: 1) Expression of a hof1 allele containing these domains (hof1ΔCT) is largely sufficient to rescue the cell size and overabundant actin cable defects of hof1Δ cells, and 2) further loss of BNR1 in hof1Δ cells does not suppress the cell size defect. Together these results imply that the F-BAR/CC2 regulates cell size and actin cable density independently of the role of the C-terminal half of Hof1 in Bnr1 regulation.
However, the exact mechanisms by which the F-BAR/CC2 might regulate mother cell size and cable levels remain elusive. In cells lacking functional tropomyosin, which is required for cable formation, mother cells grow isotropically and become enlarged, presumably due to the inability to direct secretion into the bud (Pruyne et al., 1998). Because hof1Δ cells display defects in secretory vesicle trafficking (Figure 1K), one possibility is that a portion of the misdirected vesicles in these cells are docking to the mother cortex, resulting in an increased cell size. However, we found that vesicle-trafficking defects are equally severe in a hof1ΔCT strain, in which mother cells generally have a wild type–like size, arguing against this hypothesis. Although the role of Hof1 in these processes has yet to be determined, there does appear to be a moderate correlation between cell size and total actin levels (Supplemental Figure S2E), suggesting that these two functions of Hof1 might be closely intertwined. It is worth noting that even though Hof1’s role in cytokinesis has been studied extensively, the role of the F-BAR in this process is unclear. In hof1Δ cyk3Δ cells, which arrest at cytokinesis and die (Korinek et al., 2000), expression of the C-terminal half of Hof1 partially restores viability, whereas the N-terminal half does not (Oh et al., 2013). Future work will be required to clarify the role of Hof1’s F-BAR domain in mediating both polarized cell growth and cell division.
Common roles for F-BAR proteins in regulating actin dynamics
F-BAR proteins are present in eukaryotes ranging from yeast to humans; however, the extent to which they are functionally conserved is unclear. Although it has long been known that their roles in endocytosis are shared among diverse eukaryotes (Roberts-Galbraith and Gould, 2010), it has only recently been directly shown that their functions in cytokinesis, originally characterized in yeast (Fankhauser et al., 1995; Lippincott and Li, 1998; Vallen et al., 2000; Carnahan and Gould, 2003), are also conserved in metazoans (Takeda et al., 2013). How well conserved is regulation of formin activity via F-BAR proteins? The mammalian F-BAR protein srGAP2 directly inhibits the actin filament–severing activities of the FH1 domain of FMNL1 (Mason et al., 2011). Further, the F-BAR protein Cip4 interacts with the FH1 domain of Dia during Drosophila embryogenesis and directly inhibits the formin's actin assembly activities yet promotes Arp2/3-dependent actin assembly (Yan et al., 2013). We showed that Hof1 directly inhibits formin activity, but Hof1 also interacts with yeast WASP and WIP, making it intriguing to consider whether Hof1, similar to Cip4, coordinates formin and Arp2/3 complex activities. We found that Hof1 has no effect on actin patch function during polarized growth. However, actin patches relocate to the bud neck during cytokinesis, where Hof1 may regulate both Arp2/3- and formin-mediated actin assembly activities to coordinate membrane-remodeling events underlying septation.
MATERIALS AND METHODS
Plasmids and strains
Vectors used for galactose-inducible expression of hexahistidine (His6)-C-Bni1 (pBG564; residues 1227–1953) and His6-C-Bnr1 (pBG565; residues 757–1375) in S. cerevisiae were described previously (Moseley et al., 2004; Moseley and Goode, 2005). Low-copy (CEN, URA3) plasmid for expression of GFP-SEC4 in S. cerevisiae and integration plasmid for ABP140-3GFP::LEU2 were described previously (Calero et al., 2003; Buttery et al., 2007). To construct a vector for expression and purification of His6-C-Bnr1ΔFH1 in S. cerevisiae (pBG1554), we PCR amplified sequences encoding Bnr1 residues 868–1375 from pBG565 and subcloned these into the BamHI and NotI sites of pBG564. To construct vectors for Escherichia coli expression of GST-Hof1-CT-His6 (pBG1555), GST-Hof1-SH3-His6 (pBG1014), and GST-Hof1-CTΔSH3-His6 (pBG1556), we PCR amplified sequences encoding residues 350–669, 575–699, and 350–574, respectively, from yeast genomic DNA and subcloned these into the BamHI and NotI sites of pET-GST–tobacco etch virus (TEV; Moseley et al., 2004). To construct a vector for S. cerevisiae expression of His6-SNAP-C-Bnr1 (pBG1557), we PCR amplified sequences encoding residues 757–1375 from pBG565, digested the PCR product with BclI and NotI, and subcloned into the BamHI and NotI sites of pBG1444 (Breitsprecher et al., 2012).
Unless otherwise noted, all yeast strains were isogenic to BGY1256 (MATa, ura3, leu2, his3, met15) or BGY1257 (MATα, ura3, leu2, his3, met15). Mutant yeast strains bnr1Δ::KANMX6 (BGY1271), hof1Δ::HIS3 (BGY1277), and hof1ΔCT::HIS3 (BGY3676, expressing only residues 1–349) were generated by homologous recombination using either a KANMX6 or HIS3MX6 cassette (Longtine et al., 1998). The bni1-1::HIS3 and bni1-1::HIS3 bnr1Δ::KANMX6 strains were described previously (Sagot et al., 2002). To generate the strain hof1Δ::HIS bnr1Δ::KANMX6 (BGY1311), BGY1277 was crossed to BGY1271. To generate bni1-1::HIS3 hof1Δ::HIS3 (BGY3677) and bni1-1::HIS3 hof1ΔCT::HIS3 (BGY3678), bni1-1::HIS3 was crossed to BGY1277 and BGY3676, respectively. The SLA1-GFP::HIS ABP1-mRFP::KANMX6 strain was previously described (Boettner et al., 2009). The hof1Δ::TRP1 SLA1-GFP::HIS3 ABP1-mRFP::KANMX6 (BGY3679) strain was generated by homologous recombination using a TRP1 cassette (Longtine et al., 1998). The ABP140-3GFP::LEU2 integration vector was digested with NdeI and transformed into strains BGY1257, BGY1277, and BGY3676, producing strains BGY3680, BGY3681, and BGY3682, respectively.
Protein purification
RMA was purified as previously described (Spudich and Watt, 1971). RMA was purified first by generating an acetone powder from ground muscle tissue, which was stored in aliquots at −80°C. Aliquots of acetone powder were then pulverized using a coffee grinder, resuspended in G-buffer, and cleared by low-speed centrifugation. The actin was polymerized overnight and then pelleted. Pellets were stored at 4°C until needed (up to ∼2 mo). Every 2–3 wk, fresh RMA was prepared by disrupting an RMA pellet by douncing. RMA was then dialyzed against G-buffer (3 mM Tris-HCl pH 8.0, 0.5 mM dithiothreitol [DTT], 0.2 mM ATP, 0.1 mM CaCl2) for 2–3 d and cleared by ultracentrifugation, and the supernatant was gel filtered on a 16/60 S200 column (GE Healthcare, Pittsburgh, PA). Column fractions were stored at 4°C.
For bulk actin assembly assays, RMA was fluorescently labeled with pyrenyl-iodoacetamide on cysteine 374 (Pollard and Cooper, 1984; Graziano et al., 2013). An RMA pellet stored at 4°C (prepared as described in the preceding paragraph) was dialyzed against pyrene buffer (25 mM Tris-HCl, pH 7.5, 100 mM KCl, 0.02% NaN3, 0.3 mM ATP, and 2 mM MgSO4) for 3–4 h and then diluted with pyrene buffer to 1 mg/ml (23.8 μM). A sevenfold molar excess of pyrenyl-iodoacetamide was added, the actin solution was incubated overnight at 4°C, and aggregates were cleared by low-speed centrifugation. The supernatant (containing F-actin) was centrifuged for 3 h at 4°C at 45,000 rpm in a Ti70 rotor (Beckman Coulter, Indianapolis, IN) to pellet F-actin. The actin pellets were disrupted by douncing, dialyzed against G-buffer for 1–2 d, and gel filtered on a 16/60 S200 column. Peak fractions were pooled, aliquoted, snap frozen, and stored at −80°C until use. For TIRF microscopy, RMA was labeled with Oregon green (Kuhn and Pollard, 2005; Graziano et al., 2013).
S. cerevisiae profilin was expressed in BL21(DE3) E. coli and purified as described (Moseley et al., 2004; Graziano et al., 2013). Cells were grown in Luria broth to log phase, induced with 0.4 mM isopropyl-β-d-thiogalactoside (IPTG) for 3–4 h at 37°C, and then pelleted and stored at −80°C. Frozen pellets were thawed, resuspended in 20 mM Tris-HCl, pH 8.0, with standard protease inhibitors, and lysed using lysozyme, followed by sonication. Lysates were cleared by centrifugation at 80,000 rpm at 4°C for 20 min using a TLA100.3 rotor (Beckman Coulter). The supernatant was loaded onto a 5-ml Q HiTrap column (GE Healthcare) and eluted with a 75-ml salt gradient (0–400 mM NaCl) in 20 mM Tris-HCl, pH 8.0. Peak fractions were pooled, concentrated to ∼5 ml, and loaded onto a gel filtration column (26/60 Superdex 75; GE Healthcare). Peak fractions were pooled, snap frozen in liquid N2, and stored at −80°C.
C-Bni1, C-Bnr1, C-Bnr1ΔFH1, and SNAP-C-Bnr1 were expressed as His6-fusion proteins in S. cerevisiae strain BJ2168 on 2-μm plasmids under the control of galactose-inducible promoters as previously described (Moseley et al., 2006; Graziano et al., 2013). For each formin polypeptide, 2–4 l of yeast cells were grown in synthetic medium lacking uracil with 2% raffinose to OD600 of 0.6–0.9. Then protein expression was induced by addition of galactose (2% wt/vol), and cells were grown for 8–9 h at 25–30°C. Cells were then washed in H2O, frozen under liquid N2, and mechanically lysed using a coffee grinder and liquid N2. This frozen yeast lysate powder was stored at −80°C until use. C-Bni1 was purified as described (Graziano et al., 2013). Yeast powder (∼10 g) was resuspended in one volume of buffer A (20 mM NaPO4, pH 7.4, 150 mM NaCl, 30 mM imidazole, 0.5 mM DTT, 1% NP-40, standard protease inhibitors) and cleared by ultracentrifugation at 80,000 rpm for 20 min in a TLA100.3 rotor. The supernatant was incubated with Ni–nitrilotriacetic acid (NTA) beads for ∼1.5 h at 4°C. Ni-NTA beads were washed several times with buffer B (buffer A without protease inhibitors or NP-40). Ni-NTA beads coated with SNAP-C-Bnr1 were incubated with a fivefold molar excess of BG-649-PEG-Biotin overnight at 4°C, washed with buffer B, and then eluted with buffer B plus 300 mM imidazole. For Bnr1 truncations lacking a SNAP tag, proteins were eluted with buffer B plus 300 mM imidazole immediately after the first set of washes. Eluted proteins were fractionated on a Superose 12 gel filtration column (GE Healthcare) equilibrated in buffer HEKG10D (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 7.5, 1 mM EDTA, 50 mM KCl, glycerol [10% vol/vol], 1 mM DTT). Peak fractions were pooled, aliquoted, snap frozen, and stored at −80°C. C-Bnr1 and C-Bnr1ΔFH1 were purified similarly, except that the cleared yeast lysates were filtered through a 0.45-μm syringe-driven filter unit (Millex; EMD Millipore, Billerica, MA), and the His6-fusion proteins were isolated on a Profinia purification system (Bio-Rad Laboratories, Hercules, CA) using the native immobilized metal ion affinity chromatography (IMAC) plus desalting program (1-ml IMAC column plus 5-ml desalting column). The desalting buffer used was HEKG10D. Eluted C-Bnr1 and C-Bnr1ΔFH1 polypeptides (4 ml) were concentrated using centrifugal filter units (MWCO 30 kDa; Amicon Ultra; EMD Millipore) to a final volume of 0.5–0.7 ml, snap-frozen in aliquots with liquid N2, and stored at −80°C. Yeast actin was purified as previously described (Goode, 2002).
GST-Hof1-CT, GST-Hof1-SH3, and GST-Hof1-CTΔSH3 were expressed in E. coli strain BL21(DE3). Cells were grown in terrific broth to late log phase, induced with 0.4 mM IPTG for ∼4 h at 37°C, and then pelleted and stored at −80°C. Pellets were thawed, resuspended in lysis buffer (20 mM NaPO4, pH 7.4, 150 mM NaCl, 30 mM imidazole, 0.5 mM DTT, 5% glycerol [vol/vol]) with standard protease inhibitors, and lysed by treatment with lysozyme and sonication. Lysates were cleared by centrifugation at 13,000 rpm for 15 min in an F21S-8×50y rotor (Thermo Scientific, Waltham, MA), and the supernatant was mixed with 0.5 ml of Ni-NTA beads and incubated at 4°C for ∼1.5 h. The beads were then washed repeatedly with lysis buffer plus 400 mM NaCl but lacking glycerol. Hof1 polypeptides were eluted with 1 ml of lysis buffer plus 300 mM imidazole but lacking glycerol. Eluted proteins were then diluted 10-fold in lysis buffer lacking imidazole and further purified on a Profinia purification system using the GST plus desalting program (1-ml GST column plus 5-ml desalting column). The desalting buffer was HEKG10D. The eluted GST-Hof1-His6 fusion proteins were concentrated with centrifugal filter units (MWCO 10 kDa; Amicon Ultra) to final volumes of ∼0.5 ml, snap-frozen in aliquots under liquid N2, and stored at −80°C. For Hof1-CT (lacking the GST tag), purification was the same up until elution from Ni-NTA beads. Then the protein was diluted 10-fold and incubated with 0.5 ml glutathione beads for 2 h at 4°C. The beads were washed twice with HEKG10D and incubated with TEV protease for 2 h at room temperature in 0.5 ml HEKG10D. TEV-released Hof1-CT was aliquoted, snap-frozen under N2, and stored at −80°C.
A CapZ expression vector (Soeno et al., 1998) was transformed into E. coli strain BL21(DE3) pLysS. Cells were grown in LB to mid log phase, induced with 0.4 mM IPTG for 3 h at 37°C, and then pelleted and stored at −80°C. Frozen pellets were thawed in lysis buffer (20 mM Tris pH 8.0, 1 mM EDTA, 0.1% Triton X-100, protease inhibitors) and lysed by lysozyme treatment and sonication. Lysates were cleared by centrifugation at 35,000 rpm for 30 min at 4°C in a Ti70 rotor (Beckman Coulter). Supernatants were loaded onto a 1-ml Q-HiTrap column (GE Healthcare) and eluted with a 45-ml salt gradient (0–500 mM KCl) in 20 mM Tris, pH 8.0. Peak fractions were pooled, concentrated using a centrifugal filter unit (Centiprep, MWCO 10 kDa; Millipore) to 3 ml, and loaded onto a 26/60 Superdex 75 gel filtration column (GE Healthcare) equilibrated in 50 mM KCl, 20 mM Tris, pH 8.0. Peak fractions were pooled and loaded onto a 1-ml Mono Q column (GE Healthcare) and eluted with a 30-ml salt gradient (0–500 mM KCl) in 20 mM Tris, pH 8.0. Peak fractions were pooled, dialyzed overnight at 4°C into HEK buffer (20 mM HEPES, pH 7.4, 1 mM EDTA, 50 mM KCl), aliquoted, snap-frozen under liquid N2, and stored at −80°C.
Actin assembly assays
Gel-filtered monomeric actin (in G-buffer) was cleared by centrifugation at 90,000 rpm for 1 h at 4°C in a TLA100 rotor (Beckman Coulter), and the top ∼50% of the supernatant was carefully recovered. Each assembly reaction (60 μl) contained 2 μM G-actin (5% pyrene labeled) and, where indicated, 5 μM yeast profilin. G-actin was converted to Mg2+-ATP-actin 2 min before use, and 42 μl G-actin was mixed with 15 μl of proteins/buffer, followed by 3 μl of initiation mix (40 mM MgCl2, 10 mM ATP, 1 M KCl) before the start of reactions. Pyrene–actin fluorescence was monitored in a plate reader (Infinite M200; Tecan, Männedorf, Switzerland) at excitation and emission wavelengths of 365 and 407 nm, respectively. All reactions were performed at 25°C. Normalized times to 50% polymerization (Figure 3G) were calculated as follows: 1) For each reaction condition, the time to 50% actin polymerization (T1/2) was determined. 2) T1/2 values for reactions containing Hof1 polypeptides were divided by T1/2 values obtained from identical reactions lacking Hof1. 3) These normalized T1/2 values were plotted as a function Hof1 concentration.
Cell imaging
For imaging yeast F-actin organization, cells were chemically fixed and stained with fluorescently labeled phalloidin as previously described (Graziano et al., 2013). Briefly, yeast strains were grown in yeast extract/peptone/2% glucose (YEPD) at 25°C to early/mid log phase and fixed at room temperature with 4.5% formaldehyde for 45 min. For bni1-1::HIS strains (Figure 2, D and E), cells from early/mid log-phase cultures were incubated at 34°C for 5 min before fixation. Fixed cells were stained overnight at 4°C with Alexa 488–phalloidin and washed twice with PBS before imaging. Cells were imaged in mounting media (10 mM NaPO4, pH 7.4, 75 mM NaCl, 4.3 mM p-phenylenediamine, 0.01 mg/ml 4′,6-diamidino-2-phenylindole, and 45% glycerol) at 20°C using an upright microscope (Ni-E; Nikon) equipped with a spinning disk head (CSU-W1; Yokogawa Corporation of America, Sugar Land, TX) with laser excitation at 488 nm (emission filter, 525/40 nm), a 100×/numerical aperture (NA) 1.45 Plan Apochromat objective, and an electron-multiplying charge-coupled device (EMCCD) camera (iXon 897U; Andor Technology, Belfast, Northern Ireland). Images were acquired using Elements AR software (Nikon, Tokyo, Japan). For all strains, z-series were collected (0.2-μm step size), capturing each plane with a 200-ms exposure. The number of planes in each z-series was variable but was chosen for each field of view acquired such that all the cells imaged would lie between the highest and lowest focal plane.
All image analysis of fixed cells was performed using ImageJ. To measure cell volumes, cells were assumed to be ellipsoidal, and the equation Vcell = (4/3)•π•r1•r2•r3 was used, where π•r1•r2 corresponds to the cross-sectional area of the cell at its middle focal plane and r3 corresponds to the radius of the cell in the z-direction. F-actin levels in the mother cell were calculated in tandem with the volume by summing the Alexa Fluor 488 r3 phalloidin signal present in each focal plane of the cell (Supplemental Figure S2E). To normalize F-actin fluorescence intensity with respect to cell volume (Figure 2G), total fluorescence of each cell was divided by its volume. For our comparison of F-actin cable levels in wild-type, hof1Δ, and hof1ΔCT strains, all of these strains had polarized actin patches (i.e., patches were predominantly in the bud rather than the mother). We restricted our analysis to mother cell compartments in which almost all of the F-actin intensity was specific to cables. Although some of the cells had a few patches present in the mother compartment, the intensity of each individual patch was on average only ∼0.5% of the total F-actin intensity of the mother compartment (compare the y-axes in Supplemental Figure S2, E and G). Therefore the presence of a few patches had minimal impact on the comparison of cable levels in strains.
Patch intensities (Figure 1C) were calculated with the same image sets used for determining cell volumes and F-actin levels. Maximum-intensity projections were generated from each z-series, small ellipses were drawn around individual patches, and their fluorescence intensities were measured. Each triangle in the “Actin patch intensity” plot in Figure 1C represents one patch. Approximately five to seven patches were measured per cell.
For live imaging of strains expressing Abp140-3GFP or GFP-Sec4, cells were grown in synthetic medium (2% glucose) lacking the appropriate amino acids to early/mid log phase and imaged immediately at 20–25°C. Images were acquired using the same equipment and procedures described earlier in this section, with the following exceptions. For strains expressing GFP-Sec4, 200-ms exposures of single focal planes were acquired every 250 ms for 60 s (4 frames/s). For experiments in which the angles of elongating cables were measured (Figure 1J), 30 μM latrunculin B (Enzo Life Sciences, Farmingdale, NY) was added to yeast cultures immediately before imaging, which decreased cable elongation rates by about twofold to enable accurate measurement and tracking. A single focal plane was imaged every 80 ms for 30 s (12.5 frames/s). Actin cable extension angles relative to each cell's axis of polarity (i.e., mother–bud axis) were measured using custom software written in MATLAB (MathWorks, Natick, MA).
Bnr1-GFP strains were imaged using confocal microscopy as described. The z-series of cells were acquired, and Bnr1 signal was determined by measuring the fluorescence intensity of each focal plane where GFP signal was visible at the bud neck. These intensities were then summed and divided by the total area of the bud neck. Bud sizes were scored visually.
Actin patch dynamics was monitored by live imaging using cells grown to log phase in synthetic medium and immobilized on 2% agarose in synthetic complete medium. Images were acquired as described earlier in this section with laser excitation at 488 and 561 nm. Kymographs representing single endocytic events were selected, and line profiles along each kymograph were exported as text files. Images were processed using Elements AR and ImageJ. Corrected fluorescence intensity profiles were calculated by averaging the first five time points and the last five time points in each channel and assuming that the background signal changed linearly over time as photobleaching occurred. This dynamic background value was subtracted at each time point, and the entire profile was then normalized to the maximum signal in the profile. The lifetime of each fluorescent marker was taken as the longest stretch of positive values.
TIRF microscopy
In all experiments, coverslips were first cleaned by sonication in detergent for 60 min, followed by successive rounds of sonication in 1 M KOH and 1 M HCl for 20 min each and then sonication in ethanol for at least 60 min. Coverslips were then washed extensively with double-distilled (dd) H2O, dried in an N2 stream, layered with 200 μl of 80% ethanol, pH 2.0, 2 mg/ml methoxy-poly(ethylene glycol)-silane and 2 μg/ml biotin-poly(ethylene glycol)-silane (Laysan Bio, Arab, AL), and incubated overnight at 70°C. Flow cells were assembled by rinsing poly(ethylene glycol)-coated coverslips extensively with ddH2O, followed by attachment either to a flow chamber (Ibidi, Martinsried, Germany) or glass slide. Double-sided tape (2.5 cm × 2 mm × 120 μm) and 5-min epoxy resin were used for attachment to flow chambers, whereas only double-sided tape was used for attachment to glass slides.
Immediately before each experiment, flow cells were washed with streptavidin (17 μg/ml), incubated for 3 min in HBSA (20 mM HEPES, 1 mM EDTA, 50 mM KCl, and 1% bovine serum albumin), and then washed with TIRF buffer (50 mM KCl, 1 mM MgCl2, 1 mM ethylene glycol tetraacetic acid, 0.2 mM ATP, 25 mM DTT, 15 mM glucose, and 0.5% methylcellulose [4000 cP], pH 7.4). Proteins in TIRF buffer were mixed with 0.5–1.0 μM G-actin (10% Oregon green labeled) and added to the flow cell.
For single-color TIRF microscopy experiments, images were acquired at 10-s intervals for a total of 600 s using an inverted microscope (Ti200; Nikon) with a 150-mW argon laser (Mellot Griot, Carlsbad, CA), a 60× TIRF objective, NA 1.49 (Nikon), and an EMCCD camera (iXon). Focus was maintained using the Perfect Focus System (Nikon). The temperature of the room was maintained at 20°C. Elongation rates of filaments were determined by measuring the length of each filament at 30-s intervals over a period of at least 300 s. At least two independent reactions were performed for each condition in each experiment. To determine the number of filaments nucleated, fields of view were examined 450 s after actin assembly was initiated. Filament densities were calculated for at least six fields of view from at least two separate reactions. Analysis was performed using ImageJ.
For two-color TIRF microscopy experiments, images were acquired at 5-s intervals for a total of 600 s with laser excitation (Agilent Technologies) at 488 and 640 nm. Elongation rates were determined as in the preceding paragraph. C-Bnr1 was tethered to the surface by a linkage consisting of biotin-PEG, streptavidin, and SNAP-649-PEG-Biotin-C-Bnr1 (Smith et al., 2013). Ingredients in TIRF chambers were replaced by flow-in using a Pump 11 Elite (Harvard Apparatus, Holliston, MA) at a flow rate of 10 μl/min for 2 min. Filaments were scored as “displaced” if the barbed end moved at least 1 μm from the anchored SNAP-649-PEG-Biotin-C-Bnr1 site.
Supplementary Material
Acknowledgments
We thank Richa Jaiswal and Melissa Chesarone-Cataldo for critical reading of the manuscript. We also thank Gönen Memisoglu for experimental assistance. This work was supported by a fellowship from the Leukemia and Lymphoma Society (5188-09) to H.E.Y. and grants from the National Institutes of Health (GM083137) and National Science Foundation (DMR-MRSEC-0820429) to B.L.G.
Abbreviations used:
- CC
coiled coil
- DAD
diaphanous autoregulatory domain
- F-BAR
Fer/Cip4 homology–Bin amphiphysin Rvs
- FH
formin homology
- RMA
rabbit muscle actin
- SH3
Src homology 3
- TIRF
total internal reflection fluorescence
- WASP
Wiskott–Aldrich syndrome protein
- WIP
WASP-interacting protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-03-0850) on April 9, 2014.
REFERENCES
- Asakura T, et al. Isolation and characterization of a novel actin filament-binding protein from Saccharomyces cerevisiae. Oncogene. 1998;16:121–130. doi: 10.1038/sj.onc.1201487. [DOI] [PubMed] [Google Scholar]
- Bartolini F, Ramalingam N, Gundersen GG. Actin-capping protein promotes microtubule stability by antagonizing the actin activity of mDia1. Mol Biol Cell. 2012;23:4032–4040. doi: 10.1091/mbc.E12-05-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Park HO. Cell polarization and cytokinesis in budding yeast. Genetics. 2012;191:347–387. doi: 10.1534/genetics.111.132886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner DR, et al. The F-BAR protein Syp1 negatively regulates WASp-Arp2/3 complex activity during endocytic patch formation. Curr Biol. 2009;19:1979–1987. doi: 10.1016/j.cub.2009.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D, Goode BL. Formins at a glance. J Cell Sci. 2013;126:1–7. doi: 10.1242/jcs.107250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D, Jaiswal R, Bombardier JP, Gould CJ, Gelles J, Goode BL. Rocket launcher mechanism of collaborative actin assembly defined by single-molecule imaging. Science. 2012;336:1164–1168. doi: 10.1126/science.1218062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttery SM, Kono K, Stokasimov E, Pellman D. Regulation of the formin Bnr1 by septins and a MARK/Par1-family septin-associated kinase. Mol Biol Cell. 2012;23:4041–4053. doi: 10.1091/mbc.E12-05-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttery SM, Yoshida S, Pellman D. Yeast formins Bni1 and Bnr1 utilize different modes of cortical interaction during the assembly of actin cables. Mol Biol Cell. 2007;18:1826–1838. doi: 10.1091/mbc.E06-09-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero M, Chen CZ, Zhu W, Winand N, Havas KA, Gilbert PM, Burd CG, Collins RN. Dual prenylation is required for Rab protein localization and function. Mol Biol Cell. 2003;14:1852–1867. doi: 10.1091/mbc.E02-11-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnahan RH, Gould KL. The PCH family protein, Cdc15p, recruits two F-actin nucleation pathways to coordinate cytokinetic actin ring formation in Schizosaccharomyces pombe. J Cell Biol. 2003;162:851–862. doi: 10.1083/jcb.200305012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone M, Gould CJ, Moseley JB, Goode BL. Displacement of formins from growing barbed ends by bud14 is critical for actin cable architecture and function. Dev Cell. 2009;16:292–302. doi: 10.1016/j.devcel.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev. Mol Cell Biol. 2010;11:62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- Chesarone-Cataldo M, Guerin C, Yu JH, Wedlich-Soldner R, Blanchoin L, Goode BL. The myosin passenger protein Smy1 controls actin cable structure and dynamics by acting as a formin damper. Dev Cell. 2011;21:217–230. doi: 10.1016/j.devcel.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M, Pruyne D, Amberg DC, Boone C, Bretscher A. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat Cell Biol. 2002;4:260–269. doi: 10.1038/ncb770. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Reymond A, Cerutti L, Utzig S, Hofmann K, Simanis V. The S. pombe cdc15 gene is a key element in the reorganization of F-actin at mitosis. Cell. 1995;82:435–444. doi: 10.1016/0092-8674(95)90432-8. [DOI] [PubMed] [Google Scholar]
- Feliciano D, Di Pietro SM. SLAC, a complex between Sla1 and Las17, regulates actin polymerization during clathrin-mediated endocytosis. Mol Biol Cell. 2012;23:4256–4272. doi: 10.1091/mbc.E11-12-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrezuelo F, Colomina N, Palmisano A, Gari E, Gallego C, Csikasz-Nagy A, Aldea M. The critical size is set at a single-cell level by growth rate to attain homeostasis and adaptation. Nat Commun. 2012;3:1012. doi: 10.1038/ncomms2015. [DOI] [PubMed] [Google Scholar]
- Firat-Karalar EN, Welch MD. New mechanisms and functions of actin nucleation. Curr Opin Cell Biol. 2011;23:4–13. doi: 10.1016/j.ceb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E, Chen T. A matter of life and death: self-renewal in stem cells. EMBO Rep. 2013;14:39–48. doi: 10.1038/embor.2012.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Gonczy P. Mechanisms of asymmetric cell division: flies and worms pave the way. Nat Rev. Mol Cell Biol. 2008;9:355–366. doi: 10.1038/nrm2388. [DOI] [PubMed] [Google Scholar]
- Goode BL. Purification of yeast actin and actin-associated proteins. Methods Enzymol. 2002;351:433–441. doi: 10.1016/s0076-6879(02)51862-0. [DOI] [PubMed] [Google Scholar]
- Gould CJ, Maiti S, Michelot A, Graziano BR, Blanchoin L, Goode BL. The formin DAD domain plays dual roles in autoinhibition and actin nucleation. Curr Biol. 2011;21:384–390. doi: 10.1016/j.cub.2011.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindan B, Bowser R, Novick P. The role of Myo2, a yeast class V myosin, in vesicular transport. J Cell Biol. 1995;128:1055–1068. doi: 10.1083/jcb.128.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano BR, DuPage AG, Michelot A, Breitsprecher D, Moseley JB, Sagot I, Blanchoin L, Goode BL. Mechanism and cellular function of Bud6 as an actin nucleation-promoting factor. Mol Biol Cell. 2011;22:4016–4028. doi: 10.1091/mbc.E11-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano BR, Jonasson EM, Pullen JG, Gould CJ, Goode BL. Ligand-induced activation of a formin-NPF pair leads to collaborative actin nucleation. J Cell Biol. 2013;201:595–611. doi: 10.1083/jcb.201212059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris ES, Li F, Higgs HN. The mouse formin, FRLalpha, slows actin filament barbed end elongation, competes with capping protein, accelerates polymerization from monomers, and severs filaments. J Biol Chem. 2004;279:20076–20087. doi: 10.1074/jbc.M312718200. [DOI] [PubMed] [Google Scholar]
- Higashida C, Miyoshi T, Fujita A, Oceguera-Yanez F, Monypenny J, Andou Y, Narumiya S, Watanabe N. Actin polymerization-driven molecular movement of mDia1 in living cells. Science. 2004;303:2007–2010. doi: 10.1126/science.1093923. [DOI] [PubMed] [Google Scholar]
- Jendretzki A, Ciklic I, Rodicio R, Schmitz HP, Heinisch JJ. Cyk3 acts in actomyosin ring independent cytokinesis by recruiting Inn1 to the yeast bud neck. Mol Genet Genomics. 2009;282:437–451. doi: 10.1007/s00438-009-0476-0. [DOI] [PubMed] [Google Scholar]
- Johnson JM, Jin M, Lew DJ. Symmetry breaking and the establishment of cell polarity in budding yeast. Curr Opin Genet Dev. 2011;21:740–746. doi: 10.1016/j.gde.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei T, Tanaka K, Hihara T, Umikawa M, Imamura H, Kikyo M, Ozaki K, Takai Y. Interaction of Bnr1p with a novel Src homology 3 domain-containing Hof1p. Implication in cytokinesis in Saccharomyces cerevisiae. J Biol Chem. 1998;273:28341–28345. doi: 10.1074/jbc.273.43.28341. [DOI] [PubMed] [Google Scholar]
- Korinek WS, Bi E, Epp JA, Wang L, Ho J, Chant J. Cyk3, a novel SH3-domain protein, affects cytokinesis in yeast. Curr Biol. 2000;10:947–950. doi: 10.1016/s0960-9822(00)00626-6. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci USA. 2004;101:14725–14730. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Wu JQ, Pollard TD. Profilin-mediated competition between capping protein and formin Cdc12p during cytokinesis in fission yeast. Mol Biol Cell. 2005;16:2313–2324. doi: 10.1091/mbc.E04-09-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JR, Pollard TD. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys J. 2005;88:1387–1402. doi: 10.1529/biophysj.104.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Higgs HN. The mouse formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr Biol. 2003;13:1335–1340. doi: 10.1016/s0960-9822(03)00540-2. [DOI] [PubMed] [Google Scholar]
- Li R, Albertini DF. The road to maturation: somatic cell interaction and self-organization of the mammalian oocyte. Nat Rev. Mol Cell Biol. 2013;14:141–152. doi: 10.1038/nrm3531. [DOI] [PubMed] [Google Scholar]
- Lippincott J, Li R. Dual function of Cyk2, a cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution. J Cell Biol. 1998;143:1947–1960. doi: 10.1083/jcb.143.7.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Mason FM, Heimsath EG, Higgs HN, Soderling SH. Bi-modal regulation of a formin by srGAP2. J Biol Chem. 2011;286:6577–6586. doi: 10.1074/jbc.M110.190397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F, Boehm ME, Hofmann A, Hub B, Zentgraf H, Lehmann WD, Pereira G. Phosphorylation-dependent regulation of the F-BAR protein Hof1 during cytokinesis. Genes Dev. 2011;25:875–888. doi: 10.1101/gad.622411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F, Palani S, Hub B, Pereira G. Dual function of the NDR-kinase Dbf2 in the regulation of the F-BAR protein Hof1 during cytokinesis. Mol Biol Cell. 2013;24:1290–1304. doi: 10.1091/mbc.E12-08-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley JB, Goode BL. Differential activities and regulation of Saccharomyces cerevisiae formin proteins Bni1 and Bnr1 by Bud6. J Biol Chem. 2005;280:28023–28033. doi: 10.1074/jbc.M503094200. [DOI] [PubMed] [Google Scholar]
- Moseley JB, Goode BL. The yeast actin cytoskeleton: from cellular function to biochemical mechanism. Microbiol Mol Biol Rev. 2006;70:605–645. doi: 10.1128/MMBR.00013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley JB, Maiti S, Goode BL. Formin proteins: purification and measurement of effects on actin assembly. Methods Enzymol. 2006;406:215–234. doi: 10.1016/S0076-6879(06)06016-2. [DOI] [PubMed] [Google Scholar]
- Moseley JB, Sagot I, Manning AL, Xu Y, Eck MJ, Pellman D, Goode BL. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol Biol Cell. 2004;15:896–907. doi: 10.1091/mbc.E03-08-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi SN, Feng Q, Boulton VJ, Zahn R, Munn AL. Vrp1p functions in both actomyosin ring-dependent and Hof1p-dependent pathways of cytokinesis. Traffic. 2001;2:189–201. doi: 10.1034/j.1600-0854.2001.020305.x. [DOI] [PubMed] [Google Scholar]
- Oh Y, Schreiter J, Nishihama R, Wloka C, Bi E. Targeting and functional mechanisms of the cytokinesis-related F-BAR protein Hof1 during the cell cycle. Mol Biol Cell. 2013;24:1305–1320. doi: 10.1091/mbc.E12-11-0804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Bartolini F, Deaconescu AM, Moseley JB, Dogic Z, Grigorieff N, Gundersen GG, Goode BL. Adenomatous polyposis coli protein nucleates actin assembly and synergizes with the formin mDia1. J Cell Biol. 2010;189:1087–1096. doi: 10.1083/jcb.201001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AS, Pollard TD. The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr Biol. 2008;18:9–19. doi: 10.1016/j.cub.2007.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Quantitative analysis of the effect of Acanthamoeba profilin on actin filament nucleation and elongation. Biochemistry. 1984;23:6631–6641. doi: 10.1021/bi00321a054. [DOI] [PubMed] [Google Scholar]
- Pruyne D, Legesse-Miller A, Gao L, Dong Y, Bretscher A. Mechanisms of polarized growth and organelle segregation in yeast. Annu Rev Cell Dev Biol. 2004;20:559–591. doi: 10.1146/annurev.cellbio.20.010403.103108. [DOI] [PubMed] [Google Scholar]
- Pruyne DW, Schott DH, Bretscher A. Tropomyosin-containing actin cables direct the Myo2p-dependent polarized delivery of secretory vesicles in budding yeast. J Cell Biol. 1998;143:1931–1945. doi: 10.1083/jcb.143.7.1931. [DOI] [PubMed] [Google Scholar]
- Rajmohan R, Wong MH, Meng L, Munn AL, Thanabalu T. Las17p-Vrp1p but not Las17p-Arp2/3 interaction is important for actin patch polarization in yeast. Biochim Biophys Acta. 2009;1793:825–835. doi: 10.1016/j.bbamcr.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, Gould KL. Setting the F-BAR: functions and regulation of the F-BAR protein family. Cell Cycle. 2010;9:4091–4097. doi: 10.4161/cc.9.20.13587. [DOI] [PubMed] [Google Scholar]
- Rodal AA, Manning AL, Goode BL, Drubin DG. Negative regulation of yeast WASp by two SH3 domain-containing proteins. Curr Biol. 2003;13:1000–1008. doi: 10.1016/s0960-9822(03)00383-x. [DOI] [PubMed] [Google Scholar]
- Romero S, Le Clainche C, Didry D, Egile C, Pantaloni D, Carlier MF. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004;119:419–429. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- Sagot I, Klee SK, Pellman D. Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat Cell Biol. 2002;4:42–50. doi: 10.1038/ncb719. [DOI] [PubMed] [Google Scholar]
- Schott DH, Collins RN, Bretscher A. Secretory vesicle transport velocity in living cells depends on the myosin-V lever arm length. J Cell Biol. 2002;156:35–39. doi: 10.1083/jcb.200110086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BA, Padrick SB, Doolittle LK, Daugherty-Clarke K, Correa IR, Jr, Xu MQ, Goode BL, Rosen MK, Gelles J. Three-color single molecule imaging shows WASP detachment from Arp2/3 complex triggers actin filament branch formation. eLife. 2013;2:e01008. doi: 10.7554/eLife.01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeno Y, Abe H, Kimura S, Maruyama K, Obinata T. Generation of functional beta-actinin (CapZ) in an E. coli expression system. J Muscle Res Cell Motil. 1998;19:639–646. doi: 10.1023/a:1005329114263. [DOI] [PubMed] [Google Scholar]
- Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- Sun Y, Martin AC, Drubin DG. Endocytic internalization in budding yeast requires coordinated actin nucleation and myosin motor activity. Dev Cell. 2006;11:33–46. doi: 10.1016/j.devcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Takeda T, Robinson IM, Savoian MM, Griffiths JR, Whetton AD, McMahon HT, Glover DM. Drosophila F-BAR protein syndapin contributes to coupling the plasma membrane and contractile ring in cytokinesis. Open Biol. 2013;3:130081. doi: 10.1098/rsob.130081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, et al. A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science. 2002;295:321–324. doi: 10.1126/science.1064987. [DOI] [PubMed] [Google Scholar]
- Tonikian R, et al. Bayesian modeling of the yeast SH3 domain interactome predicts spatiotemporal dynamics of endocytosis proteins. PLoS Biol. 2009;7:e1000218. doi: 10.1371/journal.pbio.1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen EA, Caviston J, Bi E. Roles of Hof1p, Bni1p, Bnr1p, and myo1p in cytokinesis in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:593–611. doi: 10.1091/mbc.11.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcarra CL, Kreutz B, Rodal AA, Toms AV, Lu J, Zheng W, Quinlan ME, Eck MJ. Structure and function of the interacting domains of Spire and Fmn-family formins. Proc Natl Acad Sci USA. 2011;108:11884–11889. doi: 10.1073/pnas.1105703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg J, Drubin DG. Clathrin-mediated endocytosis in budding yeast. Trends Cell Biol. 2012;22:1–13. [Google Scholar]
- Xu Y, Moseley JB, Sagot I, Poy F, Pellman D, Goode BL, Eck MJ. Crystal structures of a formin homology-2 domain reveal a tethered dimer architecture. Cell. 2004;116:711–723. doi: 10.1016/s0092-8674(04)00210-7. [DOI] [PubMed] [Google Scholar]
- Yang HC, Pon LA. Actin cable dynamics in budding yeast. Proc Natl Acad Sci USA. 2002;99:751–756. doi: 10.1073/pnas.022462899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Lv Z, Winterhoff M, Wenzl C, Zobel T, Faix J, Bogdan S, Grosshans J. The F-BAR protein Cip4/Toca-1 antagonizes the formin Diaphanous in membrane stabilization and compartmentalization. J Cell Sci. 2013;126:1796–1805. doi: 10.1242/jcs.118422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JH, Crevenna AH, Bettenbuhl M, Freisinger T, Wedlich-Soldner R. Cortical actin dynamics driven by formins and myosin V. J Cell Sci. 2011;124:1533–1541. doi: 10.1242/jcs.079038. [DOI] [PubMed] [Google Scholar]
- Zigmond SH, Evangelista M, Boone C, Yang C, Dar AC, Sicheri F, Forkey J, Pring M. Formin leaky cap allows elongation in the presence of tight capping proteins. Curr Biol. 2003;13:1820–1823. doi: 10.1016/j.cub.2003.09.057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.