The Ca2+-activated Cl− channel ANO1/TMEM16A is located in the primary cilium, and blocking it pharmacologically or knocking it down with shRNA interferes with ciliogenesis. Before ciliogenesis, the channel is organized into a torus-shaped structure (the “nimbus”) enriched in proteins required for ciliogenesis.
Abstract
Many cells possess a single, nonmotile, primary cilium highly enriched in receptors and sensory transduction machinery that plays crucial roles in cellular morphogenesis. Although sensory transduction requires ion channels, relatively little is known about ion channels in the primary cilium (with the exception of TRPP2). Here we show that the Ca2+-activated Cl− channel anoctamin-1 (ANO1/TMEM16A) is located in the primary cilium and that blocking its channel function pharmacologically or knocking it down with short hairpin RNA interferes with ciliogenesis. Before ciliogenesis, the channel becomes organized into a torus-shaped structure (“the nimbus”) enriched in proteins required for ciliogenesis, including the small GTPases Cdc42 and Arl13b and the exocyst complex component Sec6. The nimbus excludes F-actin and coincides with a ring of acetylated microtubules. The nimbus appears to form before, or independent of, apical docking of the mother centriole. Our data support a model in which the nimbus provides a scaffold for staging of ciliary components for assembly very early in ciliogenesis and chloride transport by ANO1/TMEM16A is required for the genesis or maintenance of primary cilia.
INTRODUCTION
The ethos of chloride ions in biology has evolved dramatically over the past two decades from one in which passive Cl− fluxes perform mundane tasks to one in which Cl− channels dynamically execute a myriad of cell biological functions, including vesicular trafficking, cell cycle regulation, cell migration, and embryonic development and morphogenesis (Hartzell, 2009; Verkman and Galietta, 2009; Duran et al., 2010; Berend et al., 2012; Stauber and Jentsch, 2013). One Cl− channel that has recently received particular attention is anoctamin-1 (ANO1; also known as TMEM16A; Hartzell et al., 2009; Ferrera et al., 2010; Duran and Hartzell, 2011; Kunzelmann et al., 2011; Huang et al., 2012a). The anoctamin gene family was identified in 2003, before its Cl− channel function was known, in a bioinformatic analysis of a region of chromosome 11 (11q13) that is amplified in many tumors (Katoh, 2003, 2005; Espinosa et al., 2008; Miettinen et al., 2009; Hwang et al., 2011; Wilkerson and Reis-Filho, 2013). In 2008, ANO1 was shown to be a plasma membrane Ca2+-activated Cl− channel (CaCC; Caputo et al., 2008; Schroeder et al., 2008; Yang et al., 2008). CaCCs are present in the apical membranes of secretory epithelial cells, where they are gated open by increases in cytosolic Ca2+ (Hartzell et al., 2005). Cl− efflux through the channel then drives fluid secretion as Na+ and water follow to maintain charge neutrality and osmotic balance. As expected for a classic CaCC, ANO1 is expressed in the apical membranes of differentiated secretory epithelial cells such as salivary gland and pancreatic acinar cells (Yang et al., 2008; Romanenko et al., 2010; Perez-Cornejo et al., 2012). Although CaCCs are best known for their role in transepithelial transport, novel functions for these channels in regulation of neuronal excitability, vascular tone, gastrointestinal motility, and nociception have been uncovered as a result of identification of the ANO1 gene (Ferrera et al., 2010; Duran and Hartzell, 2011; Kunzelmann et al., 2011; Huang et al., 2012a).
Hints that ANO1 not only drives epithelial fluid secretion but also participates in cell biological functions were first provided by its link to poor prognosis in many cancers (reviewed in Wilkerson and Reis-Filho, 2013), but awareness that ANO1 participates in cellular development and differentiation received a major boost from studies by Jason Rock and Brian Harfe, who showed that ANO1 is highly expressed in the zone of polarizing activity of the mouse limb bud (Rock et al., 2007). This anatomical region is involved in creating a gradient of the sonic hedgehog (SHH) morphogen that specifies the anterioposterior axis of the limb. A role of ANO1 in embryonic development was then established by showing that disruption of the ANO1 gene in mouse results in neonatal lethality as a result of developmental abnormalities, including defects in morphogenesis of the trachea and gut (Rock et al., 2008).
Here we report a novel observation that is likely to open a new line of investigation into the role of ANO1 in the genesis of the primary cilium. The primary cilium is an apical appendage enriched in many signaling and morphogenetic networks, including SHH (Pazour and Witman, 2003; Berbari et al., 2009; Seeley and Nachury, 2010). As cells withdraw from the cell cycle and develop apical–basal polarity, the centrosome, which in dividing cells functions to organize the mitotic spindle, migrates to the apical surface of the cell, where it becomes a basal body to nucleate the primary cilium (Kobayashi and Dynlacht, 2011). Defects in ciliogenesis are linked to mutations in a number of genes that produce a pleomorphic spectrum of diseases typified by defective tissue morphogenesis (Veland et al., 2009; Hildebrandt et al., 2011; Davis and Katsanis, 2012; Valente et al., 2014). Here we show that ANO1 forms a unique structure at the apical surface of polarizing epithelial cells before formation of the primary cilium and that ANO1 enters the cilium at early stages of ciliogenesis. This apical ANO1-containing structure is likely a hub for ciliary components that are trafficked into the cilium during early stages of ciliogenesis. We showed previously that ANO1 interacts at high stoichiometry with proteins involved in membrane–cytoskeletal organization and primary cilia (Perez-Cornejo et al., 2012), suggesting that ANO1 may serve as a component of a scaffold in this organization. Consistent with a role for ANO1 in ciliogenesis, we find that interference with ANO1 channel function or expression disrupts normal ciliogenesis.
RESULTS
Concentration of ANO1 in an apical annular structure
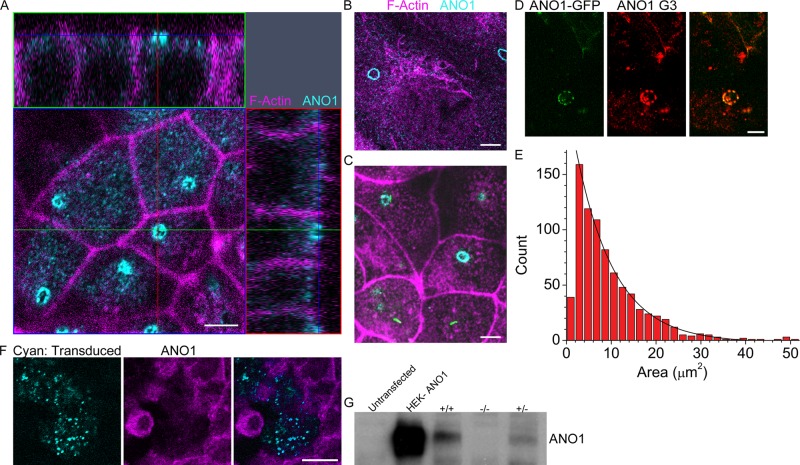
While investigating the role of the Ca2+-activated Cl− channel (CaCC) ANO1 in transepithelial transport, we immunolabeled various epithelial cell lines grown on permeable supports to determine the apical–basal distribution of ANO1. We initially concentrated on a widely used murine immortalized cortical collecting duct cell line (mpkCCD14) whose ion transport functions have been well studied (Chassin et al., 2007; Rajagopal et al., 2012). Surprisingly, we found that when cells were cultured under conditions that disfavored formation of primary cilia, ANO1 was concentrated at the apical surface in a sharply defined annulus ∼3 μm in diameter composed of smaller, discrete puncta (Figure 1A). We refer to this structure including other associated proteins as the nimbus because of its resemblance to a halo. The vast majority of cells have only one nimbus per cell. The ring of ANO1 staining circumscribes an area covering ∼6% of the apical aspect of each cell: the average area demarcated by the ring is 9.5 ± 1.2 μm2 (n = 798), compared with an average total apical membrane area of 156.9 ± 3.8 μm2. The average ANO1 nimbus is elliptically shaped, with major and minor axial radii of 2.0 and 1.4 μm, respectively. Nimbus sizes are distributed exponentially rather than in a Gaussian manner (Figure 1E), suggesting the possibility that the nimbus is a dynamic structure.
FIGURE 1:
An annulus of ANO1 is located at the apical aspect of cultured epithelial cells. (A) Confocal image of mpkCCD14 cells grown on permeable supports in the presence of serum. The xy-plane is an optical section taken near the apical surface (faint blue line in the z-sections) showing ANO1 (cyan) concentrated in an annular structure. The yz-plane (top, green line in xy image) and xz-plane (right, red line in xy image) show that the nimbus is located at the apical surface of the cell. Fluorescent phalloidin was used to label F-actin (magenta). (B) ANO1 (cyan) nimbus in RPE-J cells grown on glass coverslips. Acetylated tubulin (magenta). (C) ANO1 (cyan) nimbi in IMCD3 cells grown on permeable supports. Maximum intensity projection (MIP) of a z-stack. Acetylated tubulin (magenta). IMCD3 cells were stably transfected with SSTR3-EGFP, and some cells in the lower part of image have short cilia (green). Scale bars, 5 μm. (D) HEK cells were transfected with ANO1-EGFP (green) and stained with ANO1 antibody (red). Signal from the mANO1-EGFP overlaps with G3 antibody labeling. Scale bar, 5 μm. (E) Size distribution of ANO1 rings. MIPs were made from z-stacks using Zeiss Zen software. The MIPs were then thresholded using robust background subtraction in Cell Profiler (Materials and Methods; Jiang et al., 2012; Wang et al., 2012). The size distribution histogram was fitted to an exponential (black line, count = 223earea/8.9). For the fit, the smallest-size bin was ignored because the thresholding procedure discarded the smallest ANO1 clusters. (F) Specificity of ANO1 antibody. Representative image of mpkCCD14 cells before nimbus formation (Figure 2) treated with lentivirus encoding ANO1 shRNA and EGFP (Figure 9A). Lentivirus-infected cells are identified by EGFP expression (cyan). ANO1 G3 antibody (magenta) shows a decrease in labeling in shRNA-transduced cells. Scale bar, 10 μm. (G) Western blot showing the level of ANO1 in untransfected HEK cells, ANO1-transfected HEK cells, and embryo lysate from wild-type (+/+), ANO1 knockout (−/−), and heterozygous (+/−) mice.
The nimbus is not unique to the mpkCCD14 cell line but instead appears to be a common feature of several epithelial cells we examined. Well-defined nimbi were observed in the rat retinal pigmented epithelium cell line RPE-J (Figure 1B) and the mouse intramedullary collecting duct cell line IMCD3 (Figure 1C). In IMCD3 cells, the nimbus was often less clearly demarcated than in mpkCCD14 or RPE-J cells: in IMCD3 cells, ANO1 nimbi were regularly observed, although frequently ANO1 was concentrated at the apical surface in an amorphous patch without an annulus (Figure 1C). Nimbi were also observed in HEK cells transfected with ANO1–enhanced green fluorescent protein (EGFP; Figure 1D).
The specificity of our antibody is demonstrated by experiments showing that the antibody does not label cells in which ANO1 has been knocked down with short hairpin RNA (shRNA; Figure 1F). it recognizes a band of the expected molecular weight in Western blots that is absent in ANO1−/− mice (Figure 1G), and it colocalizes with ANO1-EGFP in transfected HEK cells (Figure 1D).
Structures that we believe are related to the nimbus have been described previously in the literature and are believed to be areas where ciliary membrane is organized for assembly into the primary cilium (see Discussion). The organization of a CaCC into a structure of such unusual and conspicuous appearance raised questions regarding its potential role in orchestrating apical membrane function. We therefore set out to characterize this structure in more detail.
The nimbus is an antecedent of ciliogenesis
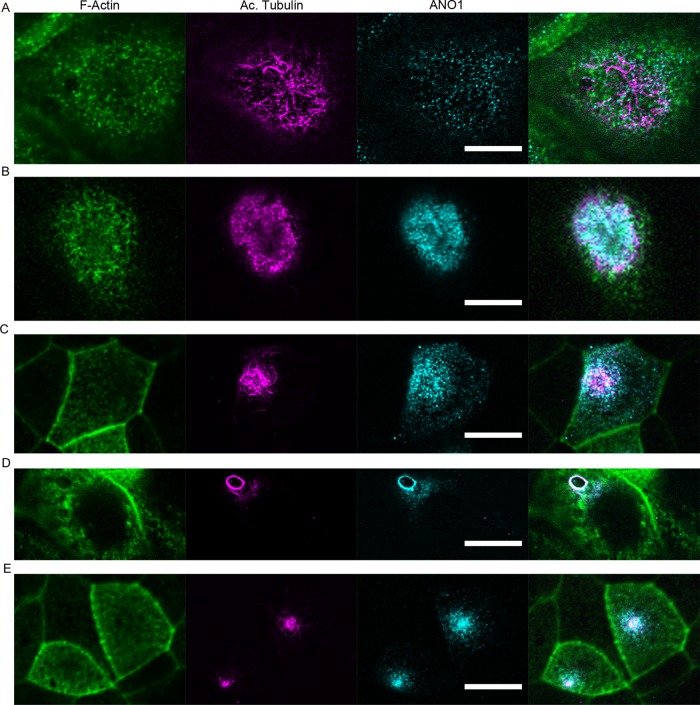
The exponential distribution of nimbus sizes observed (Figure 1E) suggests (but by no means proves) that the nimbus is a dynamic structure because the probability of finding a nimbus of a certain size would be expected to be exponentially distributed if its size were determined by kinetic processes of aggregation and disaggregation of subunits (Moffitt et al., 2010). We investigated the temporal aspects of ANO1 localization and nimbus formation in mpkCCD14 cells that were fixed and labeled at different time points after plating onto permeable supports. Before the cells reach confluency (∼2 d after plating), ANO1 is widely distributed (Figure 2A), but after the cells become confluent (>3 d after plating), ANO1 becomes progressively concentrated into an apical patch (Figure 2, B and C). The nimbus is most clearly defined after the cells reach confluence but before they develop cilia (Figure 2D). As the cells continue to polarize, ANO1 becomes localized to a highly compact apical spot that correlates with a concentration of acetylated tubulin, a marker of the primary cilium (Figure 2E).
FIGURE 2:
ANO1 localization and nimbus formation before formation of the primary cilium. mpkCCD14 cells were plated onto permeable supports, fixed at various times after plating, and stained with fluorescent phalloidin for F-actin (green) and antibodies against acetylated tubulin (magenta) and ANO1 (cyan). In this and subsequent figures, the rightmost image is a merged image of those to the left. (A) At 24 h after seeding, cells are spread out, and ANO1 is visualized as puncta throughout the cell. Acetylated tubulin is found as microtubules distributed throughout the cell. At 2 d (B) and 3 d (C) after seeding, ANO1 and acetylated tubulin become concentrated in a patch at the apical membrane. (D) At 4 d after seeding, ANO1 and acetylated tubulin are visualized in the ring structure we term the nimbus. (E) At 5 d after seeding, as cells continue to polarize and form primary cilia, ANO1 and acetylated tubulin are localized to a discrete spot at the apical portion of the cell. Scale bars, 10 μm.
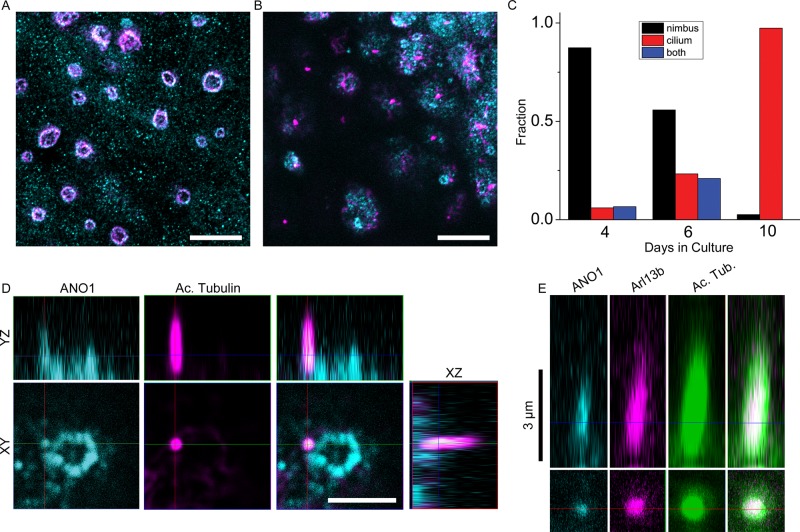
To investigate the relationship of the ANO1 nimbus to the primary cilium, we varied culture conditions (serum concentration, cell plating density, and days in culture) and quantified the fraction of cells positive for a nimbus, a primary cilium, or both. In cultures in which most cells lacked a primary cilium (4 d in culture), nearly all cells had an ANO1 nimbus (Figure 3, A and C), but in cultures in which the cells had a cilium labeled with acetylated tubulin (10 d in culture), very few cells exhibited a nimbus (Figure 3, B and C). When both a cilium and a nimbus were present in the same cell, the emerging cilium emanated from one side of the ANO1 nimbus 74% of the time (Figure 3D; n = 34 randomly selected cells having both nimbi and cilia). The emerging cilium labeled positive for ANO1 as well as acetylated tubulin and always sprouted from one side of the nimbus. The spatial proximity of the nimbus to the primary cilium in these cases and the temporal progression from nimbiated to ciliated cells support the idea that the nimbus may be involved in organization of ciliary components before or early in ciliogenesis. We also observe full-length primary cilia that label for ANO1, acetylated tubulin, and the ciliary protein Arl13b (Figure 3E).
FIGURE 3:
The ANO1 nimbus precedes primary cilium formation and localization of ANO1 in the nascent cilium. (A) Maximum intensity projection of mpkCCD14 cells grown under conditions (high serum, 4 d in culture) at which few cells develop cilia. Under these conditions most cells have a nimbus composed of both ANO1 (cyan) and acetylated tubulin (magenta). (B) Maximum intensity projection of cells grown under conditions (10 d in culture) at which most cells have cilia, labeled by acetylated tubulin (magenta), but very few nimbi (ANO1, cyan). (C) Quantification of the number of cells with well-defined nimbi (black), cilia (red), or both (blue) as a function of days in culture showing that ciliated cells rarely have a well-defined nimbus. Nimbi were defined as annular ANO1-staining structures 2–4 μm in diameter. Cilia were defined as acetylated tubulin-staining projections >2 μm in length. n = 325. (D) The primary cilium (magenta) develops as a projection from the side of a nimbus (cyan). In the few cells that have both a nimbus and a cilium, the cilium usually (74% of the time) projects from the side of the nimbus. Bottom, xy-plane. Top, yz-plane. (E) Primary cilium labeled with ANO1 (cyan), Arl13b (magenta), and acetylated tubulin (green). Scale bars, 10 μm (A, B), 2.5 μm (D), 3 μm (E).
Ciliary proteins in the nimbus
The identification of ANO1 as a component of both the nimbus and the cilium led us to test whether the nimbus contained other ciliary proteins. Assembly of primary cilia is a concerted process that involves trafficking of post-Golgi vesicles to the base of the cilium, their exocytosis by a process engineered by the exocyst complex, and transport of ciliary components by intraflagellar transport (IFT; Pedersen and Rosenbaum, 2008; Nachury et al., 2010; Ishikawa and Marshall, 2011). By immunofluorescence, we detect the exocyst complex protein Sec6 associated with the ANO1 nimbus (Figure 4A), consistent with the idea that the nimbus is associated with the establishment of a ciliary membrane compartment (Nachury et al., 2010; Das and Guo, 2011; Zuo et al., 2011). Furthermore, the ciliary protein Arl13b is also found in the nimbus at a stage in the nimbus life cycle (Figure 4B). Arl13b is a small GTPase required for proper primary cilium structure (Caspary et al., 2007; Cantagrel et al., 2008; Larkins et al., 2011), and Arl13b mutations are linked to Joubert syndrome, a human ciliopathy (Cantagrel et al., 2008). The ANO1 puncta in Arl13b-positive nimbi seemed more indistinctly defined than in Arl13b-negative nimbi. One interpretation is that Arl13b associates with nimbi that are disassembling or reorganizing at a stage just before ciliogenesis. At a later stage, some cells possessed a bleb at the apical surface that was positive for both ANO1 and Arl13b (Figure 4C). These blebs presumably are nascent procilia at an early stage of emergence from the cell. The association of ANO1 and Arl13b with the nimbus and the procilium is shown in Figure 4C, where the left cell has a fuzzy ANO1 ring associated with Arl13b puncta, whereas the right cell has a bleb that is positive for ANO1 and Arl13b that emanates from Arl13b puncta.
FIGURE 4:
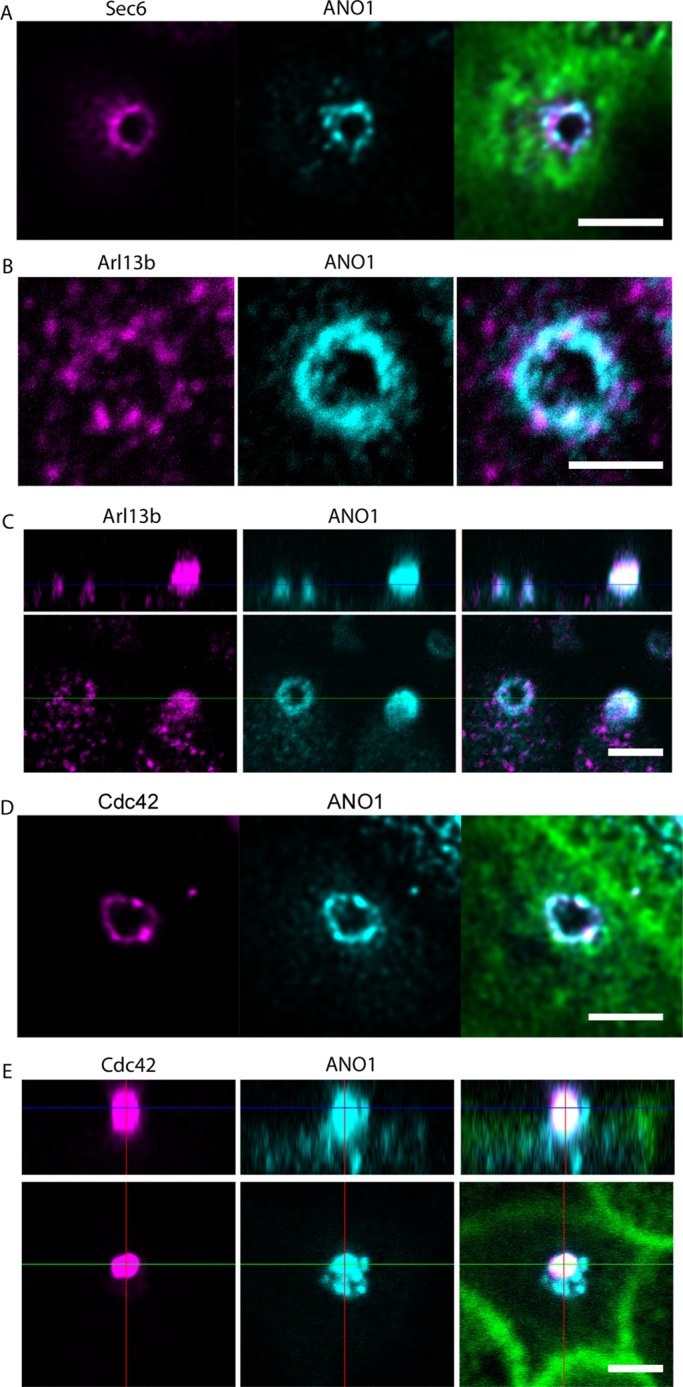
Location of key ciliary proteins in the nimbus. mpkCCD14 cells were grown on permeable supports to a nimbiated stage and labeled with the indicated antibodies. (A) Sec6 (magenta), ANO1 (cyan), and F-actin (green) labeled with Alexa Fluor 633–conjugated phalloidin. (B) Intense puncta of Arl13b (magenta) colocalize with ANO1 (cyan) in the nimbus. (C) Bottom, xy-plane. Top, yz-plane. The left cell has a nimbus containing both ANO1 (cyan) and Arl13b (magenta). The right cell contains a bleb that labels for both ANO1 and Arl13b. (D) Cdc42 (magenta), ANO1 (cyan), F-actin (green). (E) Bottom, xy-plane; top, yz-plane. A bleb of Cdc42 (magenta) also labels positive for ANO1 (cyan) and is presumably a nascent cilium. F-actin (green). Scale bars, 2.5 μm.
The nimbus also labels positive for Cdc42 (Figure 4D), a small GTPase that is essential for ciliogenesis (Choi et al., 2013; Zuo et al., 2011) and a principal determinant in establishing correct cell polarity with respect to the external environment by controlling cytoskeletal dynamics and stabilizing appropriate cellular asymmetry (de Curtis and Meldolesi, 2012; Etienne-Manneville and Hall, 2002). ANO1 and Cdc42 occupy approximately the same contour in the nimbus, and in many cases the two proteins are colocalized (Figure 4D). In addition, blebs of Cdc42 and ANO1 were seen in some cells (Figure 4E). The Cdc42 blebs are surrounded by puncta of ANO1 and are presumably nascent procilia.
Relationship of the nimbus to the centrosome
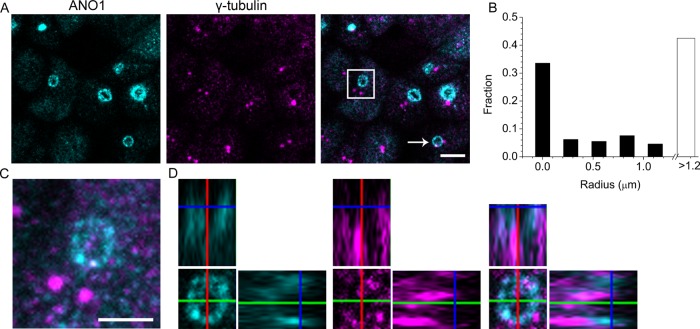
To investigate further the connection between the nimbus and the primary cilium, we examined the relationship of the centrosome to the nimbus. In dividing cells, centrosomes coordinate the formation of the microtubule spindle during mitosis, whereas in interphase cells the centrosome undergoes an incompletely understood functional transition as it migrates to the cell surface. There the mother centriole becomes a basal body that nucleates the axoneme of the primary cilium (Kobayashi and Dynlacht, 2011). Consistent with this model, we observe that each primary cilium is invariably anchored to a basal body as visualized by γ-tubulin staining. However, we believe that the nimbus forms before migration of the basal body to the apical surface, because only about half of the nimbi are associated with a basal body (Figure 5, A and B). In one typical culture, 33% of nimbi (292/870) had at least one clearly identified basal body within its perimeter (Figure 5A, white arrow), and an additional 24% of nimbi (210/870) had a basal body <1.2 μm away (Figure 5B). In the remaining 43% of the cells, the nimbus and the centrioles were clearly separated by >1.2 μm (Figure 5, A, square, and C). However, a careful examination of z-stacks of these centriole-estranged nimbi revealed significant accumulations of γ-tubulin within the nimbi, even though discrete centrioles were present at a remote location in the cell (Figure 5, C and D). Although live-cell imaging will be required to resolve this issue definitively, these data are consistent with a scenario in which nimbus formation occurs before migration of the centriole to the apical membrane (Sorokin, 1962; Joo et al., 2013).
FIGURE 5:
Relationship of the centriole to the ANO1 nimbus. (A) A maximum-intensity projection of a z-stack of confocal images of mpkCCD14 cells, showing that the spatial relationship of nimbi (ANO1, cyan) to centrioles (γ-tubulin, magenta) is variable. Some nimbi have centrioles within their boundary (white arrow), whereas others do not (white box). (B) Quantification of the distance of centrioles from ANO1 nimbi. ANO1 nimbi and centrioles were identified by thresholding using Cell Profiler. The leftmost bar indicates the fraction of ANO1 nimbi having at least one centriole completely contained within the perimeter in the maximum intensity projection (33%). Another 24% of the ANO1 nimbi had centrioles within <1.2 μm. Forty-three percent of rings did not have a centriole within 1.2 μm. (C) High-magnification image of the nimbus boxed in A. Puncta of γ-tubulin (magenta) are evident in the nimbus (ANO1, cyan), despite the remote location of the centrioles to the nimbus. (D) Individual ortho slices through the nimbus show that γ-tubulin in the nimbus partially overlaps with ANO1. Scale bar, 5 μm (A), 2.5 μm (C).
The nimbus is an interface between the microtubule and actin cytoskeletons
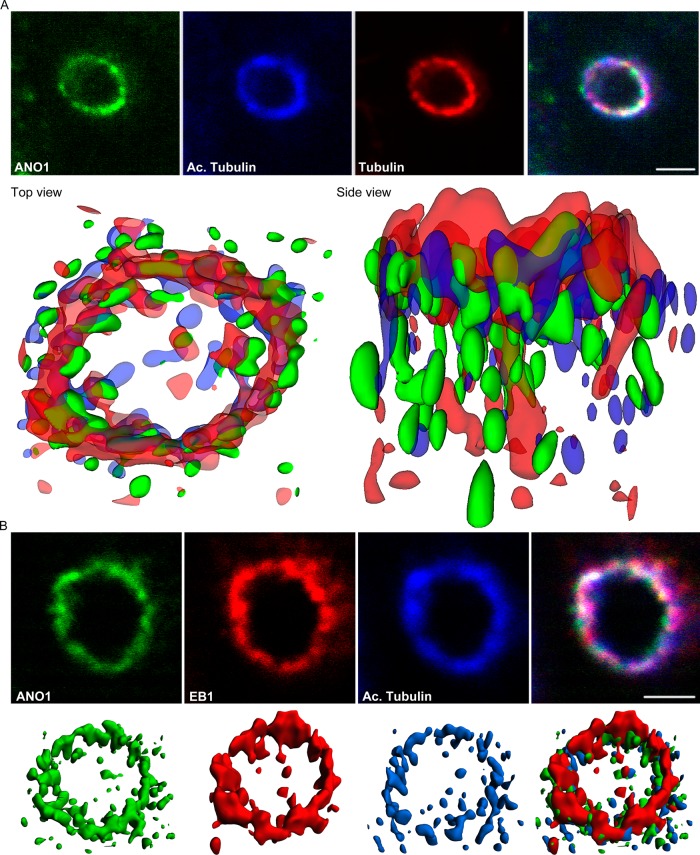
After docking of the mother centriole to the apical membrane, there is extension of the ciliary axoneme, a microtubule-based structure. To explore the spatial relationships of microtubules to the ANO1 nimbus, we deconvolved z-stacks of confocal images labeled for ANO1, acetylated tubulin, and α-tubulin and reconstructed isosurfaces (Figure 6A). This revealed that microtubules describe a torus ∼3 μm in diameter that coincides with the ring of ANO1 puncta. The torus is composed of both acetylated and nonacetylated microtubules that intermingle with puncta of ANO1. The microtubule plus end–tracking protein (+TIP), end-binding protein-1 (EB1), is also associated with the nimbus (Figure 6B). Microtubule +TIPs play an integral role in attaching and stabilizing microtubule ends at the cell cortex, and EB1 has been shown to be essential for ciliogenesis in mouse fibroblasts (Gouveia and Akhmanova, 2010; Schroder et al., 2011).
FIGURE 6:
The nimbus is a hub of the microtubule cytoskeleton. (A) The ANO1 (green) nimbus contains both acetylated (blue) and nonacetylated (red) α-tubulin. Top, representative confocal xy-planes of a z-stack of a nimbus that was subjected to deconvolution. Bottom, the z-stack was deconvolved using Huygens Essential software, and isosurfaces were then constructed from the deconvolved image using Imaris software. Top view, from the apical surface of the cell. Side view, from a plane near the apical surface. (B) EB1 (red) is located at the tips of the microtubules (blue) in the nimbus and commingles with ANO1 (green). Top, representative confocal xy-plane images of a z-stack used for deconvolution. Bottom, isosurfaces constructed from deconvolved images. Scale bars, 2.5 μm.
Recently data have emerged that branched F-actin is a negative regulator of ciliogenesis (Kim et al., 2010; Yan and Zhu, 2013). Consistent with our hypothesis that the nimbus represents a staging area for ciliogenesis, F-actin was largely excluded from the center of the nimbus, as visualized by labeling with fluorescent phalloidin (Figure 7A). The average pixel intensity of fluorescent phalloidin at the center of the ring was an average 11.9-fold less than in the adjacent cytoplasm (Figure 7D). The ezrin/radixin/moesin proteins link the underlying actin cytoskeleton to the plasma membrane (Fehon et al., 2010) and are the most highly enriched proteins in the ANO1 proteome (Perez-Cornejo et al., 2012). Labeling mpkCCD14 cells for moesin revealed that moesin is highly concentrated at the interface of the F-actin–free and F-actin–rich zones coincident with ANO1 (Figure 7, A–C). Plots of moesin and F-actin intensities revealed that the peak moesin intensity was located exactly midway between the cytoplasm and the F-actin–free zone (Figure 7D). The nimbus contained overlapping rings of moesin and ANO1 by confocal microscopy (Figure 7B), whereas structured illumination microscopy (SIM) revealed that both the ANO1 and the moesin rings each comprised 8–12 puncta (Figure 7C). Typically, the puncta of moesin and ANO1 occupy sporadically overlapping and alternating positions in the nimbus. Although moesin is best known in the context of the actin cytoskeleton, moesin also binds and stabilizes microtubules at the cell cortex (Solinet et al., 2013). This location of moesin at the interface of actin-rich and tubulin-rich zones places it in a prime location to coordinate the actin and microtubule cytoskeletons.
FIGURE 7:
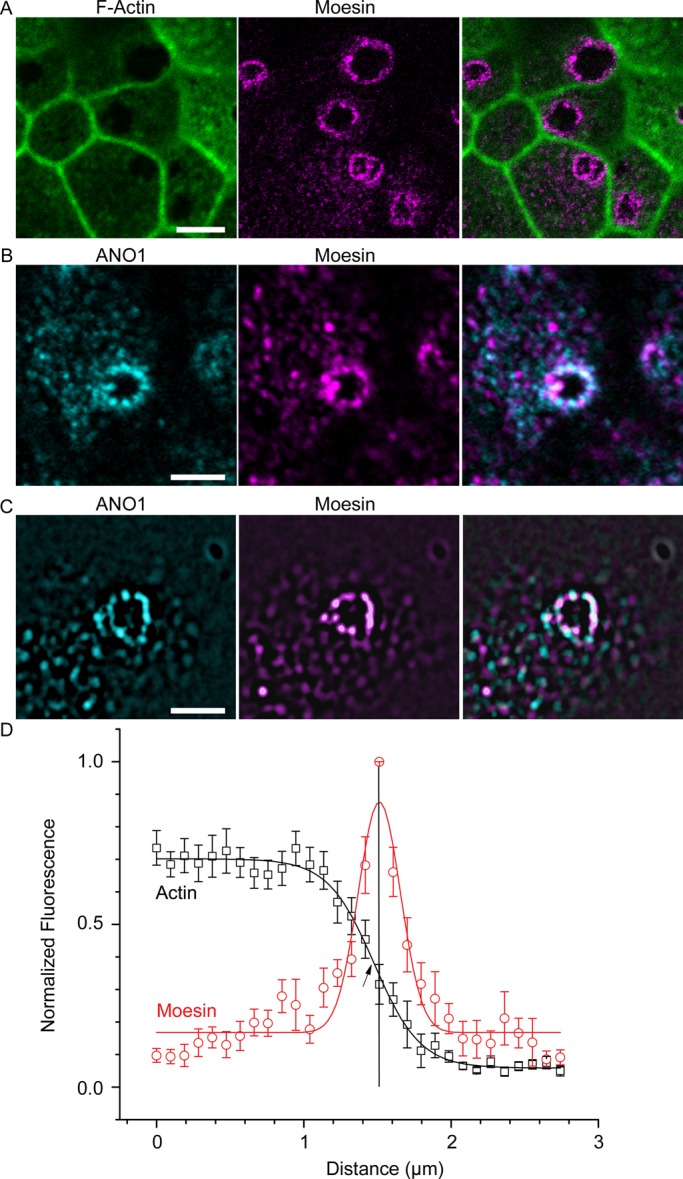
Moesin marks the boundary of an F-actin–free zone that coincides with the ANO1 nimbus. (A) F-actin (green) concentration is lower in the center of the nimbus, and the interface between F-actin–free and F-actin–rich zones is defined by moesin (magenta) labeling. (B) Confocal image of mpkCCD14 cell nimbus showing that ANO1 (cyan) and moesin (magenta) occupy overlapping locations in the nimbus. (C) Structured illumination microscopy of ANO1 (cyan) and moesin (magenta) shows that the ANO1 and moesin rings comprise distinct puncta that are irregularly superimposed. (D) Quantification of moesin distribution relative to F-actin. To measure the relationship between moesin and F-actin labeling, pixel intensities along a line starting near the center of the cell and ending at the approximate center of a nimbus were recorded and normalized to the highest intensity in each channel. These measurements for 12 nimbi were then aligned on the x-axis to the moesin pixel with the highest intensity (= 1, vertical line) and the actin (black data points) and moesin (red data points) intensities averaged at each position on the x-axis. The moesin data were fitted to a Gaussian function (red line), and the actin was fitted to a Boltzmann equation (black line). Because the moesin channels were aligned to the highest moesin pixel intensity in order to calculate the average of multiple nimbi, the peak moesin intensity is 1, whereas variation in actin intensities around each nimbus resulted in a maximum intensity <1. Scale bars, 5 μm (A), 2.5 μm (B, C).
Effects of block of ANO1 Cl− currents on ciliogenesis
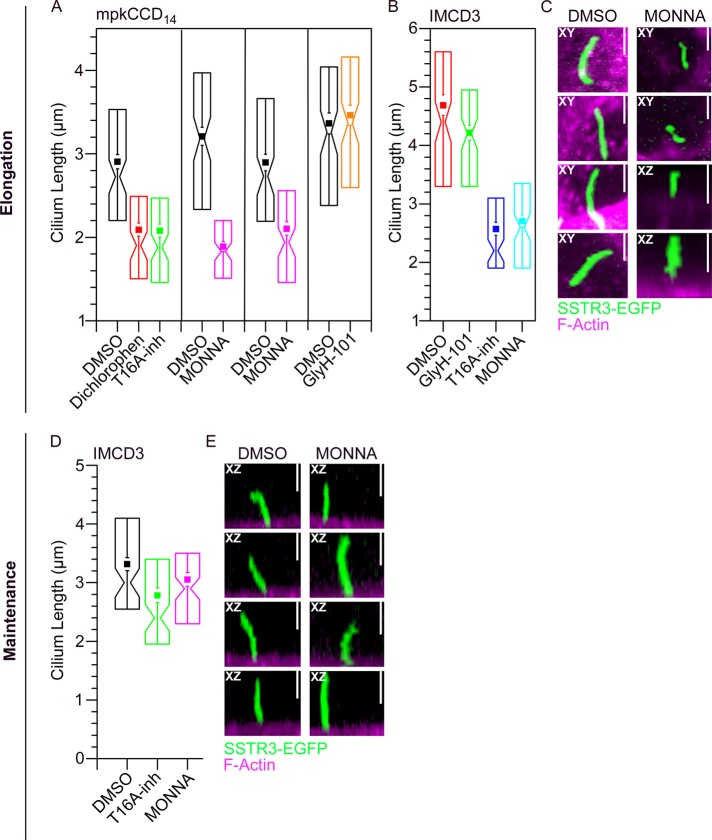
To test whether ANO1 plays a role in ciliogenesis, we measured the effects of inhibitors of ANO1 Cl− current on cilium length and percentage of ciliated cells. Although Cl− channel blockers are notoriously nonspecific (Hartzell et al., 2005), a number of selective ANO1 inhibitors have recently been described, including ANO1-blocking drug 2-[(5-ethyl-1,6-dihydro-4-methyl-6-oxo-2-pyrimidinyl)thio]-N-[4-(4-methoxyphenyl)-2-thiazolyl]acetamide (T16Ainh-A01; Namkung et al., 2011), 4-chloro-2-[(5-chloro-2-hydroxyphenyl)methyl]phenol (dichlorophen; Huang et al., 2012b), (3,5-dibromo-4-hydroxyphenyl)-(2-ethyl-1-benzofuran-3-yl)methanone (benzbromarone; Huang et al., 2012b), and N-((4-methoxy)-2-naphthyl)-5-nitroanthranilic acid (MONNA; Oh et al., 2013). We tested the effect of these drugs on mpkCCD14 cells. Cells were grown on permeable supports until a monolayer was formed (transepithelial resistance >250 Ω/cm2) and then switched to serum-free media to promote ciliogenesis and simultaneously exposed to ANO1 blockers for 24–48 h. Cells were labeled with acetylated tubulin antibody to visualize cilia, phalloidin to assess overall cell and monolayer morphology, and ANO1 antibody. Benzbromarone had the most severe effect: epithelial morphology was disrupted and no ciliated cells were observed after 24 h of treatment with drug. The effect of dichlorophen was less severe but also dramatic: it decreased the percentage of ciliated cells by 50% and the length of cilia by 28% (Figure 8A). The ANO1 inhibitors T16Ainh-A01 and MONNA did not affect the percentage of ciliated cells but consistently decreased the length of cilia by 28–42% (Figure 8A). Of the ANO1 blockers, the selectivity of MONNA has been most thoroughly examined (Oh et al., 2013). MONNA does not inhibit several other Cl− channels, including CFTR and members of the CLC and bestrophin family. In two independent experiments, MONNA decreased cilium length a similar amount. In contrast to the effects of ANO1 inhibitors, the CFTR inhibitor N-(2-naphthalenyl)-((3,5-dibromo-2,4-dihydroxyphenyl)methylene) glycine hydrazide (GlyH-101), which has no effect on ANO1 currents, has no effect on cilium length. Incubation of cells in any of the ANO1 blockers for more than several days inevitably disrupted epithelial polarity and integrity, as evidenced by large decreases in transepithelial resistance and loss of cell–cell contacts visualized by microscopy.
FIGURE 8:
Inhibitors of ANO1 channel activity have a negative effect on cilium length. (A) Quantification of the effect of ANO1 inhibitors on ciliary length in mpkCCD14 cells. (B) Quantification of the effect of ANO1 inhibitors on ciliary length in IMCD3 cells. Solid squares, mean. Whiskers, 1 SEM. Box, 25th and 75th percentiles. Notch, median and 1–99% confidence intervals of the median. *p < 0.001 by two-tailed t test compared with the matched DMSO control. Each data point is the mean of 84–110 cilia measured in randomly selected fields. (C) Representative images of DMSO (control) and MONNA-treated IMCD3 cells labeled for F-actin (magenta) and expressing EGFP-tagged somatostatin receptor 3 (SSTR3-EGFP, green). In C, MONNA was added to the medium at the same time serum starvation was initiated. This protocol tested the effect of ANO1 inhibitors on cilium formation, elongation, and maintenance (labeled “elongation”). (D) Quantification of the effect of ANO1 inhibitors added for 6 h after 24 h of serum starvation. This protocol tested the potential effect of ANO1 inhibitors on maintenance of ciliary length (labeled “maintenance”). (E) Representative image of DMSO (control) and MONNA-treated IMCD3 cells labeled for F-actin (magenta) and expressing EGFP-tagged somatostatin receptor 3 (SSTR3-EGFP, green). Under both conditions of ANO1 inhibitor exposure (C, E) the somatostatin receptor continues to localize to the primary cilium. Vertical scale bars, 2.5 μm (C, E).
To be certain that the effects of ANO1 blockers were not limited to mpkCCD14 cells, we repeated these experiments on IMCD3 cells (Figure 8, B and C). IMCD3 cells on average have longer cilia than mpkCCD14 cells. Nevertheless, both T16Ainh-A01 and MONNA caused a ∼50% decrease in average cilium length, whereas the CFTR inhibitor GlyH-101 had no effect (Figure 8B). Although overall cilia length was decreased in MONNA-treated cells, localization of the somatostatin receptor 3 tagged with EGFP (SSTR3-EGFP) to the primary cilium was not affected (Figure 8C).
The effect of ANO1 channel blockers on cilium length could be explained by an effect on the assembly of the cilium or an effect on the processes that maintain the length of an already assembled cilium. To test for an effect on maintenance, we induced IMCD3 cells stably expressing the SSTR3-EGFP to ciliate by serum starvation for 24 h and then exposed them to either T16A-inh or MONNA for 6 h. Under these conditions, the inhibitor had little effect on cilium length, as measured by SSTR3-EGFP fluorescence (Figure 8, D and E). This observation suggests that once the cilia form, their length is not dependent on ANO1 function. In contrast, ANO1 function is important when cilia are initially forming.
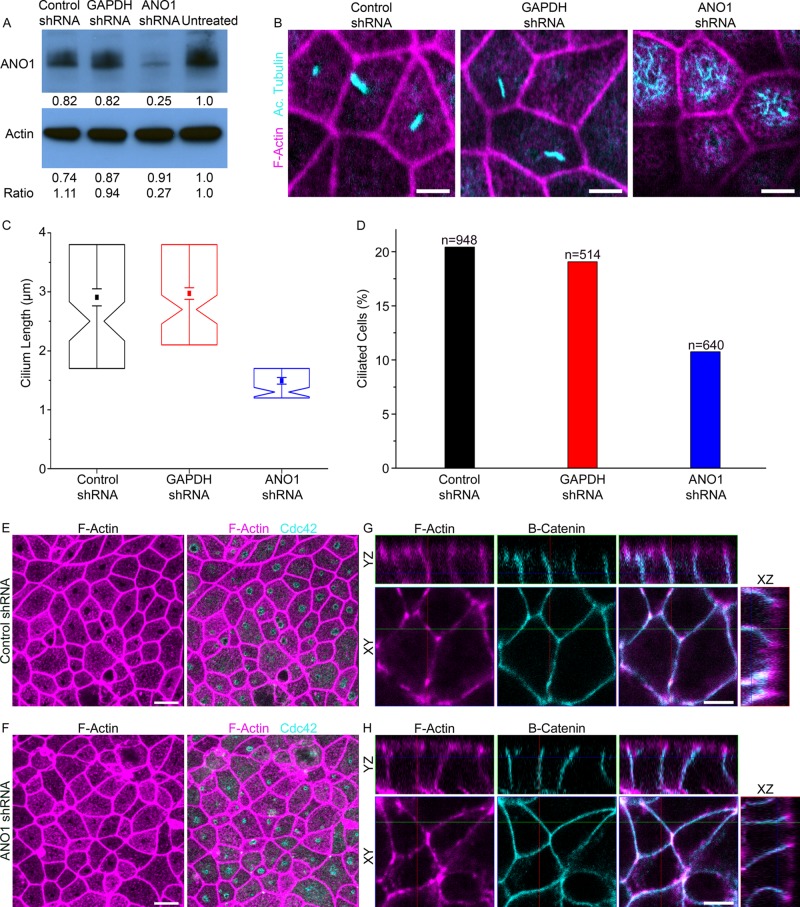
As another approach to testing the role of ANO1 in ciliogenesis, we transduced mpkCCD14 cells with lentiviral particles harboring sequences encoding shRNA against ANO1. Control cells were infected with nonsilencing negative control lentiviral particles or lentivirus having shRNA against glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Western blot showed that ANO1 shRNA reduced the level of ANO1 expression by >70% (Figure 9A). Knockdown of ANO1 caused a significant reduction in both percentage of ciliated cells and in length of cilia as measured by acetylated tubulin labeling (Figure 9, B–D). The percentage of ciliated cells was reduced by 50% (Figure 9D), and the length of the cilia was reduced by 45% (Figure 9C).
FIGURE 9:
Effects of shRNA knockdown of ANO1. (A) Western blot showing the level of ANO1 and β-actin in untreated mpkCCD14 cells and cells treated with lentiviral particles encoding shRNA against ANO1 or GAPDH or a nonsilencing control sequence as indicated. The numbers below the lanes represent the densitometric measurement of the respective bands. The row of numbers labeled “ratio” is the ratio of the ANO1 band to the β-actin band normalized to the untreated control. (B) Representative images of ciliated mpkCCD14 cells that were treated with lentivirus encoding ANO1 or GAPDH shRNA or a nonsilencing control shRNA. Cyan, acetylated tubulin. Magenta, phalloidin. Scale bars, 5 μm. (C) Cilium length in cells treated with shRNA as indicated. Box plot as in Figure 8. (D) Percentage of ciliated cells. (E, F) Nimbi of Cdc42 (cyan) are observed in cultures treated with either (E) control shRNA or (F) ANO1 shRNA. Magenta, F-actin labeled with phalloidin. Scale bars, 10 μm. (G, H) Overall cell polarity appears unperturbed and similar in cells treated with (G) control shRNA and (H) ANO1 shRNA as assessed by β-catenin (cyan) and F-actin (magenta) organization. Scale bars, 5 μm.
The observation that cilium length is shortened by knockdown or inhibition of ANO1 raises the question of whether ANO1 knockdown affects the structure of the nimbus or alters cell polarity. Although we have not investigated these issues in depth, we find that ANO1 knockdown does not affect the formation of the F-actin–free area or the nimbus as visualized by Cdc42 labeling (Figure 9, E and F). Furthermore, ANO1 knockdown does not appear to affect overall cell polarity as visualized by β-catenin labeling (Figure 9, G and H).
DISCUSSION
We describe a transient organelle, the “nimbus,” that is temporally and spatially related to the primary cilium. The nimbus is a torus-shaped structure comprising the Ca2+-activated Cl− channel ANO1 and other proteins involved in ciliogenesis, including Cdc42, Arl13b, the exocyst complex component Sec6, and acetylated tubulin (Das and Guo, 2011; Larkins et al., 2011; Zuo et al., 2011; Choi et al., 2013; Zhang et al., 2013). The nimbus is located at an interface between the actin and microtubule cytoskeletons demarcated by moesin, a protein that interacts stoichiometrically with ANO1 (Perez-Cornejo et al., 2012). The relationship of ANO1 to the primary cilium is strengthened by the composition of the ANO1 proteome (Perez-Cornejo et al., 2012), which is highly enriched in proteins known to participate in ciliogenesis and cell polarity (Hattula et al., 2002; Nachury et al., 2007; Yoshimura et al., 2007; Westlake et al., 2011; Hehnly et al., 2012), including Rab8 (stable isotope labeling by amino acids in cell culture enrichment ratio, 6.7), Rab11 (6.0), TRAPPC3 (3.4), and Cdc42 (4.0). Sixty-two percent of ANO1-interacting proteins are represented in the Ciliary Proteome Database (http://v3.ciliaproteome.org/cgi-bin/index.php; Albertson, 2006), and the highest-ranking ANO1-interacting proteins have experimentally verified functions of organization of the cytoskeleton, membrane–cytoskeletal interactions, and membrane trafficking. This suggests that ANO1 is part of a signaling network that organizes the membrane and cytoskeleton in the process of ciliogenesis.
The nimbus shares similarities to the pericentrosomal preciliary compartment (PPC) described by others (Kim et al., 2010). Although the nimbus, unlike the PPC, does not label for early endosomal markers, it seems likely that the PPC and nimbus perform related or sequential functions. The nimbus also resembles a compartment highly enriched in ceramide sphingolipids that may be involved in tubulin acetylation during ciliogenesis (He et al., 2012). Another structure that one might equate with the nimbus is the ciliary pocket, an invagination of plasma membrane around the cilium (Benmerah, 2013). The ciliary pocket might be a nimbus remnant but is smaller and collars the cilium, whereas the nimbus lies adjacent to the cilium. The ciliary vesicle that assembles at the distal end of the mother centriole (Sorokin, 1962; Joo et al., 2013) apparently forms after the nimbus, because many (43%) nimbi are not adjacent to centrioles.
The central question raised here is the role of Cl− channels in ciliogenesis. Blocking ANO1 Cl− currents or knocking down ANO1 expression results in shortened cilia, suggesting that ANO1 may participate in cilium length control. Changes in cilium length are also caused by defects in Arl13b and IFT proteins that cause polycystic kidney disease (Pazour et al., 2000; Caspary et al., 2007; Ong, 2013). Mechanisms controlling ciliary length are poorly understood but involve precursor molecule availability, intraflagellar transport, cytoskeletal dynamics, and turnover of components at the ciliary tip (Ishikawa and Marshall, 2011; Avasthi and Marshall, 2012).
The finding that ANO1 knockout in mice produces a lethal developmental phenotype supports the assertion that ANO1 is involved in cilium assembly or maintenance (Rock et al., 2008). Although ANO1−/− mice have not been reported to exhibit hallmarks of classic ciliopathies (cystic disease, polydactyly, or situs inversus; Badano et al., 2006), few ANO−/− studies have been published. It is likely that disruption of Cl− transport would have different phenotypic consequences than disruption of the BBsome or intraflagellar transport (Cardenas-Rodriguez and Badano, 2009; Bettencourt-Dias et al., 2011; Hildebrandt et al., 2011).
We propose three testable hypotheses for a role of ANO1 Cl− currents in ciliogenesis (Figure 10):
ANO1 could regulate apical membrane potential (Vm). This could affect other ciliogenic processes, such as Ca2+ signaling. The TRP ion channel polycystin-2 (PKD2, TRPP2) is a Ca2+-permeable channel responsible for the ciliopathy autosomal-dominant polycystic kidney disease (Chapin and Caplan, 2010) and left–right asymmetry in the embryo (McGrath et al., 2003; Yoshiba et al., 2012; Takao et al., 2013). Ca2+ influx through PKD2 or other Ca2+ channels could activate ANO1, which could provide feedback control on Ca2+ influx by altering Vm. Changes in Vm could affect both the driving force for Ca2+ influx and the open probability of voltage-gated channels.
ANO1 may create hydrodynamic and osmotic forces that encourage ciliary extension. In this model, membrane trafficking and cytoskeletal assembly alone are insufficient to erect a normal cilium; osmotic pressure would assist by producing a cellular bleb (Nilius et al., 1997; Hoffmann et al., 2009; Duran et al., 2010) into which ciliary components could traffic and assemble. This idea is attractive because it seems consistent with membrane blebs such as those in Figure 4, C and E.
The Cl− ion may function as a regulator of protein function (Duran et al., 2010). More than 1500 human proteins in the Protein Data Bank, including Rab11A and SHH, have Cl− as a ligand. In these structures, Cl− is frequently located at protein interfaces, hinting that anions could participate in protein–protein interaction. The observation that the enzymatic activities of many proteins, including certain G-proteins and protein kinases, are affected by Cl− in the physiological range supports a regulatory role for Cl− (Nakajima et al., 1992; Duran et al., 2010; Pacheco-Alvarez and Gamba, 2011; Lo Nostro and Ninham, 2012; Szalontai et al., 2013). Because intracellular Cl− is spatially and temporally dynamic (Kuner and Augustine, 2000; Duebel et al., 2006; Berglund et al., 2008), gradients of intracellular Cl− in the periciliary region could locally regulate protein function. For example, tubulin polymerization is affected by the species and concentration of anion (Suzaki et al., 1978; Ray et al., 1984; Wolff et al., 1996). Alternatively, Cl− concentration in intracellular vesicular compartments might alter vesicular trafficking (Faundez and Hartzell, 2004; Stauber and Jentsch, 2013).
FIGURE 10:
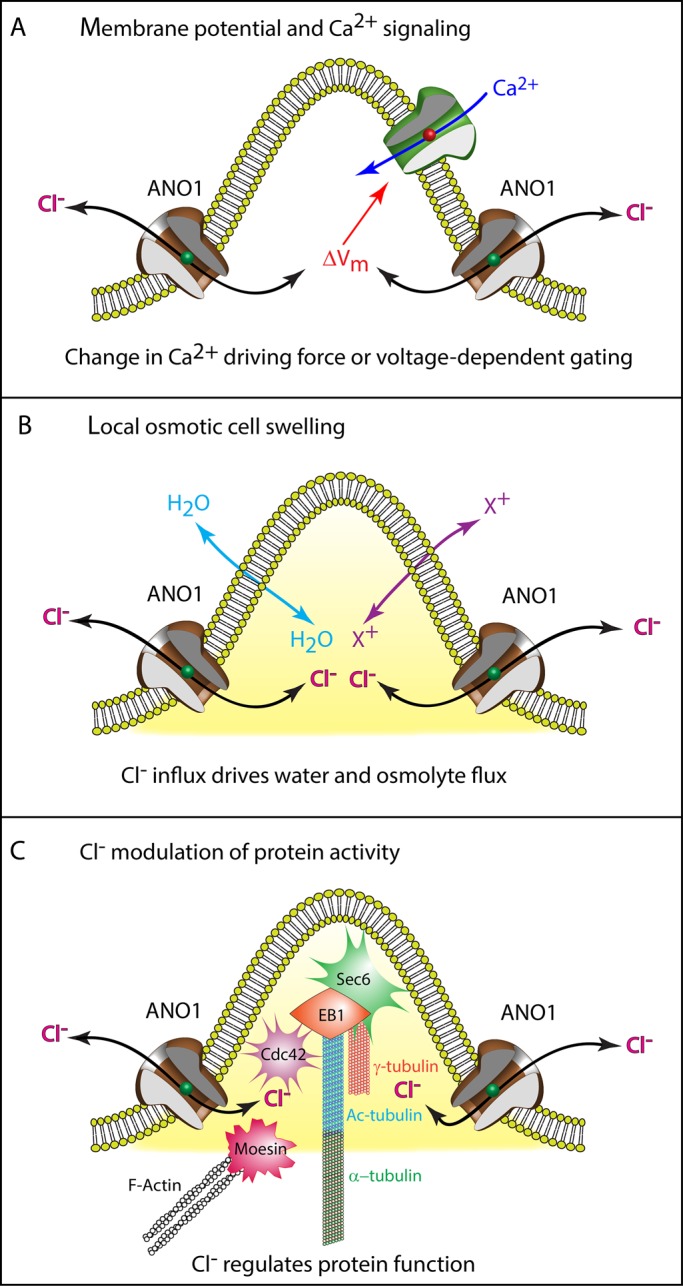
Possible mechanisms of ANO1 function in ciliogenesis. See Discussion for more information. Although the figure depicts ANO1 in the plasma membrane at the base of a nascent cilium as suggested by Figure 3E, we have no firm evidence that ANO1 is located in the plasma membrane. In addition, the proximity of Sec6, EB1, Cdc42, moesin, tubulin, and actin in the illustration is not meant to imply that they form a complex.
The ciliary membranes differ from the plasma membrane in both protein and lipid composition (Emmer et al., 2010; Maric et al., 2010; Conduit et al., 2012; He et al., 2012). Intriguingly, some members of the ANO family are required for phospholipid scrambling (Suzuki et al., 2010, 2013; Yang et al., 2012; Malvezzi et al., 2013). Although there is no evidence that ANO1 has lipid scramblase activity, the high sequence conservation among ANOs suggests that ANO1 may have important interactions with lipids (Hartzell and Ruppersburg, 2013).
It is hoped that the association of ANO1 with the primary cilium will shed light on the relationship of ANO1 to cancer (Wilkerson and Reis-Filho, 2013). Ciliary signaling has an important role in the control of cell division because of the dichotomous role of the centrosome in nucleating the cilium and organizing the mitotic spindle. Cross-talk between ciliogenesis and mitosis is evident from observations that defective cell division results in abnormal ciliogenesis and that defective ciliogenesis alters the cell cycle. For example, proliferative transformed cells typically lack a primary cilium (Wheatley, 1995), and cells having defective primary cilia can undergo inappropriate cell division, as occurs in cystic disease (Sharma et al., 2013). ANO1 is overexpressed in several cancer cell lines, and knockdown or interference with ANO1 expression or function has been reported to suppress proliferation (Stanich et al., 2011; Duvvuri et al., 2012; Mazzone et al., 2012; Britschgi et al., 2013), although results are inconsistent (Ayoub et al., 2010; Kamentsky et al., 2011; Ruiz et al., 2012; Simon et al., 2013).
In summary, we identified for the first time an anion channel that both localizes to the primary cilium and at some level appears to contribute to primary cilium formation. We believe that this novel finding will open new lines of investigation into the role of Cl− channels in primary cilium biology.
MATERIALS AND METHODS
Cell culture
The mpkCCD14 cells were maintained in modified medium (DMEM:Ham's F12 1:1 + GlutaMax; 20 μg/ml sodium selenite, 5 μg/ml apotransferrin, 1 nM triiodothyronine, 12 mM d-glucose, 2% fetal bovine serum [FBS], and 15 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4) at 37°C in a 5% CO2/95% air humidified atmosphere. Cells were seeded on semipermeable filters (Millicell, 0.4-μm pore size; Millipore, Billerica, MA) for experiments. For cell staging experiments, mpkCCD14 cells were seeded at starting concentration of 2.25 × 105 onto permeable supports (Millicell) and samples taken every 24 h for 6 d. IMCD3 cells were maintained in DMEM:F12 supplemented with 10% FBS. IMCD3 cells stably expressing SSTR3-EGFP were a generous gift from Greg Pazour, University of Massachusetts, Worcester, MA (Pazour, 2008). RPE-J cells were cultured in DMEM with 10% FBS.
Immunofluorescence and image analysis
Cells grown on permeable supports filters were fixed in 4% paraformaldehyde overnight at 4°C, washed in phosphate-buffered saline (PBS), incubated in a blocking solution of PBS containing 0.15% Triton X-100 and 2% cold-water fish gelatin at room temperature, and subsequently incubated with primary antibodies. Primary antibodies were used against the following antigens: ANO1 1:1000 (custom rabbit polyclonal genomic antibody G3 [SDIX, Newark, DE] directed against mAno1 amino acids 878–960 and G2 directed against amino acids 54–126 provided identical results), anti-moesin mouse monoclonal 1:1000 (A50007; Abcam, Cambridge, MA), anti–acetylated tubulin mouse monoclonal 1:1000 (subtype immunoglobulin G2b [IgG2b], 6-11B-1; Sigma-Aldrich, St. Louis, MO), anti-Cdc42 mouse monoclonal 1:500 (sc-8401; Santa Cruz Biotechnology, Dallas, TX), anti–α-tubulin mouse monoclonal 1:500 (subtype IgG1; T9026; Sigma-Aldrich), anti–γ-tubulin monoclonal 1:2000 (subtype IgG1; T6557; Sigma-Aldrich), anti-EB1 monoclonal 1:1000 (subtype IgG1; 610534; BD Biosciences, San Jose, CA), anti-Arl13b mouse monoclonal clone N295B 1:10 (subtype IgG2a; 73–287; UC Davis/NIH NeuroMab Facility, Davis, CA), anti–β-catenin mouse 1:200 (610153; BD Transduction Lab; gift of Michael Koval, Emory University, Atlanta, GA), and anti-Sec6 mouse monoclonal 1:5 (9H5; Charles Yeaman, University of Iowa, Iowa City, IA; Andersen and Yeaman, 2010). The cells were then washed and incubated in secondary antibodies (1:1000): Alexa Fluor 488–labeled goat anti-rabbit or anti-mouse IgG (Invitrogen, Carlsbad, CA), Daylight 549-labeled goat anti-rabbit IgG (Jackson Immunochemicals, West Grove, PA), Alexa Fluor 555–labeled goat anti–mouse IgG1 (Invitrogen), and Alexa Fluor 633–labeled goat anti-mouse IgG2b (Invitrogen). Actin filaments were stained with phalloidin conjugated to Alexa Fluor 633 (Invitrogen). The filters were mounted in ProLong Gold (Invitrogen). Confocal images were acquired at room temperature using a Zeiss LSM-510 microscope and ZEN software (Carl Zeiss, Oberkochen, Germany). Objectives used were 63×/1.4 numerical aperture (NA) and 100×/1.4 NA Plan-Apochromat oil objectives. The z-stacks were deconvolved using Huygens Essential 3.7 deconvolution software (Hilversum, Netherlands) with a signal-to-noise ratio of 15, quality of 0.1, and automatic bleaching correction. Resulting images were processed using ZEN and Imaris software. Three-dimensional SIM was performed on an inverted Nikon (Melville, NY) microscope running NIS Elements with a 100× oil objective, NA 1.49.
Measurements of ANO1 nimbus size were performed using Cell Profiler. Maximum intensity projections (MIPs) were made of z-stacks taken through the ANO1 ring using Zeiss Zen software. The MIPs were then thresholded using robust background subtraction in Cell Profiler (Carpenter et al., 2006; Kamentsky et al., 2011). The algorithm trims the brightest and dimmest 5% of pixel intensities over the image, then calculates the mean and SD of the remaining pixels and calculates the threshold as the mean plus two times the SD. The dimensions (area and axes) of the resulting thresholded structures were then automatically measured by Cell Profiler. Approximately 800 rings were identified and measured. Distances between nimbi and centrioles were also determined using Cell Profiler after identifying the structures by a similar thresholding procedure.
Cells used to assess the percentage of ciliated cells and cilium lengths were processed for immunofluorescence microscopy by staining with anti–acetylated tubulin. The ciliated cells and total cells were counted in random fields to determine the percentage of ciliated cells for each condition. Cilia lengths were measured using confocal z-stacks analyzed in ZEN software. Only cilia that projected from the apical portion of the cell were measured.
ANO1 inhibitor treatment
The mpkCCD14 cells were seeded at 5.0 × 105 and grown on permeable supports until the transepithelial resistance increased to greater than five times background (>500 Ω), usually several days. The cells were the treated with the indicated inhibitors by replacing both the apical and basal medium with serum-free medium with or without inhibitors. The inhibitors used were T16Ainh-A01 (Namkung et al., 2011; a generous gift from Alan Verkman, University of California, San Francisco, San Francisco, CA), dichlorophen (Huang et al., 2012b; Sigma-Aldrich), benzbromarone (Huang et al., 2012b; Sigma-Aldrich), MONNA (Oh et al., 2013; a generous gift from Justin Lee, Korean Institute of Science and Technology, Seoul, South Korea), and GlyH-101 (Muanprasat et al., 2004; EMD Millipore). IMCD3 cells were grown to confluency on glass-bottom dishes in the presence of serum, after which they were simultaneously switched to serum-free medium and exposed to the various inhibitors for 24 h.
The ANO1 inhibitor MONNA was used at a final concentration of 10 μM and 1% dimethyl sulfoxide (DMSO), T16Ainh-A01 at 10 μM and 0.1% DMSO, dichlorophen at 20 μM and 0.1% DMSO, and benzbromarone at 50 μM and 0.1% DMSO. The CFTR channel inhibitor GlyH-101 was used at a final concentration of 2.5 μm and 0.1% DMSO. Cells were treated with the indicated concentrations of inhibitors or matching percentage of DMSO in serum-free medium for either 24 or 48 h before fixation. Cells were labeled with an antibody against acetylated tubulin to label primary cilia. Individual cilia lengths and the percentage of ciliated cells were measured using Zeiss ZEN software.
For experiments to test the effects of ANO1 inhibitors on cilia maintenance, IMCD3 cells were cultured as described, serum starved for 24 h, and then exposed to inhibitors, still in serum-free medium, for 6 h before fixation.
Lentivirus transduction
GIPZ lentiviral shRNA particles were obtained from Thermo Fisher Scientific and targeted to mANO1 (V2LMM_92063), GAPDH (RHS4372), or negative nonsilencing control (RHS4348). For lentiviral transduction, mpkCCD14 cells were plated at 8 × 105 cells/plate in 60-mm dishes and allowed to grow overnight. After 24 h, cells were switched to serum-free medium with lentiviral particles. At 24 h after transduction, infected cells were selected by the addition of 1.5 μg/ml puromycin in serum-containing medium. At 7 d after selection, cells were plated at 5 × 105 cells/filter in serum-containing medium. After 48 h, cells were switched to serum-free medium for an additional 48 h before fixation. Only cells identified as positively transduced by expression of TurboGFP were counted in the analysis of the percentage of ciliated cells and measurements of cilia lengths.
Acknowledgments
We thank Bonnie Blazer-Yost and Stephanie Flaig for mpkCCD14 cells and advice on their culture; Alexa Mattheyses at the Integrated Cellular Imaging Core, Emory University, for advice and training in microscopy; Greg Pazour, Tamara Caspary, Joseph Besharse, and Victor Faundez for helpful advice and reagents; and Alan Verkman, Justin Lee, Charles Yeaman, and Daniel Billadeau for reagents. This work was supported by National Institutes of Health Grants GM60448 and EY11482 and a pilot grant from the Emory Center for Cystic Fibrosis Research of Children's Healthcare of Atlanta. C.C.R. was supported by National Institutes of Health Training Grant 5T32GM008367. This research project was supported in part by the Emory University Integrated Cellular Imaging Core and a SIM pilot grant from the Core.
Abbreviations used:
- ANO1
anoctamin-1
- GlyH-101
N-(2-naphthalenyl)-((3,5-dibromo-2,4-dihydroxyphenyl)methylene) glycine hydrazide
- MIP
maximum-intensity projection
- MONNA
N-((4-methoxy)-2-naphthyl)-5-nitroanthranilic acid
- SHH
sonic hedgehog
- T16Ainh-A01
ANO1-blocking drug 2-[(5-ethyl-1,6-dihydro-4-methyl-6-oxo-2-pyrimidinyl)thio]-N-[4-(4-methoxyphenyl)-2-thiazolyl]acetamide
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-10-0599) on April 2, 2014.
REFERENCES
- Albertson DG. Gene amplification in cancer. Trends Genet. 2006;22:447–455. doi: 10.1016/j.tig.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Andersen NJ, Yeaman C. Sec3-containing exocyst complex is required for desmosome assembly in mammalian epithelial cells. Mol Biol Cell. 2010;21:152–164. doi: 10.1091/mbc.E09-06-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avasthi P, Marshall WF. Stages of ciliogenesis and regulation of ciliary length. Differentiation. 2012;83:S30–S42. doi: 10.1016/j.diff.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoub C, Wasylyk C, Li Y, Thomas E, Marisa L, Robe A, Roux M, Abecassis J, de Reynies A, Wasylyk B. ANO1 amplification and expression in HNSCC with a high propensity for future distant metastasis and its functions in HNSCC cell lines. Br J Cancer. 2010;103:715–726. doi: 10.1038/sj.bjc.6605823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badano JL, Mitsuma N, Beales PL, Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genom Hum Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- Benmerah A. The ciliary pocket. Curr Opin Cell Biol. 2013;25:78–84. doi: 10.1016/j.ceb.2012.10.011. [DOI] [PubMed] [Google Scholar]
- Berbari NF, O'Connor AK, Haycraft CJ, Yoder BK. The primary cilium as a complex signaling center. Curr Biol. 2009;19:R526–R535. doi: 10.1016/j.cub.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berend K, van Hulsteijn LH, Gans ROB. Chloride: the queen of electrolytes. Eur J Internal Med. 2012;23:203–211. doi: 10.1016/j.ejim.2011.11.013. [DOI] [PubMed] [Google Scholar]
- Berglund K, Schleich W, Krieger P, Loo LS, Wang D, Cant NB, Feng G, Augustine GJ, Kuner T. Imaging synaptic inhibition in transgenic mice expressing the chloride indicator, Clomeleon. Brain Cell Biol. 2008;35:207–228. doi: 10.1007/s11068-008-9019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Hildebrandt F, Pellman D, Woods G, Godinho SA. Centrosomes and cilia in human disease. Trends Genet. 2011;27:307–315. doi: 10.1016/j.tig.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britschgi A, et al. Calcium-activated chloride channel ANO1 promotes breast cancer progression by activating EGFR and CAMK signaling. Proc Natl Acad Sci USA. 2013;110:E1026–E1034. doi: 10.1073/pnas.1217072110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantagrel V, et al. Mutations in the cilia gene ARL13B lead to the classical form of Joubert syndrome. Am J Hum Genet. 2008;83:170–179. doi: 10.1016/j.ajhg.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputo A, Caci E, Ferrera L, Pedemonte N, Barsanti C, Sondo E, Pfeffer U, Ravazzolo R, Zegarra-Moran O, Galietta LJV. TMEM16A, a membrane protein associated with calcium-dependent chloride channel activity. Science. 2008;322:590–594. doi: 10.1126/science.1163518. [DOI] [PubMed] [Google Scholar]
- Cardenas-Rodriguez M, Badano JL. Ciliary biology: understanding the cellular and genetic basis of human ciliopathies. Am J Med Genet C Semin Med Genet. 2009;151C:263–280. doi: 10.1002/ajmg.c.30227. [DOI] [PubMed] [Google Scholar]
- Carpenter AE, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Chapin HC, Caplan MJ. The cell biology of polycystic kidney disease. J Cell Biol. 2010;191:701–710. doi: 10.1083/jcb.201006173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassin C, Bens M, Vandewalle A. Transimmortalized proximal tubule and collecting duct cell lines derived from the kidneys of transgenic mice. Cell Biol Toxicol. 2007;23:257–266. doi: 10.1007/s10565-006-0169-y. [DOI] [PubMed] [Google Scholar]
- Choi SY, Chacon-Heszele MF, Huang L, McKenna S, Wilson FP, Zuo X, Lipschutz JH. Cdc42 deficiency causes ciliary abnormalities and cystic kidneys. J Am Soc Nephrol. 2013;24:1435–1450. doi: 10.1681/ASN.2012121236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit SE, Dyson JM, Mitchell CA. Inositol polyphosphate 5-phosphatases; new players in the regulation of cilia and ciliopathies. FEBS Lett. 2012;586:2846–2857. doi: 10.1016/j.febslet.2012.07.037. [DOI] [PubMed] [Google Scholar]
- Das A, Guo W. Rabs and the exocyst in ciliogenesis, tubulogenesis and beyond. Trends Cell Biol. 2011;21:383–386. doi: 10.1016/j.tcb.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis EE, Katsanis N. The ciliopathies: a transitional model into systems biology of human genetic disease. Curr Opin Genet Dev. 2012;22:290–303. doi: 10.1016/j.gde.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Curtis I, Meldolesi J. Cell surface dynamics—how Rho GTPases orchestrate the interplay between the plasma membrane and the cortical cytoskeleton. J Cell Sci. 2012;125:4435–4444. doi: 10.1242/jcs.108266. [DOI] [PubMed] [Google Scholar]
- Duebel J, Haverkamp S, Schleich W, Feng G, Augustine GJ, Kuner T, Euler T. Two-photon imaging reveals somatodendritic chloride gradient in retinal ON-type bipolar cells expressing the biosensor Clomeleon. Neuron. 2006;49:81–94. doi: 10.1016/j.neuron.2005.10.035. [DOI] [PubMed] [Google Scholar]
- Duran C, Hartzell HC. Physiological roles and diseases of TMEM16/anoctamin proteins: are they all chloride channels. Acta Pharmacol Sin. 2011;32:685–692. doi: 10.1038/aps.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran C, Thompson CH, Xiao Q, Hartzell HC. Chloride channels: often enigmatic, rarely predictable. Annu Rev Physiol. 2010;72:95–121. doi: 10.1146/annurev-physiol-021909-135811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvvuri U, et al. TMEM16A induces MAPK and contributes directly to tumorigenesis and cancer progression. Cancer Res. 2012;72:3270–3281. doi: 10.1158/0008-5472.CAN-12-0475-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmer BT, Maric D, Engman DM. Molecular mechanisms of protein and lipid targeting to ciliary membranes. J Cell Sci. 2010;123:529–536. doi: 10.1242/jcs.062968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa I, et al. A novel monoclonal antibody against DOG1 is a sensitive and specific marker for gastrointestinal stromal tumors. Am J Surg Pathol. 2008;32:210–218. doi: 10.1097/PAS.0b013e3181238cec. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- Faundez V, Hartzell HC. Intracellular chloride channels: determinants of function in the endosomal pathway. Sci STKE. 2004;2004(233):re8. doi: 10.1126/stke.2332004re8. [DOI] [PubMed] [Google Scholar]
- Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: the role of ERM proteins. Nat Rev Mol Cell Biol. 2010;11:276–287. doi: 10.1038/nrm2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrera L, Caputo A, Galietta LJ. TMEM16A protein: a new identity for Ca2+-dependent Cl channels. Physiology (Bethesda) 2010;25:357–363. doi: 10.1152/physiol.00030.2010. [DOI] [PubMed] [Google Scholar]
- Gouveia SM, Akhmanova A. Cell and molecular biology of microtubule plus end tracking proteins: end binding proteins and their partners. Int Rev Cell Mol Biol. 2010;285:1–74. doi: 10.1016/B978-0-12-381047-2.00001-3. [DOI] [PubMed] [Google Scholar]
- Hartzell HC. Chloride channels: an historical perspective. In: Alvarez-Leefmans FJDE, editor. In: Physiology and Pathology of Chloride Transporters and Channels in the Nervous System: From Molecules to Diseases. San Diego, CA: Academic Press; 2009. pp. 3–16. [Google Scholar]
- Hartzell C, Putzier I, Arreola J. Calcium-activated chloride channels. Annu Rev Physiol. 2005;67:719–758. doi: 10.1146/annurev.physiol.67.032003.154341. [DOI] [PubMed] [Google Scholar]
- Hartzell HC, Ruppersburg CC. Functional reconstitution of a chloride channel bares its soul. Proc Natl Acad Sci USA. 2013;110:19185–19186. doi: 10.1073/pnas.1319415110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzell HC, Yu K, Xiao Q, Chien LT, Qu Z. Anoctamin/TMEM16 family members are Ca2+-activated Cl- channels. J Physiol. 2009;587:2127–2139. doi: 10.1113/jphysiol.2008.163709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattula K, Furuhjelm J, Arffman A, Peranen J. A Rab8-specific GDP/GTP exchange factor is involved in actin remodeling and polarized membrane transport. Mol Biol Cell. 2002;13:3268–3280. doi: 10.1091/mbc.E02-03-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Wang G, Dasgupta S, Dinkins M, Zhu G, Bieberich E. Characterization of an apical ceramide-enriched compartment regulating ciliogenesis. Mol Biol Cell. 2012;23:3156–3166. doi: 10.1091/mbc.E12-02-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehnly H, Chen CT, Powers CM, Liu HL, Doxsey S. The centrosome regulates the Rab11-dependent recycling endosome pathway at appendages of the mother centriole. Curr Biol. 2012;22:1944–1950. doi: 10.1016/j.cub.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann EK, Lambert IH, Pedersen SF. Physiology of cell volume regulation in vertebrates. Physiol Rev. 2009;89:193–277. doi: 10.1152/physrev.00037.2007. [DOI] [PubMed] [Google Scholar]
- Huang F, et al. Calcium-activated chloride channel TMEM16A modulates mucin secretion and airway smooth muscle contraction. Proc Natl Acad Sci USA. 2012b;109:16354–16359. doi: 10.1073/pnas.1214596109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Wong X, Jan LY. International Union of Basic and Clinical Pharmacology. LXXXV: calcium-activated chloride channels. Pharmacol Rev. 2012a;64:1–15. doi: 10.1124/pr.111.005009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang DG, Qian X, Hornick JL. DOG1 antibody is a highly sensitive and specific marker for gastrointestinal stromal tumors in cytology cell blocks. Am J Clin Pathol. 2011;135:448–453. doi: 10.1309/AJCP0PPKOBNDT9LB. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Marshall WF. Ciliogenesis: building the cell's antenna. Nat Rev Mol Cell Biol. 2011;12:222–234. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- Jiang K, et al. A proteome-wide screen for mammalian SxIP motif-containing microtubule plus-end tracking proteins. Curr Biol. 2012;22:1800–1807. doi: 10.1016/j.cub.2012.07.047. [DOI] [PubMed] [Google Scholar]
- Joo K, et al. CCDC41 is required for ciliary vesicle docking to the mother centriole. Proc Natl Acad Sci USA. 2013;110:5987–5992. doi: 10.1073/pnas.1220927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamentsky L, Jones TR, Fraser A, Bray MA, Logan DJ, Madden KL, Ljosa V, Rueden C, Eliceiri KW, Carpenter AE. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics. 2011;27:1179–1180. doi: 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M. FLJ10261 gene, located within the CCND1-EMS1 locus on human chromosome 11q13, encodes the eight-transmembrane protein homologous to C12orf3, C11orf25 and FLJ34272 gene products. Int J Oncol. 2003;22:1375–1381. [PubMed] [Google Scholar]
- Katoh M. Identification and characterization of TMEM16H gene in silico. Int J Mol Med. 2005;15:353–358. [PubMed] [Google Scholar]
- Kim J, Lee JE, Heynen-Genel S, Suyama E, Ono K, Lee K, Ideker T, Aza-Blanc P, Gleeson JG. Functional genomic screen for modulators of ciliogenesis and cilium length. Nature. 2010;464:1048–1051. doi: 10.1038/nature08895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Dynlacht BD. Regulating the transition from centriole to basal body. J Cell Biol. 2011;193:435–444. doi: 10.1083/jcb.201101005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuner T, Augustine GJ. A genetically encoded ratiometric indicator for chloride: capturing chloride transients in cultured hippocampal neurons. Neuron. 2000;27:447–459. doi: 10.1016/s0896-6273(00)00056-8. [DOI] [PubMed] [Google Scholar]
- Kunzelmann K, Tian Y, Martins JR, Faria D, Kongsuphol P, Ousingsawat J, Thevenod F, Roussa E, Rock J, Schreiber R. Anoctamins. Pflugers Arch. 2011;462:195–208. doi: 10.1007/s00424-011-0975-9. [DOI] [PubMed] [Google Scholar]
- Larkins CE, Aviles GD, East MP, Kahn RA, Caspary T. Arl13b regulates ciliogenesis and the dynamic localization of Shh signaling proteins. Mol Biol Cell. 2011;22:4694–4703. doi: 10.1091/mbc.E10-12-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Nostro P, Ninham BW. Hofmeister phenomena: an update on ion specificity in biology. Chem Rev. 2012;112:2286–2322. doi: 10.1021/cr200271j. [DOI] [PubMed] [Google Scholar]
- Malvezzi M, Chalat M, Janjusevic R, Picollo A, Terashima H, Menon AK, Accardi A. Ca2+-dependent phospholipid scrambling by a reconstituted TMEM16 ion channel. Nat Commun. 2013;4:2367. doi: 10.1038/ncomms3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maric D, Epting CL, Engman DM. Composition and sensory function of the trypanosome flagellar membrane. Curr Opin Microbiol. 2010;13:466–472. doi: 10.1016/j.mib.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzone A, Eisenman ST, Strege PR, Yao Z, Ordog T, Gibbons SJ, Farrugia G. Inhibition of cell proliferation by a selective inhibitor of the Ca(2+)-activated Cl(-) channel. Ano1. Biochem Biophys Res Commun. 2012;427:248–253. doi: 10.1016/j.bbrc.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J, Somlo S, Makova S, Tian X, Brueckner M. Two populations of node monocilia initiate left-right asymmetry in the mouse. Cell. 2003;114:61–73. doi: 10.1016/s0092-8674(03)00511-7. [DOI] [PubMed] [Google Scholar]
- Miettinen M, Wang ZF, Lasota J. DOG1 antibody in the differential diagnosis of gastrointestinal stromal tumors: a study of 1840 cases. Am J Surg Pathol. 2009;33:1401–1408. doi: 10.1097/PAS.0b013e3181a90e1a. [DOI] [PubMed] [Google Scholar]
- Moffitt JR, Chemla YR, Bustamante C. Methods in statistical kinetics. Methods Enzymol. 2010;475:221–257. doi: 10.1016/S0076-6879(10)75010-2. [DOI] [PubMed] [Google Scholar]
- Muanprasat C, Sonawane ND, Salinas D, Taddei A, Galietta LJ, Verkman AS. Discovery of glycine hydrazide pore-occluding CFTR inhibitors: mechanism, structure-activity analysis, and in vivo efficacy. Gen J Physiol. 2004;124:125–137. doi: 10.1085/jgp.200409059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- Nachury MV, Seeley ES, Jin H. Trafficking to the ciliary membrane: how to get across the periciliary diffusion barrier. Annu Rev Cell Dev Biol. 2010;26:59–87. doi: 10.1146/annurev.cellbio.042308.113337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima T, Sugimoto T, Kurachi Y. Effects of anions on the G protein-mediated activation of the muscarinic K+ channel in the cardiac atrial cell membrane. Intracellular chloride inhibition of the GTPase activity of GK. Gen J Physiol. 1992;99:665–682. doi: 10.1085/jgp.99.5.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namkung W, Yao Z, Finkbeiner WE, Verkman AS. Small-molecule activators of TMEM16A, a calcium-activated chloride channel, stimulate epithelial chloride secretion and intestinal contraction. FASEB J. 2011;25:4048–4062. doi: 10.1096/fj.11-191627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilius B, Eggermont J, Voets T, Buyse G, Manolopoulos V, Droogmans G. Properties of volume-regulated anion channels in mammalian cells. Prog Biophys Mol Biol. 1997;68:69–119. doi: 10.1016/s0079-6107(97)00021-7. [DOI] [PubMed] [Google Scholar]
- Oh SJ, Hwang SJ, Jung J, Yu K, Kim J, Choi JY, Hartzell HC, Roh EJ, Lee CJ. MONNA, a potent and selective blocker for transmembrane protein with unknown function 16/anoctamin-1. Mol Pharmacol. 2013;84:726–735. doi: 10.1124/mol.113.087502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong ACM. Primary cilia and renal cysts: does length matter. Nephrol Dial Transplant. 2013;28:2661–2663. doi: 10.1093/ndt/gft354. [DOI] [PubMed] [Google Scholar]
- Pacheco-Alvarez D, Gamba G. WNK3 is a putative chloride-sensing kinase. Cell Physiol Biochem. 2011;28:1123–1134. doi: 10.1159/000335848. [DOI] [PubMed] [Google Scholar]
- Pazour GJ, Bloodgood RA. Targeting proteins to the ciliary membrane. Curr Top Dev Biol. 2008;85:115–149. doi: 10.1016/S0070-2153(08)00805-3. [DOI] [PubMed] [Google Scholar]
- Pazour GJ, Dickert BL, Vucica Y, Seeley ES, Rosenbaum JL, Witman GB, Cole DG. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene Tg737, are required for assembly of cilia and flagella. J Cell Biol. 2000;151:709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Witman GB. The vertebrate primary cilium is a sensory organelle. Curr Opin Cell Biol. 2003;15:105–110. doi: 10.1016/s0955-0674(02)00012-1. [DOI] [PubMed] [Google Scholar]
- Pedersen LB, Rosenbaum JL. Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Curr Top Dev Biol. 2008;85:23–61. doi: 10.1016/S0070-2153(08)00802-8. [DOI] [PubMed] [Google Scholar]
- Perez-Cornejo P, Gokhale A, Duran C, Cui Y, Xiao Q, Hartzell HC, Faundez V. Anoctamin 1 (Tmem16A) Ca2+-activated chloride channel stoichiometrically interacts with an ezrin-radixin-moesin network. Proc Natl Acad Sci USA. 2012;109:10376–10381. doi: 10.1073/pnas.1200174109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal M, Kathpalia PP, Widdicombe JH, Pao AC. Differential effects of extracellular ATP on chloride transport in cortical collecting duct cells. Am J Physiol Renal Physiol. 2012;303:F483–F491. doi: 10.1152/ajprenal.00062.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K, Bhattacharyya B, Biswas BB. Anion-induced increases in the affinity of colcemid binding to tubulin. Eur J Biochem. 1984;142:577–581. doi: 10.1111/j.1432-1033.1984.tb08325.x. [DOI] [PubMed] [Google Scholar]
- Rock JR, Futtner CR, Harfe BD. The transmembrane protein TMEM16A is required for normal development of the murine trachea. Dev Biol. 2008;321:141–149. doi: 10.1016/j.ydbio.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Rock JR, Lopez MC, Baker HV, Harfe BD. Identification of genes expressed in the mouse limb using a novel ZPA microarray approach. Gene Expr Patterns. 2007;8:19–26. doi: 10.1016/j.modgep.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Romanenko VG, Catalan MA, Brown DA, Putzier I, Hartzell HC, Marmorstein AD, Gonzalez-Begne M, Rock JR, Harfe BD, Melvin JE. Tmem16A encodes the Ca2+-activated Cl- channel in mouse submandibular salivary gland acinar cells. J Biol Chem. 2010;285:12990–13001. doi: 10.1074/jbc.M109.068544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz C, et al. Enhanced expression of ANO1 in head and neck squamous cell carcinoma causes cell migration and correlates with poor prognosis. PLoS One. 2012;7:e43265. doi: 10.1371/journal.pone.0043265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder JM, et al. EB1 and EB3 promote cilia biogenesis by several centrosome-related mechanisms. J Cell Sci. 2011;124:2539–2551. doi: 10.1242/jcs.085852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder BC, Cheng T, Jan YN, Jan LY. Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell. 2008;134:1019–1029. doi: 10.1016/j.cell.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley ES, Nachury MV. The perennial organelle: assembly and disassembly of the primary cilium. J Cell Sci. 2010;123:511–518. doi: 10.1242/jcs.061093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N, Malarkey EB, Berbari NF, O'Connor AK, Vanden Heuvel GB, Mrug M, Yoder BK. Proximal tubule proliferation is insufficient to induce rapid cyst formation after cilia disruption. J Am Soc Nephrol. 2013;24:456–464. doi: 10.1681/ASN.2012020154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S, et al. DOG1 regulates growth and IGFBP5 in gastrointestinal stromal tumors. Cancer Res. 2013;73:3661–3670. doi: 10.1158/0008-5472.CAN-12-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinet S, Mahmud K, Stewman SF, Ben El Kadhi K, Decelle B, Talje L, Ma A, Kwok BH, Carreno S. The actin-binding ERM protein moesin binds to and stabilizes microtubules at the cell cortex. J Cell Biol. 2013;202:251–260. doi: 10.1083/jcb.201304052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin S. Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J Cell Biol. 1962;15:363–377. doi: 10.1083/jcb.15.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanich JE, Gibbons SJ, Eisenman ST, Bardsley MR, Rock JR, Harfe BD, Ordog T, Farrugia G. Ano1 as a regulator of proliferation. Am J Physiol Gastrointest Liver Physiol. 2011;301:G1044–G1051. doi: 10.1152/ajpgi.00196.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber T, Jentsch TJ. Chloride in vesicular trafficking and function. Annu Rev Physiol. 2013;75:453–477. doi: 10.1146/annurev-physiol-030212-183702. [DOI] [PubMed] [Google Scholar]
- Suzaki T, Sakai H, Endo S, Kimura I, Shigenaka Y. Effects of various anions, glutamate and GTP on microtubule assembly in vitro. J Biochem. 1978;84:75–81. doi: 10.1093/oxfordjournals.jbchem.a132121. [DOI] [PubMed] [Google Scholar]
- Suzuki J, Fujii T, Imao T, Ishihara K, Kuba H, Nagata S. Calcium-dependent phospholipid scramblase activity of TMEM16 protein family members. J Biol Chem. 2013;288:13305–13316. doi: 10.1074/jbc.M113.457937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki J, Umeda M, Sims PJ, Nagata S. Calcium-dependent phospholipid scrambling by TMEM16 F. Nature. 2010;468:834–838. doi: 10.1038/nature09583. [DOI] [PubMed] [Google Scholar]
- Szalontai B, Nagy G, Krumova S, Fodor E, Pali T, Taneva SG, Garab G, Peters J, Der A. Hofmeister ions control protein dynamics. Biochim Biophys Acta. 2013;1830:4564–4572. doi: 10.1016/j.bbagen.2013.05.036. [DOI] [PubMed] [Google Scholar]
- Takao D, Nemoto T, Abe T, Kiyonari H, Kajiura-Kobayashi H, Shiratori H, Nonaka S. Asymmetric distribution of dynamic calcium signals in the node of mouse embryo during left-right axis formation. Dev Biol. 2013;376:23–30. doi: 10.1016/j.ydbio.2013.01.018. [DOI] [PubMed] [Google Scholar]
- Valente EM, Rosti RO, Gibbs E, Gleeson JG. Primary cilia in neurodevelopmental disorders. Nat Rev Neurol. 2014;10:27–36. doi: 10.1038/nrneurol.2013.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veland IR, Awan A, Pedersen LB, Yoder BK, Christensen ST. Primary cilia and signaling pathways in mammalian development, health and disease. Nephron Physiol. 2009;111:39–53. doi: 10.1159/000208212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkman AS, Galietta LJ. Chloride channels as drug targets. Nat Rev Drug Discov. 2009;8:153–171. doi: 10.1038/nrd2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, et al. Downregulation of TMEM16A calcium-activated chloride channel contributes to cerebrovascular remodeling during hypertension by promoting basilar smooth muscle cell proliferation. Circulation. 2012;125:697–707. doi: 10.1161/CIRCULATIONAHA.111.041806. [DOI] [PubMed] [Google Scholar]
- Westlake CJ, et al. Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc Natl Acad Sci USA. 2011;108:2759–2764. doi: 10.1073/pnas.1018823108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley DN. Primary cilia in normal and pathological tissues. Pathobiology. 1995;63:222–238. doi: 10.1159/000163955. [DOI] [PubMed] [Google Scholar]
- Wilkerson PM, Reis-Filho JS. The 11q13-q14 amplicon: clinicopathological correlations and potential drivers. Genes Chromosomes Cancer. 2013;52:333–355. doi: 10.1002/gcc.22037. [DOI] [PubMed] [Google Scholar]
- Wolff J, Sackett DL, Knipling L. Cation selective promotion of tubulin polymerization by alkali metal chlorides. Protein Sci. 1996;5:2020–2028. doi: 10.1002/pro.5560051008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YD, et al. TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature. 2008;455:1210–1215. doi: 10.1038/nature07313. [DOI] [PubMed] [Google Scholar]
- Yang H, et al. TMEM16F forms a Ca2+-activated cation channel required for lipid scrambling in platelets during blood coagulation. Cell. 2012;151:111–122. doi: 10.1016/j.cell.2012.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Zhu X. Branched F-actin as a negative regulator of cilia formation. Exp Cell Res. 2013;319:147–151. doi: 10.1016/j.yexcr.2012.08.009. [DOI] [PubMed] [Google Scholar]
- Yoshiba S, et al. Cilia at the node of mouse embryos sense fluid flow for left-right determination via Pkd2. Science. 2012;338:226–231. doi: 10.1126/science.1222538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S, Egerer J, Fuchs E, Haas AK, Barr FA. Functional dissection of Rab GTPases involved in primary cilium formation. J Cell Biol. 2007;178:363–369. doi: 10.1083/jcb.200703047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Hu J, Ling K. Molecular views of Arf-like small GTPases in cilia and ciliopathies. Exp Cell Res. 2013;319:2316–2322. doi: 10.1016/j.yexcr.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Fogelgren B, Lipschutz JH. The small GTPase Cdc42 is necessary for primary ciliogenesis in renal tubular epithelial cells. J Biol Chem. 2011;286:22469–22477. doi: 10.1074/jbc.M111.238469. [DOI] [PMC free article] [PubMed] [Google Scholar]