Abstract
Background
Carboxypeptidase N (CPN) plays an important role in regulating vasoactive peptide hormones, growth factors, and cytokines by specifically cleaving their C-terminal basic residues. Herein we demonstrated that the circulating peptides specifically cleaved by CPN in the tumor microenvironment can indeed be stage-specific indicators of breast cancer.
Methods
The activity of CPN was evaluated using an ex-vivo peptide cleavage assay, in which synthesized C3f peptide (His6-C3f_S1304-R1320-His6) is incubated in interstitial fluids of breast tumor and adjacent normal breast tissues in mice with orthotopic implantation of the human cell line MDA-MB-231. Fragment profiling, by an approach combining nanopore fractionation and mass spectrometry, revealed the nature and extent of cleavage by CPN. These results correlated with the level of CPN-catalyzed peptides in blood specimens taken from the tumor-bearing mice, healthy women and breast cancer patients. CPN expression in the same set of samples was further examined by immunohistochemistry and immunoblotting.
Results
We showed that generation of C3f_R1310-L1319 specifically correlated with the CPN expression level. In both the mouse and clinical patient samples, the amount of CPN was clearly increased in tumor tissues compared to that seen in normal breast tissue, while its counterpart in blood remained constant. The amount of 6 CPN-catalyzed peptides predominantly increased in sera taken from the mice (n=8) at 2 weeks after orthotropic implantation. Six homologous peptides displayed the significantly higher expression in the patients’ plasma as early as the first pathologic stage of breast cancer.
Conclusions
The amount of circulating CPN-catalyzed peptides reported here reflects the extent of this enzyme’s activity in tumors, and our results clearly indicate their strong potential as biomarkers for non-invasive and early diagnosis of breast cancer.
Keywords: Circulating peptides, Nanopore-based assay, Breast cancer, Early detection, Carboxypeptidase N
Introduction
Perhaps in preparation for or as a result of disease progression, cells in the tumor microenvironment typically secrete several kinds of proteins, such as cytokines, growth factors, proteases, peptidases, to name a few (1, 2). In theory, any or all of these secreted proteins can serve as the biomarkers of tumor progression (3, 4). However, the reality is much more challenging to monitor because of the large degree of fluctuation in abundance and localization of these tumor-secreted proteins, especially in the early stage of tumor development and/or metastasis (5, 6). As such, it seems feasible that we might take advantage of the fact that secreted proteases/peptidases in the tumor microenvironment generate proteolytic products, also referred to as “circulating peptides”, and these are continuously released into interstitial fluid, lymph, eventually making their way into the bloodstream (7). Consequently, blood samples can provide ample information about the body, “coded” in the patterns and quantity of these peptides (8). An additional advantage of scouring blood samples for this type of information is its accessibility, while being less prone to selection bias compared to tissue samples. Indeed, several studies have presented comprehensive qualitative and quantitative profiling of circulating peptides, demonstrating their enormous potential use as a rapid, cost-effective, and accurate platform for cancer diagnosis and therapeutic evaluation, even despite the complexity cancer pathophysiology landscape (9–13). However, only a few of them have shown a direct correlation between the presence (and/or quantity) of these circulating peptides, the proteases/peptidases that generate them, or the roles these play in cancer progression (12, 14–16).
Carboxypeptidase N (CPN), also known as an arginine/lysine carboxypeptidase, kininase I, or anaphyla-toxin inactivator, is a member of a larger family of zinc metallopeptidases (17). Many metallopeptidases cleave the carboxy (C)-terminal arginine or lysine of endogenous peptides, thus changing the peptide substrates’ activity and receptor binding-capability (18, 19). CPN is known to act on the peptides, bradykinin (BK), anaphylatoxins (C3a, C4a, and C5a), and fibrinopeptides A and B (20). The nona-peptide BK9, whose amino acid sequence “RPPGFSPFR” remains identical in the human and mouse genomes, enlarges blood vessels and causes the blood pressure to decrease as a result. Removal of its C-terminal arginine by CPN, and thus loss function of binding with its receptor, is an important regulatory step for the inactivation of BK9 (19). C3f (C3f_S1304-R1320), a heptadecapeptide (amino acid sequence “SSKITHRIHWESASLLR” in human and “SSATTFRLLWENGNLLR” in mouse) that is generated from the proteolysis of C3b by complement factor I in blood (21). C3f_S1304-R1320 has also been reported as a substrate of CPN in an in-vitro study (22). Villanuva and Profumo have both attributed several daughter fragments of C3f_S1304-R1320 and BK9 to human bladder cancer, prostate cancer and breast cancer (12, 13). Given these results, the only that we could identify that attempts to draw a relationship between CPN and cancer is study conducted decades ago in 1983, where the investigators showed that increased CPN activity was observed in the sera of lung cancer patients (23). In this paper, we clearly link the catalytic activity of CPN to the patterns of its proteolytic products during tumor progression in a breast cancer mouse model and in clinical samples from breast cancer patients. Our results strongly indicate that indeed circulating peptides generated by CPN, as a representative member of the metallopeptidases, can serve as clear signatures of early disease onset and progression.
Materials and Methods
BREAST CANCER MOUSE MODEL AND BLOOD SAMPLE COLLECTION
All animal study protocols have been approved by the Institutional Animal Care and Use Committee of the Methodist Hospital Research Institute. Female mice [age-matched between 6 and 8 weeks, homozygous for the severe combined immune deficiency] were purchased from Charles River Laboratories. Eight mice were injected subcutaneously into the right flank with 1 × 107 MDA-MB-231 cells suspended in 100 μl of phosphate buffered saline (PBS). Tumor volume was calculated using the formula π/6 × length × width2 every 2 weeks. Blood samples were collected from 8 mice by retro orbital bleeding at five different time points (before injection, 2, 4, 6, and 8 weeks after injection). For each time point, 100 μl of blood per mouse were collected. The blood samples were kept at 25 °C for 1 hour and then centrifuged at 4, 000 g for 15 minutes at 25 °C. The sera were collected and stored at -80 °C until use. After the mice were sacrificed at the 8th week, adjunct normal breast tissue and tumor tissue from mice were collected and then stored at −80 °C until use.
EXTRATION OF INTERSTITIAL FLUID
Five milligrams of tissue (normal or tumor) were homogenized in 300 μl of PBS on ice by an OMNI homogenizer (Kennesaw, GA). The interstitial fluid was obtained by centrifugation of homogenized tissue samples at 3,000 g for 30 min at 4 °C. Protein concentration was determined by Bradford assay (BioRad, USA). Aliquots of the interstitial fluid were stored at −80 °C until use.
CPN DEPLETION FROM INTERSTITIAL FLUID
The anti-CPN antibody (Abcam) was immobilized on Dynal magnetic beads (Life Technologies, USA) according to the manufacturer’s instructions. Aliquots (1 mg/ml) of the interstitial fluid or conditioned cell-culture medium (CM) were incubated with 0.4 mg of antibody-coated beads for 1 hour at 25 °C. After five washes with 0.5 ml of PBS, CPN was eluted from the beads with 100 μl of 0.1 mol/L glycine (pH 3). The flow-through and eluate were analyzed by immunoblotting to evaluate the depletion efficiency.
EX-VIVO PEPTIDE CLEAVAGE ASSAY
Aliquots (1mg/ml) of the interstitial fluid were preincubated at 37 °C for 15 min with 10 mmol/L EDTA for an inhibition test. His-tagged C3f peptide (His6-C3f_S1304-R1320-His6), synthesized with 95% purity by GLbiochem (Shanghai, China), was used at a final concentration of 100 μmol/L for this assay. Cleaved peptides were extracted from the reaction solution by nanopore fractionation and detected by matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS) according to protocols described in our previous publication (19).
NANOPORE FRACTIONATION
Five microliters of each serum sample, collected from either mice or clinical breast cancer patients at five different stages (see details in supplemental information), were pipetted into individual wells arrayed on the 4″ nanoporous silica (NPS) chip. After incubation at 25 °C in a humidified chamber for 30 minutes, the residual samples were removed, and the wells were washed with 10 μl of deionized water four times. For elution, 5 μl of a solution containing 50% acetonitrile (ACN) and 0.1% trifluoroacetic acid was applied to each well for 90 seconds, allowing the peptides to be extracted from the NPS chip. The eluates were then sent to MALDI-TOF MS profile and peptide sequencing (see protocols in supplemental information).
STATISTICAL ANALYSIS
MALDI-TOF MS data were processed by MarkerView software v. 1.2.1 (AB SCIEX, Concord, Canada). The data were normalized by the internal standard peptide ACTH 18–39 (Sigma-Aldrich, St. Louis, MO). A Student’s t-test was also performed by comparing control and case samples using MarkerView.
Results
WORK FLOW OF PEPIDOMICS STUDY ON CIRCULATING PEPTIDE BIOMARKERS
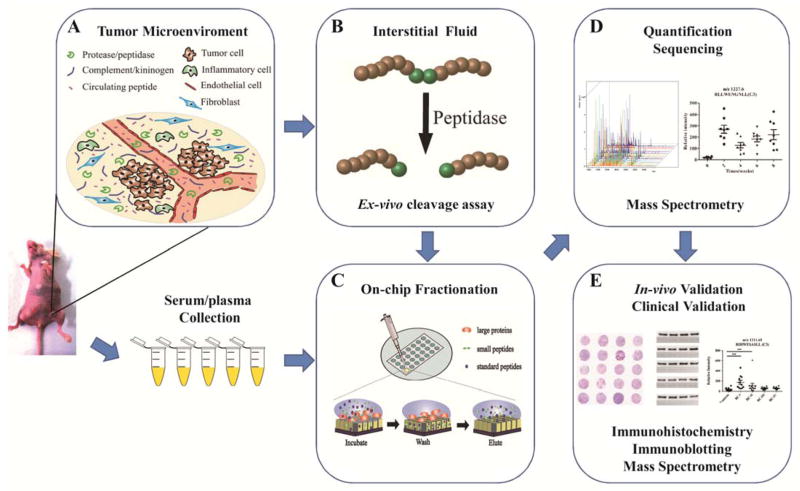
To verify our hypothesis that CPN-catalyzed peptide products found in circulation can indeed be strong indicators of disease, we designed and implemented a systematic workflow to culminate in a set of signatures that could inform disease diagnosis (Fig. 1), overviewed as follows. Some of the foundational experiments involved inducing a breast cancer mouse model by injecting MDA-MB-231 human breast cancer cells into the animals. It was important to first evaluate the activity of CPN in the microenvironment by an ex vivo cleavage assay. Here, interstitial fluids extracted from the animals were incubated with synthesized C3f peptides (His6-C3f_S1304-R1320-His6), as the substrate of CPN, with the cleavage sites predicted by bioinformatics tools (e.g. PROSPER (14), http://lightning.med.monash.edu/PROSPER/webserver.html), and marked in Fig. 2A. The peptides were isolated using a nanopore fractionation method previously developed by our group (24–29). Briefly, low-molecular weight peptides are captured and preserved on NPS thin films, separating them from often overwhelming, high-molecular weight proteins/peptides prior to mass spectrometry analysis. Analysis by mass spectrometry can provide not only the quantitative information for multiple peptides species in a single sample, but also helps to identify their sequence information (24–26, 28–30). The in vivo expression of CPN in tissues (evaluated by immunohistochemistry (IHC)) or blood (by immunoblotting) is correlated to the presence or quantity (see protocols in supplemental information). Taking these results, we then perform clinical validation, where the peptide biomarker candidates are examined in plasma samples collected from patient cohorts at different stage of cancer. Expression of CPN in human tumor tissue and blood is also evaluated and correlated to that of its peptide substrates.
Figure 1. Workflow of peptidomics study on circulating peptide biomarkers.
A. Circulating peptides are produced in the tumor microenviroment and released into the bloodstream. B. The activity of tumor specific peptidase (CPN, for example) in interstitial fluid is evaluated by ex-vivo cleavage assay. C. Nanopore fractionation is used to extract peptides from serum/plasma samples or the reaction solution from the ex-vivo cleavage assay. D. Quantification and sequence identification of peptides are determined by mass spectrometry. E. In-vivo validation and clinical validation are performed by immunohistochemistry, immunoblotting and mass spectrometry.
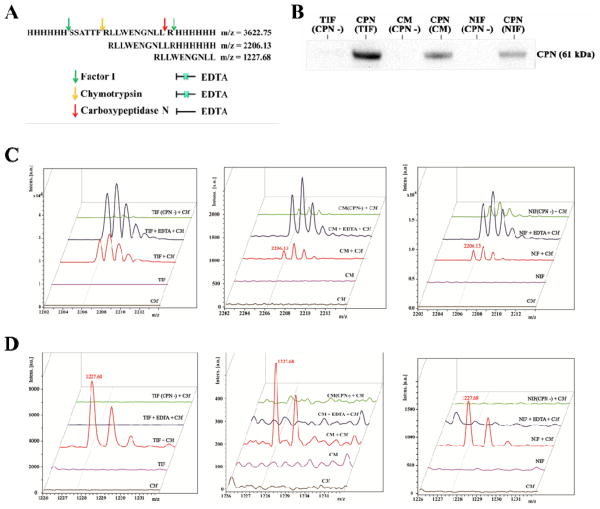
Figure 2. Ex vivo peptide cleavage assay on the synthetic C3f peptide.
A. Specific cleavage sites are indicated on the C3f peptide, a substrate of the enzymes factor I, chymotrypsin and CPN. B. Detecting CPN in different assay conditions. CPN-depletion in normal [NIF (CPN -)] or tumor interstitial [TIF (CPN -)] fluid; CPN protein pull-down from normal [CPN (NIF)] or tumor interstitial [CPN (TIF)] fluid. C & D. Normal interstitial fluid (NIF) and tumor interstitial fluid (TIF) (20 μl of 1 mg/ml) from breast cancer mouse model or conditioned medium (CM) of MB-MDA 231 cells were incubated with synthetic C3f peptides (Final concentration is 100 μmol/L) at 37 °C for 1 hour, followed by nanopore fractionation and MALDI-TOF MS. C3f, synthetic peptide His6-C3f_S1304-R1320-His6 only; NIF or TIF, normal or tumor interstitial fluid only; EDTA, an inhibitor of CPN; NIF (CPN -) and TIF (CPN -), CPN-depleted normal and tumor tissue interstitial fluid; CM (CPN -), CPN-depleted conditioned medium.
EX-VIVO PEPTIDE CLEAVAGE ASSAY AND IN VIVO VALIDATION
As indicated in Fig. 2A, “FR” marked on the peptide sequence is a well-known cleavage site of chymotrypsin, a serine protease that cannot be inhibited by EDTA. If cleaved by chymotrypsin, a peptide peak can be detected in a MS spectrum at m/z of 2206.13. After Factor I removes the His6-tag on both termini of the synthetic peptide, CPN cleaves the C-terminal “R” of C3f_R1310-R1320 to produce C3f_R1310-L1319 with m/z of 1227.68. The addition of EDTA inactivates CPN by chelating the zinc ion in its catalytic domain. For further ex vivo validation of these cleavage events, we conducted the peptide cleavage assay by incubating synthetic peptides with tumor interstitial fluid (TIF) and conditioned cell-culture medium (CM) of MDA-MB-231 cells. Results indicated that production of C3f_R1310-L1319 in TIF was six-fold higher than in normal interstitial fluid (NIF). As shown in Fig. 2C and 2D, the addition of EDTA clearly inhibits cleavage of C3f_R1310-L1319, however it could not prevent the generation of the peak at m/z = 2206.13 in samples containing TIF, NIF and CM, suggesting the likelihood that C3f_R1310-L1319 was finally generated by a metallopeptidase, which was proven to be CPN in this study. Most notably, the generation of C3f_R1310-L1319 with m/z of 1227.68, but not the peptide with m/z of 2206.13, was inhibited if CPN is removed by immunodepletion (2-fold, confirmed by immunoblotting, Fig. 2B) from the interstitial fluids or CM. Expression of CPN, validated by anti-CPN immunoblotting and IHC staining, was increased in tumor tissue compared to that in normal breast tissue (Fig. 3A and 3B). In fact, the adipocytes and glandular cells in normal mouse breast tissue, expressed CPN weakly or none at all, whereas the myoepithelial cells exhibited an elevated level of CPN. As shown in Fig. 3C, the concentration of CPN in mouse sera remained constant during tumor growth.
Figure 3. CPN expression in mouse tissues and sera.
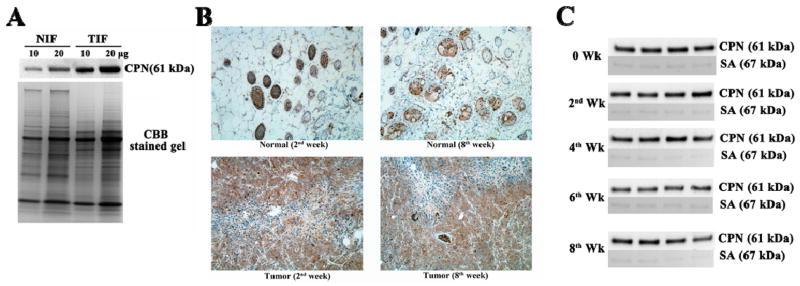
A. CPN immunoblotting expression in normal breast interstitial fluid (NIF) and tumor interstitial fluid (TIF). Coomassie Brilliant Blue (CBB) stained gel was used for the evaluation of equal loading. Both immunoblot and CBB stained gel were subjected to quantitative image analysis by software Image J, and the result was presented by histogram. B. Immunohistochemistry analysis of CPN in normal breast tissue and tumor tissue collected at the 2nd and 8th week. C. Immunoblotting of CPN in mouse sera. Ponceau S-stained serum albumin (SA) serves as internal control for loading. (Groups: 0, 2nd, 4th, 6th, 8th week; n = 4/group).
SERUM PEPTIDOMIC PROFILING IN MOUSE MODEL
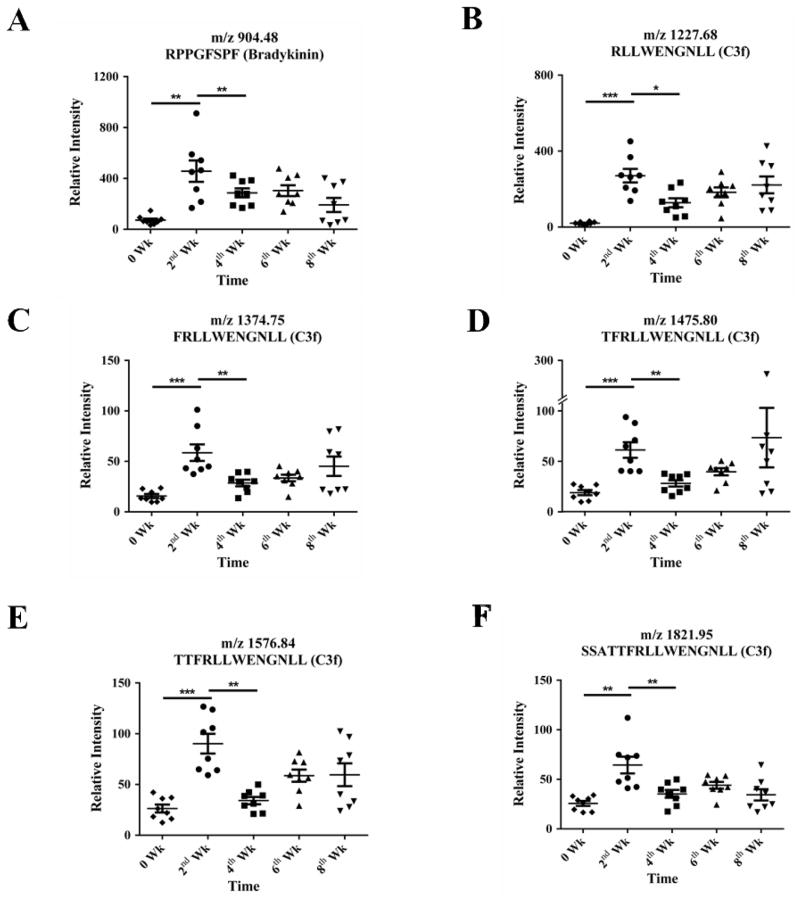
We introduced MDA-MB-231 cells into nude mice by orthotopic implantation to induce tumor growth. Tumor volume was measured at 2-week intervals for a total of four time points, with average tumor sizes across 8 mice at 69.8 ± 30.9 mm3, 190.3 ± 91.9 mm3, 718.9 ± 380.9 mm3 and 1548.0 ± 804.6 mm3, respectively (Fig. S1). After collecting mouse sera, we processed a total of 120 sera samples (triplicates of 40 serum samples, including sera collected from 8 mice over the course of 5 time points) on-NPS chip fractionation, and then tested all of the eluates on one 192-wells MALDI-TOF MS plate. Each MALDI-TOF MS spectrum contained about 150 monoisotopic peaks in the range of 800 ~ 3,500 Da (Fig. S2A). The data were imported into the MarkerView software for t-test analysis. Compared to the normal controls collected before tumor implantation, different peaks were found in the diseased samples, after applying the primary (p < 0.05) and secondary (fold change > 2) criteria (Fig. S2B). Sequence information obtained from high-performance liquid chromatography-tandem mass spectrometry revealed six differential peptides, including five from C3f and one from bradykinin, and their appearance correlated with the CPN expression. These peptides were thus chosen for further study (Table S1). It is important to note that all six peptides exhibited a similar trend during the first four weeks after tumor induction (Fig. 4), with the quantity peaking at week 2 (the first tumor growth measurement) and then decreasing dramatically by week 4. Interestingly, the quantity of nearly all C3f peptides increased again slightly at week 6 and week 8. Exceptions included the peptide C3f_R1304-L1319 with m/z of 1821.95, which increased again only at week 6 before declining for the second time at week 8, and the BK8 peptide which remained steady at week 6 before decreasing at week 8. The absolute intensities (indicator of concentration) of C3f_R1310-L1319 and, more prominently BK8, were higher than the other fragments.
Figure 4. The vertical scattering plot of the normalized peak intensities in MALDI TOF MS for the potential peptide signatures in serum samples collected from tumor-bearing animals (breast cancer mouse model).
The mass-to-charge ratio and sequence identification is listed in each panel. Mouse number: n = 8; Student’s t-test, * p < 0.05, ** p < 0.01, *** p < 0.001.
CLINICAL VALIDATION
We expected our findings in the breast cancer mouse model to be reflected in clinical samples collected from patients with breast cancer, positioning our approach for clinical translation. To provide unequivocal evidence, we obtained commercial tissue array slide that contains 110 human breast tissue sections, including 100 breast cancer cases (pathologic stage I-III) and 10 adenosis/cancer adjacent normal breast tissues as controls (see array details at http://www.biomax.us/tissue-arrays/Breast/BC081116a). Analysis by IHC using these commercial arrays revealed that indeed CPN expression was generally increased in samples taken from breast cancer patients regardless of disease stage (Fig. 5A). When we scored these 110 sections based on a scoring system reported by Zhang et al.(31), 100% of the controls (n = 10) received a score of 1+ or less, which meant that CPN was weakly or not expressed in controls. However, across all stages of breast cancer represented on the array, 85% of the samples received scores higher than 1+, and 70% received scores higher than 2+ (Fig. 5B and Fig. S3). Conversely, the amount of CPN in plasma remained constant between breast cancer patient and that of the control cohort (Fig. 5C).
Figure 5. CPN expression in human tissues and plasma.
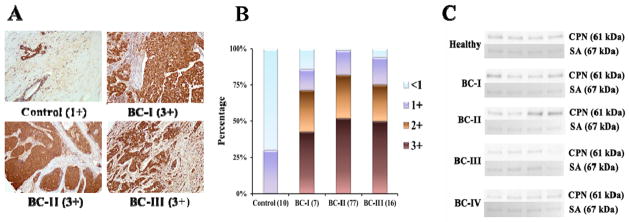
A. IHC stains for CPN in human control cohort (cancer adjunct tissue or adenosis) and tumor tissues. B. The histogram shows the percentage of the scoring results of IHC stains for CPN (n = 10, 7, 77 and 16 for control, BC-I, BC-II and BC-III slides, respectively). See Material and Method for scoring method of IHC staining pattern. C. CPN expression by immunoblotting in human plasma from healthy controls and breast cancer (BC) patients at different stages of disease (I, II, III and IV). Ponceau S-stained serum albumin serves as internal control. n = 4/group.
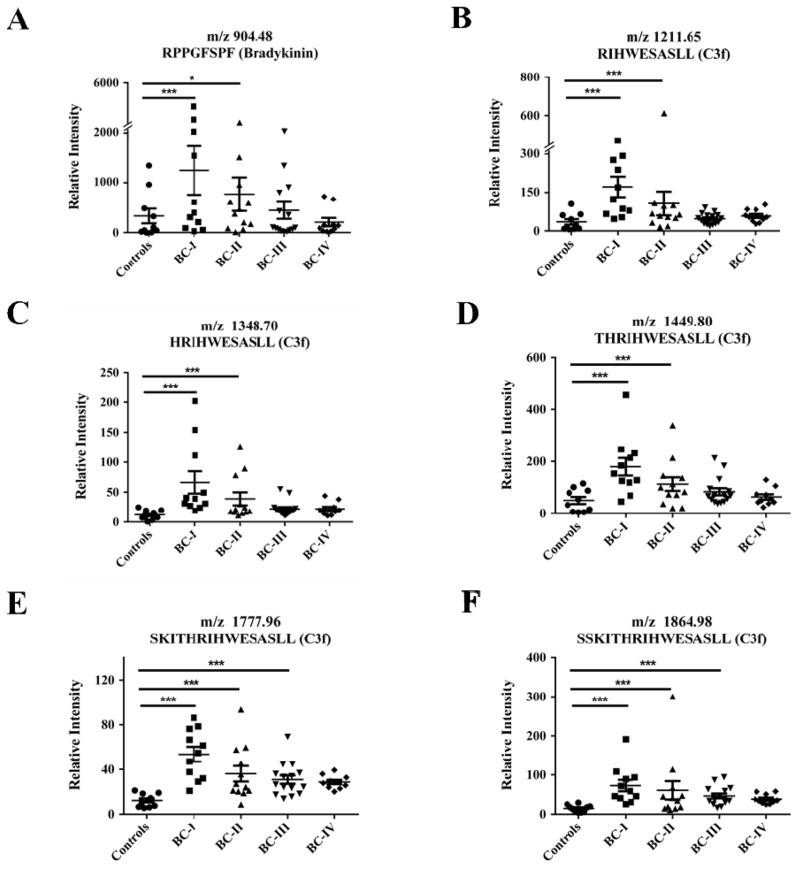
For further validation, circulating peptides of interests were profiled in 58 human plasma specimens, including healthy controls (n = 10), stage I breast cancer (BC-I, n = 11), stage II breast cancer (BC-II, n = 12), stage III breast cancer (BC-III, n = 15) and stage IV breast cancer (BC-IV, n = 10). MALDI-TOF MS data showed that the BK8 specie appeared more abundant in stage I and II breast cancer (BC-I and BC-II) compared to healthy controls (BC-I vs control p < 0.001, BC-II vs control p < 0.05) (Fig. 6 and Table S2). Perhaps most striking, the peptide species C3f_R1310-L1319, C3f_R1309-L1319, C3f_R1308-L1319, C3f_R1305-L1319, and C3f_R1304-L1319 presented prominently in samples of BC-I and BC-II, compared to the healthy cohort (p < 0.001) (Fig. 6), consistent with the report by Profumo et al (13). Overall, the patterns of these CPN-catalyzed products in the clinical samples from breast cancer patients matched strongly with our observations in the breast cancer mouse model. As shown in our results, the expression levels of the 6 CPN-specific peptides showed no significant difference between mouse serum and plasma before we processed the validation experiment on human plasma specimens (Fig S4).
Figure 6. Profiles of the potential peptide signatures in healthy woman plasma and woman patients’ plasma with breast cancer of different pathological stages.
Sample number: n = 10, healthy controls; n = 11, stage I breast cancer (BC-I); n = 12, stage II breast cancer (BC-II); n = 15, stage III breast cancer (BC-III); n = 10, stage IV breast cancer (BC-IV); Student’s t-test, * p < 0.05, *** p < 0.001.
Discussion
There have been numerous studies to find biomarkers for early disease diagnosis, but frustratingly we still lack definitive candidates that can be used to predict disease onset – early and with great precision and specificity. The often poor diagnosis and prognosis is due, in part, to the absence of an approach that can identify markers with such features, in a manner that is non-invasive, reproducible, and conducive to rapid therapeutic processes (32). Because it is known that certain proteases/peptidases play an important role in several cancers, a deeper understanding of certain proteolytic events, particularly in the tumor microenvironment, will facilitate the identification of tumor-derived products that are eventually secreted into circulation (8, 12, 33, 34). Profiling the serum proteome then provides a “map” that we can use to trace cancer-specific metabolic or immunological signatures indicative of early-stage tumor progression (6, 13, 35, 36).
In this study, we examined the activity of CPN, a protease known to play a role in several types of cancer, within the tumor microenvironment as well as in the blood circulation of mice (breast cancer mouse model) and patients diagnosed with breast cancer (different stages). The peptide fragments generated by tumor-resident CPN and released into the bloodstream – their quantities and identities presented in our comprehensive analyses here – strongly correlate with enzymatic expression and reflect the organism’s pathophysiological state. Therefore, we advocate their use as distinct and precise biomarkers of early breast cancer diagnosis and risk assessment, certainly to be detected and identified before metastasis and perhaps even before the tumor presents with any observable characteristics commonly used in the clinic.
A previous in-vitro study has shown that CPN can cleave the C-terminal arginine of the C3f_S1304-R1320; however it was unclear whether CPN was the only specific peptidase able to do so in that particular assay condition (22). To make such a conclusion or otherwise, we depleted CPN activity in protease assays that contain NIF, TIF and CM, before adding the synthetic C3f peptide. The resulting MALDI-TOF MS spectra revealed that cleavage to C3f_R1310-L1319 was indeed terminated in both interstitial fluid and CM and confirmed the specificity of the CPN to the C-terminal arginine of C3f. It was CPN but not other proteases, for example carboxypeptidase E (CPE), who was the specific enzyme for this cleavage event because both the previous study has shown MDA-MB-231 cells can secret both CPN and CPE (37). In addition, the strong m/z intensities of C3f_R1310-L1319 fragments observed in TIF demonstrate the higher activity of CPN in tumor microenvironment than its counterpart in the normal breast tissue. The immunoblotting study displayed two folds higher of CPN activity in TIF than in NIF, further confirmed by IHC analysis of normal and tumor tissues. Therefore it is very clear that CPN is highly expressed in tumor tissue and secreted into TIF in early stage of the breast cancer. The physiological and oncological significance of this event needs to be further studied. Although CPN is weakly expressed in the glandular cells and highly expressed in myoepithelial cells, both cell types are considered the minority population in normal breast tissue. In adipocytes, the majority population in normal breast tissue, we did not observe any detectable expression of CPN. The cell line MDA-MB-231 used here belongs to the epithelial cell lineage, as are myoepithelial cells. Besides its high expression and secretion of CPN, MDA-MB-231 cells had a rapid growth rate and caused the poor differentiation of the tumor tissue, which probably explain the elevated CPN activity observed in TIF compared to NIF.
It was somewhat surprising to find that the level of CPN expression did not differ significantly in the sera of control and diseased subjects, whether mouse or human. This observation suggests that any additional leakage of CPN from the tumor microenvironment may not be readily detectable above the background level of CPN in the serum due to dilution. As described by other investigators, active CPN is secreted into circulation as a 280 kDa molecule, comprised of two regulatory 83 kDa-subunits and two catalytic 50 kDa-subunits (17, 20, 23). We concluded that the increased levels of circulating CPN-catalyzed peptides presented here resulted from cleavage by CPN resident in the tumor microenvironment, rather than enzymes circulating in blood. We firmly believe it is precisely this observation that makes the idea of using CPN-catalyzed circulating peptides as disease biomarkers much more appealing and feasible.
As the MALDI-TOF MS spectra featured the peptide signatures being expressed more prominently in the blood specimens of animals and patients with breast cancer, one would expect to see some changes (increases) of these peptides in TIF compared to NIF in tumor-bearing mouse. In fact, their signals or peaks were not detectable by MALDI-TOF MS. The low abundance of these peptides in interstitial fluid is likely due to their continuous release from the tumor microenvironment to accumulate in the blood. Nevertheless, whether this explanation reflects the actual physiological process remains to be proven.
The expression levels of circulating peptides can be balanced by the activities of multiple proteases/peptidases, some of which promote the proteolytic production, but others simultaneously inhibit the production of degraded peptides (19, 22, 38). In the breast cancer mouse model examined here, the quantities of all 6 circulating peptides had dramatically decreased at week 4, after peaking at week 2. Some studies have reported that angiotensin-converting enzyme and neutral endopeptidase inhibit the cleavage of bradykinin in human plasma (19). We speculate that a peptidase other than CPN may be more active in either tissue or blood as the tumor matures, leading to a decline in peptides cleaved by CPN. Based on predictions using the program PROSPER (14), we selected cathepsin G as a peptidase that could be responsible for cleaving C3f fragments at the site of “LW”. Other studies have suggested an important role for cathepsin G in tumor progression, especially in metastasis (39). As such, cathepsin G seems likely to be involved in the declining abundance of the C3f fragments in blood at mid-stage tumor progression. As the peptides reported in this study are not directly released from the tumor, their abundance in blood may be regulated by other enzymes in addition to CPN, such as cathepsin G. Thus, these peptides may not correlate entirely with tumor burden.
Cumulatively, our results represent a first demonstration, to our knowledge, that clearly links the proteolytic activity of CPN, particularly at tumor sites, to the cleavage patterns of its catalytic substrates C3f and BK in the blood, by means of a rapid, reproducible, sensitive, precise and non-invasive approach. It would certainly be naïve to think that CPN-catalyzed peptides and the enzyme itself are the only biomarkers specific for breast cancer diagnosis. We do however argue that they can be used as a complementary measure for very early diagnosis of tumor growth, particularly because CPN activity within the tumor at this point is already detectable by blood sampling. Such sensitivity together with all of the features mentioned above lend well to their translation to the clinic, for breast cancer and perhaps other pathologies.
Supplementary Material
Acknowledgments
The work was primarily supported by research funding provided by DOD innovator award (DoD W81XWH-09-1-0212). Y.H., H.S. and M.F. also acknowledge the partial support from the following grants: U54CA143837, NIH U54CA151668 and DoD W81XWH-11-2-0168. F.J.E. wants to thank the support from the Breast Cancer Research Foundation. All authors thank the support of clinical samples from Department of Breast Cancer Oncology at the University of Texas M. D. Anderson Cancer Center. We specially acknowledge the contribution from Pamela Mcshane at Biorepository Core of the Methodist Hospital (TMH) and funding support from Department of Pathology at TMH.
Abbreviation
- CPN
carboxypeptidase N
- PBS
Phosphate buffered saline
- BK
bradykinin
- CM
conditioned cell-culture medium
- IHC
immunohistochemistry
- NPS
nanoporous silica
- ACN
acetonitrile
- MALDI-TOF MS
matrix-assisted laser desorption/ionization time of flight mass spectrometry
References
- 1.Holzel M, Bovier A, Tuting T. Plasticity of tumour and immune cells: A source of heterogeneity and a cause for therapy resistance? Nature reviews Cancer. 2013;13:365–76. doi: 10.1038/nrc3498. [DOI] [PubMed] [Google Scholar]
- 2.Sounni NE, Noel A. Targeting the tumor microenvironment for cancer therapy. Clinical chemistry. 2013;59:85–93. doi: 10.1373/clinchem.2012.185363. [DOI] [PubMed] [Google Scholar]
- 3.Chen J, Xi J, Tian Y, Bova GS, Zhang H. Identification, prioritization and evaluation of glycoproteins for aggressive prostate cancer using quantitative glycoproteomics and antibody-based assays on tissue specimens. Proteomics. 2013 doi: 10.1002/pmic.201200541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marimuthu A, Chavan S, Sathe G, Sahasrabuddhe NA, Srikanth SM, Renuse S, et al. Identification of head and neck squamous cell carcinoma biomarker candidates through proteomic analysis of cancer cell secretome. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbapap.2013.04.029. [DOI] [PubMed] [Google Scholar]
- 5.Meng R, Gormley M, Bhat VB, Rosenberg A, Quong AA. Low abundance protein enrichment for discovery of candidate plasma protein biomarkers for early detection of breast cancer. J Proteomics. 2011;75:366–74. doi: 10.1016/j.jprot.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 6.Misek DE, Kim EH. Protein biomarkers for the early detection of breast cancer. Int J Proteomics. 2011;2011:343582. doi: 10.1155/2011/343582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okwan-Duodu D, Landry J, Shen XZ, Diaz R. Angiotensin converting enzyme and the tumor microenvironment: Mechanisms beyond angiogenesis. Am J Physiol Regul Integr Comp Physiol. 2013 doi: 10.1152/ajpregu.00544.2012. [DOI] [PubMed] [Google Scholar]
- 8.Rubin H. Systemic effects of cancer: Role of multiple proteases and their toxic peptide products. Medical science monitor: international medical journal of experimental and clinical research. 2005;11:RA221–8. [PubMed] [Google Scholar]
- 9.Ardill JE, Erikkson B. The importance of the measurement of circulating markers in patients with neuroendocrine tumours of the pancreas and gut. Endocrine-related cancer. 2003;10:459–62. doi: 10.1677/erc.0.0100459. [DOI] [PubMed] [Google Scholar]
- 10.Petricoin EF, Belluco C, Araujo RP, Liotta LA. The blood peptidome: A higher dimension of information content for cancer biomarker discovery. Nature reviews Cancer. 2006;6:961–7. doi: 10.1038/nrc2011. [DOI] [PubMed] [Google Scholar]
- 11.Liotta LA, Petricoin EF. Serum peptidome for cancer detection: Spinning biologic trash into diagnostic gold. The Journal of clinical investigation. 2006;116:26–30. doi: 10.1172/JCI27467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Villanueva J, Shaffer DR, Philip J, Chaparro CA, Erdjument-Bromage H, Olshen AB, et al. Differential exoprotease activities confer tumor-specific serum peptidome patterns. The Journal of clinical investigation. 2006;116:271–84. doi: 10.1172/JCI26022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Profumo A, Mangerini R, Rubagotti A, Romano P, Damonte G, Guglielmini P, et al. Complement c3f serum levels may predict breast cancer risk in women with gross cystic disease of the breast. Journal of proteomics. 2013;85:44–52. doi: 10.1016/j.jprot.2013.04.029. [DOI] [PubMed] [Google Scholar]
- 14.Kudo T, Kigoshi H, Hagiwara T, Takino T, Yamazaki M, Yui S. Cathepsin g, a neutrophil protease, induces compact cell-cell adhesion in mcf-7 human breast cancer cells. Mediators of inflammation. 2009;2009:850940. doi: 10.1155/2009/850940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villanueva J, Martorella AJ, Lawlor K, Philip J, Fleisher M, Robbins RJ, Tempst P. Serum peptidome patterns that distinguish metastatic thyroid carcinoma from cancer-free controls are unbiased by gender and age. Molecular & cellular proteomics: MCP. 2006;5:1840–52. doi: 10.1074/mcp.M600229-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Fiedler GM, Leichtle AB, Kase J, Baumann S, Ceglarek U, Felix K, et al. Serum peptidome profiling revealed platelet factor 4 as a potential discriminating peptide associated with pancreatic cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15:3812–9. doi: 10.1158/1078-0432.CCR-08-2701. [DOI] [PubMed] [Google Scholar]
- 17.Keil C, Maskos K, Than M, Hoopes JT, Huber R, Tan F, et al. Crystal structure of the human carboxypeptidase n (kininase i) catalytic domain. Journal of molecular biology. 2007;366:504–16. doi: 10.1016/j.jmb.2006.11.025. [DOI] [PubMed] [Google Scholar]
- 18.Skidgel RA, Erdos EG. Angiotensin converting enzyme (ace) and neprilysin hydrolyze neuropeptides: A brief history, the beginning and follow-ups to early studies. Peptides. 2004;25:521–5. doi: 10.1016/j.peptides.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 19.Kuoppala A, Lindstedt KA, Saarinen J, Kovanen PT, Kokkonen JO. Inactivation of bradykinin by angiotensin-converting enzyme and by carboxypeptidase n in human plasma. American journal of physiology Heart and circulatory physiology. 2000;278:H1069–74. doi: 10.1152/ajpheart.2000.278.4.H1069. [DOI] [PubMed] [Google Scholar]
- 20.Levin Y, Skidgel RA, Erdos EG. Isolation and characterization of the subunits of human plasma carboxypeptidase n (kininase i) Proceedings of the National Academy of Sciences of the United States of America. 1982;79:4618–22. doi: 10.1073/pnas.79.15.4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riley-Vargas RC, Lanzendorf S, Atkinson JP. Targeted and restricted complement activation on acrosome-reacted spermatozoa. The Journal of clinical investigation. 2005;115:1241–9. doi: 10.1172/JCI23213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ganu VS, Muller-Eberhard HJ, Hugli TE. Factor c3f is a spasmogenic fragment released from c3b by factors i and h: The heptadeca-peptide c3f was synthesized and characterized. Molecular immunology. 1989;26:939–48. doi: 10.1016/0161-5890(89)90112-0. [DOI] [PubMed] [Google Scholar]
- 23.Schweisfurth H, Reinhart E, Heinrich J, Brugger E. A simple spectrophotometric assay of carboxypeptidase n (kininase i) in human serum. J Clin Chem Clin Biochem. 1983;21:605–9. doi: 10.1515/cclm.1983.21.10.605. [DOI] [PubMed] [Google Scholar]
- 24.Fan J, Deng X, Gallagher JW, Huang H, Huang Y, Wen J, et al. Monitoring the progression of metastatic breast cancer on nanoporous silica chips. Philosophical Transactions of the Royal Society a-Mathematical Physical and Engineering Sciences. 2012;370:2433–47. doi: 10.1098/rsta.2011.0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan J, Gallagher JW, Wu H-J, Landry MG, Sakamoto J, Ferrari M, Hu Y. Low molecular weight protein enrichment on mesoporous silica thin films for biomarker discovery. Journal of visualized experiments: JoVE. 2012 doi: 10.3791/3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan J, Huang Y, Finoulst I, Wu H-J, Deng Z, Xu R, et al. Serum peptidomic biomarkers for pulmonary metastatic melanoma identified by means of a nanopore-based assay. Cancer letters. 2013;334:202–10. doi: 10.1016/j.canlet.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu Y, Bouamrani A, Tasciotti E, Li L, Liu X, Ferrari M. Tailoring of the nanotexture of mesoporous silica films and their functionalized derivatives for selectively harvesting low molecular weight protein. Acs Nano. 2010;4:439–51. doi: 10.1021/nn901322d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Y, Gopal A, Lin K, Peng Y, Tasciotti E, Zhang X, Ferrari M. Microfluidic enrichment of small proteins from complex biological mixture on nanoporous silica chip. Biomicrofluidics. 2011:5. doi: 10.1063/1.3528237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu Y, Peng Y, Lin K, Shen H, Brousseau LC, III, Sakamoto J, et al. Surface engineering on mesoporous silica chips for enriching low molecular weight phosphorylated proteins. Nanoscale. 2011;3:421–8. doi: 10.1039/c0nr00720j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bouamrani A, Hu Y, Tasciotti E, Li L, Chiappini C, Liu X, Ferrari M. Mesoporous silica chips for selective enrichment and stabilization of low molecular weight proteome. Proteomics. 2010;10:496–505. doi: 10.1002/pmic.200900346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang F, Ren G, Lu Y, Jin B, Wang J, Chen X, et al. Identification of trak1 (trafficking protein, kinesin-binding 1) as mgb2-ag: A novel cancer biomarker. Cancer letters. 2009;274:250–8. doi: 10.1016/j.canlet.2008.09.031. [DOI] [PubMed] [Google Scholar]
- 32.Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, et al. American society of clinical oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25:5287–312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 33.Shen Y, Tolic N, Liu T, Zhao R, Petritis BO, Gritsenko MA, et al. Blood peptidome-degradome profile of breast cancer. PLoS One. 2010;5:e13133. doi: 10.1371/journal.pone.0013133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diamandis EP. Oncopeptidomics: A useful approach for cancer diagnosis? Clinical chemistry. 2007;53:1004–6. doi: 10.1373/clinchem.2006.082552. [DOI] [PubMed] [Google Scholar]
- 35.Diamandis EP. Peptidomics for cancer diagnosis: Present and future. Journal of proteome research. 2006;5:2079–82. doi: 10.1021/pr060225u. [DOI] [PubMed] [Google Scholar]
- 36.Koomen JM, Li D, Xiao LC, Liu TC, Coombes KR, Abbruzzese J, Kobayashi R. Direct tandem mass spectrometry reveals limitations in protein profiling experiments for plasma biomarker discovery. Journal of proteome research. 2005;4:972–81. doi: 10.1021/pr050046x. [DOI] [PubMed] [Google Scholar]
- 37.Hu Y, Bouamrani A, Tasciotti E, Li L, Liu X, Ferrari M. Tailoring of the nanotexture of mesoporous silica films and their functionalized derivatives for selectively harvesting low molecular weight protein. ACS nano. 2010;4:439–51. doi: 10.1021/nn901322d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozik A, Moore RB, Potempa J, Imamura T, Rapala-Kozik M, Travis J. A novel mechanism for bradykinin production at inflammatory sites. Diverse effects of a mixture of neutrophil elastase and mast cell tryptase versus tissue and plasma kallikreins on native and oxidized kininogens. J Biol Chem. 1998;273:33224–9. doi: 10.1074/jbc.273.50.33224. [DOI] [PubMed] [Google Scholar]
- 39.Wilson TJ, Nannuru KC, Futakuchi M, Sadanandam A, Singh RK. Cathepsin g enhances mammary tumor-induced osteolysis by generating soluble receptor activator of nuclear factor-kappab ligand. Cancer research. 2008;68:5803–11. doi: 10.1158/0008-5472.CAN-07-5889. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.