Abstract
The Warburg effect is a tumorigenic metabolic adaptation process characterized by augmented aerobic glycolysis, which enhances cellular bioenergetics. In normal cells, energy homeostasis is controlled by AMPK; however, its role in cancer is not understood, as both AMPK-dependent tumor-promoting and -inhibiting functions were reported. Upon stress, energy levels are maintained by increased mitochondrial biogenesis and glycolysis, controlled by transcriptional coactivator PGC-1α and HIF, respectively. In normoxia, AMPK induces PGC-1α, but how HIF is activated is unclear. Germline mutations in the gene encoding the tumor suppressor folliculin (FLCN) lead to Birt-Hogg-Dubé (BHD) syndrome, which is associated with an increased cancer risk. FLCN was identified as an AMPK binding partner, and we evaluated its role with respect to AMPK-dependent energy functions. We revealed that loss of FLCN constitutively activates AMPK, resulting in PGC-1α–mediated mitochondrial biogenesis and increased ROS production. ROS induced HIF transcriptional activity and drove Warburg metabolic reprogramming, coupling AMPK-dependent mitochondrial biogenesis to HIF-dependent metabolic changes. This reprogramming stimulated cellular bioenergetics and conferred a HIF-dependent tumorigenic advantage in FLCN-negative cancer cells. Moreover, this pathway is conserved in a BHD-derived tumor. These results indicate that FLCN inhibits tumorigenesis by preventing AMPK-dependent HIF activation and the subsequent Warburg metabolic transformation.
Introduction
Kidney cancer is a metabolic disease, because renal cancer genes, including VHL, MET, TSC1, TSC2, FH, and SDH, are involved in metabolic sensing and adaptation to fluctuations in oxygen, energy, and nutrient status (1). Intriguingly, aberrant HIF-driven gene expression is a shared feature observed upon deregulation of these pathways. To maintain cellular homeostasis under energy stress, HIF mediates transcription of genes that favor glycolysis (hexokinase 2 [HK2] and lactate dehydrogenase A [LDHA]) and energy supply (SLC2A1, encoding GLUT1, and VEGF). HIF induction occurs via protein stabilization of its regulatory α subunit and/or an increase in transcriptional activity of the HIF complex (encompassing α and β subunits). While HIF activation in response to hypoxia has been described extensively, its role in normoxia and tumorigenic metabolic adaptation is poorly defined (2, 3).
Germline mutations in the folliculin (FLCN) gene lead to Birt-Hogg-Dubé (BHD) syndrome, characterized by lung cysts, pneumothorax susceptibility, renal cell carcinoma, and skin tumors (4). We reported recently that loss of the tumor suppressor FLCN leads to increased HIF transcriptional activity and a higher glycolytic rate in human kidney cancer cells, which is a recurrent characteristic of cancer metabolic adaptation referred to as the Warburg effect (5, 6). HIF-α protein levels are regulated by oxygen-dependent prolyl hydroxylation, sequential VHL binding, and degradation of HIF-α via the proteasome (7). Interestingly, it has been demonstrated that AMP-activated protein kinase (AMPK) increased the transcriptional activity of HIF without affecting its stability via an unknown mechanism (8). AMPK is a heterotrimeric enzyme that monitors the energy status and maintains energy homeostasis under metabolic stress by activating catabolic processes and inhibiting anabolic processes (9). However, the role of AMPK in metabolic transformation is still unclear, as both oncogenic and tumor suppressor functions have been reported (10). Interestingly, FLCN and its binding partner FLCN-interacting protein (FNIP) are known to interact with AMPK (11, 12). However, it is unknown how FLCN and AMPK interact mechanistically, how they regulate HIF transcriptional activity and metabolic adaptation in normoxia, and whether this effect is linked to tumor suppression.
Here, we used untransformed cells to investigate the effect of FLCN on cellular metabolism and signaling pathways linked to AMPK. We show that loss of FLCN binding to AMPK results in AMPK activation and uncover a novel signaling pathway that directly couples AMPK-dependent mitochondrial biogenesis to HIF-driven aerobic glycolysis through mitochondrial ROS production in physiological conditions. The concomitant induction of mitochondrial oxidative phosphorylation (OXPHOS) with aerobic glycolysis results in excess production of ATP and metabolic intermediates, a cancer adaptation mechanism also known as the Warburg effect. We reveal that this metabolic transformation is conserved in human cancer cells and renal tumors from patients with BHD and that this mechanism confers a tumorigenic advantage upon loss of FLCN.
Results
Loss of FLCN increases ATP levels through enhanced HIF-dependent aerobic glycolysis.
To study the cellular role of FLCN, we generated syngenic paired Flcn+/+ (WT) and Flcn–/– (KO) mouse embryonic fibroblast (MEF) lines and used a stably rescued FLCN cell line derived from the KO MEFs (Resc) (Figure 1A). Consistent with our earlier work, we observed that Flcn KO cells displayed a 2-fold increase in HIF activity when compared with Flcn WT or Resc MEFs in hypoxia using a HIF reporter assay (Figure 1B) (5). Under normoxic conditions, HIF-induced target gene (HK2, LDHA, SLC2A1, and VEGF) expression was increased by 2-fold upon loss of FLCN (Figure 1C). Loss of FLCN in MEFs did not affect the levels of HIF-1α or HIF-2α protein, which we reported previously in human renal cancer cells (Figure 1D) (5). Moreover, we tested whether the HIF-1α subunit drives the increased HIF target gene expression in Flcn KO MEFs under normoxia by using 2 independent shRNAs for HIF-1α (Figure 1, E and F). Together with our previous results, we concluded that under both hypoxic and normoxic conditions FLCN controls HIF transcriptional activity. However, we observed that, in hypoxia, the augmented HIF-α protein levels potentiate the HIF transcriptional activity and thus intensify the difference observed between FLCN-positive and FLCN-negative cells (5).
Figure 1. Loss of FLCN stimulates HIF-dependent glycolysis and ATP production.
(A, D, and E) Western blot analysis of FLCN, HIF-1α, and HIF-2α expression levels in the indicated MEF cell lines. Actin was used as loading control. Results are representative of 3 independent experiments. (B) Fold HIF activity assessed under hypoxic conditions using HIF reporter assay. (C and F) Relative mRNA expression of HIF target genes determined by qRT-PCR (C) in the indicated MEFs or (F) in KO MEFs downregulated (shHIF-1α cell lines) or not (shNT) for HIF-1α. (G) Fold change in glucose uptake (Glu up), lactate production (Lact), and extracellular acidification rate (ECAR) in the indicated MEFs. (H) Glucose uptake and ATP levels measured in the indicated MEF cell lines. Data in B, C, and F–H represent the mean ± SD of 4 independent experiments performed in triplicate. *P < 0.05 **P < 0.01, ***P < 0.001.
Interestingly, Flcn KO MEFs exhibited an increased rate of aerobic glycolysis characterized by an augmentation of glucose uptake and lactate production, leading to an increased extracellular acidification rate (Figure 1G). Consistently, the ATP levels were increased in Flcn KO MEFs compared with those in WT MEFs (Figure 1H). Stable downregulation of HIF-1α confirmed that the increased aerobic glycolysis and ATP levels observed in Flcn KO cells depend on HIF-1α activation (Figure 1H). Altogether, our data strongly suggest that Flcn deficiency enhances HIF-transcriptional activity, which drives aerobic glycolysis, increasing cellular ATP levels in normoxic conditions.
Increased mitochondrial biogenesis enhances ROS production and activates HIF.
In aerobic conditions, cells preferentially use oxygen to efficiently produce ATP through mitochondrial OXPHOS. It was thought initially that aerobic glycolysis associated with the Warburg effect was accompanied by impaired mitochondrial activity (13). However, recent reports have shown that most cancer cells have normal mitochondrial function and that OXPHOS is not dispensable and actively contributes to energy and biosynthetic precursor production, which constitute a tumorigenic advantage (14–16). It has been reported recently that conditional loss of FLCN in mouse kidney and muscle resulted in increased mitochondrial function (17). To confirm this finding in our cellular model, we acutely measured the rate of mitochondrial respiration in Flcn KO MEFs compared with that in WT and Resc cells and show that loss of FLCN significantly increased total mitochondrial respiration (Figure 2A). Using oligomycin, an ATP synthase inhibitor that suppresses mitochondrial ATP turnover, we determined the amount of proton leak over total mitochondrial respiration. Interestingly, we did not observe a significant difference in the percentage of proton leak to total ATP turnover (Figure 2A). Next, we examined the mitochondrial abundance and efficiency and observed that loss of FLCN resulted in a 1.2-fold increase in mitochondrial abundance and potential using mitochondrial dyes (Figure 2B). These effects were reversed by FLCN reexpression in KO cells. Strikingly, the mitochondrial membrane potential per mitochondrial mass was unchanged in cells devoid of FLCN (Figure 2B), suggesting that the increase in mitochondrial respiration observed in Flcn KO MEFs is partly caused by an increase in mitochondrial mass. To determine whether FLCN functions similarly in other cell lines, we measured the relative mitochondrial levels in the UOK257 kidney cancer devoid of FLCN. We confirmed the increased mitochondrial abundance upon loss of FLCN by determining the percentage of mitochondrial DNA normalized to the nuclear DNA content in UOK257 and MEF lines by quantitative PCR (Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI71749DS1). This observation is in agreement with the fact that some patients with BHD syndrome are predisposed to develop renal oncocytomas, which are tumors characterized by abundant mitochondria, suggesting an extensive mitochondrial accumulation due to years of FLCN deficiency in this kind of BHD-derived tumors (18).
Figure 2. Enhancement of mitochondrial biogenesis upon loss of FLCN activates HIF in MEFs.
(A) Fold changes in total mitochondrial respiration, divided in ATP turnover and proton leak, determined following inhibition of ATP synthase by oligomycin treatment. (B) Fold changes in mitochondrial load and potential determined using MitoTracker Green FM and MitoTracker Red CMXROS, respectively. Pot, mitochondrial membrane potential; Load, mitochondrial mass. (C and F) Fold ROS levels assessed using the general oxidative stress indicator CM-H2DCFDA incubated with cells for (C) 30 minutes or (F) the indicated time points in a time course experiment. Data are expressed as fold ROS levels normalized to (C) WT or as (F) the KO/WT ROS ratio. (D) Extent of ROS-dependent protein and (E) DNA damage quantified using the OxyBlot Protein Oxidative Detection Kit and the OxiSelect Oxidative DNA Damage Kit, respectively. Data are representative of 3 independent experiments. (G) Relative mRNA expression of HIF target genes in MEFs treated with 10 mM of the antioxidant NAC for 24 hours. Data in A–C and E–G represent the mean ± SD of 4 independent experiments performed in triplicate. *P < 0.05, **P < 0.01, ***P < 0.001.
Since enhanced mitochondrial load is likely to increase the production of ROS, a side product of mitochondrial respiration, we quantified the relative levels of cellular ROS using the CM-H2DCFDA general oxidative stress indicator. We observed that loss of FLCN is associated with a significant 1.7-fold increase of intracellular ROS levels, which was rescued by FLCN reexpression (Figure 2C). Consistently, we observed a 3-fold increase of ROS levels in UOK257 cells devoid of FLCN (empty vector [EV]) when compared with UOK257 cells rescued for FLCN expression (Supplemental Figure 1B). ROS accumulation causes DNA and protein damage, which affects cellular functions. Therefore, we hypothesized that the increased ROS levels conferred by loss of FLCN could lead to cellular damage. We measured protein carbonylation and 8-hydroxydeoxyguanosine (8-OHdG) levels, 2 major oxidized forms of ROS-dependent protein and DNA damage, respectively. We did not observe a difference in the levels of protein carbonylation (Figure 2D) or 8-OHdG (Figure 2E) between WT and Flcn KO cells, suggesting that the increased ROS levels are not causing long-term damage in Flcn KO cells. Since the ROS assay was performed on live cells using a probe irreversibly oxidized upon contact with ROS, one attractive explanation to interpret the lack of ROS-induced damages would be that a higher rate of mitochondrial ROS production is measured by this assay and does not represent ROS accumulation. To test this hypothesis, we incubated the cells with the CM-H2DCFDA dye in a time course experiment, measured the ROS levels, and determined the ratio between KO and WT ROS signal (Figure 2F). Interestingly, while we observed an increase in ROS signal over time in both Flcn WT and KO cells, the ROS level ratio between KO and WT cells was constantly elevated over time, suggesting an enhanced ROS production in KO cells rather than an accumulation.
Whereas ROS were described initially as harmful to the cell, beneficial and physiological roles of ROS as signaling molecules are now widely acknowledged (19). Interestingly, increased ROS production upon mitochondrial dysfunction is well known to activate HIF under hypoxic conditions (20, 21). Moreover, heightened mitochondrial ROS levels are also known to drive HIF activation in normoxia. Indeed, a recent study in Caenorhabditis elegans showed an upregulation of HIF activity in response to mitochondrial ROS production promoting longevity (22). In addition, mitochondrial ROS-dependent HIF activation has been shown to modulate the immune response in long-lived Mclk1+/– mouse mutants (23), and HIF activation was reported to be both AMPK- and ROS-dependent in prostate cancer cells (8). Finally, HIF activation by mitochondrial ROS caused invasive growth and angiogenesis in melanoma cells (24). Therefore, we hypothesized that the increased production of mitochondrial ROS observed in Flcn-deficient cells might be responsible for the HIF transcriptional activation. To test this possibility, we treated WT and Flcn KO MEFs with the antioxidant N-acetyl cysteine (NAC) and determined the effect on the transcriptional activation of HIF target genes (Figure 2G). In Flcn KO MEFs, NAC reduced mRNA expression of HIF target genes to a level equivalent to that in WT MEFs treated with NAC. We also confirmed a similar effect using the HIF reporter assay in hypoxic conditions (Supplemental Figure 1C). Collectively, these data reveal that ROS enhances HIF transcriptional activation in Flcn KO MEFs.
Increased PGC-1α expression in FLCN-null cells enhances ROS production.
Mitochondrial biogenesis is largely controlled by a transcriptional network dependent on the peroxisome proliferator-activated receptor γ coactivator 1 (PGC-1α, encoded by PPARGC1) (25). It has been suggested that through mitochondrial biogenesis, PGC-1α indirectly stimulates HIF target gene expression (26). Therefore, we quantified PGC-1 expression and observed a 3-fold increase in both the PGC-1α and PGC-1β transcripts upon loss of FLCN, which was restored by FLCN reexpression (Resc), suggesting that the increased mitochondrial biogenesis is driven by PGC-1 in FLCN-null cells (Figure 3A). Consistently, we observed a substantial increase in PGC-1α protein levels by Western blot (Figure 3B), and a corresponding increase in PGC-1α target genes (COX7a, ATP5j, NDUFB5) and coactivators (TFAM, NRF1, NRF2) (Figure 3C). We also observed a reduction of PGC-1α expression after FLCN reintroduction in the FLCN-deficient (EV) UOK257 kidney cell line (Supplemental Figure 2). However, we could not detect PGC-1β protein using commercially available antibodies. In line with this work, it was previously reported that tumors from patients with BHD harbor an induced PGC-1α transcription profile associated with a high expression of mitochondria and OXPHOS-associated genes (17, 27). Furthermore, PGC-1α and its target genes are consistently upregulated in 30 thyroid oncocytomas when compared with normal tissue (28). Mitochondria-rich oncocytic tumors, which are observed in kidneys of some patients with BHD, might severely affect organ functionality and are also observed in other organs, such as the thyroid gland, suggesting that the PGC-1α–dependent mitochondrial biogenesis observed upon loss of FLCN is not restricted to BHD syndrome.
Figure 3. ROS-mediated HIF activation depends on PGC-1α upregulation in FLCN-null cells.
(A) PPARGC1A and PPARGC1B relative mRNA expression measured by qRT-PCR in the indicated MEFs. (B) Western blot analysis of the PGC-1α and FLCN expression levels. Actin was blotted as loading control. Results are representative of 3 independent experiments. (C) Relative mRNA expression of PGC-1α target genes and coactivators determined by qRT-PCR. (D) PGC-1α protein and relative mRNA expression levels in KO MEFs downregulated using shRNA for PGC-1α (shPGC-1α-A and -B) or control (shNT), as measured by Western blot and qRT-PCR. (E) Relative ROS levels and (F) HIF target gene expression determined in the indicated cell lines. Data represent the (A and D–F) mean ± SD or (C) mean ± SEM of 4 independent experiments performed in triplicate. *P < 0.05, **P < 0.01, ***P < 0.001.
It is accepted that PGC-1α increases ROS production through its effect on mitochondrial biogenesis. However, PGC-1α simultaneously enhances cellular antioxidant defense mechanisms to neutralize the damaging effect caused by ROS, and generally PGC-1α upregulation is not associated with ROS accumulation (29, 30). To determine whether PGC-1α upregulation was responsible for the increased production of ROS in our cell lines, we downregulated PGC-1α levels in Flcn KO MEFs (Figure 3D) and quantified ROS levels (Figure 3E). In contrast with control, the knockdown of PGC-1α isoform in Flcn KO MEFs significantly reduced ROS levels, suggesting PGC-1α–dependent ROS production (Figure 3E). To confirm that PGC-1α–dependent ROS production mediates HIF activation in Flcn KO MEF cells, we determined the levels of expression of HIF target genes upon PGC-1α knockdown and observed a significant decrease (Figure 3F). Together, these data suggest that PGC-1α is upregulated upon loss of FLCN and stimulates mitochondrial biogenesis and ROS production, which drive HIF transcriptional activation.
AMPK activation in FLCN-deficient cells promotes PGC-1α upregulation.
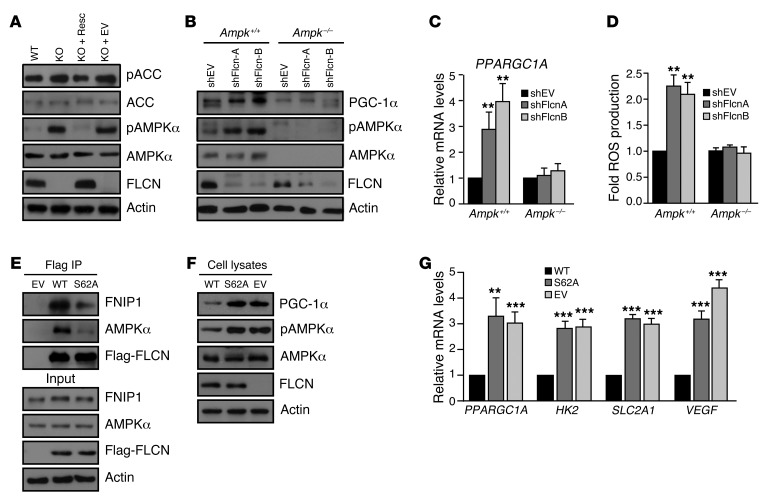
FLCN has been linked to the AMPK signaling pathway, as FLCN binds to AMPK through FNIP (5, 11, 12). The FLCN-AMPK interaction is controlled by the phosphorylation of FLCN on 2 sites (serine 62 and 302), suggesting possible regulatory roles in AMPK-dependent energy sensing and maintenance of energy homeostasis (31, 32). Through allosteric interactions with AMP and ADP, AMPK acutely monitors adenylate charge fluctuations. Under energetic or cellular stress conditions, AMPK enzymatic function is activated by phosphorylation of its threonine 172 catalytic site by the upstream kinase, LKB1 (9). Active AMPK is known to upregulate PGC-1α gene expression and to directly phosphorylate PGC-1α (33). The impact of FLCN loss on mitochondrial biogenesis, accompanied with high PGC-1α expression levels, made us speculate that AMPK is involved in the PGC-1-ROS–dependent activation of HIF. By Western blot, we confirmed that loss of FLCN activates AMPK and increases AMPK target gene phosphorylation in normoxia and in the absence of energetic stress (Figure 4A). To test whether AMPK activation upon Flcn loss is responsible for increased PGC-1α expression and activity, we downregulated FLCN in AMPKα-null (Ampk–/–) cells using shRNA or EV control (Figure 4B). While Flcn knockdown resulted in an increased in AMPKα activation and elevated PGC-1α protein (Figure 4B) and transcript (Figure 4C) levels in control cells (Ampk+/+), the expression levels of PGC-1α remained unchanged in the Ampk–/– cells. Similarly, ROS production was increased in the Ampk+/+ cells upon Flcn downregulation, while ROS levels remain unchanged in the Ampk–/– cells (Figure 4D). Our data reveal that, in the absence of FLCN, AMPK drives a sequential chain of events leading to upregulation of PGC-1α, increased mitochondrial biogenesis, ROS production, and HIF activation, which stimulates aerobic glycolysis.
Figure 4. AMPK activation upon loss of FLCN binding drives the PGC-1α-ROS–mediated HIF induction.
(A) Western blot analysis of AMPK expression (AMPKα) and activation (pT172 AMPKα [pAMPKα]) levels and acetyl-CoA carboxylase (ACC) expression and activation (pS79 ACC [pACC]) levels in the indicated MEFs. Actin was used as loading control. (B) Western blot analysis of Ampk–/– or Ampk+/+ MEFs downregulated (shFlcn) for FLCN or not (shEV). Actin was used as loading control. (C) Relative PPARGC1A mRNA expression and (D) ROS production levels in the indicated MEFs. (E–G) Flcn KO MEFs were rescued with FLCN WT, FLCN S62A mutant (S62A), or EV constructs, and (E) the extent of FLCN binding to AMPKα and FNIP1 was determined by coimmunoprecipitation. The effect of the S62A mutation on (F and G) PGC-1α, (G) HIF target gene expression, and (F) AMPK activation (pT172 AMPKα) was assayed by (F) Western blot and (G) qRT-PCR. Results in A, B, E, and F are representative of 3 independent experiments and data in C, D, and G represent the mean ± SD of 4 independent experiments performed in triplicate. **P < 0.01, ***P < 0.001.
To further substantiate the notion that FLCN binding to AMPK prevents its activation, we rescued FLCN expression in KO MEFs with an EV (Flag-EV), Flag-FLCN-WT, or the nonphosphorylatable Flag-FLCN-S62A mutant. This point mutation has been reported previously to reduce the affinity of FLCN to AMPK in a HEK293T overexpression system (32). To validate this observation, we immunoprecipitated FLCN in MEF lines described above. As expected, we observed reduced binding of the FLCN S62A mutant to AMPK and FNIP1 when compared with FLCN-WT (Figure 4E). Strikingly, reduced interaction of FLCN-S62A mutant with AMPK enhanced the phosphorylation of AMPK (Figure 4F), suggesting that binding of FLCN to the AMPK-FNIP1 complex blocks AMPK activation. To test this notion further, we asked whether the FLCN-dependent regulation of PGC-1α expression was dependent on the association of FLCN and AMPK. Consequently, the FLCN-S62A mutant failed to restore the levels of PGC-1α protein expression (Figure 4F) and transcript levels (Figure 4G) to a level observed with Flag-FLCN WT. Similarly, the S62A mutant failed to repress HIF target gene activation (HK2, SLC2A1, VEGF) (Figure 4G), revealing a loss-of-function phenotype of the S62A mutant. Our data uncover a new FLCN function as a negative regulator of AMPK and show that loss of FLCN binding to AMPK is a critical signaling event that leads to AMPK/PGC-1α/HIF activation.
Loss of FLCN leads to metabolic transformation.
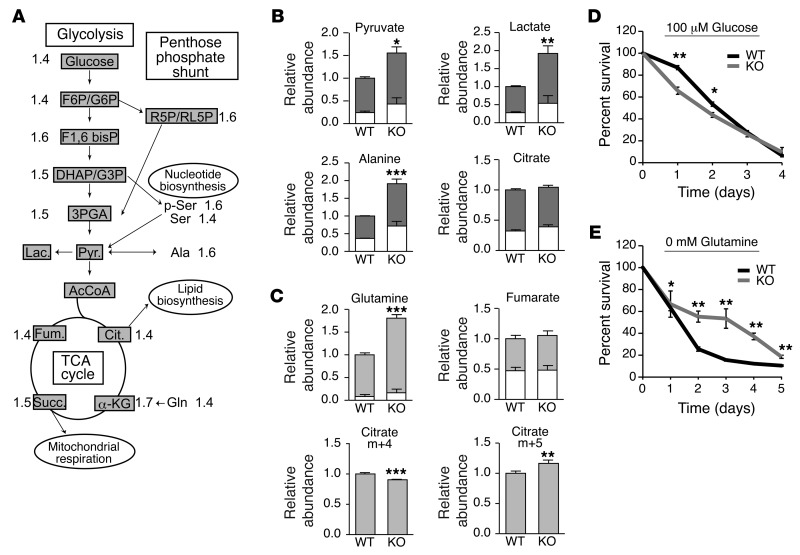
Our data reveal that loss of FLCN in untransformed MEFs induces metabolic reprogramming, characterized by PGC-1α–dependent mitochondrial biogenesis and HIF-driven aerobic glycolysis. However, while FLCN-deficient MEFs exhibit increased amounts of ATP, they do not acquire a proliferative advantage in vitro (Supplemental Figure 3) or spontaneous transformation features, such as anchorage-independent growth or in vivo tumorigenic potential per se (M.-C. Gingras, unpublished observations). To further characterize the metabolic profile associated with loss of FLCN, we used an approach combining gas chromatography and mass spectrometry (GC/MS) and liquid chromatography tandem mass spectrometry (LC/MS) analyses to monitor the intracellular steady-state levels of metabolites (Figure 5A and Supplemental Table 1). As expected, the glycolysis intermediates (glucose, fructose 6-phosphate/glucose 6-phosphate, fructose 1, 6-bisphosphate, dihydroxyacetone phosphate/glyceraldehyde 3-phosphate, 3-phosphoglyceric acid) were augmented in the Flcn KO MEFs. Consistently, the intermediates of the pentose phosphate pathway (ribose 5-phosphate/ribulose 5-phosphate) and the levels of serine as well as phosphorylated serine, both known as critical intermediates for nucleotide biosynthesis, were also significantly increased in the Flcn KO MEFs. In agreement with the observed increase in mitochondrial number, respiration, and OXPHOS, the tricarboxylic acid (TCA) cycle metabolites were increased (citrate, α-ketoglutarate, succinate, and fumarate) in Flcn KO MEFs. It has been reported that glycolytic cancer cells increase their consumption of glutamine and use it as a source of carbon to feed the TCA cycle and facilitate the production of biosynthetic precursors (34). Consistently, we observed an increase in glutamine levels in Flcn KO MEFs. Surprisingly, the metabolic signature of the Flcn KO cells suggested a highly proliferative profile, while their proliferation rates were similar to those of WT MEFs in cell culture (Supplemental Figure 3).
Figure 5. FLCN-null cell survival depends on glucose-derived biosynthetic precursor production.
(A) Metabolic signature of Flcn KO cells determined by GC/MS and LC/MS. Data are expressed as fold of the WT MEF metabolite quantification and are indicated in the schematic representation of the metabolic pathways. (B and C) Mass isotopomer labeling of MEFs pulsed with (B) 13C glucose or (C) 13C glutamine. Relative incorporation of (B) 13C glucose (dark gray) and (C) 13C glutamine (light gray) to total metabolite pool are indicated, and the metabolite abundance relative to WT condition was measured. (D and E) Percentage cell survival under (D) glucose and (E) glutamine starvation relative to cell number at day 0. Data represent the mean ± SD of (A) 8 or (B–E) 3 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
Next, we defined the profile of glucose and glutamine consumption by performing mass isotopomer labeling experiments in cells pulsed with [13C]-labeled glucose (Figure 5B and Supplemental Table 2) or glutamine (Figure 5C and Supplemental Table 2) and examined the amount of 13C contribution of these carbon fuels to metabolite pools. Consistent with the metabolic signature, we observed an accumulation of glycolytic intermediates derived from the labeled glucose in the Flcn KO MEFs, suggesting an increased glycolytic rate. Indeed, the total abundance of pyruvate, lactate, and alanine derived from the labeled glucose was increased by 1.5- to 2-fold in Flcn-null cells, a characteristic feature of the Warburg effect. Our data further reveals a significant amount of [13C]-citrate derived from the labeled glucose, which was not increased in Flcn KO MEFs, while the other TCA intermediates are mainly unlabeled (M.-C. Gingras, unpublished observations). These results might be explained by the utilization of citrate for the generation of lipid precursors and membrane synthesis, which would imply that a different source of carbon may fuel the TCA cycle. Consistent with this notion, we observed a significant level of [13C]-glutamine uptake in WT MEFs, which was drastically increased in Flcn KO MEFs (Figure 5C). However, this uptake was not associated with a higher TCA cycle utilization of glutamine, since labeled fumarate (Figure 5C), α-ketoglutarate, and succinate (M.-C. Gingras, unpublished observations) were similar in WT and Flcn KO MEFs (Figure 5C). Importantly, the level of m+5 citrate (5 carbons labeled), which could only be derived from reductive carboxylation of the labeled glutamine, was significantly increased in Flcn KO MEFs compared with that in WT. Conversely, the m+4 citrate derived from the classical TCA cycle reaction was decreased in Flcn KO MEFs (Figure 5C). This characteristic is frequent in glycolytic cancer cells, which sometimes decrease their glucose-derived replenishment of the TCA cycle that must rely on alternative source of metabolites, such as glutamine (35, 36). Strikingly, it has been reported recently that HIF is necessary and sufficient to induce reductive carboxylation of glutamine in renal cell carcinoma to maintain lipogenesis and cell growth (37). Our findings reveal that the increased uptake of glucose drives the glycolytic pathway to favor the generation of additional ATP, nucleotide precursors through the pentose phosphate and serine pathway, and enhanced lactate and alanine production. While the glutamine uptake is significantly increased upon loss of FLCN, the glutamine flux into the TCA cycle is not enhanced. Rather, glutamine uptake might serve to produce citrate and lipids through glutamine reductive carboxylation.
Our data strongly suggest that glucose is the major source of energy in Flcn KO MEFs and that glutamine might be an alternative carbon source used for lipogenesis. Therefore, we investigated the dependency of Flcn KO MEFs on both sources of energy by evaluating their ability to survive under either glucose or glutamine deprivation. Under low glucose concentration (100 μM), Flcn KO MEFs exhibited a diminution of cell survival compared with WT MEFs (Figure 5D), while complete glutamine withdrawal lead to an increase in survival in Flcn KO MEFs compared with WT MEFs (Figure 5E). Collectively, our data reveal that FLCN-deficient cells are “metabolically transformed” to favor aerobic glycolysis and depend on glucose to survive.
Loss of FLCN-mediated metabolic transformation confers tumorigenic advantage.
AMPK has been proposed to be a contextual oncogene that may confer a survival advantage under selection pressure, suggesting an involvement in tumor progression rather than in tumor initiation (10). To determine whether the metabolic transformation induced upon loss of FLCN would promote a tumorigenic advantage in an established cancer cell line, we used the human follicular thyroid cancer cell line FTC-133, which is naturally deficient for FLCN expression, that we stably rescued with an EV or FLCN expression vector (Figure 6A and refs. 38, 39). In agreement with our previous results, FTC-133 control cells exhibited an increase in AMPK activation (Figure 6A), PGC-1α levels (Figure 6, A and E), mitochondrial biogenesis (Figure 6, B and C), and ROS production (Figure 6D) compared with FLCN-rescued FTC-133 cells. Moreover, the FTC-133 control cells displayed transcriptional upregulation of HIF target genes (Figure 6E) in absence of HIF-1α stabilization (Figure 6A). Consistently, we observed an increase in aerobic glycolysis (glucose uptake) and ATP levels in FLCN-null FTC-133 control cells (stably rescued with an EV and infected with a scramble sequence [shScram]) compared with FTC-133 cells reexpressing FLCN (Resc) or downregulated for HIF-1α expression (shHIF-1α) (Figure 6F and Supplemental Figure 4A). Strikingly, these data suggest that the AMPK/PGC-1α/ROS/HIF pathway identified in FLCN-null MEFs and UOK257 cells is conserved in the human FTC-133 cancer cell line. Similar to what we observed in Flcn KO MEFs, the metabolic advantage conferred by loss of FLCN did not lead to a proliferative advantage in FTC-133 cells, as the proliferation rate and cell size were comparable between FTC-133 control and FLCN-rescued cells (M.-C. Gingras, unpublished observations). However, we observed a significant increase in the number of soft agar colonies in FTC-133 control cells (EV + shScram) when compared with that in rescued cells (Figure 6G and Supplemental Figure 4B). Moreover, downregulation of HIF-1α completely abolished the increased colony numbers observed in control cells, which were reduced to a similar level as that observed in the rescued cells (Figure 6G and Supplemental Figure 4B). In agreement with these data, it has been reported that while FLCN reintroduction in FLCN-deficient UOK257 cells does not affect cell growth, it significantly decreased the anchorage-independent and in vivo tumorigenic potential (40). Using UOK257 cells, we confirmed a similar decrease in anchorage-independent cell growth upon HIF-1α downregulation (Supplemental Figure 4, C and D). In addition, FLCN deficiency in FTC-133 cells was associated with an increased tumor volume and growth rate when compared with that in FLCN-rescued cells or HIF-1α–downregulated cell lines (Figure 6H and Supplemental Figure 4E).
Figure 6. HIF-dependent metabolic advantage drives tumorigenesis in FLCN-deficient cancer cells.
(A) Western blot analysis of the AMPK expression (AMPKα) and activation (pT172 AMPKα) and PGC1-α and HIF-1α expression levels in FTC-133 cells deficient (EV) or rescued (Resc) for FLCN expression. (B–D) Fold (B) mitochondrial respiration, (C) load and potential, and (D) ROS production in FTC-133 cells. (E) Fold mRNA levels of the indicated genes relative to FTC-133 cells rescued for FLCN expression. (F) Glycolysis (glucose uptake) and ATP levels quantified in the FLCN-null (EV) cells downregulated (shHIF-1α) or not (shScram) for HIF-1α expression and compared with FTC-133 cells rescued for FLCN expression (Resc). (G) Fold change in soft agar colony number and (H) xenograft tumor volume 42 days after subcutaneous tumor cell injection in nude mice using the indicated FTC-133 cell lines. Data represent the mean ± SD of (A–G) 4 independent experiments or (H) 5 tumors per group. *P < 0.05, **P < 0.01, ***P < 0.001.
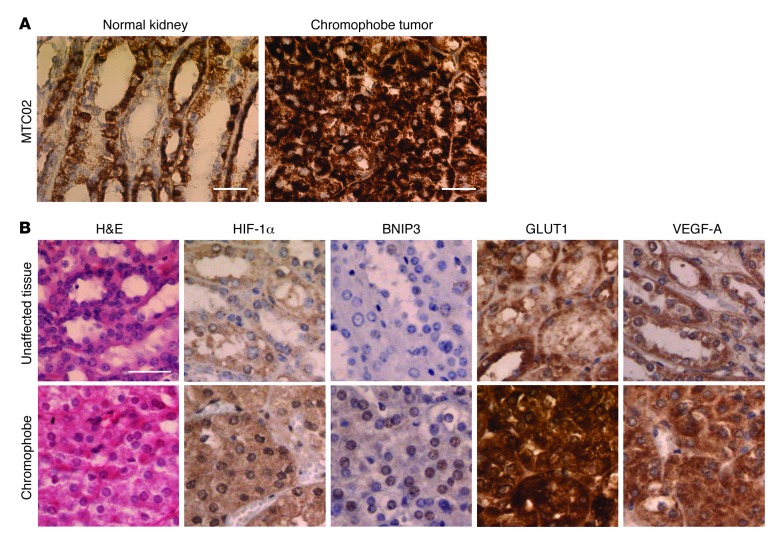
To test the relevancy of our finding for human BHD disease, we performed immunohistochemistry staining on normal kidney tissues and a chromophobe tumor isolated from a patient with BHD. We observed an increase in the mitochondrial marker (MTC02) (Figure 7A). Moreover, we detected an increase in HIF-1α nuclear staining associated with a significant heightening in HIF targets staining (BNIP3, GLUT1, VEGF-A) (Figure 7B), suggesting that loss of FLCN stimulates mitochondrial biogenesis and HIF-1 signaling during renal tumor progression in patients with BHD. Altogether, our data strongly suggest that loss of FLCN induces an AMPK-HIF–dependent metabolic reprogramming, resulting in an energetic advantage, which favors tumor progression in vivo.
Figure 7. Increased mitochondrial content and HIF target gene expression in a BHD tumor.
(A and B) Representative images of immunohistochemistry staining performed on a BHD kidney chromophobe tumor and (A) normal kidney or (B) adjacent unaffected tissues. Scale bar: 50 μm (A); 100 μm (B).
Discussion
In accordance with our results, it was shown recently that upregulation of PGC-1α expression resulted in an increase of mitochondrial biogenesis and ATP production in a muscle-specific Flcn KO mouse model (17). Interestingly, all features observed in our FLCN-deficient cells have been linked independently with cancer-promoting metabolic adaptations. Indeed, AMPK activation (10, 41–43), PGC-1 induction and mitochondrial biogenesis (44), increased ROS production (45), and HIF activation (46) have been described as steps leading to cellular metabolic reprogramming that favor tumor development in different cellular systems. Interestingly, while some AMPK-dependent signaling pathways inducing HIF have been reported, the exact mechanism of this was still unclear (10). Our work identifies an additional mechanism for AMPK to activate HIF and couples this activation to an AMPK downstream target: PGC-1α.
While HIF activation has been associated with tumor development in mammals, it leads to extension of life span in nematodes. Indeed, a loss-of-function mutation of the VHL tumor suppressor gene in C. elegans promotes a HIF-α–dependent increase in life span (47). Interestingly, promotion of C. elegans longevity upon transient AMPK-dependent increase of ROS or upon ROS-mediated HIF-1 activation has been reported independently (22, 48, 49). Together with these studies, our results suggest the existence of an AMPK-driven pathway activating HIF-mediated metabolic adaptation through ROS, which appears conserved in lower organisms.
Classically, ROS is known to oxidize and inhibit the hydroxylases responsible for HIF degradation, leading to HIF stabilization and activation (46). However, we reveal that ROS production can drive HIF transcriptional activation without affecting the protein levels of HIF-α. In accordance with our data, it has been shown in prostate cancer cells that H2O2 activates HIF beyond what would be solely expected through increased HIF-1α stability (for example, compared with MG132 treatment) (8). Indeed, the use of a reporter system measuring transactivation of HIF revealed that ROS but not MG132 stimulates HIF activity. Moreover, AMPK inhibition blocked this activation, suggesting that ROS could stimulate HIF-1 transactivation via AMPK, which is in addition to any effect on HIF stabilization. In addition to the various transcriptional cofactors that might influence HIF activity, several HIF posttranslational modifications, such as phosphorylation, S-nitrosylation, and sumoylation, have been reported as potent modulators of HIF transcriptional activity (50). Moreover, the MgcRacGAP cytoskeleton regulator has been identified recently as a novel inhibitor of HIF dimerization affecting HIF transcriptional activity (51). Further investigations will be required to decipher this mechanism, which is not within the scope of this study.
Since increased ROS and HIF activation has been shown independently to promote tumorigenesis, it would be worthwhile to further determine whether this signaling connection from ROS to HIF is relevant to other types of cancers. Interestingly, it was shown recently that FLCN expression negatively correlates with PGC-1α activation in numerous tumors, such as those of the breast, cervix, colon, kidney, lung, lymph, ovary, pancreas, prostate, stomach, thyroid, and vulva (27). Specifically, the PGC-1α gene set and other OXPHOS gene sets correlated negatively with FLCN expression across all tumor types examined. Of interest, we also observed enhancement of PGC-1α expression, ROS production, and HIF activation in a follicular thyroid carcinoma cell line that innately lacks FLCN and was not derived from a patient with BHD. Our analogous observations in this cell line importantly confirm that our research findings presented in this manuscript have wider implications to other cancers, rather than only being restricted to tumors developed by patients with BHD. Together, our study suggests that loss of FLCN expression leads to activation of the AMPK/PGC-1α/OXPHOS/HIF signaling axis, which is an important driver of sporadic tumors in multiple organs.
AMPK has been shown to drive antitumorigenic functions, revealing an AMPK metabolic tumor suppressor function (52, 53). This notion is compatible with the fact that AMPK activates catabolic processes and inhibits anabolic processes, an unfavorable context for cell proliferation (10). In contrast, several recent reports have also shown that gain of function of AMPK and PGC-1α is a driver of tumorigenesis via maintenance of metabolic homeostasis, promotion of metastasis, and support of cancer cell survival (41–43, 54–58). This concept is supported by the fact that loss of AMPK or its upstream activator LKB1 is associated with apoptosis under bioenergetically stressful conditions (54). Based on these reports, it was proposed that AMPK tumor suppressor function might prevent tumor initiation through its ability to restrict cell proliferation, whereas AMPK activation permits metabolic adaptation and survival under energetic stress conditions and is beneficial for tumor progression and metastasis (10). In agreement with this concept, the central regulatory role of LKB1 in progrowth metabolism and tumor initiation has been reported recently (59). Results presented here are consistent with an AMPK oncogenic function and uncover a signaling pathway that links AMPK activity to HIF via PGC-1α upregulation, mitochondrial biogenesis, and ROS generation. Our study also reveals how loss of FLCN mediates aerobic glycolysis to drive a metabolic advantage in cancer cells when oxygen and nutrients are plentiful. Our findings have wider implications for cancers and human disease in which HIF activity is known to be induced and shed further light on the tumor suppressor mechanism of FLCN.
Methods
See the Supplemental Methods for details regarding plasmids, antibodies, and reagents; for description of the cell lines and culture methods; and for descriptions of extended procedures used for RNA extraction, quantitative PCR, real-time PCR (qRT-PCR), protein extraction, immunoprecipitation, Western blotting, immunohistochemistry, proliferation, soft agar, and xenografts assays. See Supplemental Table 3 for primers.
HIF luciferase reporter assays.
MEFs were transfected with the firefly luciferase HIF activity reporter pGL2-TK-HRE plasmid (gifted by G. Melillo; NCI, Frederick, Maryland, USA), and the assays were carried out under hypoxia (1% oxygen) as described previously (60). Briefly, 20 μl cleared lysed cell supernatant was injected with 50 μl luciferase assay reagent II (Promega) into a luminometer. Luciferase levels were measured at 2- and 10-second intervals after initial injection. Results were normalized to TK-renilla reporter control. When indicated, the antioxidant, NAC, was added to the cells for 16 hours prior to transfection and maintained after transfection.
Glucose uptake and lactate production assays.
Glucose uptake was determined using the 2-(N-[7-nitrobenz-2-oxa-1,3-diazol-4-yl]amino)-2-deoxyglucose (2-NBDG) fluorescent glucose analog (Invitrogen). Briefly, cells were incubated with 100 nM 2-NBDG for 1 hour, and the mean fluorescent intensity was detected using a FACSCalibur flow cytometer (BD Biosciences). Lactate production was determined in media collected from 48-hour seeded cells using the l-Lactate Assay Kit (Eton Bioscience). Absorbance measurements were read at 490 nm using a Spectramax spectrophotometer microplate reader (Molecular Devices), and data were normalized to the cell number.
Seahorse XF24 Extracellular Flux analysis.
The extracellular acidification rate was determined using the XF24 Extracellular Flux Analyzer (Seahorse Bioscience). Briefly, 40,000 cells per well were seeded in 625 μl nonbuffered DMEM containing 25 mM glucose, 2 mM glutamine, and 10% FBS and were incubated in a CO2-free incubator at 37°C for 1 hour. XF24 assays consisted of sequential mix (3-minute), pause (3-minute), and measurement (5-minute) cycles, and assays were performed 3 time in triplicates.
Protein and DNA damage assays.
The extent of oxidative protein damage was measured using the OxyBlot Protein Oxidation Detection Kit (Millipore) using 20 μg total protein extract. Carbonylated proteins were detected by Western blot, and α-tubulin was used as loading control. The amount of DNA oxidative damage (8-OHdG) was quantified on 16 μg purified genomic DNA using the OxiSelect Oxidative DNA Damage ELISA Kit (Cell Biolabs Inc.).
Respiration assay.
Respiration assays were performed as described previously (57, 61). Briefly, 1 × 106 cells were resuspended in PBS supplemented with 25 mM glucose, 1 mM pyruvate, and 2% (w/v) BSA. Oxygen consumption was measured at 37°C using a Clark-type oxygen electrode. The ATP synthase inhibitor oligomycin (2.5 μg per 106 cells) was used to determine the amount of proton leak over ATP turnover.
ROS, mitochondrial load, and potential.
Cellular ROS levels were determined using the general oxidative stress indicator CM-H2DCFDA (Invitrogen), and the mitochondrial mass and potential were determined using the MitoTracker Green FM and MitoTracker Red CMXRos (Invitrogen), respectively. Briefly, subconfluent adherent cells were incubated for 30 minutes at 37°C or in time course for 5, 15, or 30 minutes in serum-free media supplemented with the dyes. Then, cells were incubated for 10 minutes in complete DMEM and resuspended in 500 μl PBS supplemented with 2% (v/v) FBS. Fluorescence intensity was detected using the FACSCalibur flow cytometer (BD Biosciences). Cells not incubated with the dye or pretreated with 100 μM H2O2 were used as negative and ROS-positive controls, respectively.
ATP quantification.
ATP levels were determined by luminescence using the CellTiter-Glo Luminescent Cell Viability Assay (Promega). Briefly, 5,000 subconfluent cells were loaded in a 96-well plate in triplicates. After addition of equal volume of CellTiter-Glo reagent, relative luminescence units (RLUs) was measured using the FLUOstar Omega microplate reader (BMG LabTech), and results were expressed as fold changes.
Metabolism.
Metabolites from subconfluent cells cultured for 48 hours were extracted and quantified by GC/MS (TCA intermediates) or by LC/MS (glycolytic and pentose phosphate intermediates and amino acids) as described previously (41, 53). Metabolite levels were normalized to the cell number, and data are expressed as fold of the control WT cells. For the mass isotopomer experiment, 5 × 105 cells were cultured for 48 hours and pulsed using uniformly labeled 13C-glucose (30 minutes) or 13C-glutamine (1 hour). Metabolites were extracted and GC/MS analyses were performed as described previously (41).
Statistics.
Statistical analyses were performed using unpaired, 1-tailed Student’s t tests using Excel software (Microsoft). A P value of less than 0.05 was considered significant. Results are expressed as fold difference relative to the control conditions and are shown as mean ± SD or SEM of values obtained in at least 3 independent experiments.
Study approval.
Maintenance and experimental manipulation of animals were performed in facilities managed by the McGill University Animal Resources Centre. All animal experiments were conducted under animal use protocol approved by McGill University and according to the guidelines and regulations of the McGill University Research and Ethic Animal Committee and the Canadian Council on Animal Care.
Supplementary Material
Acknowledgments
We are indebted to D. Avizonis and G. Bridon from the Goodman Cancer Research Center Metabolomics Core Facility and Ken McDonald from the McGill Core Flow Cytometry Facility (McGill University) for their technical support and expertise. We thank J. St-Pierre and I. Topisirovic for critical reading of this manuscript and V. Chénard for technical support. M. Yan was supported by a McGill Integrated Cancer Research Training Program (MICRTP) studentship award, and M. Yan and B. Faubert are supported by a Canadian Institutes of Health Research (CIHR) Doctoral Research Award. This project is funded by the Kidney Foundation of Canada (to A. Pause: KFOC100021), Association for International Cancer Research Career Development Fellowship (to A.R. Tee: 06-914/915), the Myrovlytis Trust (to A.R. Tee, E.A. Dunlop, and A. Pause), the Cancer Research UK development fund (to A.R. Tee), the Terry Fox Foundation team grant on Oncometabolism (to A. Pause and R. Jones: 116128), the Dutch Cancer Society (to M.A.M. van Steensel: KWF U2009-4352), and the Annadal Foundation and GROW (to M.A.M. van Steensel and B.J. Coull). A. Pause was a recipient of the Canada Research Chair in Molecular Oncology. The Goodman Cancer Research Centre Metabolomics Core Facility is supported by the New Innovation Fund CFI 21875 (V. Giguere), The Dr. John R. and Clara M. Fraser Memorial Trust, the Terry Fox Foundation team grant on Oncometabolism (116128), and McGill University.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article:J Clin Invest. 2014;124(6):2640–2650. doi:10.1172/JCI71749.
References
- 1.Linehan WM, Srinivasan R, Schmidt LS. The genetic basis of kidney cancer: a metabolic disease. Nat Rev Urol. 2010;7(5):277–285. doi: 10.1038/nrurol.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keith B, Johnson RS, Simon MC. HIF1α and HIF2α: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2012;12(1):9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40(2):294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt LS, et al. Germline BHD-mutation spectrum and phenotype analysis of a large cohort of families with Birt-Hogg-Dube syndrome. Am J Hum Genet. 2005;76(6):1023–1033. doi: 10.1086/430842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Preston RS, et al. Absence of the Birt-Hogg-Dube gene product is associated with increased hypoxia-inducible factor transcriptional activity and a loss of metabolic flexibility. Oncogene. 2011;30(10):1159–1173. doi: 10.1038/onc.2010.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 7.Greer SN, Metcalf JL, Wang Y, Ohh M. The updated biology of hypoxia-inducible factor. EMBO J. 2012;31(11):2448–2460. doi: 10.1038/emboj.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jung SN, et al. Reactive oxygen species stabilize hypoxia-inducible factor-1 alpha protein and stimulate transcriptional activity via AMP-activated protein kinase in DU145 human prostate cancer cells. Carcinogenesis. 2008;29(4):713–721. doi: 10.1093/carcin/bgn032. [DOI] [PubMed] [Google Scholar]
- 9.Oakhill JS, Scott JW, Kemp BE. AMPK functions as an adenylate charge-regulated protein kinase. Trends Endocrinol Metab. 2012;23(3):125–132. doi: 10.1016/j.tem.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 10.Liang J, Mills GB. AMPK: a contextual oncogene or tumor suppressor? Cancer Res. 2013;73(10):2929–2935. doi: 10.1158/0008-5472.CAN-12-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baba M, et al. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc Natl Acad Sci U S A. 2006;103(42):15552–15557. doi: 10.1073/pnas.0603781103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466(7302):68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11(5):325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 14.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pollak M. Targeting oxidative phosphorylation: why, when, and how. Cancer Cell. 2013;23(3):263–264. doi: 10.1016/j.ccr.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 16.Schulze A, Harris AL. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 2012;491(7424):364–373. doi: 10.1038/nature11706. [DOI] [PubMed] [Google Scholar]
- 17.Hasumi H, et al. Regulation of mitochondrial oxidative metabolism by tumor suppressor FLCN. J Natl Cancer Inst. 2012;104(22):1750–1764. doi: 10.1093/jnci/djs418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pavlovich CP, et al. Renal tumors in the Birt-Hogg-Dube syndrome. Am J Surg Pathol. 2002;26(12):1542–1552. doi: 10.1097/00000478-200212000-00002. [DOI] [PubMed] [Google Scholar]
- 19.Finkel T. Signal transduction by reactive oxygen species. J Cell Biol. 2011;194(1):7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brunelle JK, et al. Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metab. 2005;1(6):409–414. doi: 10.1016/j.cmet.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 21.Guzy RD, et al. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 2005;1(6):401–408. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 22.Lee SJ, Hwang AB, Kenyon C. Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr Biol. 2010;20(23):2131–2136. doi: 10.1016/j.cub.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang D, Malo D, Hekimi S. Elevated mitochondrial reactive oxygen species generation affects the immune response via hypoxia-inducible factor-1α in long-lived Mclk1+/– mouse mutants. J Immunol. 2010;184(2):582–590. doi: 10.4049/jimmunol.0902352. [DOI] [PubMed] [Google Scholar]
- 24.Comito G, et al. HIF-1α stabilization by mitochondrial ROS promotes Met-dependent invasive growth and vasculogenic mimicry in melanoma cells. Free Radic Biol Med. 2011;51(4):893–904. doi: 10.1016/j.freeradbiomed.2011.05.042. [DOI] [PubMed] [Google Scholar]
- 25.Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18(4):357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 26.O’Hagan KA, et al. PGC-1alpha is coupled to HIF-1alpha-dependent gene expression by increasing mitochondrial oxygen consumption in skeletal muscle cells. Proc Natl Acad Sci U S A. 2009;106(7):2188–2193. doi: 10.1073/pnas.0808801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klomp JA, et al. Birt-Hogg-Dube renal tumors are genetically distinct from other renal neoplasias and are associated with up-regulation of mitochondrial gene expression. BMC Med Genomics. 2010;3:59. doi: 10.1186/1755-8794-3-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savagner F, et al. PGC-1-related coactivator and targets are upregulated in thyroid oncocytoma. Biochem Biophys Res Commun. 2003;310(3):779–784. doi: 10.1016/j.bbrc.2003.09.076. [DOI] [PubMed] [Google Scholar]
- 29.St-Pierre J, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127(2):397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 30.Finkel T. Cell biology: a clean energy programme. Nature. 2006;444(7116):151–152. doi: 10.1038/444151a. [DOI] [PubMed] [Google Scholar]
- 31.Piao X, et al. Regulation of folliculin (the BHD gene product) phosphorylation by Tsc2-mTOR pathway. Biochem Biophys Res Commun. 2009;389(1):16–21. doi: 10.1016/j.bbrc.2009.08.070. [DOI] [PubMed] [Google Scholar]
- 32.Wang L, et al. Serine 62 is a phosphorylation site in folliculin, the Birt-Hogg-Dube gene product. FEBS Lett. 2010;584(1):39–43. doi: 10.1016/j.febslet.2009.11.033. [DOI] [PubMed] [Google Scholar]
- 33.Jager S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1α. Proc Natl Acad Sci U S A. 2007;104(29):12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeBerardinis RJ, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104(49):19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Metallo CM, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481(7381):380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mullen AR, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481(7381):385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gameiro PA, et al. In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell Metab. 2013;17(3):372–385. doi: 10.1016/j.cmet.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu X, et al. Therapeutic targeting the loss of the Birt-Hogg-Dube suppressor gene. Mol Cancer Ther. 2011;10(1):80–89. doi: 10.1158/1535-7163.MCT-10-0628. [DOI] [PubMed] [Google Scholar]
- 39.Reiman A, et al. Gene expression and protein array studies of folliculin-regulated pathways. Anticancer Res. 2012;32(11):4663–4670. [PubMed] [Google Scholar]
- 40.Hong SB, et al. Tumor suppressor FLCN inhibits tumorigenesis of a FLCN-null renal cancer cell line and regulates expression of key molecules in TGF-β signaling. Mol Cancer. 2010;9:160. doi: 10.1186/1476-4598-9-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leprivier G, et al. The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation. Cell. 2013;153(5):1064–1079. doi: 10.1016/j.cell.2013.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jeon SM, Chandel NS, Hay N. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature. 2012;485(7400):661–665. doi: 10.1038/nature11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu L, et al. Deregulated MYC expression induces dependence upon AMPK-related kinase 5. Nature. 2012;483(7391):608–612. doi: 10.1038/nature10927. [DOI] [PubMed] [Google Scholar]
- 44.Deblois G, St-Pierre J, Giguere V. The PGC-1/ERR signaling axis in cancer. Oncogene. 2013;32(30):3483–3490. doi: 10.1038/onc.2012.529. [DOI] [PubMed] [Google Scholar]
- 45.Haigis MC, Deng CX, Finley LW, Kim HS, Gius D. SIRT3 is a mitochondrial tumor suppressor: a scientific tale that connects aberrant cellular ROS, the Warburg effect, and carcinogenesis. Cancer Res. 2012;72(10):2468–2472. doi: 10.1158/0008-5472.CAN-11-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Curr Opin Genet Dev. 2010;20(1):51–56. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, Shao Z, Zhai Z, Shen C, Powell-Coffman JA. The HIF-1 hypoxia-inducible factor modulates lifespan in C. elegans. PLoS One. 2009;4(7):e6348. doi: 10.1371/journal.pone.0006348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, Ristow M. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6(4):280–293. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 49.Zarse K, et al. Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal. Cell Metab. 2012;15(4):451–465. doi: 10.1016/j.cmet.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lisy K, Peet DJ. Turn me on: regulating HIF transcriptional activity. Cell Death Differ. 2008;15(4):642–649. doi: 10.1038/sj.cdd.4402315. [DOI] [PubMed] [Google Scholar]
- 51.Lyberopoulou A, et al. MgcRacGAP, a cytoskeleton regulator, inhibits HIF-1 transcriptional activity by blocking its dimerization. Biochim Biophys Acta. 2013;1833(6):1378–1387. doi: 10.1016/j.bbamcr.2013.02.025. [DOI] [PubMed] [Google Scholar]
- 52.Luo Z, Zang M, Guo W. AMPK as a metabolic tumor suppressor: control of metabolism and cell growth. Future Oncol. 2010;6(3):457–470. doi: 10.2217/fon.09.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Faubert B, et al. AMPK is a negative regulator of the Warburg effect and suppresses tumor growth in vivo. Cell Metab. 2013;17(1):113–124. doi: 10.1016/j.cmet.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kottakis F, Bardeesy N. LKB1-AMPK axis revisited. Cell Res. 2012;22(12):1617–1620. doi: 10.1038/cr.2012.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Girnun GD. The diverse role of the PPARγ coactivator 1 family of transcriptional coactivators in cancer. Semin Cell Dev Biol. 2012;23(4):381–388. doi: 10.1016/j.semcdb.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhalla K, et al. PGC1α promotes tumor growth by inducing gene expression programs supporting lipogenesis. Cancer Res. 2011;71(21):6888–6898. doi: 10.1158/0008-5472.CAN-11-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klimcakova E, et al. PGC-1α promotes the growth of ErbB2/neu-induced mammary tumors by regulating nutrient supply. Cancer Res. 2012;72(6):1538–1546. doi: 10.1158/0008-5472.CAN-11-2967. [DOI] [PubMed] [Google Scholar]
- 58.Tennakoon JB, et al. Androgens regulate prostate cancer cell growth via an AMPK-PGC-1α-mediated metabolic switch. Oncogene. doi: 10.1038/onc.2013.463. [published online ahead of print November 4, 2013]. doi: 10.1038/onc.2013.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dupuy F, et al. LKB1 is a central regulator of tumor initiation and pro-growth metabolism in ErbB2-mediated breast cancer. Cancer Metab. 2013;1(1):18. doi: 10.1186/2049-3002-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rapisarda A, et al. Identification of small molecule inhibitors of hypoxia-inducible factor 1 transcriptional activation pathway. Cancer Res. 2002;62(15):4316–4324. [PubMed] [Google Scholar]
- 61.Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9(6):425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.