Abstract
Activation of the ER stress response is associated with malignant progression of B cell chronic lymphocytic leukemia (CLL). We developed a murine CLL model that lacks the ER stress–associated transcription factor XBP-1 in B cells and found that XBP-1 deficiency decelerates malignant progression of CLL-associated disease. XBP-1 deficiency resulted in acquisition of phenotypes that are disadvantageous for leukemic cell survival, including compromised BCR signaling capability and increased surface expression of sphingosine-1-phosphate receptor 1 (S1P1). Because XBP-1 expression requires the RNase activity of the ER transmembrane receptor IRE-1, we developed a potent IRE-1 RNase inhibitor through chemical synthesis and modified the structure to facilitate entry into cells to target the IRE-1/XBP-1 pathway. Treatment of CLL cells with this inhibitor (B-I09) mimicked XBP-1 deficiency, including upregulation of IRE-1 expression and compromised BCR signaling. Moreover, B-I09 treatment did not affect the transport of secretory and integral membrane-bound proteins. Administration of B-I09 to CLL tumor–bearing mice suppressed leukemic progression by inducing apoptosis and did not cause systemic toxicity. Additionally, B-I09 and ibrutinib, an FDA-approved BTK inhibitor, synergized to induce apoptosis in B cell leukemia, lymphoma, and multiple myeloma. These data indicate that targeting XBP-1 has potential as a treatment strategy, not only for multiple myeloma, but also for mature B cell leukemia and lymphoma.
Introduction
The functional role of the ER stress response in mature B cell leukemia or lymphoma has been largely overlooked because leukemia and lymphoma cells do not expand their ER as do multiple myeloma (MM) cells. Recently, chronic lymphocytic leukemia (CLL), the most common adult leukemia, was shown to require activation of the ER stress response for survival (1).
The IRE-1/XBP-1 pathway represents the most conserved ER stress-response pathway. IRE-1 contains a luminal stress-sensor domain and a cytoplasmic kinase/RNase domain (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI73448DS1). The RNase domain is critical for the function of IRE-1 because it splices 26 nucleotides from the XBP1 mRNA, causing a frame shift in translation (2–4). The spliced XBP1 mRNA encodes a functional 54-kDa XBP-1s transcription factor. The role of XBP-1 in cancer has not been validated by genetic deletion of the Xbp1 gene in mice. Thus, we deleted the Xbp1 gene from B cells of Eμ-TCL1 transgenic mice (Cd19-Cre Xbp1flox/flox Eμ-TCL1, herein referred to as XBP-1KO/Eμ-TCL1), arguably the best CLL mouse model to date (5, 6). The Eμ-TCL1 mouse model is clinically relevant because TCL1 expression is found in 90% of human CLL cases (1, 7). Eμ-TCL1 mice develop leukemia with all clinical features of aggressive human CLL (6, 8) and have been used repeatedly for preclinical drug tests (9–16). Using XBP-1KO/Eμ-TCL1 mice, we examine the role of the IRE-1/XBP-1 pathway in tumor progression.
While most transcription factors remain undruggable, the specific activation mechanism of XBP-1 renders IRE-1 an attractive target for therapeutic intervention. Although chemical screens have led to the identification of inhibitors of the IRE-1 RNase activity (17–20), there is a need to develop novel small molecules with improved cellular and in vivo efficacy. We synthesized and evaluated novel tricyclic chromenone inhibitors of IRE-1 RNase activity that potently suppress the expression of XBP-1 and induce apoptosis. We also determined the bioavailability and pharmacokinetics of our lead inhibitor, B-I09, and showed that B-I09, when administered as a single agent, effectively induces leukemic regression without causing systemic toxicity in CLL-bearing Eμ-TCL1 mice. Since the inhibition of the IRE-1/XBP-1 pathway compromises B cell receptor (BCR) signaling, we tested for a potential synergistic effect between B-I09 and the Bruton’s tyrosine kinase (BTK) inhibitor ibrutinib. Our results demonstrate the effectiveness of targeting both the IRE-1/XBP-1 and BCR signaling pathways to induce apoptosis in human B cell leukemia, lymphoma, and MM cells.
Results
XBP-1KO/Eμ-TCL1 mice develop leukemia significantly more slowly than XBP-1WT/Eμ-TCL1 mice.
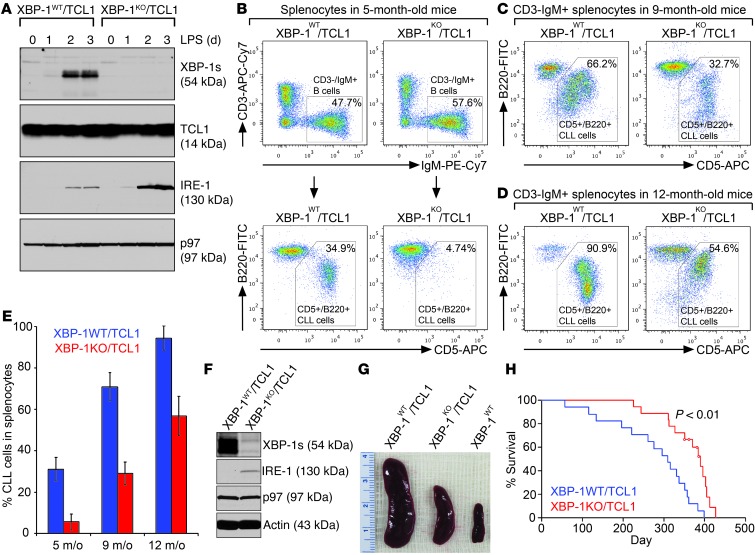
To investigate how the loss of XBP-1 can counter malignant progression of leukemia, we crossed B cell–specific XBP-1KO mice (Cd19-Cre Xbp1flox/flox; the expression of Cre recombinase is under the control of the Cd19 promoter) with Eμ-TCL1 mice to create a B cell–specific XBP-1–deficient CLL mouse model, XBP-1KO/Eμ-TCL1. To show that B cells produced by this new mouse model do not produce the 54-kDa spliced XBP-1 protein (XBP-1s), we isolated B cells from spleens of 6-week-old XBP-1KO/Eμ-TCL1 and XBP-1WT/Eμ-TCL1 mice (Supplemental Figure 2A), stimulated them with LPS, and confirmed no expression of XBP-1s in XBP-1KO/Eμ-TCL1 B cells (Figure 1A). When XBP-1s is missing, the elevated expression of IRE-1 is observed in XBP-1KO/Eμ-TCL1 B cells (Figure 1A), consistent with previous XBP-1 knockout and inhibition data in WT B cells (1, 21). We monitored leukemic progression in 5-, 9-, and 12-month-old XBP-1KO/Eμ-TCL1 mice by analyzing CD5+B220+ CLL cells on gated CD3–IgM+ B cell populations in the spleens (Figure 1B) and found that XBP-1KO/Eμ-TCL1 mice develop leukemia significantly more slowly than their age-matched XBP-1WT/Eμ-TCL1 littermates (Figure 1, B–E). We also confirmed that indeed XBP-1s is expressed by CD3–IgM+CD5+B220+ CLL cells isolated from the spleens of 12-month-old XBP-1WT/Eμ-TCL1 mice but not by those of the age-matched XBP-1KO/Eμ-TCL1 littermates (Figure 1F). As a result, spleens isolated from XBP-1KO/Eμ-TCL1 mice are significantly smaller than those from their control littermates (Figure 1G). XBP-1KO/Eμ-TCL1 mice also survive longer than their WT counterparts (Figure 1H).
Figure 1. XBP-1 deficiency decelerates leukemic progression in Eμ-TCL1 mice.
(A) CD5–/B220+ B cells purified from 6-week-old XBP-1WT/Eμ-TCL1 (XBP-1WT/TCL1) and XBP-1KO/Eμ-TCL1 (XBP-1KO/TCL1) mice were stimulated with LPS for a course of 3 days and lysed for analysis of indicated proteins by immunoblots. Data shown in immunoblots are representative of 3 independent experiments. (B–D) Splenocytes isolated from XBP-1WT/Eμ-TCL1 and XBP-1KO/Eμ-TCL1 mice at the ages of 5, 9, and 12 months were stained with CD3-APC-Cy7, IgM-PE-Cy7, B220-FITC, CD5-APC, and DAPI. Gated live CD3–IgM+ B cell populations were analyzed for the expression of B220 and CD5. (E) The percentages of CD5+B220+ CLL cells in splenocytes of XBP-1WT/Eμ-TCL1 and XBP-1KO/Eμ-TCL1 mice at the ages of 5, 9, and 12 months were plotted as mean ± SEM (n = 5 in each age group). (F) CD5+B220+ CLL cells purified from spleens of XBP-1WT/Eμ-TCL1 and XBP-1KO/Eμ-TCL1 mice were lysed to analyze for the expression of indicated proteins. Data shown in immunoblots are representative of 3 independent experiments. (G) Spleens from 12-month-old age-matched XBP-1WT/Eμ-TCL1 and XBP-1KO/Eμ-TCL1 littermates and a WT mouse. (H) Kaplan-Meier analysis of overall survival of XBP-1KO/Eμ-TCL1 mice (n = 18). Four mice from the XBP-1KO/Eμ-TCL1 group were censored (circled in red), as they were removed for other studies.
XBP-1–deficient Eμ-TCL1 CLL cells exhibit compromised BCR signaling.
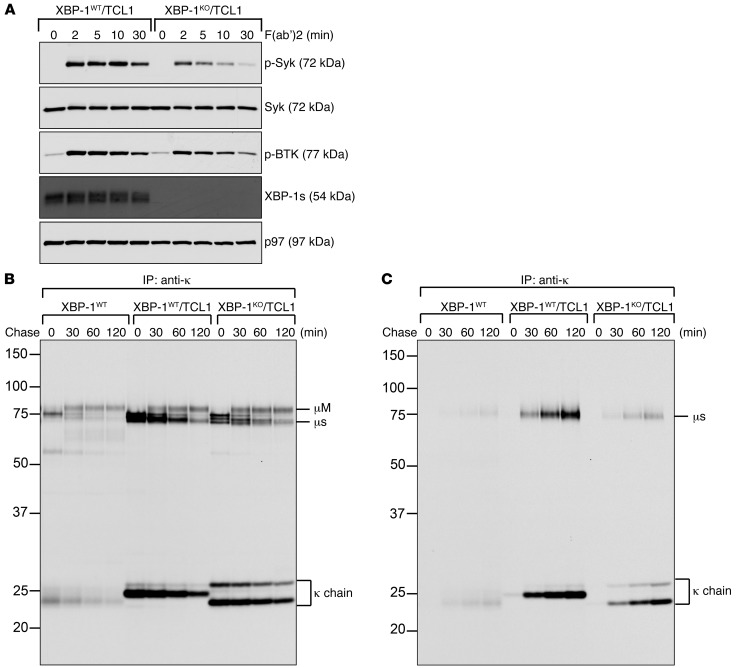
Constitutive BCR activation is a critical survival signal for CLL cells (22, 23). To understand how the loss of XBP-1 may contribute to the slower progression of leukemia in Eμ-TCL1 mice, we purified CLL cells from XBP-1WT/Eμ-TCL1 and XBP-1KO/Eμ-TCL1 littermates (Supplemental Figure 2, B and C), cultured them in LPS for 3 days, activated the BCR using F(ab′)2 anti-mouse IgM, and lysed the cells. Cell lysates were immunoblotted for phospho-Syk and phospho-BTK because Syk and BTK are critical BCR signaling molecules for CLL survival (22, 23). Compared with XBP-1WT/Eμ-TCL1 CLL cells, XBP-1KO/Eμ-TCL1 CLL cells are defective in Syk and BTK phosphorylation upon activation of the BCR (Figure 2A). Unlike naive normal B cells, XBP-1WT/Eμ-TCL1 CLL cells synthesize significantly increased amounts of secretory forms of IgM and release them into culture medium in the absence of any stimulation (Figure 2, B and C). The lack of XBP-1 leads to dramatically decreased synthesis of secretory μ heavy chains, resulting in decreased secretion of IgM into culture medium. Both XBP-1WT/Eμ-TCL1 and XBP-1KO/Eμ-TCL1 CLL cells produce comparable amounts of membrane-bound μ heavy chains (Figure 2B). The μ heavy chains can be assembled with κ light chains in the ER to form membrane-bound IgM (mIgM), which can be delivered to the cell surface (Figure 2B). While CLL cells isolated from XBP-1WT/Eμ-TCL1 mice acquire monoclonality, as evidenced by the use of a unique κ light chain, XBP-1–deficient CLL cells undergo slower clonal selection (Figure 2, B and C). The synthesis, assembly, and transport of the class I and class II MHC molecules are similar when comparing XBP-1WT/Eμ-TCL1 and XBP-1KO/Eμ-TCL1 CLL cells (Supplemental Figure 3).
Figure 2. XBP-1 deficiency compromises activation of the BCR and synthesis of secretory IgM in Eμ-TCL1 B cells.
(A) XBP-1KO/Eμ-TCL1 B cells respond ineffectively to activation via the BCR. XBP-1WT/Eμ-TCL1 and XBP-1KO/Eμ-TCL1 B cells were treated with LPS for 3 days, stimulated with F(ab′)2 anti-mouse IgM to crosslink the BCR for indicated times, and lysed for analysis of indicated proteins by immunoblots. (B and C) WT B cells and CLL cells were isolated from 12-month-old WT, XBP-1WT/Eμ-TCL1 and XBP-1KO/Eμ-TCL1 mice. Purified cells were radiolabeled for 15 minutes, chased for indicated times, and lysed. Intracellular and extracellular IgM were immunoprecipitated from lysates (B) and culture medium (C), respectively, using an anti-κ antibody. Immunoprecipitates were analyzed on an SDS-PAGE gel. Data are representative of 3 independent experiments.
XBP-1 deficiency leads to increased surface expression of S1P1, but not other critical B cell markers in Eμ-TCL1 CLL cells.
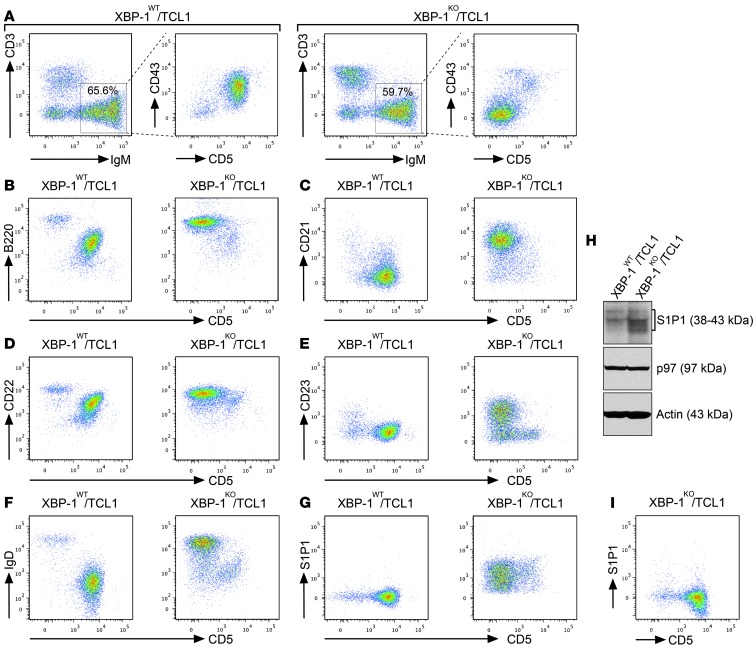
In addition to the BCR, CLL cells express critical surface proteins that contribute to their survival. To determine how XBP-1 deficiency contributes to the surface expression of B cell markers on Eμ-TCL1 CLL cells, we chose to investigate age-matched XBP-1WT/Eμ-TCL1 and XBP-1KO/Eμ-TCL1 mice, the spleens of which still contain CD5– precancerous B cells and CD5+ CLL cells. When Eμ-TCL1 CD5– B cells turned into CD5+ CLL cells, we observed the significantly increased surface expression of CD43 and decreased expression of B220, CD21, CD22, CD23, and IgD (Figure 3). There was little change in the surface expression of CD1d, CD49b, CD20, CD24, CD38, CD184, MHC class II molecules, CD25, and GL7 (Supplemental Figure 4). In contrast to MM cells, these CLL cells do not express CD138 (Supplemental Figure 4). Notably, sphingosine-1-phosphate receptor 1 (S1P1) was synthesized and expressed at the elevated levels on the surface of XBP-1KO/Eμ-TCL1 B cells (Figure 3, G and H). Although XBP-1KO/Eμ-TCL1 B cells eventually express similar levels of S1P1 when they turn into CD5+ CLL cells (Figure 3I), the initial increased expression of S1P1 as a result of XBP-1 deficiency may contribute to opposing homing signals and facilitate the egress of CLL cells from spleens and lymph nodes, leading to delayed leukemic progression (24).
Figure 3. XBP-1 deficiency delays immunophenotypic changes in leukemia.
(A) Splenocytes isolated from approximately 9-month-old XBP-1WT/Eμ-TCL1 and XBP-1KO/Eμ-TCL1 mice were stained with monoclonal antibodies against CD3, IgM, CD5, and CD43. The expression of CD43 on the surface of CD5– B cells and CD5+ CLL cells were analyzed on gated CD3–IgM+ B cell populations of the spleens. Data are representative of 3 independent experiments. (B–G) Splenocytes isolated from approximately 9-month-old XBP-1WT/Eμ-TCL1 and XBP-1KO/Eμ-TCL1 mice were stained with monoclonal antibodies against CD3, IgM, CD5, and an additional marker indicated in each panel: (B) B220, (C) CD21, (D) CD22, (E) CD23, (F) IgD, and (G) S1P1. The expressions of the indicated markers on the surface of CD5– B cells and CD5+ CLL cells were analyzed on gated CD3–IgM+ B cell populations of the spleens. Data are representative of 3 independent experiments. (H) B cells purified from XBP-1WT/Eμ-TCL1 and XBP-1KO/Eμ-TCL1 mice were stimulated with LPS for 3 days and lysed for analysis of the expression of S1P1, p97, and actin by immunoblots. (I) Splenocytes isolated from a 14-month-old XBP-1KO/Eμ-TCL1 mouse were stained with monoclonal antibodies against CD3, IgM, CD5, and S1P1 and similarly analyzed.
A potent IRE-1 inhibitor, B-H09, derived from chemical synthesis of 4μ8C analogs, interacts with IRE-1 in B cells.
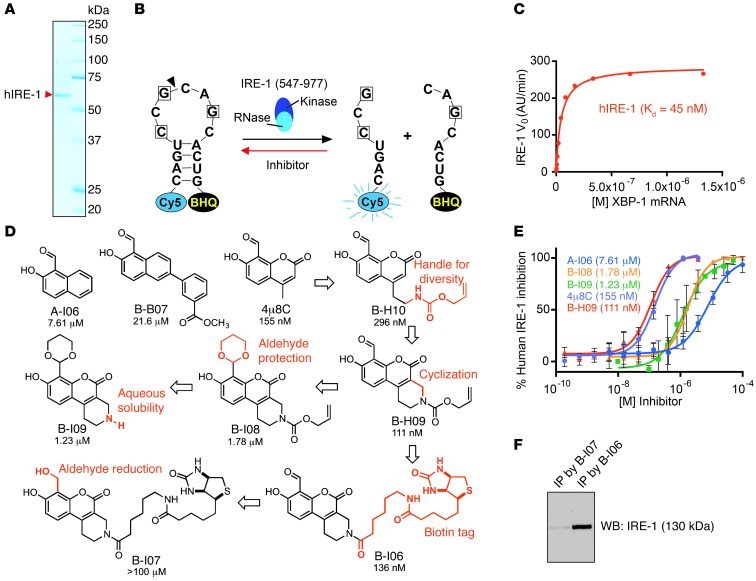
In an effort to develop novel IRE-1 RNase inhibitors with improved potency and cellular efficacy, we carried out the expression and purification of recombinant human IRE-1 for use in an in vitro fluorescence resonance energy transfer (FRET) suppression assay (25). The cytoplasmic kinase/RNase domain (aa 547–977) of IRE-1 was expressed as a puritin-His–tagged IRE-1 fusion protein in SF21 insect cells and purified by Ni-NTA affinity chromatography (Figure 4A). We evaluated the activity of our recombinant IRE-1 using a synthetic mRNA stem loop corresponding to the XBP1 substrate sequence. This stem loop incorporates a Cy5 fluorophore on its 5′ end and the black hole quencher (BHQ) on its 3′ end, resulting in fluorescence only upon site-specific cleavage by IRE-1 (Figure 4B). IRE-1 exhibits functional RNase activity with a Km value of 45 nM (Figure 4C).
Figure 4. Development of potent inhibitors to target the IRE-1/XBP-1 pathway.
(A) Recombinant human IRE-1 (hIRE-1) was expressed in insect cells and purified using Ni-NTA column chromatography. Purified hIRE-1 was analyzed by SDS-PAGE and stained with Coomassie Brilliant Blue G-250. (B) A diagram depicting the mini-XBP1 stem-loop RNA and its cleavage by hIRE-1. IRE-1 inhibitors block hIRE-1 from cleaving the XBP1 stem-loop RNA substrate. (C) The Michaelis-Menten curve for hIRE-1 showing catalytic RNase activity in a FRET assay. Initial reaction rates are plotted as a function of different XBP1 stem-loop RNA concentrations in the presence of 5 nM hIRE-1. (D) Structures of IRE-1 inhibitors with in vitro IC50 values obtained from FRET-suppression assays. (E) Dose-response curves were generated from FRET-suppression assays. Dose-response experiments were carried out a minimum of 3 times on different days, and data were plotted as mean ± SEM. The IC50 values were calculated from the mean inhibition value at each concentration. Shown here are representative curves for selected IRE-1 inhibitors. (F) LPS-stimulated XBP-1–deficient B cells were treated with B-I06 or B-I07 (control) for 24 hours and lysed for immunoprecipitations using an anti-biotin antibody and protein G–conjugated agarose. Immunoprecipitates were analyzed by SDS-PAGE and immunoblotted for IRE-1.
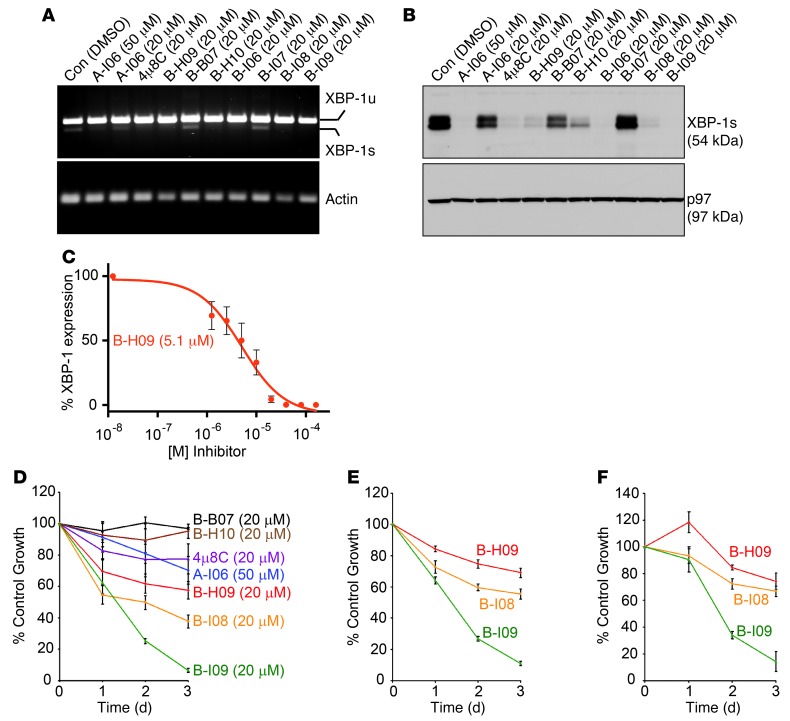
We first assayed salicylaldehyde-based compounds A-I06 and 4μ8C, which are known to potently inhibit XBP1 mRNA splicing by IRE-1 (1, 19). The IC50 value of each compound was calculated by the fluorescence readout, which was inversely correlated with the capability of a compound to inhibit IRE-1 from cleaving the XBP1 stem-loop substrate. The coumarin derivative 4μ8C exhibited an IC50 of 155 nM in the FRET suppression assay, while A-I06 displayed weaker activity in vitro. We then conducted the chemical synthesis of a library of A-I06 and 4μ8C analogs (26), of which B-H09 stands out as the most potent inhibitor, with an IC50 of 111 nM (Figure 4, D and E). The aldehyde moiety of each of these inhibitors is believed to be critical for inhibition of RNase function, allowing the highly specific formation of a Schiff base with lysine 907 in the RNase domain of IRE-1 (19, 20). Indeed, protection of the aldehyde group of B-H09 as an acid-labile 1,3-dioxane acetal (as in B-I08) resulted in a 16-fold drop in inhibitory activity.
We next prepared a biotin-tagged derivative, B-I06, in order to carry out direct binding experiments with endogenously expressed IRE-1 in B cells. In our FRET suppression assay, we confirmed that B-I06 essentially retains the potency of its parent compound in vitro (IC50 = 136 nM), while the reduced (primary alcohol) derivative B-I07 was found to be inactive. As confirmation that B-H09 interacts with IRE-1, we showed that B-I06, but not the negative control B-I07, can pull down IRE-1 from mouse B cell lysates via the use of a monoclonal anti-biotin antibody immobilized to protein G–conjugated agarose beads (Figure 4F).
IRE-1 inhibitors with masked aldehyde moieties exert stronger effects in suppressing XBP-1s and leukemic cell growth.
We and others have previously shown that A-I06 induces cell death in MM and CLL cells without imposing toxicity to normal B cells or to mice (1, 18). Consistent with the IC50 values derived from in vitro FRET suppression assay (Figure 4E), B-H09 is significantly more effective than A-I06 in inhibiting the splicing of XBP1 mRNA in human WaC3 CLL cells, as determined by RT-PCR (Figure 5A). At the protein level, B-H09 effectively blocks the expression of XBP-1s in LPS-stimulated B cells, while A-I06 requires a higher concentration to achieve complete inhibition (Figure 5B). We have also assessed the dose-dependent efficacy of B-H09 in suppressing XBP-1s expression in LPS-stimulated B cells and determined an approximate IC50 value of 5.1 μM by immunoblots followed by quantitation using densitometry (Figure 5C). To establish growth-inhibitory effects, we treated Eμ-TCL1 CLL cells with these inhibitors (20 μM) and found that B-H09 is more potent than A-I06 and the closely related coumarin derivative 4μ8C in inhibiting mouse CLL cell growth (Figure 5D). To optimize the cellular efficacy of B-H09, we tested whether the 1,3-dioxane group in B-I08 may serve as a putative prodrug moiety, thus avoiding potential interactions of the aldehyde with serum proteins in culture medium prior to cell entry. The aqueous solubility of B-I08 was also enhanced through removal of the allyloxycarbonyl group to generate B-I09. Although B-I08 and B-I09 exhibited weaker activity in the FRET suppression assay (Figure 4E), both were highly effective in inhibiting splicing of XBP1 mRNA in human WaC3 cells and the expression of XBP-1s in LPS-stimulated B cells (Figure 5, A and B). The water-soluble 1,3-dioxane derivative B-I09 was more effective than B-H09 and 4μ8C in suppressing the growth of mouse Eμ-TCL1 CLL cells (Figure 5D). B-I09 was the most potent inhibitor of the growth of CLL cells freshly isolated from human patients (Figure 5, E and F). We used LC-MS to confirm that the 1,3-dioxane–protecting group of B-I09 remains completely intact after 48-hour exposure to the FRET suppression assay buffer (pH 7.4) and remains greater than 50% intact after incubation in cell culture medium at 37°C for 24 hours (Supplemental Figure 5, A and B). These results suggest that the 1,3-dioxane moiety improves cellular uptake and may decompose to reveal the bioactive aldehyde once inside the cell. This decomposed compound, C-B06, has an IC50 of 248 nM and can inhibit splicing of XBP1 mRNA and expression of XBP-1s in human MEC1 and MEC2 CLL cell lines (Supplemental Figure 5, C–E).
Figure 5. IRE-1 inhibitors with masked aldehyde moieties potently suppress XBP-1s expression and leukemic growth.
(A) WaC3 CLL cells were treated with indicated compounds for 24 hours and lysed for RNA extraction. Human unspliced XBP1 (XBP1u), spliced XBP1 (XBP1s), and actin were detected by RT-PCR using specific primers. Data are representative of 3 experiments. (B) Mouse B cells were stimulated with LPS for 48 hours to allow for XBP-1s expression, and then treated with indicated inhibitors for 24 hours. Lysates were immunoblotted for XBP-1s and p97. Data are representative of 3 experiments. (C) LPS-stimulated B cells were treated with 0, 1.25, 2.5, 5, 10, 20, 40, 80, or 160 mM B-H09 for 24 hours. Equal amounts of lysates were immunoblotted for XBP-1s. The XBP-1s protein band intensity was determined using ImageJ. The percentage of inhibition was determined by comparing with the untreated group. Data from 3 experiments were plotted as mean ± SEM. (D) Mouse CD3–IgM+CD5+ Eμ-TCL1 CLL cells were treated with DMSO or indicated inhibitors for 3 days and subjected to XTT assays. Percentages of growth were determined by comparing inhibitor-treated with DMSO-treated groups. Data from 4 identical experimental groups were plotted as mean ± SD. Results are representative of 3 experiments. (E and F) Primary CLL cells from 2 human patients were treated with DMSO or indicated inhibitors (20 μM), subjected to XTT assays, and similarly analyzed. Data from 4 identical experimental groups were plotted as mean ± SD. Results are representative of 3 experiments.
Protein trafficking pathways are unaffected by inhibiting the IRE-1/XBP-1 pathway using B-I09.
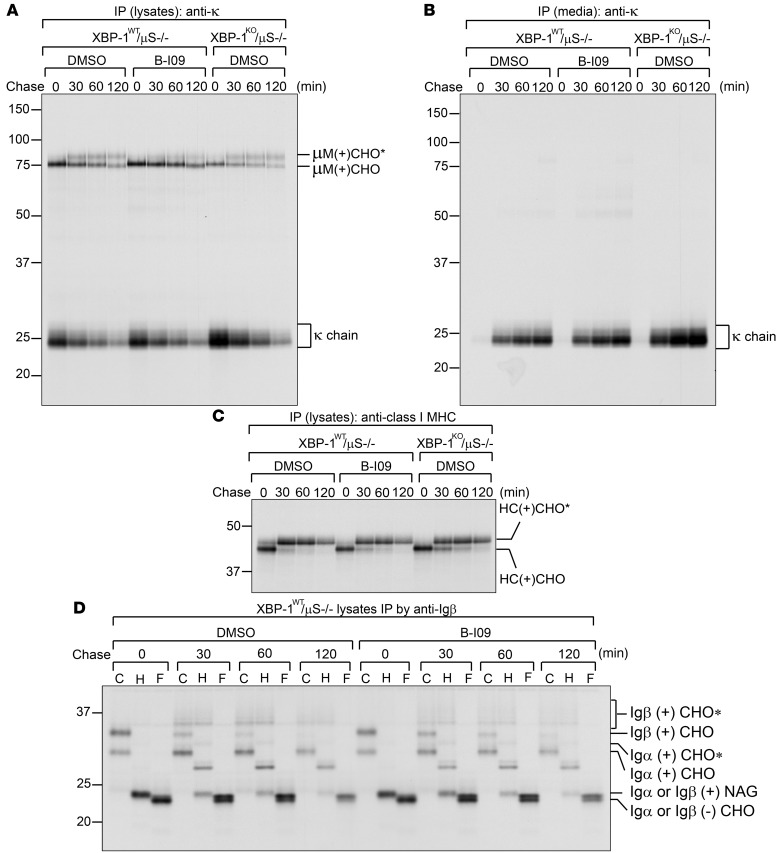
Secretory protein transport in mammalian cells posits an intricate process, as it requires coordination among chaperones, glycosyltransferases, GTPases, and vesicular transport systems (27). Minor defects in posttranslational modifications, folding, or assembly of a membrane protein can stall transport and lead to proteolysis. Thus, we tested whether B-I09 can impose adverse effects on this process. To investigate the trafficking of mIgM to the cell surface, we took advantage of μS KO B cells, which have been genetically manipulated to allow for the expression of only membrane-bound μ heavy chain (28). We stimulated B cells with LPS for 2 days to allow the expression of XBP-1s, treated these B cells for an additional 24 hours with B-I09 (20 μM) to inhibit the expression of XBP-1s, performed pulse-chase experiments, and immunoprecipitated IgM from cell lysates and culture medium using an anti-κ antibody. The surface display of mIgM is clearly not affected by treatment with B-I09, as evidenced by successful acquisition of complex glycans on membrane-bound μ heavy chains (Figure 6A). The B-I09–treated μS KO B cells also produce comparable amounts of membrane-bound μ chains and κ light chains, and the latter can be secreted into culture medium (Figure 6B). In addition, B-I09–treated μS KO and WT B cells synthesize, assemble, and present class I MHC molecules, Igα, and Igβ to their surface normally (Figure 6, C and D, and Supplemental Figure 6, C and D). These data suggest that B-I09 does not target critical cellular mechanisms involved in protein transport. Similar to genetic XBP-1–deficient CLL cells, B-I09–treated B cells are ineffective in synthesizing secretory μ chains (Supplemental Figure 6, A and B, and Figure 2, B and C). B-I09 also phenocopies genetic Xbp1 knockout by upregulating the expression levels of IRE-1 (Supplemental Figure 7 and Figure 1, A and F).
Figure 6. B-I09 does not affect synthesis, assembly and transport of critical B cell integral membrane proteins.
(A and B) To reveal the effect of B-I09 on membrane-bound IgM (mIgM), XBP-1WT/μS KO B cells were stimulated with LPS for 2 days and subsequently treated with DMSO (control) or B-I09 (20 μM) for 1 additional day. DMSO-, or B-I09–treated XBP-1WT/μS KO B cells and DMSO-treated XBP-1KO/μS KO B cells were radiolabeled for 15 minutes, chased for indicated time, and lysed. Intracellular mIgM and κ light chain were immunoprecipitated from lysates using an anti-κ antibody (A). Secreted free κ chains were immunoprecipitated from culture medium using an anti-κ antibody (B). Immunoprecipitates were analyzed by SDS-PAGE. Data are representative of 3 independent experiments. (C) Lysates similar to those in A were immunoprecipitated using an antibody against the class I MHC heavy chain (HC), and immunoprecipitates were analyzed by SDS-PAGE. Data are representative of 3 independent experiments. (D) Using lysates similar to those in A, we performed immunoprecipitations using an anti-Igβ antibody to retrieve the Igα/Igβ heterodimers. Immunoprecipitated Igα/Igβ proteins were eluted from the beads and treated with endo-H or PNGase F before being analyzed by SDS-PAGE. μM, membrane-bound μ chain; CHO, high mannose-type glycans; CHO*, complex-type glycans; NAG, N-acetylglucosamines. Data are representative of 3 independent experiments.
B-I09 suppresses leukemic progression without imposing systemic toxicity in mice.
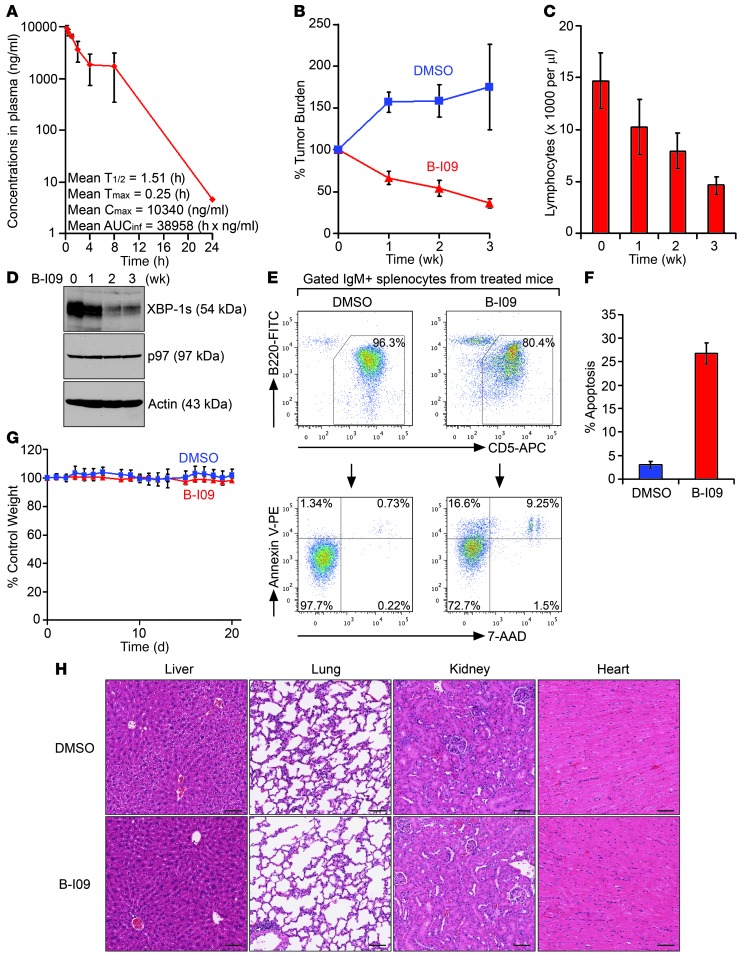
To determine appropriate dosing of B-I09 in vivo, mice were intraperitoneally injected with B-I09 (50 mg/kg). B-I09 has a half-life of approximately 1.5 hours and reaches its peak concentration of approximately 39 μM in mouse plasma serum 15 minutes after administration (Figure 7A). We selected tumor-bearing Eμ-TCL1 mice in which CLL cells comprise the majority of the lymphocytes in the peripheral blood (Supplemental Figure 8), treated these mice with 50 mg/kg B-I09 on the first 5 days of each week for 3 weeks, and observed clear CLL regression in the peripheral blood (Figure 7B). After treatment with B-I09 for 3 weeks, the numbers of lymphocytes in the peripheral blood dropped close to the normal range of 500∼5000 cells/μl (Figure 7C). We also confirmed by immunoblots that B-I09 indeed inhibits the expression of XBP-1s in CLL cells collected from the peripheral blood of B-I09–injected Eμ-TCL1 mice (Figure 7D). Since CLL cells proliferate in secondary lymphoid organs and are protected from cell death through interactions with microenvironments, we examined whether B-I09 has an effect on CLL cells residing in the spleen. When comparing mice injected with DMSO or B-I09 (50 mg/kg) for 3 weeks, we observed significantly increased apoptotic CD5+ CLL cells in the spleens of B-I09–injected mice (Figure 7, E and F). Injection of B-I09 has no apparent adverse effects or toxicity to mice, as evidenced by no significant weight loss or apparent histological changes to vital organs, such as liver, lungs, kidney, and heart, after 3 weeks of treatment (Figure 7, G and H).
Figure 7. B-I09 suppresses leukemic growth and is well tolerated in mice.
(A) Pharmacokinetic analysis of B-I09 is described in Methods (n = 3; mean ± SEM). The terminal half-life (T1/2), time of peak concentration (Tmax), maximum concentration (Cmax), and AUC versus time calculated using zero to infinity (AUCinf) of B-I09 in mouse plasma are indicated. (B) CLL-bearing Eμ-TCL1 mice were intraperitoneally injected with DMSO (n = 3) or B-I09 (50 mg/kg in DMSO, n = 8) daily for the first 5 days of each of 3 weeks. Blood was collected to measure lymphocyte numbers using a HemaTrue Hematology Analyzer (HESKA). Data were compared with lymphocyte counts prior to B-I09 injections and plotted as mean ± SEM. (C) Lymphocyte counts in the peripheral blood of B-I09–treated Eμ-TCL1 mice (n = 8) were plotted as mean ± SEM. (D) PBMCs from B-I09 mice, before and after injections, were lysed for analysis of indicated proteins. (E) Splenocytes from DMSO- or B-I09–injected Eμ-TCL1 mice were stained with IgM-PE-Cy7, B220-FITC, CD5-APC, annexin V–PE, and 7-AAD. Gated IgM+B220+CD5+ splenic CLL cells were analyzed for annexin V– and/or 7-AAD–positive populations. (F) Percentages of apoptotic cells in gated IgM+B220+CD5+ CLL populations from spleens of DMSO-injected (n = 3) or B-I09–injected (n = 8) Eμ-TCL1 mice were plotted as mean ± SEM. (G) Weight of DMSO-injected (n = 3) or B-I09–injected (n = 8) Eμ-TCL1 mice was plotted as mean ± SD. (H) Paraffin-embedded sections of indicated organs from Eμ-TCL1 mice receiving 3 weeks of injections with DMSO or B-I09 were stained with H&E. Scale bars: 80 μm.
Inhibition of the IRE-1/XBP-1 pathway compromises BCR signaling and synergizes with ibrutinib to induce apoptosis in B cell cancer.
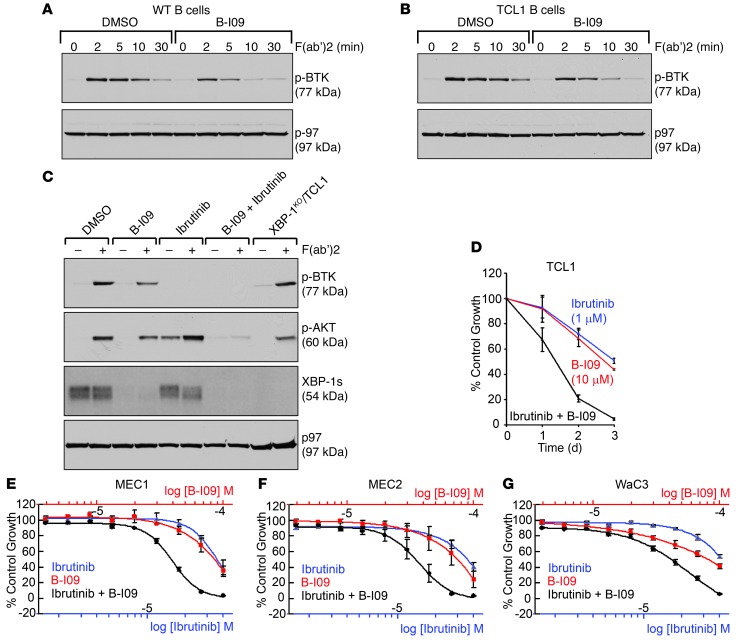
Genetic XBP-1 deficiency is known to compromise BCR signaling (ref. 21 and Figure 2A), which is crucial for the survival of CLL (22, 23). To examine whether B-I09 can phenocopy genetic Xbp1 knockout in compromising BCR signaling, we cultured B cells from WT and Eμ-TCL1 mice in LPS (20 μg/ml) and B-I09 (20 μM) for 48 hours. These cells were subsequently stimulated with F(ab′)2 anti-mouse IgM and analyzed for the phosphorylation of BTK, a central BCR-signaling molecule serving as a promising target for the treatment of CLL. Similar to XBP-1–deficient B cells, B-I09–treated WT and Eμ-TCL1 B cells exhibited compromised BCR signaling, as evidenced by reduced phosphorylation of BTK (Figure 8, A–C, and Figure 2A). In recent studies, the treatment of mouse TCL1 CLL and human CLL with ibrutinib (a specific inhibitor of BTK) has led to significantly improved prognosis (11, 29–31). Since inhibition of the IRE-1/XBP-1 pathway results in reduced phosphorylation of BTK, we tested whether B-I09 can aggrandize the effect of ibrutinib. While mouse Eμ-TCL1 CLL cells were treated with B-I09 in combination with ibrutinib, we observed enhanced growth inhibition, possibly because such a combination can effectively block phosphorylation of AKT upon BCR activation on LPS-stimulated Eμ-TCL1 B cells (Figure 8, C and D). Of note, TCL1 drives leukemia via activation of AKT (32, 33).
Figure 8. B-I09 mimics genetic Xbp1 knockout in compromising BCR signaling, and exerts a strong synergistic effect with ibrutinib.
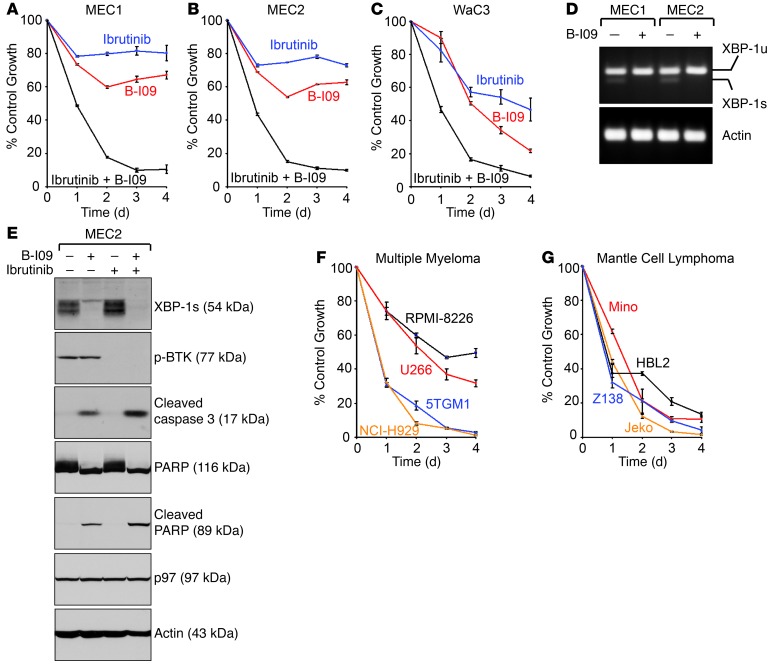
(A) WT and (B) Eμ-TCL1 B cells were treated with DMSO or B-I09 (20 μM) in the presence of LPS (20 μg/ml) for 2 days, stimulated with F(ab′)2 anti-mouse IgM for indicated times to activate the BCR, and lysed for analysis of indicated proteins by immunoblots. Data are representative of 3 experiments. (C) Eμ-TCL1 B cells were stimulated with LPS for 2 days and subsequently treated with DMSO, B-I09 (20 μM), ibrutinib (10 μM), or B-I09 in combination with ibrutinib for another day. LPS-stimulated XBP-1KO/Eμ-TCL1 B cells serve as controls. After stimulation with F(ab′)2 anti-mouse IgM for 5 minutes, cells were lysed for analysis of indicated proteins by immunoblots. Data are representative of 3 experiments. (D) Eμ-TCL1 CLL cells were treated with DMSO (control), B-I09 (10 μM), ibrutinib (1 μM), or a combination of both for 3 days and subjected to XTT assays. Percentages of growth were determined by comparing inhibitor-treated groups with control groups. Data from 4 identical experimental groups were plotted as mean ± SD. Results are representative of 3 experiments. (E–G) Dose-dependent growth inhibition curves of MEC1, MEC2, and WaC3 human CLL cells treated for 48 hours with B-I09, ibrutinib, or a combination were determined by CellTiter Blue assays. The concentration ranges for B-I09 and ibrutinib were 3.9 μM∼100 μM and 1.56 μM∼40 μM, respectively. Data from 2 experimental repeats were plotted as mean ± SEM.
When ibrutinib was combined with B-I09 (dual serial dilution) to treat human MEC1, MEC2, and WaC3 CLL cell lines for 48 hours, we determined GI50 (50% growth inhibition) concentrations for ibrutinib (between 10∼20 μM) and B-I09 (between 30∼40 μM) and observed a strong synergistic effect in suppressing human CLL cell growth (Figure 8, E–G, and Supplemental Table 1). When treating these human CLL cell lines with 10 μM ibrutinib, 20 μM B-I09, or a combination of the 2 for a course of 4 days, we observed that more than 80% growth inhibition occurs within the first 2 days of combined treatment (Figure 9, A–C). Although MEC1 and MEC2 cells become resistant to treatment with ibrutinib or B-I09 alone, both cell lines are highly sensitive to the combined treatment (Figure 9, A and B). WaC3 cells are sensitive to ibrutinib, B-I09, and a combination (Figure 9C). We confirmed that the expression of XBP-1s is indeed suppressed by B-I09 in all 3 human CLL cell lines (Figure 9D and Figure 5A) and that the growth inhibition is a result of apoptosis, as evidenced by proteolytic cleavage of procaspase-3 and PARP in B-I09–treated human CLL cells (Figure 9E).
Figure 9. B-I09 synergizes with ibrutinib to induce apoptosis in human CLL, MM, and MCL cell lines.
(A–C) MEC1, MEC2 and WaC3 human CLL cells were treated with DMSO (control), B-I09 (20 μM), ibrutinib (10 μM), or a combination of both for 4 days, and subjected to XTT assays. Percentages of growth were determined by comparing inhibitor-treated groups with control groups. Data from 4 identical experimental groups were plotted as mean ± SD. Results are representative of 3 independent experiments. (D) MEC1 and MEC2 human CLL cells were treated with DMSO (control) or B-I09 (20 μM) for 48 hours and lysed for RNA extraction and RT-PCR. Human XBP1u, XBP1s, and actin were detected using specific primers. Results are representative of 3 independent experiments. (E) Human MEC2 CLL cells were treated for 72 hours with DMSO (control), B-I09 (20 μM), ibrutinib (10 μM), or the combination of B-I09 and ibrutinib. Cells were lysed for analysis of indicated proteins by immunoblots. Data are representative of 3 independent experiments. (F and G) MM cell lines (F) and MCL cell lines (G) were treated with DMSO or the combination of B-I09 (20 μM) and ibrutinib (10 μM) for a course of 4 days and subjected to XTT assays at the end of each day. Percentages of growth were determined by comparing treated groups with control groups. Each data point derived from 4 independent groups receiving exactly the same treatment was plotted as mean ± SD. Results are representative of 3 independent experiments.
The IRE-1/XBP-1 pathway is important for the survival of MM malignancies derived from plasma cells (18, 34). We examined several MM cell lines (mouse 5TGM1, human RPMI-8226, human U266, and human NCI-H929) and showed that B-I09 (20 μM) can effectively suppress the expression of XBP-1s (Supplemental Figure 9A). The combination of B-I09 with ibrutinib exerts a synergistic cytotoxic effect against all 4 MM cell lines (Figure 9F, Supplemental Figure 9, B–E, and Supplemental Table 1). Mantle cell lymphoma (MCL) is an incurable non-Hodgkin lymphoma derived from mature B cells in the mantle zone. Since the role of the IRE-1/XBP-1 pathway in MCL is completely unknown, we examined 4 human MCL cell lines (HBL2, Jeko, Mino, and Z138) for the expression of XBP-1s and discovered that XBP-1s is constitutively expressed by all 4 human MCL cell lines (Supplemental Figure 10A). Treatment with 20 μM B-I09 for 24 hours effectively inhibited the expression of XBP-1s in these MCL cells (Supplemental Figure 10A). The combination of B-I09 with ibrutinib similarly exerted synergistic cytotoxicity against all 4 human MCL cell lines (Figure 9G, Supplemental Figure 10, B–E, and Supplemental Table 1). The synergistic cytotoxicity is a result of apoptosis in both MM and MCL cells (Supplemental Figure 11, A and B).
Discussion
Overexpression of XBP-1s in mouse B cells leads to monoclonal gammopathy of undetermined significance (MGUS) (35), a precursor syndrome of MM. Inhibitors that block the splicing of XBP1 mRNA by IRE-1 were discovered through screening chemical libraries (17–19). While the inhibitor STF-083010 (or A-I06) shows promising results in the treatment of MM and CLL (1, 18), we show here that B cell–specific deletion of the Xbp1 gene can decelerate leukemic progression in mice. These data provide the strongest validation of the IRE-1/XBP-1 pathway as a target for therapeutic intervention in B cell cancer (Figure 1). By chemical synthesis, we developed a “prodrug” inhibitor, B-I09, which is highly effective in suppressing the expression of XBP-1s and leukemic cell growth in vitro and in mice (Figures 4, 5, and 7).
The expression of XBP-1s is crucial for the survival of B cell leukemia and lymphoma (Figure 5, D–F, Figures 7–9, and Supplemental Figures 9–11). Notably, CLL cells freshly purified from spleens of Eμ-TCL1 mice produced significantly increased amounts of secretory IgM relative to B cells isolated from spleens of WT mice (Figure 2, B and C). Deletion of the Xbp1 gene from Eμ-TCL1 CLL cells specifically inhibited synthesis of secretory IgM but not membrane-bound IgM or class I and class II MHC molecules (Figure 2, B and C, and Supplemental Figure 3). Eμ-TCL1 CLL cells thus require the expression of XBP-1s to support the production of secretory IgM.
Deletion or inhibition of the IRE-1/XBP-1 pathway can compromise BCR signaling (Figure 2A and Figure 8, A–C). Because the survival of mature B cell cancer relies on activation of the BCR (22, 36), this makes targeting the IRE-1/XBP-1 pathway even more attractive for the therapy of B cell leukemia and lymphoma. Targeting BCR signaling in CLL and MCL using ibrutinib is considered to be one of the most exciting breakthroughs in B cell cancer therapy (29–31, 37). The pharmacological synergism between B-I09 and ibrutinib in inducing apoptosis of human CLL, MCL, and MM cells suggests a promising treatment strategy for B cell cancer (Figures 8 and 9, Supplemental Figures 9–11, and Supplemental Table 1).
Methods
Mice.
The XBP-1–deficient CLL mouse model (XBP-1KO/Eμ-TCL1) was generated by crossing CD19-Cre Xbp1flox/flox mice (21) with Eμ-TCL1 mice (6). These colonies, together with μS KO mice (with B cells lacking sIgM) (28), were maintained at our animal facility strictly following the guidelines provided by the University of South Florida and the H. Lee Moffitt Cancer Center Committees on Animal Care.
Immunofluorescent staining and flow cytometric analysis of mouse splenocytes and purified B and CLL cells.
Splenocytes were obtained from mice by mashing the spleens through cell strainers followed by RBC lysis (QIAGEN). Mouse B cells, μS KO B cells, and Eμ-TCL1 CLL cells were purified from mouse spleens by negative selection using Pan-B Magnetic Beads (Miltenyi Biotech). After nonspecific blocking for 30 minutes using FBS, cell-surface staining was achieved by incubating cells at 4°C for 30 minutes with the following anti-mouse antibodies: CD3 (145-2C11; Biolegend), IgM (e-Bioscience), B220 (RA3-6B2; BD Pharmingen), CD5 (53-7.3; eBioscience), CD1d (1B1; Biolegend), CD20 (A1SB12; e-Bioscience), CD21 (7E9; Biolegend), CD22 (OX-97; Biolegend), CD23 (B3B4; Biolegend), CD24 (M1/69; Biolegend), CD25 (PC61; Biolegend), CD38 (90; Biolegend), CD43 (eBioR2/60; e-Bioscience), CD49b (DX5; Biolegend), CD138 (281-2; BD Pharmingen), CD184 (2B11; e-Bioscience), MHC II (M5/114; Biolegend), S1P1 (713412; R&D), GL7 (GL7; Biolegend), and IgD (11-26c.2a; Biolegend). Viability staining was accomplished using DAPI exclusion during acquisition. Acquisition of B cell and CLL cell populations was performed on a LSRII Cytometer (BD Biosciences) harboring a custom configuration for the H. Lee Moffitt Cancer Center and Research Institute. Mid-range Spherotech FL1 Fluorescent Rainbow Beads (BD Biosciences) were used to maintain consistent gains for all parameters across different time points. Cytometry data was analyzed using FlowJo software, version 7.6.1 (Tree Star Inc.).
Antibodies and reagents.
Antibodies to TCL1 (Cell Signaling), IRE-1 (Cell Signaling), XBP-1 (Santa Cruz Biotechnology Inc.), Syk (Cell Signaling), phospho-Syk (Tyr525/526) (Cell Signaling), phospho-BTK (Cell Signaling), S1P1 (Santa Cruz Biotechnology Inc.), phospho-AKT (Invitrogen), cleaved caspase-3 (Cell Signaling), PARP (Cell Signaling), cleaved PARP (Cell Signaling), p97 (Fitzgerald), actin (Sigma-Aldrich), μ (SouthernBiotech), and κ (SouthernBiotech) were obtained commercially. Polyclonal antibodies against Igβ, class I MHC, and class II MHC molecules were generated in rabbits. LPS was procured from Sigma-Aldrich.
Cell culture.
Primary human CLL cells were obtained from patients at the H. Lee Moffitt Cancer Center and Research Institute by Pinilla-Ibarz. Primary mouse B cells, mouse Eμ-TCL1 CLL cells, primary human CLL cells, human CLL cell lines (MEC1, MEC2 and WaC3), MM cell lines (mouse 5TGM1, human RPMI-8226, human U266, and human NCI-H929), and human MCL cell lines (Mino, Jeko, and HBL2) were all cultured in the RPMI 1640 medium (Gibco; Invitrogen) supplemented with 10% heat-inactivated FBS, 2 mM l-glutamine, 100 U/ml penicillin G sodium, 100 μg/ml streptomycin sulfate, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, and 0.1 mM β-mercaptoethanol (β-ME). Human MCL cell line Z138 was cultured in the IMDM medium (Gibco; Invitrogen) with the same supplemental nutrients.
Protein isolation and immunoblotting.
Cells were lysed using RIPA buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA) supplemented with protease inhibitors (Roche). Protein concentrations were determined by BCA assays (Pierce). Samples were boiled in SDS-PAGE sample buffer (62.5 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 0.1% bromophenol blue) with β-ME and analyzed by SDS-PAGE. Proteins were transferred to nitrocellulose membranes, blocked in 5% nonfat milk (wt/vol in PBS), and immunoblotted with indicated primary antibodies and appropriate horseradish peroxidase–conjugated secondary antibodies. Immunoblots were developed using Western Lighting Chemiluminescence Reagent (PerkinElmer).
BCR activation and phosphorylation assay.
XBP-1WT/Eμ-TCL1 B cells, XBP-1KO/Eμ-TCL1 B cells, B-I09–treated WT B cells, or B-I09–treated Eμ-TCL1 B cells were suspended in RPMI serum-free medium supplemented with 25 mM Hepes, stimulated with F(ab′)2 fragments of the goat anti-mouse IgM antibody (20 μg/ml) (SouthernBiotech) for 2 minutes, and lysed immediately by adding ice-cold lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA) supplemented with protease inhibitor cocktail (Roche), 4 mM sodium pyrophosphate, 2 mM sodium vanadate, and 10 mM sodium fluoride. The lysates were analyzed by SDS-PAGE. Phosphorylated proteins of interest were detected by immunoblots using phospho-specific antibodies.
Pulse-chase experiments, immunoprecipitation, protein deglycosylation, and SDS-PAGE.
Eμ-TCL1 CLL cells and primary mouse B cells were starved in methionine- and cysteine-free medium containing dialyzed serum for 1 hour, then pulse-labeled with 250 μCi/ml [35S]-methionine and [35S]-cysteine (PerkinElmer) for 15 minutes. After labeling, cells were incubated in chase medium containing unlabeled methionine (2.5 mM) and cysteine (0.5 mM). At the end of each chase interval, cells were lysed in RIPA buffer containing protease inhibitors. Precleared lysates were incubated with a primary antibody and Protein G–agarose beads (Sigma-Aldrich). Bead-bound proteins were eluted using glycoprotein-denaturing buffer (0.5% SDS, 1% β-ME) or reducing Laemmli SDS-PAGE sample buffer. Enzymatic deglycosylation of proteins was achieved by denaturation of the immunoprecipitates in glycoprotein-denaturing buffer at 95°C for 10 minutes, followed by addition of sodium citrate (pH 5.5) to a final concentration of 50 mM and incubation with Endo H (New England Biolabs) at 37°C for 2 hours. Alternatively, sodium phosphate (pH 7.5) and NP-40 were added to the denatured cell lysates to a final concentration of 50 mM and 1%, respectively, and the mixture was incubated with PNGase F (New England Biolabs) at 37°C for 2 hours. Protein samples were then analyzed by SDS-PAGE followed by fluorography.
RT-PCR.
Total RNA was isolated using TRIzol reagent (Invitrogen). Complementary DNA was synthesized from RNA using Superscript II Reverse Transcriptase (Invitrogen). The following sets of primers were used together with Platinum Taq DNA polymerase (Invitrogen) in PCR to detect the expression of the following: human XBP1 (GAGTTAAGACAGCGCTTGGG and ACTGGGTCCAAGTTGTCCAG); human actin (CTGAGCGTGGCTACTCCTTC and GGCATACAGGTCCTTCCTGA); mouse Xbp1 (GATCCTGACGAGGTTCCAGA and ACAGGGTCCAACTTGTCCAG); and mouse actin (AGCCATGTACGTAGCCATCC and CTCTCAGCTGTGGTGGTGAA).
Recombinant human IRE-1 expression and purification.
Expression of 59.2 kD polyhistidine-tagged puritin–hIRE-1 fusion protein was carried out in SF21 cells using the Bac to Bac expression system (Invitrogen) according to manufacturer specifications. An 8X-His-puritin sequence was fused to the N-terminal end of the cytoplasmic kinase/RNase domain of human IRE-1 (aa. 547-977) in the pFastbacDual-PBL expression vector and included a PreScission protease cleavage site in the linker. Frozen insect cell paste (1 g) was suspended in 8 ml lysis buffer (50 mM Tris/HCl pH 8.0, 300 mM NaCl, 5 mM BME, 10 mM imidazole) containing a protease inhibitor tablet and lysed using sonication. After removal of the cell debris via centrifugation, the supernatant was applied to an Ni (NTA) column (5 ml). After washing untagged protein by flushing with 10 column volumes of lysis buffer, the target protein was eluted using a linear imidazole gradient (15 column volumes, 10–300 mM). Fractions were analyzed via SDS-PAGE. Pooled protein-containing fractions were concentrated and rebuffered into 50 mM Tris, pH 8.0, 150 mM NaCl, and 1 mM DTT via ultrafiltration. Typically, 1 liter of insect cell culture yielded 3 mg of recombinant 8x-His-puritin-hIRE-1 following Ni(NTA) column purification.
In vitro FRET-suppression assay.
The endoribonuclease activity of recombinant hIRE-1 was assayed by incubation of 50 μl of 10 nM hIRE-1 and 50 μl of various concentrations (0.01–1 μM) of fluorescently tagged XBP-1 RNA stem loop (5′-Cy5-CAGUCCGCAGCACUG-BHQ-3′, obtained from Sigma-Aldrich) in assay buffer (20 mM HEPES, pH 7.5, 50 mM KOAc, 0.5 mM MgCl2, 3 mM DTT, 0.4% PEG, and 5% DMSO) for up to 2 hours at room temperature in a black 96-well plate. Fluorescence was read at various time points using a Biotek Synergy H1 plate reader with excitation and emission at 620 nm and 680 nm, respectively. The Km of purified recombinant hIRE-1 was determined to be 45 nM using the Michaelis-Menten kinetic model. Inhibition of RNA cleavage by small molecules was determined by preincubation of 40 μl of 15 nM hIRE-1 with various concentrations of compounds (40 μl) in assay buffer for 30 minutes at room temperature. A 150 nM solution of fluorescent XBP-1 RNA (40 μl) was then added to each well and the reaction allowed to proceed for 2 hours before reading fluorescence as described above. Final concentrations of hIRE-1 and XBP-1 RNA were 5 nM and 50 nM, respectively. All fluorescence readings were corrected using background values from wells containing only 120 μl of 50 nM XBP-1 RNA. Dose-response experiments were carried out a minimum of 3 times on different days and IC50 values calculated from the mean inhibition value at each concentration.
Chemical inhibitors.
A-I06 (2-hydroxynaphthaldehyde) and 4μ8C (7-hydroxy-4-methyl-2-oxo-2H-chromene-8-carbaldehyde) are commercially available and were obtained from Sigma-Aldrich and Matrix Scientific, respectively. All other IRE-1 inhibitors were chemically synthesized in-house as described in the Supplemental Information. Ibrutinib was prepared from commercially available 4-aminopyrazolo-(3,4-d)pyrimidine (Sigma-Aldrich) using a modification to the known synthetic route (38).
Pharmacokinetics.
In vivo pharmacokinetics studies were carried out at Agilux Laboratories Inc. using male CD-1 mice (Charles River Laboratories). B-I09 was administered intraperitoneally at 50 mg/kg as a solution in DMSO to a group of 3 male CD-1 mice. Compound blood was collected via tail snip into calibrated microvette tubes containing K2EDTA at 0.25, 0.5, 1, 2, 4, 8, and 24 hours after dosing. Blood samples were stored on wet ice until processed to plasma by centrifugation within 1 hour of collection. All plasma samples were transferred to 96-well plates and stored at –80°C until analyzed for B-I09 concentration via LC/MS/MS using a C18 RP-HPLC column (2.1 × 50 mm) and an acetonitrile/water (with 0.1% formic acid) mobile phase elution gradient. B-I09 was quantified by analysis with API 5500 TurboIonSpray–positive ion MS detection. A noncompartmental model was applied to calculate pharmacokinetic parameters using WinNonlin 4.2 software.
Compound synergism.
The activity levels of compounds alone and in combination were determined by a high-throughput CellTiter-Blue (Promega Corp.) cell viability assay. Cell viability measurement is based on the ability of living cells to convert resazurin dye into the fluorescent resorufin. Cells (3 × 103) were plated in each well of 384-well plates using a Precision XS liquid handling station (Bio-Tek Instruments Inc.). A liquid handling station was then used to serially dilute all drugs in medium, and 5 μl was added to 4 replicate wells; an additional 4 control wells received a diluent control without drug. At the end of the incubation period with drugs, 5 μl of CellTiter-Blue reagent were added to each well. The fluorescence of the product of viable cells’ bioreduction, resorufin (579 nm excitation/584 nm emission), was measured with a Synergy 4 microplate reader (Bio-Tek Instruments Inc). The fluorescence data were transferred to Microsoft Excel to calculate the percentage of viability. We determined IC50 values using a sigmoidal equilibrium model regression and XLfit version 5.2 (ID Business Solutions Ltd.). The IC50 values obtained from single-drug cell viability assays were used to design subsequent drug-combination experiments.
For drug-combination experiments, the cell viability assays were performed as described above, and the results were analyzed for synergistic, additive, or antagonistic effects using the combination index (CI) method developed by Chou (39). For the application of this method, the drug concentration dilutions were used at fixed dose molar ratios based on the IC50 levels of each drug obtained from preliminary experiments. The dose-effect curve for each drug alone was determined based on experimental observations using the median-effect principle and then compared with the effect achieved with a combination of the 2 drugs to derive a CI value. This method involves plotting dose-effect curves for each agent and their combination using the following median-effect equation: fa/fu = (D/Dm)m, where D is the dose of the drug, Dm is the dose required for a 50% effect (equivalent to IC50), fa and fu are the affected and unaffected fractions, respectively (fa = 1 – fu), and m is the exponent signifying the sigmoidicity of the dose-effect curve. XLfit software was used to calculate the values of Dm and m. The CI used for the analysis of the drug combinations was determined by the isobologram equation for mutually nonexclusive drugs that have different modes of action: CI = (D)1/(Dx)1 + (D)2 /(Dx)2 + (D)1(D)2/(Dx)1(Dx)2, where (Dx)1 and (Dx)2 in the denominators are the doses (or concentrations) for D1 (drug 1) and D2 (drug 2) alone that gives 75%, 90%, and 95% inhibition, whereas (D)1 and (D)2 in the numerators are the doses of drug 1 and drug 2 in combination that also inhibited 75%, 90%, and 95% (i.e., isoeffective). CI < 1, CI = 1, and CI > 1 indicate synergism, additive effects, and antagonism, respectively.
Cell proliferation XTT assays.
Appropriate numbers of cells were suspended in phenol red–free culture medium, seeded in 96-well cell culture plates, and treated with indicated IRE-1 inhibitors (20 μM, unless indicated otherwise), PCI-32765 (ibrutinib; 10 μM), or a combination. Every 24 hours after the treatment, cells were spun down and proliferation was assessed by XTT assays (Roche) according to the manufacturer’s instructions. Briefly, 50 μl XTT labeling reagent, 1 μl electron-coupling reagent, and 100 μl phenol red–free culture medium was combined and applied to each well of the 96-well plates. Cells were then incubated for 4 hours in a CO2 incubator to allow for the yellow tetrazolium salt XTT to be cleaved by mitochondrial dehydrogenases of metabolic active cells to form the orange formazan dye, which can be quantified at 492 nm using a BioTek Synergy H1 MicroPlate Reader.
Statistics.
Kaplan-Meier analysis was used to evaluate mouse survival data. A P value of less than 0.05 was considered significant.
Study approval.
All experiments involving the use of mice were performed following protocols approved by the Institutional Animal Care and Use Committee at the University of South Florida. Patients providing cells gave informed consent per IRB guidelines.
Supplementary Material
Acknowledgments
This study was partially supported by awards from the Miles for Moffitt Foundation (to C.C.A. Hu and J.R. Del Valle), the Leukemia Research Foundation (to C.C.A. Hu), the American Cancer Society (to C.C.A. Hu), and the NIH/NCI (grant 1R01CA163910 to C.C.A. Hu). We thank Carlo M. Croce for providing us with Eμ-TCL1 mice, and Eduardo M. Sotomayor for useful discussion.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article:J Clin Invest. 2014;124(6):2585–2598. doi:10.1172/JCI73448.
References
- 1.Kriss CL, et al. Overexpression of TCL1 activates the endoplasmic reticulum stress response: a novel mechanism of leukemic progression in mice. Blood. 2012;120(5):1027–1038. doi: 10.1182/blood-2011-11-394346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen X, et al. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 2001;107(7):893–903. doi: 10.1016/S0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- 3.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107(7):881–891. doi: 10.1016/S0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 4.Calfon M, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415(6867):92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 5.Bertilaccio MT, et al. Xenograft models of chronic lymphocytic leukemia: problems, pitfalls and future directions. Leukemia. 2013;27(3):534–540. doi: 10.1038/leu.2012.268. [DOI] [PubMed] [Google Scholar]
- 6.Bichi R, et al. Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proc Natl Acad Sci U S A. 2002;99(10):6955–6960. doi: 10.1073/pnas.102181599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herling M, et al. TCL1 shows a regulated expression pattern in chronic lymphocytic leukemia that correlates with molecular subtypes and proliferative state. Leukemia. 2006;20(2):280–285. doi: 10.1038/sj.leu.2404017. [DOI] [PubMed] [Google Scholar]
- 8.Yan XJ, et al. B cell receptors in TCL1 transgenic mice resemble those of aggressive, treatment-resistant human chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2006;103(31):11713–11718. doi: 10.1073/pnas.0604564103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson AJ, et al. Characterization of the TCL-1 transgenic mouse as a preclinical drug development tool for human chronic lymphocytic leukemia. Blood. 2006;108(4):1334–1338. doi: 10.1182/blood-2005-12-011213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suljagic M, et al. The Syk inhibitor fostamatinib disodium (R788) inhibits tumor growth in the Emu- TCL1 transgenic mouse model of CLL by blocking antigen-dependent B-cell receptor signaling. Blood. 2010;116(23):4894–4905. doi: 10.1182/blood-2010-03-275180. [DOI] [PubMed] [Google Scholar]
- 11.Ponader S, et al. The Bruton tyrosine kinase inhibitor PCI-32765 thwarts chronic lymphocytic leukemia cell survival and tissue homing in vitro and in vivo. Blood. 2012;119(5):1182–1189. doi: 10.1182/blood-2011-10-386417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lapalombella R, et al. Selective inhibitors of nuclear export show that CRM1/XPO1 is a target in chronic lymphocytic leukemia. Blood. 2012;120(23):4621–4634. doi: 10.1182/blood-2012-05-429506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hertlein E, et al. 17-DMAG targets the nuclear factor-κB family of proteins to induce apoptosis in chronic lymphocytic leukemia: clinical implications of HSP90 inhibition. Blood. 2010;116(1):45–53. doi: 10.1182/blood-2010-01-263756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamblin TJ. The TCL1 mouse as a model for chronic lymphocytic leukemia. Leuk Res. 2010;34(2):135–136. doi: 10.1016/j.leukres.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Lucas DM, et al. The novel plant-derived agent silvestrol has B-cell selective activity in chronic lymphocytic leukemia and acute lymphoblastic leukemia in vitro and in vivo. Blood. 2009;113(19):4656–4666. doi: 10.1182/blood-2008-09-175430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zanesi N, et al. Effect of rapamycin on mouse chronic lymphocytic leukemia and the development of nonhematopoietic malignancies in Emu-TCL1 transgenic mice. Cancer Res. 2006;66(2):915–920. doi: 10.1158/0008-5472.CAN-05-3426. [DOI] [PubMed] [Google Scholar]
- 17.Volkmann K, et al. Potent and selective inhibitors of the inositol-requiring enzyme 1 endoribonuclease. J Biol Chem. 2011;286(14):12743–12755. doi: 10.1074/jbc.M110.199737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papandreou I, et al. Identification of an Ire1α endonuclease specific inhibitor with cytotoxic activity against human multiple myeloma. Blood. 2011;117(4):1311–1314. doi: 10.1182/blood-2010-08-303099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cross BC, et al. The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proc Natl Acad Sci U S A. 2012;109(15):E869–E878. doi: 10.1073/pnas.1115623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomasio SM, Harding HP, Ron D, Cross BC, Bond PJ. Selective inhibition of the unfolded protein response: targeting catalytic sites for Schiff base modification. Mol Biosyst. 2013;9(10):2408–2416. doi: 10.1039/c3mb70234k. [DOI] [PubMed] [Google Scholar]
- 21.Hu CC, Dougan SK, McGehee AM, Love JC, Ploegh HL. XBP-1 regulates signal transduction, transcription factors and bone marrow colonization in B cells. EMBO J. 2009;28(11):1624–1636. doi: 10.1038/emboj.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zenz T, Mertens D, Kuppers R, Dohner H, Stilgenbauer S. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev Cancer. 2010;10(1):37–50. doi: 10.1038/nrc2764. [DOI] [PubMed] [Google Scholar]
- 23.Woyach JA, Johnson AJ, Byrd JC. The B-cell receptor signaling pathway as a therapeutic target in CLL. Blood. 2012;120(6):1175–1184. doi: 10.1182/blood-2012-02-362624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capitani N, et al. S1P1 expression is controlled by the pro-oxidant activity of p66Shc and is impaired in B-CLL patients with unfavorable prognosis. Blood. 2012;120(22):4391–4399. doi: 10.1182/blood-2012-04-425959. [DOI] [PubMed] [Google Scholar]
- 25.Wiseman RL, et al. Flavonol activation defines an unanticipated ligand-binding site in the kinase-RNase domain of IRE1. Mol Cell. 2010;38(2):291–304. doi: 10.1016/j.molcel.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ranatunga S, et al. Synthesis of novel tricyclic chromenone-based inhibitors of IRE-1 RNase activity. J Med Chem. doi: 10.1021/jm5002452. [published online ahead of print April 21, 2014]. doi: 10.1021/jm5002452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2003;4(3):181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- 28.Boes M, Esau C, Fischer MB, Schmidt T, Carroll M, Chen J. Enhanced B-1 cell development, but impaired IgG antibody responses in mice deficient in secreted IgM. J Immunol. 1998;160(10):4776–4787. [PubMed] [Google Scholar]
- 29.Burger JA, Buggy JJ. Bruton tyrosine kinase inhibitor ibrutinib (PCI-32765). Leuk Lymphoma. 2013;54(11):2385–2391. doi: 10.3109/10428194.2013.777837. [DOI] [PubMed] [Google Scholar]
- 30.Advani RH, et al. Bruton tyrosine kinase inhibitor ibrutinib (PCI-32765) has significant activity in patients with relapsed/refractory B-cell malignancies. J Clin Oncol. 2013;31(1):88–94. doi: 10.1200/JCO.2012.42.7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Byrd JC, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2013;369(1):32–42. doi: 10.1056/NEJMoa1215637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pekarsky Y, et al. Tcl1 enhances Akt kinase activity and mediates its nuclear translocation. Proc Natl Acad Sci U S A. 2000;97(7):3028–3033. doi: 10.1073/pnas.97.7.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laine J, Kunstle G, Obata T, Sha M, Noguchi M. The protooncogene TCL1 is an Akt kinase coactivator. Mol Cell. 2000;6(2):395–407. doi: 10.1016/S1097-2765(00)00039-3. [DOI] [PubMed] [Google Scholar]
- 34.Mimura N, et al. Blockade of XBP1 splicing by inhibition of IRE1α is a promising therapeutic option in multiple myeloma. Blood. 2012;119(24):5772–5781. doi: 10.1182/blood-2011-07-366633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carrasco DR, et al. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell. 2007;11(4):349–360. doi: 10.1016/j.ccr.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gururajan M, Jennings CD, Bondada S. Cutting edge: constitutive B cell receptor signaling is critical for basal growth of B lymphoma. J Immunol. 2006;176(10):5715–5719. doi: 10.4049/jimmunol.176.10.5715. [DOI] [PubMed] [Google Scholar]
- 37.Wang ML, et al. Targeting BTK with ibrutinib in relapsed or refractory mantle-cell lymphoma. N Engl J Med. 2013;369(6):507–516. doi: 10.1056/NEJMoa1306220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan ZY, et al. Discovery of selective irreversible inhibitors for Bruton’s tyrosine kinase. ChemMedChem. 2007;2(1):58–61. doi: 10.1002/cmdc.200600221. [DOI] [PubMed] [Google Scholar]
- 39.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58(3):621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.