Abstract
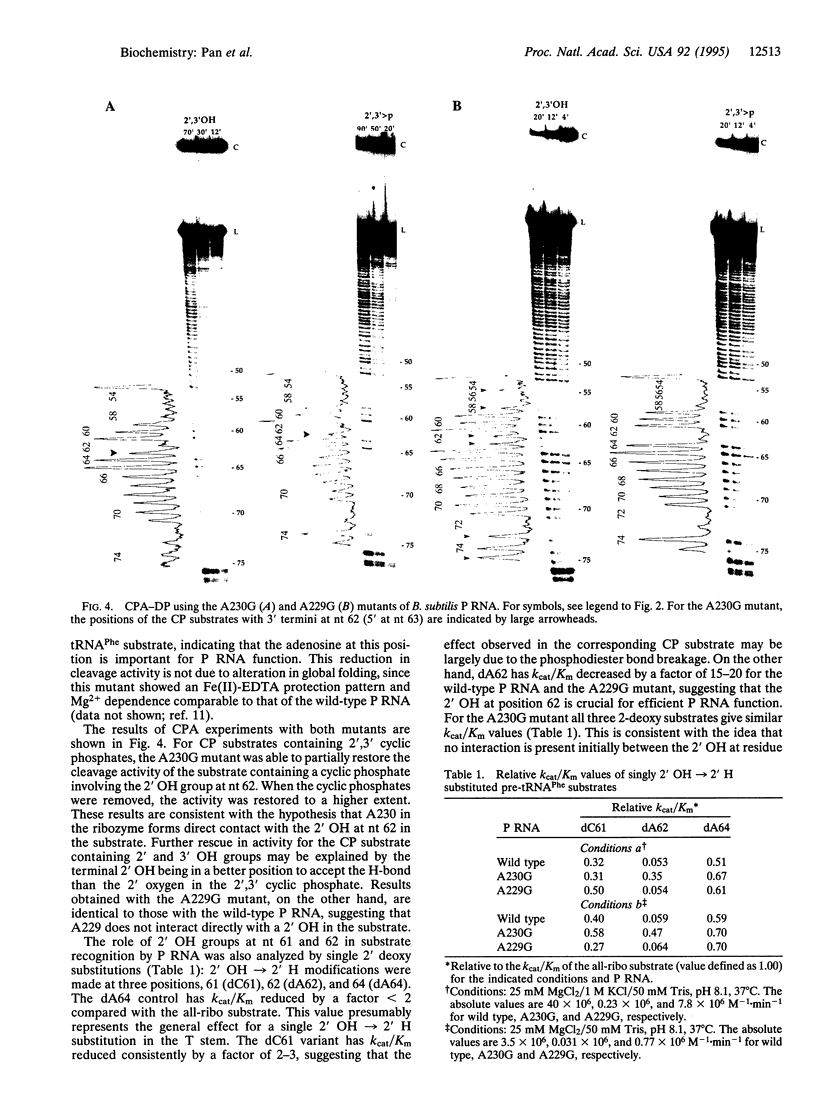
A general method has been developed to analyze all 2' hydroxyl groups involved in tertiary interactions in RNA in a single experiment. This method involves comparing the activity of populations of circularly permuted RNAs that contain or lack potential hydrogen-bond donors at each position. The 2' hydroxyls of the pre-tRNA substrate identified as potential hydrogen bond donors in intermolecular interactions with the ribozyme from eubacterial RNase P (P RNA) are located in the T stem and T loop, acceptor stem, and 3' CCA regions. To locate the hydrogen-bond acceptors for one of those 2' hydroxyls in the P RNA, a phylogenetically conserved adenosine was mutated to a guanosine. When this mutant P RNA was used, increased cleavage activity of a single circularly permuted substrate within the population was observed. The cleavage efficiency (kcat/Km) of a singly 2'-deoxy-substituted substrate at this position in the T stem was also determined. For the wild-type P RNA, the catalytic efficiency was significantly decreased compared with that of the all-ribo substrate, consistent with the notion that this 2' hydroxyl plays an important role. For the P RNA mutant, no additional effect was found upon 2'-deoxy substitution. We propose that this particular 2' hydroxyl in the pre-tRNA interacts specifically with this adenosine in the P RNA. This method should be useful in examining the role of 2' hydroxyl groups in other RNA-RNA and RNA-protein complexes.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown J. W., Pace N. R. Ribonuclease P RNA and protein subunits from bacteria. Nucleic Acids Res. 1992 Apr 11;20(7):1451–1456. doi: 10.1093/nar/20.7.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrara G., Calandra P., Fruscoloni P., Tocchini-Valentini G. P. Two helices plus a linker: a small model substrate for eukaryotic RNase P. Proc Natl Acad Sci U S A. 1995 Mar 28;92(7):2627–2631. doi: 10.1073/pnas.92.7.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gott J. M., Pan T., LeCuyer K. A., Uhlenbeck O. C. Using circular permutation analysis to redefine the R17 coat protein binding site. Biochemistry. 1993 Dec 14;32(49):13399–13404. doi: 10.1021/bi00212a004. [DOI] [PubMed] [Google Scholar]
- Harris M. E., Nolan J. M., Malhotra A., Brown J. W., Harvey S. C., Pace N. R. Use of photoaffinity crosslinking and molecular modeling to analyze the global architecture of ribonuclease P RNA. EMBO J. 1994 Sep 1;13(17):3953–3963. doi: 10.1002/j.1460-2075.1994.tb06711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschlag D., Eckstein F., Cech T. R. Contributions of 2'-hydroxyl groups of the RNA substrate to binding and catalysis by the Tetrahymena ribozyme. An energetic picture of an active site composed of RNA. Biochemistry. 1993 Aug 17;32(32):8299–8311. doi: 10.1021/bi00083a034. [DOI] [PubMed] [Google Scholar]
- Kirsebom L. A., Svärd S. G. Base pairing between Escherichia coli RNase P RNA and its substrate. EMBO J. 1994 Oct 17;13(20):4870–4876. doi: 10.1002/j.1460-2075.1994.tb06814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaGrandeur T. E., Hüttenhofer A., Noller H. F., Pace N. R. Phylogenetic comparative chemical footprint analysis of the interaction between ribonuclease P RNA and tRNA. EMBO J. 1994 Sep 1;13(17):3945–3952. doi: 10.1002/j.1460-2075.1994.tb06710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Binding of tRNA to the ribosomal A and P sites protects two distinct sets of nucleotides in 16 S rRNA. J Mol Biol. 1990 Jan 5;211(1):135–145. doi: 10.1016/0022-2836(90)90016-F. [DOI] [PubMed] [Google Scholar]
- Nolan J. M., Burke D. H., Pace N. R. Circularly permuted tRNAs as specific photoaffinity probes of ribonuclease P RNA structure. Science. 1993 Aug 6;261(5122):762–765. doi: 10.1126/science.7688143. [DOI] [PubMed] [Google Scholar]
- Noller H. F. Ribosomal RNA and translation. Annu Rev Biochem. 1991;60:191–227. doi: 10.1146/annurev.bi.60.070191.001203. [DOI] [PubMed] [Google Scholar]
- Oh B. K., Pace N. R. Interaction of the 3'-end of tRNA with ribonuclease P RNA. Nucleic Acids Res. 1994 Oct 11;22(20):4087–4094. doi: 10.1093/nar/22.20.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T., Gutell R. R., Uhlenbeck O. C. Folding of circularly permuted transfer RNAs. Science. 1991 Nov 29;254(5036):1361–1364. doi: 10.1126/science.1720569. [DOI] [PubMed] [Google Scholar]
- Pan T. Higher order folding and domain analysis of the ribozyme from Bacillus subtilis ribonuclease P. Biochemistry. 1995 Jan 24;34(3):902–909. doi: 10.1021/bi00003a024. [DOI] [PubMed] [Google Scholar]
- Pan T., Zhong K. Selection of circularly permuted ribozymes from Bacillus subtilis RNAse P by substrate binding. Biochemistry. 1994 Nov 29;33(47):14207–14212. doi: 10.1021/bi00251a032. [DOI] [PubMed] [Google Scholar]
- Perreault J. P., Altman S. Important 2'-hydroxyl groups in model substrates for M1 RNA, the catalytic RNA subunit of RNase P from Escherichia coli. J Mol Biol. 1992 Jul 20;226(2):399–409. doi: 10.1016/0022-2836(92)90955-j. [DOI] [PubMed] [Google Scholar]
- Perreault J. P., Wu T. F., Cousineau B., Ogilvie K. K., Cedergren R. Mixed deoxyribo- and ribo-oligonucleotides with catalytic activity. Nature. 1990 Apr 5;344(6266):565–567. doi: 10.1038/344565a0. [DOI] [PubMed] [Google Scholar]
- Pyle A. M., Cech T. R. Ribozyme recognition of RNA by tertiary interactions with specific ribose 2'-OH groups. Nature. 1991 Apr 18;350(6319):628–631. doi: 10.1038/350628a0. [DOI] [PubMed] [Google Scholar]
- Pyle A. M., Murphy F. L., Cech T. R. RNA substrate binding site in the catalytic core of the Tetrahymena ribozyme. Nature. 1992 Jul 9;358(6382):123–128. doi: 10.1038/358123a0. [DOI] [PubMed] [Google Scholar]
- Quigley G. J., Rich A. Structural domains of transfer RNA molecules. Science. 1976 Nov 19;194(4267):796–806. doi: 10.1126/science.790568. [DOI] [PubMed] [Google Scholar]
- Smith D., Pace N. R. Multiple magnesium ions in the ribonuclease P reaction mechanism. Biochemistry. 1993 May 25;32(20):5273–5281. doi: 10.1021/bi00071a001. [DOI] [PubMed] [Google Scholar]
- Strobel S. A., Cech T. R. Tertiary interactions with the internal guide sequence mediate docking of the P1 helix into the catalytic core of the Tetrahymena ribozyme. Biochemistry. 1993 Dec 14;32(49):13593–13604. doi: 10.1021/bi00212a027. [DOI] [PubMed] [Google Scholar]
- Westhof E., Altman S. Three-dimensional working model of M1 RNA, the catalytic RNA subunit of ribonuclease P from Escherichia coli. Proc Natl Acad Sci U S A. 1994 May 24;91(11):5133–5137. doi: 10.1073/pnas.91.11.5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Altman S. Substrate recognition by human RNase P: identification of small, model substrates for the enzyme. EMBO J. 1995 Jan 3;14(1):159–168. doi: 10.1002/j.1460-2075.1995.tb06986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Hwang E. S., Altman S. Targeted cleavage of mRNA by human RNase P. Proc Natl Acad Sci U S A. 1992 Sep 1;89(17):8006–8010. doi: 10.1073/pnas.89.17.8006. [DOI] [PMC free article] [PubMed] [Google Scholar]