Abstract
Feline immunodeficiency virus (FIV)-infected cats enter a clinically asymptomatic phase during chronic infection. Despite the lack of overt clinical disease, the asymptomatic phase is characterized by persistent immunologic impairment. In the peripheral blood obtained from cats experimentally infected with FIV-C for approximately 5 years, we identified a persistent inversion of the CD4/CD8 ratio. We cloned and sequenced the FIV-C long terminal repeat containing the viral promoter from cells infected with the inoculating virus and from in vivo-derived peripheral blood mononuclear cells and CD4 T cells isolated at multiple time points throughout the asymptomatic phase. Relative to the inoculating virus, viral sequences amplified from cells isolated from all of the infected animals demonstrated multiple single nucleotide mutations and a short deletion within the viral U3, R and U5 regions. A transcriptionally inactivating proviral mutation in the U3 promoter AP-1 site was identified at multiple time points from all of the infected animals but not within cell-associated viral RNA. In contrast, no mutations were identified within the sequence of the viral dUTPase gene amplified from PBMC isolated at approximately 5 years post-infection relative to the inoculating sequence. The possible implications of these mutations to viral pathogenesis are discussed.
Keywords: Latency, Promoter, LTR, Lentivirus, Feline immunodeficiency virus, FIV
1. Introduction
Feline immunodeficiency virus (FIV) is a lentivirus that infects cats, resulting in an acute infection syndrome followed by a prolonged asymptomatic period during which the CD4/CD8 T cell ratio is inverted (Ackley et al., 1990; Barlough et al., 1991; Joshi et al., 2004; Kohmoto et al., 1998; Pedersen and Barlough, 1991; Torten et al., 1991). FIV causes progressive immunologic impairment, culminating in an AIDS-like syndrome and death, akin to HIV-infected humans (Ikeda et al., 1996; Joshi et al., 2004; Kohmoto et al., 1998). The FIV-infected cat is the only naturally-occurring, outbred, large animal model of lentivirus-induced immunodeficiency and like HIV, FIV is capable of infecting both CD4 T cells and monocytes in the susceptible host (Bendinelli et al., 1995; Burkhard and Dean, 2003; Joshi et al, 2004).
Our laboratory has established a model of lentiviral cellular latency in experimentally FIV-infected specific pathogen free (SPF) cats during the asymptomatic phase of infection (Murphy et al., 2012). SPF cats infected with a biological isolate of FIV clade C (Pgmr) for approximately 3 years have an estimated proviral load of 1 infected peripheral CD4 T cell in approximately 103 peripheral CD4 T cells (McDonnel et al., 2012b). Our laboratory has experimentally determined that of those infected CD4 T cells, there is approximately 1 copy of viral DNA per cell and 1 in 10 proviral copies appear capable of transcription after ex vivo activation. In latently infected peripheral CD4 T cells, the integrated and transcriptionally inactive FIV promoter is physically associated with deacetylated, methylated histone proteins, consistent with a restrictive chromatin environment (McDonnel et al., 2012b). The latent provirus can readily be reactivated in vitro with exposure to histone deacetylase inhibitors such as suberoylanilide hydroxamic acid (SAHA), which result in histone acetylation at the integration site of the proviral promoter and transcriptional activation of the provirus (McDonnel et al., 2012a).
A single nucleotide mutation within the FIV-C U3 AP-1 site was previously shown to abrogate transcription in a β galactosidase reporter gene assay (Murphy et al., 2012). This AP-1 mutation was found to be present in the proviral DNA of CD4 T cells isolated from all of the FIV-infected cats. Lentiviral latency has been defined as a reversible low-productive state of infection, where infected cells retain the capacity to produce new viral particles (Eisele and Siliciano, 2012). Although we have previously demonstrated that latency is associated with a restrictive chromatin environment, we wondered whether the AP-1 mutation might be associated with an additional mechanism of viral latency. Although our in vitro experiment indicated transcriptional abrogation, in vivo viral latency mechanisms may be more complex (e.g. AP-1 mutation associated with leaky or low-level viral transcription in certain cellular states). We hypothesized that the FIV-C proviral U3 AP-1 mutation is associated with intermittent/low-level viral transcription and therefore, latency.
For the study reported here, serial peripheral blood samples were obtained from FIV-infected cats and mock-infected control cats throughout the asymptomatic phase and were systematically analyzed for detectable plasma virus and the enumeration of total white blood cells and cellular subsets using surface antigen-specific antibodies (anti-CD4, CD8, MHC II, CD11b, CD21 and CD25). Nucleic acids isolated from peripheral blood mononuclear cells (PBMC) and peripheral CD4 T cells were analyzed for detectable viral promoters via nested PCR; amplified viral promoters were subsequently cloned and sequenced. Since lentiviral latency is likely mechanistically attributable to the host/viral promoter interface, we focused our sequencing efforts on the viral promoter. During the study, multiple G to A transition mutations were identified in the proviral LTR. Since it has previously been demonstrated that FIV lacking a functional dUTPase gene is prone to G to A transition mutations, the FIV-C dUTPase gene was also amplified and sequenced.
2. Materials and methods
2.1. Animals
Six FIV SPF kittens were purchased from the breeding colony of the Feline Nutrition and Pet Care Center, University of California at Davis (UC Davis). At time of purchase, the kittens ranged in age from 4 to 5 months and were housed in the Feline Research Laboratory of the Center for Companion Animal Health, UC Davis. Four kittens were intramuscularly inoculated with FIV-C-Pgmr viral inoculums (kittens 165, 184, 187 and 186) and monitored as described previously (Murphy et al, 2012). Two control kittens (183 and 185) were mock-inoculated with 1 ml of sterile culture media. The FIV-C-Pgmr biological isolate was provided by Drs. E. Hoover (Colorado State University) and N. Pedersen (University California, Davis). This study spans the time of inoculation to approximately 253 weeks post-infection (5 years). Blood samples were obtained approximately once a month throughout this time period. The study protocol was approved by the UC Davis Institutional Animal Care and Use Committee.
2.2. Plasma virus
Whole blood was collected from FIV-infected and uninfected cats every 2–4 weeks via jugular venipuncture in EDTA-containing tubes and centrifuged at 500 × g for 5 min. Plasma was subsequently transferred and centrifuged at 17,000 × g for five additional minutes. Viral RNA was isolated from clarified plasma using a commercially available kit (QIAmp Viral RNA Minikit, Qiagen). Isolated vRNA was DNase treated (Turbo DNase, Ambion) and reverse transcribed using the First-Strand cDNA Synthesis System for Quantitative RT-PCR (OriGene). A control reaction excluding reverse transcriptase was included for each set of reverse transcribed cDNA.
Isolated RNA was then assayed for the presence of FIV gag via real-time PCR utilizing the primers FIVQT gag for and FIVQT gag rev, as described previously (Murphy et al, 2012). Real-time PCR was performed in triplicate with Real Mastermix SYBR Rox (5 PRIME) on an Applied Biosystems 7300 Real-Time PCR System with the following cycling conditions: 50 °C for 2 min, 95 °C for 5 min followed by 40 cycles of 95 °C for 15 s, 58 °C for 30 s, 68 °C for 30 s and a final elongation step at 72 °C for 5 min as previously described (Murphy et al, 2012). The real-time PCR assay has a detection limit of approximately 10 copies of FIV gag (data not shown). All real-time PCR assays were followed with a dissociation step (melt curve) to assess amplicon validity.
2.3. Leukocytes
The total WBC concentration was determined from whole blood collected in EDTA-containing tubes with a commercially available system (LeukoChek, Biomedical Polymers, Inc.) coupled with hemocytometer enumeration. The relative proportion of specific leukocyte subsets was assessed utilizing the following antigen-specific antibodies: anti-feline CD4 (clone FE1.7B12), anti-feline CD8 (clone FE1.10E9), anti-canine CD21 (clone CA2.1D6), anticanine 11b (clone CA16.3E10), anti-feline MHC II (clone 42.3), and anti-feline CD25 (clone 9F23). All of the antibodies were obtained from Dr. Peter Moore (UC Davis) with the exception of anti-feline CD25, which was a gift of Koichi Ohno, (University of Tokyo). The proportion of cells expressing a specific marker was determined by flow cytometry, using a procedure described previously (Murphy et al, 2012). Briefly, 100μl of whole blood was incubated at room temperature with each of the antibodies listed above. The cells were subsequently incubated with eythrocyte lysis buffer (154mM ammonium chloride, 82 mM potassium bicarbonate and 18mM EDTAtetrasodium salt in phosphate buffered saline (PBS), pH 7.2), washed with flow buffer (2 mM EDTA trisodium salt, 2.55 mM ETA disodium salt and 15 mM sodium azide in PBS, pH 7.2), stained with a secondary fluoroscein isothiocyanate (FITC)-conjugated horse anti-mouse IgG (Vector Laboratories, Inc.) and incubated in the dark for 15 min. Cells were washed a final time with flow buffer and centrifuged at 500 × g for 5 min. Pelleted cells were resuspended in flow buffer and analyzed using a FACScan flow cytometer (Becton Dickinson); resulting data was analyzed with FlowJo v8.6.3 (Treestar). CD4 and CD8 cell counts were derived from the absolute lymphocyte counts (multiplying the absolute lymphocyte number by the percent of cells expressing the CD4 or CD8 marker); the CD4/CD8 ratio was determined by division of the percent expression of the two markers. Viable peripheral CD4 T cells were purified from whole blood as described previously using feline antigen-specific antibodies and magnetic columns (Murphy et al., 2012).
2.4. FIV LTR amplification, cloning and sequencing
The proviral LTR was amplified from nucleic acid isolated from cryogenically stored preparations of single passage feline PBMC infected with the inoculating virus along with FIV-infected cat-derived peripheral blood mononuclear cells (PBMC) and column-isolated CD4 T cells obtained throughout the infection. PBMC were isolated from peripheral blood using Ficoll-Hypaque (Sigma), DNA was extracted (DNA Mini Kit, Qiagen), the proviral LTR was amplified via nested PCR (described below) and cloned using a commercial system (pCR2.1, TA cloning system, Invitrogen). Plasmid DNA was purified via a commercial kit (Promega) and the inserted DNA was sequenced by a local vendor (Davis Sequencing). Nucleotide sequences were aligned and compared using the AlignX function of Vector NTI software (Invitrogen).
Freshly column-isolated CD4 T cells were co-cultured at a ratio of 1:1 with allogeneic SPF feline PBMC at a final concentration of 1 × 106 cells/ml for 7–23 days using previously described protocols (Sparger et al., 1994) with the addition of mitogens (phorbol myristate acetate, 0.5 μg/ml, Sigma-Aldrich) and Concanavalin A (5 μg/ml, ThermoFisher Scientific). Cell-associated DNA and RNA were co-isolated (AllPrep DNA/RNA Mini kit, Qiagen) at incubation days 0, 7 and 23. RNA was DNase treated (TURBO DNase, Ambion) and reverse transcribed as described above. Viral transcription in CD4 T cells was assessed via real-time PCR for the presence of a 100 bp product for FIV gag using the primers FIVQT gag for and FIVqT gag rev, as described previously (Murphy et al., 2012).
The proviral 3′ LTR derived from vDNA isolated from FIV-infected cells was amplified by semi-nested PCR, cloned and sequenced. To prevent inadvertent DNA contamination during the nested PCR procedures, the reactions were set up in UV Sterilizing PCR Workstations (UVP) pre-treated with UV light exposure for 30 min followed by treatment with DNAse (DNAzap, Ambion). The initial PCR reaction utilized the primers FIVenv for and U5rev (Fig. 1) and was run with the following cycling conditions: 95 °C for 5 min, followed by 30 cycles of 95 °C for 15 s, 60 °C for 30 s, 72 °C for 30 s, and a final elongation step of 72 °C for 5 min. Ten microliters of primary PCR product provided the template for the secondary PCR reaction utilizing the primers FIVenv nest for and U5rev and was run with the following cycling conditions: 95 °C for 2 min, followed by 30 cycles of 95 °C for 15 s, 62 °C for 30 s, 72 °C for 60s, and a final elongation step of 72 °C for 5 min. The resulting PCR amplicons were cloned into pCR2.1 (Invitrogen) and sequenced as described above. In order to determine the sequence diversity of the proviral 3′ LTR derived from the inoculating virus, amplification procedure was utilized that was identical to the initial PCR amplification described above; a secondary PCR reaction was not necessary and was not performed. Resulting amplicons were cloned and sequenced as described above.
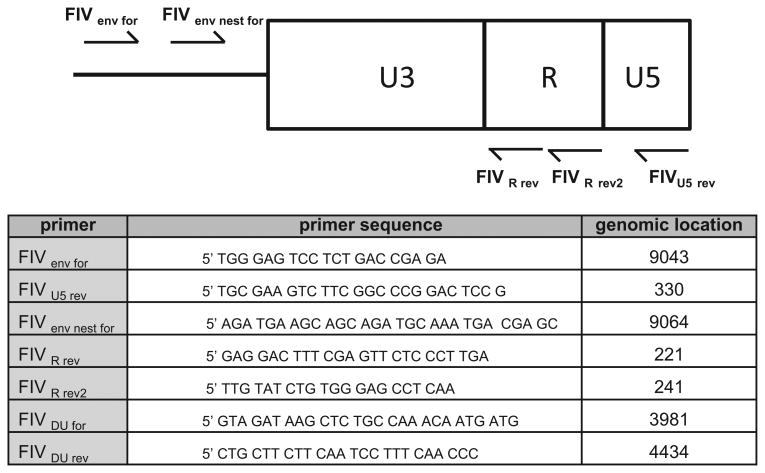
Fig. 1.
Primers used to clone FIV LTR and dUTPase gene. Schematic of the genomic location and sequence of primers utilized to amplify and clone the FIV-C LTR and dUTPase gene. The genomic location refers to the locus of the 5′ nucleotide (genome loci are derived from GenBank FIV-C reference strain AF474246.1).
The FIV U3-R region derived from cell-associated vRNA was amplified using either a nested or semi-nested PCR strategy. For nested PCR, the initial reaction utilized the primers FIVenv for and FIVR rev2 (Fig. 1) and was run with the following cycling conditions: 95 °C for 5 min, followed by 30 cycles of 95 °C for 15 s, 58 °C for 15 s, 72 °C for 60s, and a final elongation step of 72 °C for 5 min. Ten microliters of primary PCR product provided the template for the secondary PCR reaction using FIVenv nest for and FIVR rev with the following conditions: 95 °C for 5 min, followed by 30 cycles of 95 °C for 15 s, 60 °C for 30 s, 72 °C for 30 s, and a final elongation step of 72 °C for 5 min. For semi-nested PCR, the FIVR rev primer served as the downstream primer for both reactions. PCR amplicons were cloned and sequenced as described above.
2.5. FIV dUTPase gene amplification and sequencing
The FIV-C dUTPase gene (nt 4018-4409) was amplified from within the enzyme cassette encoded by the pol gene (nt 1857–5249). The amplifying primers, FIVDU for and FIVDu rev, were based upon the FIV-C sequence in GenBank (AF474246.1) and were designed to flank the dUTPase gene (Fig. 1). FIV-C dUTPase was PCR amplified (Invitrogen Taq polymerase) from the inoculating FIV-C proviral DNA and from PBMC harvested at approximately 43/4 years PI from all of the FIV-infected cats. PCR cycling conditions were as follows: 95 °C for 2 min, followed by 40 cycles of 95 °C for 15s then 55 °C for 30s and 72 °C for 60s, a final elongation at 72 °C for 5 min. PCR products were run on 1% agarose gels with ethidium bromide. PCR amplicons of ∼450 base pairs were visualized and recorded using a Protein Simple imaging system. Attempts to clone the dUTPase amplicon using the pCR 2.1 TA cloning system (Invitrogen) were not successful. Therefore, the PCR amplicons were sequenced directly. Amplicons were purified using Amicon 100k centrifugal filters following the manufacturer's protocol. Sequencing of the PCR products was done by a local vendor (Davis Sequencing, Davis, CA) in both the 5′ and 3′ directions.
2.6. Statistics
The graphical numerical data is the mean of three or more values with the standard deviation represented by error bars. For data sets with more than two mean values, analysis of variance (ANOVA) was determined. In data sets where global differences were identified, the Tukey-Kramer multiple comparisons test was used for select pair-wise comparisons of the mean response between treatment groups. A p value < 0.05 was considered to be statistically significant. Statistics were performed with InStat software (GraphPad Software, Inc., La Jolla, CA).
3. Results
3.1. Viremia
Plasma viremia was monitored by RT-PCR in 4 FIV-C and 2 mock-infected SPF cats from 0 to 242 weeks post-inoculation (PI, approximately 5 years). The total number of vRNA assays determined per FIV-infected cat was 37, 35, 35 and 37, for cats 165, 184, 187 and 186, respectively. All of the FIV-infected cats demonstrated an undulating positive viremia for the first 5–10 months of infection, but no vRNA was detected in the plasma of any of the cats after 44 weeks PI (data not shown). Mock- infected control cats were negative for plasma vRNA at all time points (data not shown). FIV-infected cats exhibited transient signs of peripheral lymphadenopathy which were generally maintained for approximately 10 months, after which the cats were clinically asymptomatic. Thus plasma viremia was below the limit of detection during the chronic, asymptomatic phase of FIV-C infection using these methods.
3.2. Leukocyte counts and immunophenotype
Total leukocyte concentrations were serially determined from peripheral blood samples obtained from FIV-infected and uninfected cats approximately once a month over 5 years of infection. The cumulative average leukocyte count in the infected cats was significantly less than in uninfected control animals ((6.60 ± 3.22) × 106 cells/ml versus (12.01 ± 3.19) × 106 cells/ml, respectively, p < 0.05). The percentage of cells expressing CD4 and CD8 T-cell antigens was monitored over the same period using flow cytometry and antigen-specific antibodies. The total number of peripheral CD4 and CD8 enumerations determined per cat was 29, 22, 29 and 23 for FIV infected cats 165, 184, 186 and 187; 20 and 21 total enumerations were performed for control cats 183 and 185, respectively. The CD4/CD8 ratio was inverted (less than 1) relative to the mock-infected control cats in 3 out of 4 FIV-infected cats during the chronic phase of infection (Fig. 2a). When the CD4/CD8 ratio was determined for progressive intervals of time post-infection, a gradual decrease in the ratio was identified for all 4 of the FIV infected cats relative to control animals (Fig. 2b). At 2 and 4 weeks post-infection, the CD4/CD8 ratio was not significantly different between infected and uninfected animals, while at 6 weeks post-infection onward, the ratio was significantly reduced for infected animals. The cumulative CD4/CD8 ratio averaged over all time points was significantly less in the FIV-infected relative to uninfected animals (0.75 ± 0.47 vs. 1.65 ± 0.38, respectively, p < 0.05). When peripheral CD4 and CD8 T-cell subsets were analyzed individually, the absolute CD4 T cell counts were significantly reduced for the FIV-infected cats relative to the uninfected control cats ((1.27 ± 1.15) × 106 cells/ml vs. (2.56 ± 1.01) × 106 cells/ml, respectively, p < 0.05). There was no significant difference between infected and uninfected cats in terms of cumulative absolute CD8 T-cell count ((1.69 ± 0.92) × 106 cells/ml vs. (1.60 ± 0.78) × 106 cells/ml, respectively, p > 0.05). Though a higher percentage of leukocytes expressed the CD8 T-cell marker in infected relative to uninfected cats (not shown), the total concentration of these cells was not significantly different due the relative leukopenia in infected animals.
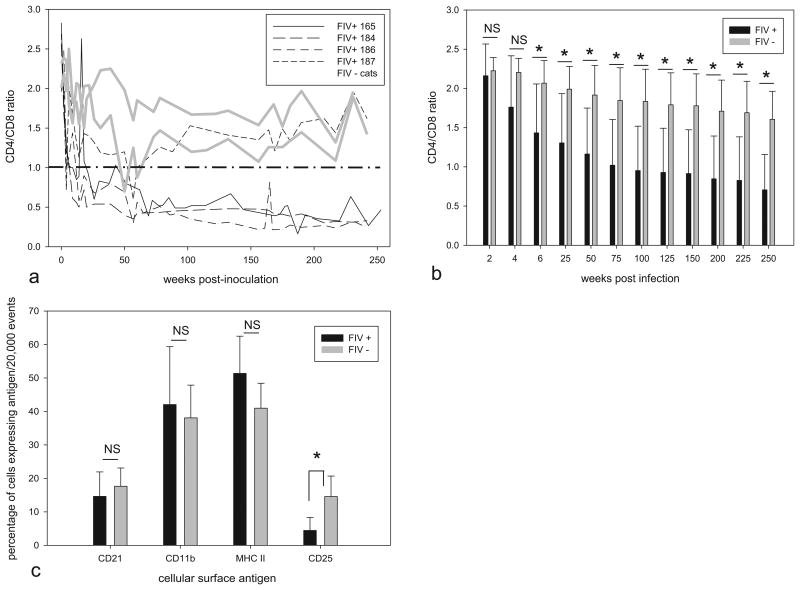
Fig. 2.
CD4/CD8 ratios and leukocyte subsets. (a) CD4/CD8 T-cell ratios were determined by flow cytometry in 4 FIV C-infected and 2 mock-infected cats over the first 253 weeks PI. Individual animals are represented by dashed black lines (FIV+ cats 165, 184, 186, and 197) or solid gray lines (uninfected control cats 183 and 185). (b) Cumulative CD4/CD8 T-cell ratios for FIV-infected (black bars) and uninfected (gray bars) cats from 2 to 250 weeks post-infection. For infected vs. uninfected animals, the ratios are not significantly different (NS) for 2 and 4 weeks post-infection. Infected animals have reduced ratios relative to controls for ratios 6 weeks post-infection and greater (*p < .05). (c) Percentage of peripheral leukocytes expressing the cell surface antigens CD21, CD11b, MHC II, and CD25 assessed by flow cytometry at 4 time points for the infected (black bars) and uninfected (gray bars) cats during the chronic phase of FIV infection (175–253 weeks PI). The percentage of CD25 cells in infected cats was less than uninfected control cats, respectively (*p < 0.05). Statistical significance is denoted by an asterisk for selected pair wise comparisons (*p < 0.05); error bars denote standard deviation. No difference in the percentage of CD21, CD11b and MHC II cells was identified between infected and uninfected cats (NS, not significant).
The percentages of peripheral leukocytes expressing the antigens CD21, CD11b, MHC II, and CD25 were assessed in all of the cats at 4 time points from 175 to 253 weeks PI. The percentage of cells expressing the B-cell marker CD21, MHC II and monocyte/granulocyte marker CD11b was not significantly different between groups (p > 0.05, Fig. 2c). However, CD25 expression was greater on leukocytes from FIV– cats (4.44 ±3.86% infected versus 14.58 ±6.12% uninfected, p < 0.05, Fig. 2c). Samples serially obtained from individual cats demonstrated minimal variation in the percentage of the leukocyte subsets.
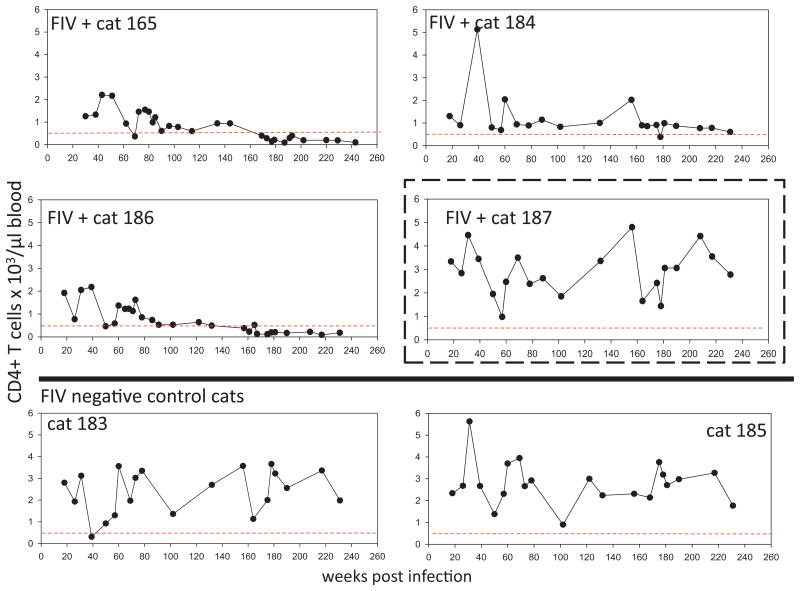
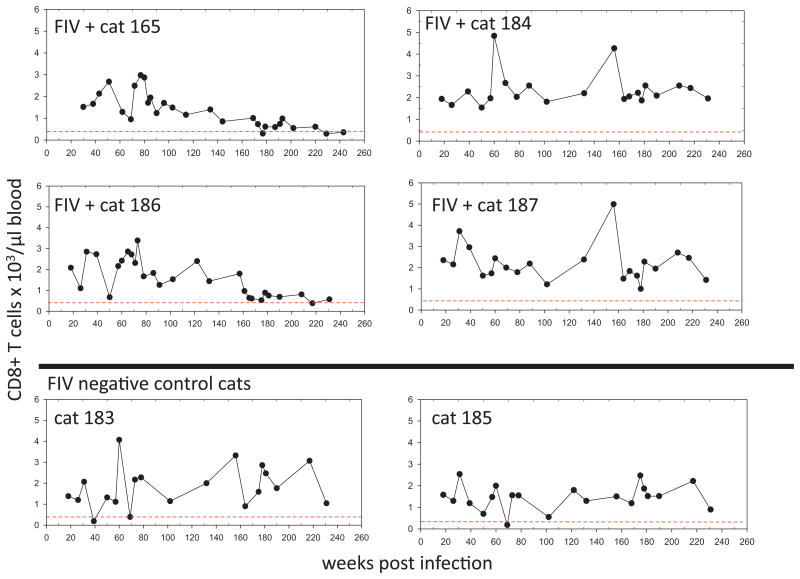
Total peripheral CD4 and CD8 T cell counts over the course of infection in all cats are displayed in Figs. 3 and 4. Although the T-cell counts oscillated from week to week, two of the FIV+ cats (165 and 186) demonstrated persistent CD4 T cell counts of less than 500 cells/μl blood starting at approximately 3 years of infection (Fig. 3). For both of these animals, The CD4 T cell count was determined to be less than 500 cells/μl at 12 different time points. These same two animals demonstrated persistently reduced numbers of CD8 T-cells relative to uninfected controls during this time period (Fig. 4). Throughout this time, the FIV-infected cats were clinically normal (as determined by periodic complete physical examinations, feeding/drinking habits and lack of behavioral changes).
Fig. 3.
CD4 T cell counts. Total number of peripheral CD4T cells over the first 240 weeks post-inoculation in 4 FIV-infected cats (165, 184, 186 and 187) and 2 mock-infected cats (183 and 185) as determined by flow cytometry. For each graph, a dotted line is shown at ∼500cells/μl blood, the lower boundary limit for normal adult cats (Dean et al., 1991). A dashed box is drawn around the data for FIV-infected animal 187, who demonstrates a persistently normal CD4/CD8 ratio.
Fig. 4.
CD8 T-cell counts. Total number of peripheral CD8+ cells over the first 240 weeks post-inoculation in 4 FIV-infected cats (165, 184, 186 and 187) and 2 mock-infected cats (183 and 185) as determined by flow cytometry. For each graph, a dotted line is shown at ∼350cells/μl blood, the lower boundary limit for normal adult cats (Dean et al., 1991).
3.3. Viral promoter variants
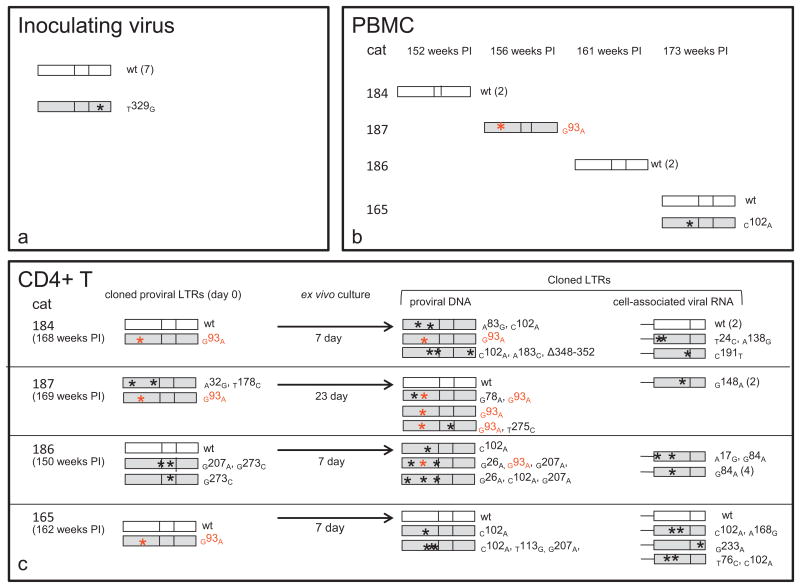
The 3′ FIV proviral LTR was amplified, cloned and sequenced from feline PBMC freshly infected with the inoculating virus. In 8 examined LTR clones, the sequences of 7 of these were identical; a single SNP mutation was identified within the U5 region (T329G) in one LTR clone (Fig. 5a). This SNP mutation was not subsequently identified.
Fig. 5.
Viral promoter variants. LTR sequences were amplified, cloned, and then sequenced from the inoculating provirus and from PBMC or CD4 T cells isolated from the infected cats during the asymptomatic phase of infection. LTRs shown in white are identical to the inoculating virus sequence (wild type, wt) while shaded LTRs had at least one point mutation, approximate genomic position denoted by an asterisk. The number in parentheses next to the cloned sequences represents the number of copies sequenced of that exact same clone. The U3 point mutation in red (G93A) has been previously shown to be transcriptionally inactivating, whereas the C102A mutation had no effect on transcriptional activity of the viral promoter (Murphy et al., 2012). (a) Sequenced clones (8) of the inoculating provirus were identical except for a single SNP within the U5 region (T329G). (b) Mutations relative to the inoculating virus in the FIV proviral LTR at various time points PI in PBMC isolated from 4 FIV-infected cats. (c) Column-isolated, latently infected CD4T cells from 4 FIV-infected cats were cultured ex vivo for 7–23 days until viral reactivation was detected by RT-PCR. FIV gag amplicons were not detected by real-time RT-PCR assays in cDNA obtained from freshly isolated cells (day 0), but were detected on day 7 (cats 184, 186 and 165) or 23 (cat 187) of ex vivo culture. The FIV proviral LTR was cloned and sequenced before and after ex vivo reactivation. The U3/R region at the 3′ end of cell-associated vRNA was also cloned and sequenced after ex vivo reactivation. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
During weeks 152–173 PI, PBMC were obtained from the FIV infected cats, DNA was isolated and the proviral LTR was amplified, cloned and sequenced (7 clones) (Fig. 5b). Previously, we amplified and sequenced 13 promoter regions from PBMC isolated from the FIV-infected cats between 2 and 173 weeks post-infection (Murphy et al., 2012). Collectively, within this group of 20 promoter sequences, only 2 mutations were identified relative to the sequence of the inoculating virus; however, a transcriptionally inactivating (Murphy et al., 2012) mutation in the U3 region AP-1 binding site (G93A) was present in sequences from the PBMC of one FIV-infected animal (cat 187).
Viable CD4 T cells were isolated using magnetic column enrichment from each FIV-infected cat between 150 and 169 weeks (approximately 3 years) post-infection as previously described (Murphy et al., 2012). Freshly isolated CD4 T cells had undetectable cell-associated vRNA, consistent with the concept of viral latency (Pace et al., 2011). The CD4 T cells were then co-cultured with SPF allogeneic feline PBMC and mitogens for 7 to 23 days in an attempt to reactivate viral transcription ex vivo. Viral reactivation was detected by real time RT-PCR for cell-associated vRNA (FIV gag, data not shown) after 7 days for cells isolated from three of the cats, but not until 23 days post-culture in cells isolated from cat 187 (Fig. 5c). The proviral LTR (U3/R/U5) was cloned and sequenced from cellular DNA before and after ex vivo culture and the U3/R region of cell-associated vRNA was also cloned and sequenced at the end of the reactivation period (Fig. 5c). For each sample, 5 clones were selected for amplification and sequencing, yielding 2–5 sequences per sample. A variety of point mutations and one 5 base pair deletion were identified within the LTR both before and after ex vivo culture, though wild-type (inoculating FIV-C virus) sequences were isolated from cells from all FIV infected cats. The same transcriptionally inactivating AP-1 binding site mutation (G93A) found in the PBMC of cat 187 was present in proviral sequences isolated from all four cats’ CD4 T cells. Importantly, none of the sequenced U3/R regions from cell-associated vRNA had this mutation, consistent with the transcriptional silencing of this SNP. A summary of the mutations discovered in the viral LTR relative to known FIV U3 transcription factor binding sites is presented in Fig. 6.
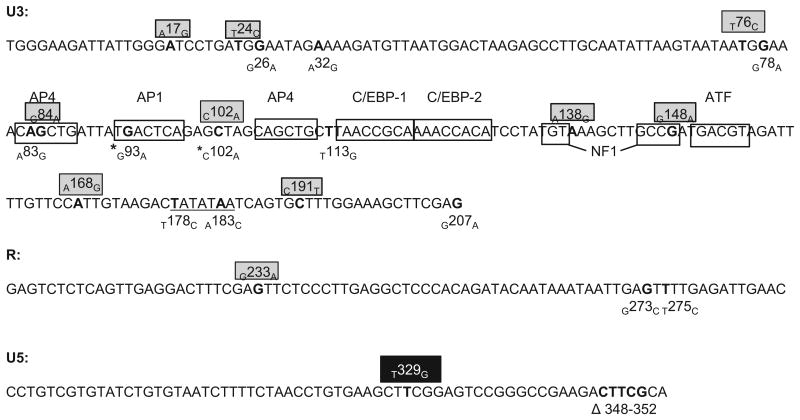
Fig. 6.
Mutations in the FIV LTR. Schematic of the inoculating FIV LTR sequence (FIV-C pgmr) including the genomic location of all mutations discovered overtime in PBMC or in CD4 T cells before and after ex vivo reactivation in all 4 FIV-infected cats. The single SNP identified in the U5 region of the inoculating virus is depicted by a black box. Mutations within gray boxes represent those found in cell-associated vRNA; unboxed mutations were discovered in proviral DNA. The location of various transcription factor binding sites in the U3 region are denoted by unshaded boxes. Mutations denoted by asterisks have been previously cloned into expression constructs, and their promoter activity has been assessed. The TATA box is underlined.
3.4. dUTPase gene stability
The FIV-C dUTPase gene derived from the inoculating provirus and from proviral DNA isolated from PBMC from FIV-infected cats 165, 187 and 184 at 217–230 weeks PI was PCR amplified, sequenced and compared (454 nucleotide amplicon). The dUTPase gene sequence for the inoculating virus was identical to the sequences amplified from the FIV-infected cats at 217–230 weeks PI. In all of the dUTPase gene sequences, 4 SNPs were present relative to the FIV-C AF474246.1 GenBank sequence.
4. Discussion
In our chronically-infected FIV positive cats, a persistent immunologic impairment (CD4/CD8 ratio less than control animals) was demonstrated from 6 to 253 weeks PI. This phenomenon of CD4/CD8 inversion has been previously reported many times and has become a marker of an infection with an immunodeficiency-causing lentivirus (Pedersen and Barlough, 1991). However, for experimentally FIV-infected cats in the asymptomatic phase, serial chronological determination of the CD4/CD8 ratio has only rarely been published (Ackley et al., 1990; Kohmoto et al., 1998; Torten et al., 1991), and these previous reports are limited in detail. In this study, three FIV-infected cats (165, 184, 186) demonstrate a persistent inversion of the CD4/CD8 ratio while a single infected cat (187) is indistinguishable from the uninfected control animals (140 total CD4/CD8 ratio calculations were determined during the study period). This data indicates that the CD4/CD8 ratio is a persistently stable signature of individualized infection. It has been suggested that a CD4/CD8 ration of <1.0 might serve as a useful discriminator for FIV infection (Barlough et al., 1991). The CD4/CD8 ratios of the infected cats progressively decreased as the time post-infection increased.This phenomenon has been identified previously (Ackley et al, 1990; Kohmoto et al., 1998) and supports the concept that the immune dysfunction associated with FIV infection is gradually progressive over time (Torten et al, 1991). In our study, a significant decrease in the CD4/CD8 ratio was first detectable at 6 weeks PI. In a study by Ackley et al. (1990), a significant decrease in the CD4/CD8 ratio of infected cats relative to uninfected cats was first detected at 18 months (72 weeks) PI. Interestingly, during the current study, CD4/CD8 ratios continue to decline despite the fact that plasma viremia becomes undetectable at approximately 42 weeks PI (132 total plasma vRNA assays performed).
It has not been determined how the integrated and transcriptionally inactive provirus results in a reduction of peripheral CD4 T cell number. It remains possible that central lymphoid lentiviral reservoirs (e.g. lymph nodes, gut-associated lymphatic tissue, spleen or thymus) could harbor ongoing viral transcription and virion assembly at low levels. Such low-level viral replication might remain tissue or cell-associated and therefore undetectable by the plasma RT-PCR assays, resulting in central CD4 T cell destruction/senescence/apoptosis, culminating in a reduced peripheral CD4 T cell number. Recent results from our laboratory indicate that latently infected peripheral CD4 T cells contain abundant short promoter-proximal vRNA transcripts (R region, +1 to +66) but lack detectable viral gag RNA (McDonnel et al., 2012b). It is possible that the intracellular presence of these viral transcripts could result in cell death by apoptosis via innate pattern recognition receptors or other mechanism.
The percentage of peripheral WBC expressing the IL2 receptor CD25 was found to be reduced in FIV-infected cats relative to uninfected controls. Preferential replication of FIV in CD4+ CD25+T cells relative to CD4+ CD25– T cells might explain this phenomenon (Joshi et al., 2004, 2005). A severe depletion of CD4+ CD25+ T cells has been identified within the intestinal lamina propria, but not the peripheral blood, during acute infection with simian immunodeficiency virus in macaques (Chase et al., 2007). FIV is capable of infecting not only CD4 T cells, but also B cells and monocytes (Beebe et al., 1994). Much less has been published about the B cell tropism of FIV. Here we found no difference in the number of peripheral CD21+ B cells between asymptomatic infected and uninfected cats. This result is supported by previous data from another group identifying no difference in peripheral B cells between infected and uninfected cats (Ackley et al., 1990). In our study, the infection status of the peripheral B cells was not determined and remains an area of future exploration. We have previously documented a dual CD4 T cell/monocyte tropism for the inoculating virus described here (FIV-C-Pgmr) (Murphy et al., 2012). Interestingly, we found that the number of peripheral CD11b+ and MHC II+ cells was similar, regardless of infection status. Whether FIV-C results in a latent infection in peripheral monocytes remains to be determined.
Although FIV has been intensively researched for over 27 years, there are relatively few detailed chronological determinations of peripheral CD4 and CD8 T cell numbers for FIV-infected cats in the asymptomatic phase. In a study by Barlough et al. (1991), chronically FIV-infected cats (25–40 months PI) have a total average number of peripheral leukocytes and CD4 T cells of (10.377 ± 1.060) × 106 cells/ml and (0.631 ± 0.124) × 106/ml, respectively (n = 19 animals). These values contrast with the findings reported here – (6.61 ± 3.36) × 106 peripheral leukocytes/ml and (1.29 ± 1.01)× 106 CD4 T cells/ml. Importantly, the enumerations reported here were determined from samples obtained throughout infection, from 2 weeks PI to 240 weeks PI and include data from all 4 infected cats (including the outlier cat 187). In a study by Torten et al. (1991), serial CD4 T cell enumerations of FIV-infected cats over a 42-month infection period are graphed but numerical data are not distilled into summary statistics. In the Torten et al. paper, the data are grouped as FIV positive or negative, while individual data from infected cats are not provided. In another study, averaged peripheral CD4 and CD8 T cell enumerations are reported over a 24-month period, but for only three time points (Ackley et al., 1990).
Previous researchers have observed that the longer a cat is infected with FIV, the more likely it is to have a lower CD4+ T cell number (Barlough et al., 1991) and that immune dysfunction in FIV is gradually progressive over time (Torten et al., 1991). In a rare 8 year long observation of FIV-infected SPF cats, a cat exhibiting a terminal FAIDS like syndrome had a terminal CD4/CD8 ratio of 0.075 and an absolute peripheral CD4 count of 21 cells/μl (Kohmoto et al, 1998). In the study reported here, two of the FIV-infected cats (cats 165 and 186) demonstrated persistent peripheral CD4 cell numbers less than 500 cells/μl of blood from 167 to 242 weeks PI (the nadir was 90 cells/μl, for both cats 165 and 186). In the same time period, these two cats demonstrate persistent peripheral CD8 T cell counts at or less than 1 × 103 cells/μl of blood. This is markedly reduced from the mean peripheral CD8 T cell count in the uninfected animals ((1.57 ± 0.77) × 103 cells/μl blood). Although the peripheral CD4 cell number is consistent with a CDC Stage II AIDS (200–499 cells/μl) or Stage III AIDS (<200cells/μl) (Center for Disease Control and Prevention Surveillance Brief, http://www.cdc.gov/hiv/library/factsheets/index.html), these cats maintained clinical normalcy throughout this 23-week period. It is possible that the protected environment in which the cats are housed prevented them from thus far succumbing to opportunistic infections. This hypothesis has been advanced previously by others (Ackley et al., 1990; Barlough et al., 1991). Alternatively, unlike in lentivirus-infected humans, the cat immune system might maintain functionality at this level of peripheral CD4 T cell reduction. Careful clinical observations of naturally FIV-infected client-owned cats suggest that low peripheral CD4 T cell counts appear to be tolerated (personal communication, Dr. K. Hartmann, University of Munich).
Our group previously reported an FIV-associated increase in the absolute number of peripheral CD8 T cells (Murphy et al., 2012); here, with progressively more complete data, we report no difference between FIV-infected and uninfected cats in regards to cumulative absolute CD8 T cell number. In other studies, FIV infection in cats has been associated with either an increased peripheral CD8 T cell number (Ackley et al., 1990) or no statistically significant difference relative to control animals (Barlough et al, 1991; Torten et al., 1991). It would seem that with FIV-disease progression, the absolute peripheral CD8 T cell numbers may decrease, and that CD8 T cell elevation therefore cannot be absolutely relied upon as a signature of FIV-infection.
Multiple single nucleotide mutations were identified in cloned proviral promoters (U3 and R regions) isolated from PBMC and peripheral CD4 T cells. A total of 11, 9, 13 and 11 LTR clones were sequenced from nucleic acids derived from the PBMC or CD4 T cells from cats 184, 187, 186 and 165. Of these 44 clones, 13 were identical to the wt sequence. Importantly, a single mutation in the U3 AP-1 binding site (g93a) was identified in freshly isolated proviral DNA from PBMC from 1 animal (187) and in freshly isolated CD4 T cells from 3 of 4 animals. We have previously demonstrated that this point mutation is transcriptionally inactivating in an in vitro system (Murphy et al, 2012). In order to determine the effect of this naturally arising U3 mutation on viral transcription ex vivo, we coincubated the column-isolated CD4T cells with SPF feline PBMC and assayed them for cell-associated vRNA (to assess viral transcriptional activity). CD4 T cells from all 4 FIV+ cats produced detectable vRNA in 7 to 23 days of incubation. The cell-associated vRNA was converted to cDNA, cloned and sequenced. Interestingly (and perhaps predictably) none of the sequenced U3/R regions derived from vRNA demonstrated the G93A AP-1 mutation, supporting the transcriptional lethality of this point mutation. This suggests that this promoter mutation, although common, does not serve as an additional mechanism of latency in our model. These findings are in agreement with previous in vitro studies in which a chloramphenicol acetyltransferase (CAT) reporter gene construct of a FIV AP-1 deletion (FIV-UK8) (Rigby et al, 1993) caused a 10-fold to 25-fold drop in CAT activity (Thompson et al, 1994). However, in other studies, mutant FIV proviruses derived from FIV-PPR (FIV subtype A) with deletion of the U3 AP-1 site demonstrated a negligible effect on viral expression and replication while mutant proviruses encoding deletions of both the AP-1 and ATF sites were severely restricted for virus expression (Bigornia et al., 2001). An infectious molecular clone of FIV with a 31 bp deletion of the U3 region including the AP-1 and AP-4 binding sequences demonstrated replication rates and cytopathology almost the same as wild type virus (Miyazawa et al., 1994). Thus, mutations within or around the AP-1 site may have varied consequences for the transcriptional activity of the LTR depending upon the particular strain and mutation.
It is interesting to note that the single cat (187) that required longer than 7 days for ex vivo activation of CD4 T cell-associated FIV transcription is the only animal from which no wild type FIV promoter was isolated on ex vivo culture day 0. Interestingly, cat 187 persistently demonstrated a CD4/CD8 ratio indistinguishable from the uninfected control animals, had a peripheral CD4 T cell number that never dropped below 1 × 103 cells/μl blood and had CD4 and CD8 T cell profiles similar to the two uninfected control cats (cats 183 and 185). In addition, this cat (187) is the only animal from which we were unable to amplify a wild type promoter from PBMC. In the 2 proviral clones obtained from cat 187's freshly isolated CD4+ T cells, one clone has the transcriptionally inactivating G93A mutation while the other has a mutation in the TATA box (T178C). This suggests the possibility that the existing viral promoter clones present in CD4T cells isolated from cat 187 may have reduced transcriptional fitness relative to wild type virus. It is therefore possible that the reduced fitness of cat 187's integrated CD4T cell proviral clones may be associated with the persistently normal CD4/CD8 ratio (>1.0) and the relatively normal chronological profiles of CD4 and CD8 T cells. Importantly, all 3 of the FIV-infected cats with inverted CD4/CD8 ratios (cats 165, 184 and 186) have readily detectable wild type viral promoters in freshly isolated CD4 T cells. For cat 187, the source of the “re-emergent” wild type promoter detected on day 23 of ex vivo CD4 T cell culture was not determined. We speculate that this wild type promoter represents a reversion mutation from a less fit precursor. Alternatively, the wild type promoter was present at low copy number and was therefore undetected in the freshly isolated CD4 T cells and PBMC. These cloning and sequencing results should be interpreted with some caution, as the relatively small number of sequenced promoters may not precisely reflect the actual in vivo diversity of the proviral promoter genotypes.
The FIV promoter with the C102A mutation was previously introduced into an expression construct and found to have transcriptional efficiency similar to the wild type promoter (Murphy et al, 2012). Unlike the G93A mutation, the C102A mutation lies between transcription factor binding sites. The transcriptional efficiencies of the other mutated promoters identified here were not determined in this study. Most of the documented viral RNA point mutations in the U3 region fall outside of the known transcription factor binding sites. No vRNA mutations were identified in binding sites for AP-1, the second AP4, C/EBP-1, C/EBP-2, ATF and the TATA box, suggesting that these promoter loci may be important for transcriptional competence. Proviral DNA mutations in the TATA box (T178C and A183C) and the short deletion in U5 (Δ348–352) were not identified in any of the vRNA clones, suggesting that these LTR mutants may not be transcriptionally competent. Interestingly, vRNA mutations were found in both the first AP-4 and NF-1 binding sites, indicating that these mutants were at least transcriptionally competent. The NF-1 mutant, G148A, is interesting in that the consensus binding sequence for NF1 is 5′ TGT (N6-7) GCCA (Thompson et al., 1994). The FIV-C wild type sequence is 5′ TGT (N6-7) GCCG, therefore the G148A mutation reverts the NF1 site back to the consensus sequence. Other U3 mutations identified in the vRNA (A17G, T24C, T76C, c102A, A138G, A168G and C191T) were not located within described transcription factor binding sites, suggesting that these regions of U3 may have a greater tolerance for mutation.
Five of the SNPs identified within U3 represent G to A transition mutations. It has previously been demonstrated that FIV lacking a functional dUTPase gene is prone to G to A transition mutations (Lerner et al., 1995; Payne and Elder, 2001). For this reason, the FIV-C dUTPase gene was amplified and sequenced both from the initial inoculating provirus and from PBMC DNA derived from the chronically-infected cats. All of these dUTPase sequences were identical, indicating that mutations within this gene had not taken place during the in vivo infection. It would seem, relative to the viral LTR region, sequence mutations within the FIV dUTPase gene are uncommon and that this gene product may be under strong selection pressure to maintain function.
Multiple features of the FIV-infected cat model demonstrate parallelism with HIV-infected people. Falling CD4 T cell numbers are a hallmark of HIV infection (Fauci et al., 1996) and most HIV-infected human patients have a reduced CD4/CD8 ratio prior to initiation of antiretroviral therapy (ART) (Serrano-Villar et al., 2014). Recent 2013 WHO guidelines advise initiation of ART at 500 (or less) CD4 T cells/μl of blood (Doherty et al., 2013). Well established documentation also suggests that low CD8 T cell counts predict the onset of AIDS in HIV-infected individuals (Karim et al., 2013). As mentioned previously, two cats in our study demonstrate persistently reduced numbers of peripheral CD4 (less than 500 cells/μl) and CD8 T cells. Remarkably, these animals seem to be clinically normal. Sequence variation has been documented within the HIV-1 LTR, including the AP-1 binding site, in virus-infected patients (Ait-Khaled and Emery, 1994). In 3 of 12 HIV-1 infected individuals, mutations within the U3 and TAR region of the HIV LTR seem to be responsible for enhanced promoter transcription, possibly leading to accelerated disease progression (Hiebenthal-Millow et al., 2003).
5. Conclusions
FIV-infected cats during the asymptomatic phase have a persistently inverted CD4/CD8 ratio, a decreased number of peripheral CD4 T cells, and a decreased percentage of CD25+ cells relative to uninfected control animals. The infected cats demonstrated multiple mutations within the proviral DNA isolated from PBMC and peripheral CD4+ T-cells. These U3 promoter mutations may have contributed to transcriptional latency; however, since provirus with a wild type U3 promoter can also apparently achieve latency, promoter mutation is apparently not required to create a latent state.
Acknowledgments
The authors are grateful for the excellent animal care provided by Monica Durden and the staff of the University of California, Davis Feline Research Laboratory. The authors would like to acknowledge Drs. Ed Hoover and Niels Pedersen for providing the biological isolate of FIV-C-Pgmr and editing assistance provided by Dr. Kirsten Murphy. Funding for this study was provided, in part, by the Center for Companion Animal Health, School of Veterinary Medicine, University of California, Davis. SM was supported by NIH T32 training grants #5TL1RR024145 and #5T32AI060555.
References
- Ackley CD, Yamamoto JK, Levy N, Pedersen NC, Cooper MD. Immunologic abnormalities in pathogen-free cats experimentally infected with feline immunodeficiency virus. J Virol. 1990;64(11):5652–5655. doi: 10.1128/jvi.64.11.5652-5655.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ait-Khaled M, Emery VC. Phylogenetic relationship between human immunodeficiency virus type 1 (HIV-1) long terminal repeat natural variants present in the lymph node and peripheral blood of three HIV-1-infected individuals. J Gen Virol. 1994;75(Pt 7):1615–1621. doi: 10.1099/0022-1317-75-7-1615. [DOI] [PubMed] [Google Scholar]
- Barlough JE, Ackley CD, George JW, Levy N, Acevedo R, Moore PF, Rideout BA, Cooper MD, Pedersen NC. Acquired immune dysfunction in cats with experimentally induced feline immunodeficiency virus infection: comparison of short-term and long-term infections. J Acquir Immune Defic Syndr. 1991;4(3):219–227. [PubMed] [Google Scholar]
- Beebe AM, Dua N, Faith TG, Moore PF, Pedersen NC, Dandekar S. Primary stage of feline immunodeficiency virus infection: viral dissemination and cellular targets. J Virol. 1994;68(5):3080–3091. doi: 10.1128/jvi.68.5.3080-3091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendinelli M, Pistello M, Lombardi S, Poli A, Garzelli C, Matteucci D, Ceccherini-Nelli L, Malvaldi G, Tozzini F. Feline immunodeficiency virus: an interesting model for AIDS studies and an important cat pathogen. Clin Microbiol Rev. 1995;8(1):87–112. doi: 10.1128/cmr.8.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigornia L, Lockridge KM, Sparger EE. Construction and in vitro characterization of attenuated feline immunodeficiency virus long terminal repeat mutant viruses. J Virol. 2001;75(2):1054–1060. doi: 10.1128/JVI.75.2.1054-1060.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhard MJ, Dean GA. Transmission and immunopathogenesis of FIV in cats as a model for HIV. Curr HIV Res. 2003;1(1):15–29. doi: 10.2174/1570162033352101. [DOI] [PubMed] [Google Scholar]
- Center for Disease Control and Prevention Surveillance Brief. http://www.cdc.gov/hiv/library/factsheets/index.html.
- Chase AJ, Sedaghat AR, German JR, Gama L, Zink MC, Clements JE, Siliciano RF. Severe depletion of CD4+ CD25+ regulatory T cells from the intestinal lamina propria but not peripheral blood or lymph nodes during acute simian immunodeficiency virus infection. J Virol. 2007;81(23):12748–12757. doi: 10.1128/JVI.00841-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean GA, Quackenbush SL, Ackley CD, Cooper MD, Hoover EA. Flow cytometric analysis of T-lymphocyte subsets in cats. Vet Immunol Immunopathol. 1991;28(3–4):327–335. doi: 10.1016/0165-2427(91)90124-u. [DOI] [PubMed] [Google Scholar]
- Doherty M, Ford N, Vitoria M, Weiler G, Hirnschall G. The 2013 WHO guidelines for antiretroviral therapy: evidence-based recommendations to face new epidemic realities. Curr Opin HIV AIDS. 2013;8(6):528–534. doi: 10.1097/COH.0000000000000008. [DOI] [PubMed] [Google Scholar]
- Eisele E, Siliciano RF. Redefining the viral reservoirs that prevent HIV-1 eradication. Immunity. 2012;37(3):377–388. doi: 10.1016/j.immuni.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauci AS, Pantaleo G, Stanley S, Weissman D. Immunopathogenic mechanisms of HIV infection. Ann Intern Med. 1996;124(7):654–663. doi: 10.7326/0003-4819-124-7-199604010-00006. [DOI] [PubMed] [Google Scholar]
- Hiebenthal-Millow K, Greenough TC, Bretttler DB, Schindler M, Wildum S, Sullivan JL, Kirchhoff F. Alterations in HIV-1 LTR promoter activity during AIDS progression. Virology. 2003;317(1):109–118. doi: 10.1016/j.virol.2003.08.034. [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Kawaguchi Y, Tomonaga K, Inoshima Y, Kohmoto M, Miyazawa T, Mikami T. Regulatory properties of the integrated long terminal repeat of the feline immunodeficiency virus. Virus Res. 1996;41(2):201–207. doi: 10.1016/0168-1702(96)01284-1. [DOI] [PubMed] [Google Scholar]
- Joshi A, Garg H, Tompkins MB, Tompkins WA. Different thresholds of T cell activation regulate FIV infection of CD4+ CD25+ and CD4+ CD25– cells. Virology. 2005;335(2):212–221. doi: 10.1016/j.virol.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Joshi A, Vahlenkamp TW, Garg H, Tompkins WA, Tompkins MB. Preferential replication of FIV in activated CD4(+)CD25(+)T cells independent of cellular proliferation. Virology. 2004;321(2):307–322. doi: 10.1016/j.virol.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Karim R, Mack WJ, Stiller T, Operskalski E, Frederick T, Landay A, Young MA, Tien PC, Augenbraun M, Strickler HD, Kovacs A. Association of HIV clinical disease progression with profiles of early immune activation: results from a cluster analysis approach. AIDS. 2013;27(9):1473–1481. doi: 10.1097/QAD.0b013e3283601bad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohmoto M, Uetsuka K, Ikeda Y, Inoshima Y, Shimojima M, Sato E, Inada G, Toyosaki T, Miyazawa T, Doi K, Mikami T. Eight-year observation and comparative study of specific pathogen-free cats experimentally infected with feline immunodeficiency virus (FIV) subtypes A and B: terminal acquired immunodeficiency syndrome in a cat infected with FIV petaluma strain. J Vet Med Sci. 1998;60(3):315–321. doi: 10.1292/jvms.60.315. [DOI] [PubMed] [Google Scholar]
- Lerner DL, Wagaman PC, Phillips TR, Prospero-Garcia O, Henriksen SJ, Fox HS, Bloom FE, Elder JH. Increased mutation frequency of feline immunodeficiency virus lacking functional deoxyuridine-triphosphatase. Proc Natl Acad Sci U S A. 1995;92(16):7480–7484. doi: 10.1073/pnas.92.16.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnel SJ, Sparger EE, Luciw PA, Murphy BG. Pharmacologic reactivation of latent feline immunodeficiency virus ex vivo in peripheral CD4+ T-lymphocytes. Virus Res. 2012a;170:174–179. doi: 10.1016/j.virusres.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnel SJ, Sparger EE, Luciw PA, Murphy BG. Transcriptional regulation of latent feline immunodeficiency virus in peripheral CD4+ T-lymphocytes. Viruses. 2012b;4(5):878–888. doi: 10.3390/v4050878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa T, Inoshima Y, Kohmoto M, Ikeda Y, Mikami T. Growth properties of a feline immunodeficiency virus mutant which lacks an AP-1 binding site in primary peripheral blood mononuclear cells. J Vet Med Sci. 1994;56(5):869–872. doi: 10.1292/jvms.56.869. [DOI] [PubMed] [Google Scholar]
- Murphy B, Vapniarsky N, Hillman C, Castillo D, McDonnel S, Moore P, Luciw PA, Sparger EE. FIV establishes a latent infection in feline peripheral blood CD4+ T lymphocytes in vivo during the asymptomatic phase of infection. Retrovirology. 2012;9:12. doi: 10.1186/1742-4690-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace MJ, Agosto L, Graf EH, O'Doherty U. HIV reservoirs and latency models. Virology. 2011;411(2):344–354. doi: 10.1016/j.virol.2010.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne SL, Elder JH. The role of retroviral dUTPases in replication and virulence. Curr Protein Pept Sci. 2001;2(4):381–388. doi: 10.2174/1389203013381008. [DOI] [PubMed] [Google Scholar]
- Pedersen NC, Barlough JE. Clinical overview of feline immunodeficiency virus. J Am Vet Med Assoc. 1991;199(10):1298–1305. [PubMed] [Google Scholar]
- Rigby MA, Holmes EC, Pistello M, Mackay A, Brown AJ, Neil JC. Evolution of structural proteins of feline immunodeficiency virus: molecular epidemiology and evidence of selection for change. J Gen Virol. 1993;74(Pt 3):425–436. doi: 10.1099/0022-1317-74-3-425. [DOI] [PubMed] [Google Scholar]
- Serrano-Villar S, Moreno S, Fuentes-Ferrer M, Sanchez-Marcos C, Avila M, Sainz T, de Villar N, Fernandez-Cruz A, Estrada V. The CD4:CD8 ratio is associated with markers of age-associated disease in virally suppressed HIV-infected patients with immunological recovery. HIV Med. 2014;15:40–49. doi: 10.1111/hiv.12081. [DOI] [PubMed] [Google Scholar]
- Sparger EE, Beebe AM, Dua N, Himathongkam S, Elder JH, Torten M, Higgins J. Infection of cats with molecularly cloned and biological isolates of the feline immunodeficiency virus. Virology. 1994;205(2):546–553. doi: 10.1006/viro.1994.1677. [DOI] [PubMed] [Google Scholar]
- Thompson FJ, Elder J, Neil JC. Cis- and trans-regulation of feline immunodeficiency virus: identification of functional binding sites in the long terminal repeat. J Gen Virol. 1994;75(Pt 3):545–554. doi: 10.1099/0022-1317-75-3-545. [DOI] [PubMed] [Google Scholar]
- Torten M, Franchini M, Barlough JE, George JW, Mozes E, Lutz H, Pedersen NC. Progressive immune dysfunction in cats experimentally infected with feline immunodeficiency virus. J Virol. 1991;65(5):2225–2230. doi: 10.1128/jvi.65.5.2225-2230.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]