Abstract
Candida albicans is a leading pathogen in infections of central venous catheters, which are frequently infused with heparin. Binding of C. albicans to medically relevant concentrations of soluble and plate-bound heparin was demonstrable by confocal microscopy and enzyme-linked immunosorbent assay (ELISA). A sequence-based search identified 34 C. albicans surface proteins containing ≥1 match to linear heparin-binding motifs. The virulence factor Int1 contained the most putative heparin-binding motifs (n = 5); peptides encompassing 2 of 5 motifs bound to heparin-Sepharose. Alanine substitution of lysine residues K805/K806 in 804QKKHQIHK811 (motif 1 of Int1) markedly attenuated biofilm formation in central venous catheters in rats, whereas alanine substitution of K1595/R1596 in 1593FKKRFFKL1600 (motif 4 of Int1) did not impair biofilm formation. Affinity-purified immunoglobulin G (IgG) recognizing motif 1 abolished biofilm formation in central venous catheters; preimmune IgG had no effect. After heparin treatment of C. albicans, soluble peptides from multiple C. albicans surface proteins were detected, such as Eno1, Pgk1, Tdh3, and Ssa1/2 but not Int1, suggesting that heparin changes candidal surface structures and may modify some antigens critical for immune recognition. These studies define a new mechanism of biofilm formation for C. albicans and a novel strategy for inhibiting catheter-associated biofilms.
Heparin is a highly sulfated, nonbranched, anionic polysaccharide composed of uronic acid (predominantly iduronic acid) in 1,4 linkage with glucosamine [1]. Doses of heparin up to 1000 U/mL are frequently used for anticoagulation in central venous catheters [2]; hence, the interactions between heparin and microorganisms may have important implications for catheter-associated infections.
Human immunodeficiency virus type 1 gp120 and Tat are known to bind to heparin [3, 4], and many more microbial proteins bind to heparan sulfate, an identical polysaccharide with reduced sulfation [reviewed in 1]. In one study, four intracellular DNA-binding proteins from C. albicans (Gcf1, Nhp6, Htb1, Hta1) bound to heparin-Sepharose [5]. However, the amino acids that mediate microbial binding to heparin have not been identified.
Eukaryotic proteins that bind heparin express conformational or linear heparin-binding motifs (HBMs). Conformational motifs include the recently described CPC clip motif, a structural signature in which 2 cationic residues surround 1 polar residue [6, 7]. Linear HBMs are conserved sequences of basic (B) and hydropathic (X) amino acids in specific patterns identified by Cardin (XBBXBX), Weintraub (XBBBXXBX), and Sobel (XBBBXXBBBXXBBX) [1, 8, 9]. Basic amino acids, such as lysine, arginine, and, rarely, histidine, are critical for interaction with anionic sulfate or carboxylate groups in heparin through electrostatic and hydrogen bonds. For example, substitution of alanine for basic amino acids in linear HBMs in the morphogen Sonic Hedgehog abolished binding to heparin [10]. Because of the widespread use of heparin in central venous catheters, we identified linear HBMs in C. albicans surface proteins and characterized their functions in heparin binding and biofilm formation.
METHODS
Sequence-Based Search for Determination of Linear HBMs
Nonredundant proteins were identified from Gene Ontology functional annotations in the Candida Genome Database (www.candidagenome.org; Assembly 21) and relevant references [11, 12] and were mapped to Assembly 21. In the search for HBMs, basic amino acids were H, K, and R. Hydropathic amino acids were W, F, Y, L, I, C, M, G, V, S, T, A, N, P, and Q. The 159 proteins that met the search criteria were manually curated to confirm surface localization by prediction of a glycosylphosphatidylinositol anchoring sequence [13–16] or by assignment to the cell surface in the Candida Genome Database.
Strains, Media, and Chemicals
C. albicans laboratory strains used in this study were BWP17wt [17] and derivatives, as described in Supplementary Table 1. All strains were maintained at −80°C in 20% glycerol. Working cultures were plated on yeast peptone dextrose (YPD) plates (10 g of yeast extract, 20 g of peptone, 20 g of glucose, and 1.5% agar per liter) at 30°C for 48 hours then stored at 4°C.
Buffers, including Roswell Park Memorial Institute medium (RPMI)–1% HEPES and RPMI–5% MOPS (3-morpholinopropane-1-sulfonic acid), were obtained from Life Technologies. Formaldehyde was purchased from Fisher, dimethyl sulfoxide and calcofluor white from Sigma, filipin from Polysciences, and Fluoromount G from SouthernBiotech. Commercial-grade heparin was purchased from Sigma, pharmaceutical heparin (1000 U/mL preservative free or 20 000 U/mL with 1.5 mg/mL methylparaben and 0.15 mg/mL propylparaben) from AAP Pharmaceuticals, and desulfated heparin analogs from Neoparin.
Binding of Heparin-Alexa Fluor 488 to C. albicans
Heparin was labeled with Alexa Fluor 488 by a method modified from Osmond et al [18]. After purification on a PD-10 desalting column (GE Healthcare) equilibrated in autoclaved nanopure water, fractions containing the labeled material detected at 490 nm were combined and dried overnight on a SpeedVac concentrator (Savant) with heating. The resulting solid was redissolved in autoclaved nanopure water to 10 mg/mL and stored at 4°C.
After overnight growth at 30°C in YPD, 2 × 107 C. albicans cells (BWP17wt) were labeled with a PKH26 Red Fluorescent Cell staining kit (Sigma), according to manufacturer's instructions, followed by Heparin-Alexa Fluor 488 (0.1 mL of 10 mg/mL solution). Organisms were incubated at 30°C with shaking (225 rpm) for 30 minutes, at which time a 0.4-mL aliquot was removed, pelleted (10 000 rpm for 3 minutes), and washed twice with phosphate-buffered saline (PBS). After reconstitution with 0.5 mL of PBS, 4′,6-diamidino-2-phenylindole dihydrochloride (Sigma, stock solution of 5 mg/mL) was added to a final concentration of 1 µg/mL and the solution allowed to stand at room temperature for 10 minutes. Cells were pelleted, washed twice with PBS, and then mounted on a microscope slide using Fluoromount G.
ELISA Techniques
Sigma heparin (fresh solution made daily), diluted to 25 U/mL in autoclaved, sterile-filtered PBS, was added as 0.1-mL aliquots (2.5 U) to each well of an allylamine-coated 96-well heparin-binding microtiter plate (BD Biosciences). In indicated experiments, equimolar amounts of desulfated heparins were added in lieu of heparin. The plate was incubated at room temperature overnight in the dark [19]. In the morning, the plate was washed with acetate buffer (100 mmol/L sodium chloride [NaCl], 50 µmol/L sodium acetate, 0.2% Tween 20; pH 7.2), incubated with 3% bovine serum albumin in PBS at 30°C for 1 hour, and then washed with PBS. Overnight cultures of C. albicans BWP17wt grown at 30°C in YPD with shaking at 225 rpm were diluted to an optical density at 600 nm (OD600) of 0.2 in 25 mL of YPD and grown at 30°C to midlog phase (OD600, 0.6–0.7). Cells were pelleted (3000 rpm for 7 minutes), washed twice with PBS, and diluted in RPMI-HEPES to 4 × 105, 2 × 105, and 1 × 105 colony-forming units (CFU)/mL, respectively, before application of 100 µL per well.
After incubation at 30°C for 1 hour and washing with PBS, 0.1 mL of biotinylated rabbit anti–C. albicans IgG recognizing soluble proteins in a C. albicans lysate (Meridian Life Science), diluted 1:2500 in assay buffer (0.3% bovine serum albumin in PBS with 0.05% Tween 20), was added to each well. After incubation at 30°C for 1 hour and washing with PBS-Tween (0.05% Tween 20 in PBS), 0.1 mL of streptavidin alkaline phosphatase (Biolegend) diluted 1:10 000 in assay buffer was added, and the plate was incubated at 30°C for 30 minutes. After washing with PBS-Tween and alkaline phosphatase buffer (100 mmol/L Tris base, 50 µmol/L magnesium chloride, 100 mmol/L NaCl; pH 9.5.), 0.1 mL of alkaline phosphatase substrate (KPL) was added to each well for 45 minutes, and then absorbance at 595 nmol/L was read on a Beckman Coulter DTX 880. Experiments were performed in quadruplicate.
Construction of Mutants
Candida albicans genomic DNA was isolated from saturated overnight cultures using glass beads, as described elsewhere [20]. A lithium acetate method was used to transform C. albicans [17]. Plasmids and polymerase chain reaction (PCR) products were purified using kits (Fermentas) or established methods [21]. Pfu enzyme (New England Biolabs) with High Fidelity buffer was employed for all amplifications. Products were sequenced to affirm fidelity before use. Primers are described in Supplementary Table 2. A single copy of INT1, including 1450 base pairs upstream and 548 base pairs downstream from the INT1 open reading frame (www.candidagenome.org; Assembly 21), was integrated into the hisG locus of the int1−/− strain VBIDM2 [22] to produce the reconstituted strain KO509. Briefly, a copy of INT1 was generated by PCR, using primers 1 and 2 with BWP17wt DNA as template and cloned into the SacI/MluI sites of pGEMHIS [17] to create pKO509. pKO509 was digested with SwaI and transformed into VBIDM2 to create the reconstituted strain KO509.
PCR-mediated overlap extension mutagenesis [23] was used to produce copies of INT1 mutated at putative heparin-binding domains. Briefly, primer pairs 1 + 3 and 2 + 4 (or 1 + 5 and 2 + 6) were used to produce 2 overlapping fragments of INT1 in which putative HBMs were mutated (FKKRFFKL → FKAAFFKL or KQKKHQ →KQAAHQ), and a full-length mutated sequence was generated in a third PCR using primers 1 + 2 with the fragments as template. The mutated sequences were cloned into the SacI/MluI sites of pGEMHIS to create pKO503 and pKO507, respectively. A construct mutated at both sites (pKO508) was produced using primers 1 + 5 and 2 + 6 with plasmid DNA pKO503 as template. Full-length mutated products cloned into pGEMHIS were used to transform VBIDM2 as described above, producing strains KO503, KO507, and KO508. The correct insertion and orientation of all constructs were confirmed by PCR. In growth curves performed with and without 100 U/mL heparin at 30° C in RPMI-HEPES, there was no difference in doubling times between the wild type, the double disruptant, the INT1 reintegrant, and the reintegrants containing alanine substitutions in motif 1, motif 4, or motif 1&4.
Identification of Peptides From C. albicans by Mass Spectrometry
Ten million (1 × 107) C. albicans were incubated with 250 U of pharmaceutical heparin (Hep+) or without heparin (Hep−) in RPMI-HEPES for 1 hour at 37°C on a rotator. Organisms were pelleted and discarded, and supernatants were removed. Next, 300 µL of Hep+ and Hep− supernatants were incubated with 100 µL of avidin agarose beads (Thermo Scientific) for 60 minutes at room temperature on a rotator in the presence of 50 U heparin (Hep+); an equal volume of RPMI-HEPES was substituted for heparin in the Hep− supernatants. Beads were pelleted; 100 µL of beads were incubated with 100 µL of 3.0 mol/L NaCl for 30 minutes at room temperature on a rotator. Beads were pelleted, and the supernatants were withdrawn, precipitated with an equal volume of trichloroacetic acid (TCA), incubated on ice overnight at 4°C, and stored at −80°C until analysis by mass spectroscopy.
Six biological replicates of TCA-precipitated proteins from equal cellular equivalence of heparin-treated (Hep+) and untreated (Hep−) conditioned medium from cultures of C. albicans were solubilized in 50 µL of Laemmli buffer. Samples were subjected to buffer exchange and concentration using an Amicon Ultra 3-kDa microfuge filtration cartridge at 14 000 g for 15 minutes with 5 subsequent additions of ×1 Laemmli buffer (50 µL) between spins. The resulting retained proteins (6 Hep+ and 6 Hep−) were subsequently prepared for sodium dodecyl sulfate polyacrylamide gel electrophoresis by combining into 2 pools of 3 samples for the medium conditioned with Hep+ and Hep−. The replicate sample pools were loaded onto 2 4%–12% mini gels and separated using the MOPS buffer system followed by silver staining (Sigma Proteosilver). Gel lanes from the replicates of Hep+ and Hep− samples were gridded into 11 equal regions, followed by in gel trypsin digestion and extraction of peptides, as described elsewhere [24].
The recovered peptides from the gridded gel sections were analyzed by liquid chromatography coupled nanoelectrospray mass spectrometry on a TripleTOF 5600 (AB Sciex) attached to an Eksigent NanoLC ultra nanoflow system. Only proteins with a minimum of 2 peptides with Mascot peptide score indicating a peptide identity and a false discovery rate against an inverse database at <1% were reported. Semiquantitative measurements between the Hep+ and Hep− proteins were generated by using a minimum of 2 tryptic peptides from each protein as surrogates for the amount of proteins from the 2 groups. This was accomplished by capturing extracted ion profiles for each peptide and comparing the monoisotopic peak intensity at the apex of the signal for the M + 2H or M + 3H signal for each peptide.
Antibody Production
A peptide corresponding to amino acids 799HKQEKQKKHQIHKV812 (motif 1 of Int1 underlined) was conjugated to KLH via an N-terminal cysteine residue (Pacific Immunology; www.pacificimmunology.com; National Institutes of Health animal welfare assurance No. A41820-01; US Department of Agriculture license 93-R-283). Two NZW rabbits were immunized once with conjugated peptide in a proprietary formulation of Freund's complete adjuvant and boosted 3 times with conjugated peptide in Freund's incomplete adjuvant. The same peptide was conjugated to cyanogen bromide–Sepharose and used for affinity purification of epitope-specific IgG. The final serum titer for both animals was >1:100 000 by ELISA. Preimmune serum and serum from bleed 3 were chromatographed on a 2-mL Protein A column (Thermo Scientific) to yield pre- and postimmune IgG.
Rat Model of Biofilm Formation
Polyethylene catheters were inserted into the jugular veins of female Sprague-Dawley rats, heparinized with 100 U/mL heparin, and clamped for 24 hours, as described elsewhere [25]. After removal of 0.5 mL for culture, the desired C. albicans strain was instilled and allowed to dwell for 24 hours before animals were euthanized. Catheters were removed aseptically, as described elsewhere [25], and prepared for scanning electron microscopy at ×100 and ×2000 to assess biofilm formation on the intraluminal surface of the catheter. For antibody experiments, 1 × 107 C. albicans yeast cells (BWP17wt) were preincubated with a 1:10 dilution of preimmune IgG or affinity-purified IgG (both at 1.0 mg/mL) for 1 hour at 30°C, before injection into the catheter and retention for 24 hours.
Ethics Statement
All animal procedures were approved by the Institutional Animal Care and Use Committee at the University of Wisconsin, according to the guidelines of the Animal Welfare Act, the Institute of Laboratory Animal Resources Guide for the Care and Use of Laboratory Animals, and Public Health Service Policy.
Statistical Analyses
Statistical analyses were performed using the GraphPad Prism 6 statistical software package. The Bonferroni correction was applied whenever multiple comparisons were made.
RESULTS
Linear HBMs in C. albicans Surface Proteins
Approximately 400 C. albicans proteins have been identified in cell wall fractions from yeast or hyphae, depending upon the extraction technique [11]. The 34 proteins in Table 1 include only those with one of the following listed as the cellular component in the Candida Genome Database: cell surface, fungal cell wall, yeast cell wall, or hyphal cell wall. Three of these proteins—Als7, Pga4, and Rbt1—have glycosylphosphatidylinositol anchors. The genes encoding 11 of 34 proteins—Als7, Cat1, Dot4, Eno1, Gph1, Ino1, Rbt1, Sam2, Srb1, Ssa2, and Ugp1—are located in newly defined transcriptionally active regions that are involved in biofilm formation [26].
Table 1.
Putative Linear Heparin-Binding Motifs in Candida albicans Surface Proteins
| Gene ID (Assembly 21) | Protein | Referencea | No. of Motifs | Motif 1 | Motif 2 | Motif 3 | Motif 4 | Motif 5 |
|---|---|---|---|---|---|---|---|---|
| orf19.4257 | Int1 | C | 5 | 804QKKHQIHKb | 1383THKGRFc | 1530MKRGKPc | 1593FKKRFFKLb | 1612SHKTRAc |
| orf19.1738 | Ugp1 | B, D | 3 | 176SHRIRVc | 310IKKFKYc | 368IRHFKGc | … | … |
| orf19.3370 | Dot4 | B | 3 | 442NKKGKSc | 519CHKCHNc | 635FKRFKFc | … | … |
| orf19.3651 | Pgk1 | A, B, D | 3 | 136GKKVKAc | 146VKKFRQc | 168AHRAHSc | … | … |
| orf19.4660 | Rps6A | B, D | 2 | 185QRKRALKAb | 192AKKVKNc | … | … | … |
| orf19.5107 | Not5 | B | 2 | 114QKRSRFc | 333VKKLKPc | … | … | … |
| orf19.5130 | Pdi1 | D | 2 | 210NKKFKNc | 301GKKYRGc | … | … | … |
| orf19.6387 | Hsp104 | D | 2 | 53VKRARYc | 199ARRSKSc | … | … | … |
| orf19.1065 | Ssa2 | B, D | 1 | 258LRRLRTc | … | … | … | … |
| orf19.1067 | Gpm2 | B | 1 | 45IKKNHLc | … | … | … | … |
| orf19.1327 | Rbt1 | A, B, C | 1 | 121GKKVKQc | … | … | … | … |
| orf19.2762 | Ahp1 | B, D | 1 | 171LKRIHNc | … | … | … | … |
| orf19.2803 | Hem13 | B, D | 1 | 257IRRGRYc | … | … | … | … |
| orf19.3590 | Ipp1 | B, D | 1 | 74TKKGKLc | … | … | … | … |
| orf19.377 | Phr3 | B, C | 1 | 115PHHHLNRYb | … | … | … | … |
| orf19.395 | Eno1 | B, D | 1 | 141AKKGKFc | … | … | … | … |
| orf19.3966 | Crh12 | A, B, C | 1 | 420TKHIHNc | … | … | … | … |
| orf19.4035 | Pga4 | B, C, D | 1 | 259AKRPRPc | … | … | … | … |
| orf19.4152 | Cef3 | B | 1 | 623LRKYKGc | … | … | … | … |
| orf19.4304 | Gap1 | B | 1 | 72QRKLKTc | … | … | … | … |
| orf19.4980 | Ssa1 | B, D | 1 | 259LRRLRTc | … | … | … | … |
| orf19.5188 | Chs1 | B, C | 1 | 126PKRQKTc | … | … | … | … |
| orf19.5788 | Eft2 | B, D | 1 | 581NKHNRIc | … | … | … | … |
| orf19.6190 | Srb1 | B, D | 1 | 123FHKAHGc | … | … | … | … |
| orf19.6214 | Atc1 | A, B | 1 | 959PKRVKVc | … | … | … | … |
| orf19.6229 | Cat1 | B | 1 | 82GKKTRIc | … | … | … | … |
| orf19.6367 | Ssb1 | B, D | 1 | 263LRRLRTc | … | … | … | … |
| orf19.657 | Sam2 | B, D | 1 | 380PKKLKFc | … | … | … | … |
| orf19.6814 | Tdh3 | B, D | 1 | 70GHKIKVc | … | … | … | … |
| orf19.7021 | Gph1 | B, D | 1 | 656TKHHIPKAb | … | … | … | … |
| orf19.7400 | Als7 | B, C | 1 | 1482SKRNKNc | … | … | … | … |
| orf19.7585 | Ino1 | B, D | 1 | 151MKRAKVc | … | … | … | … |
| orf19.7676 | Xyl2 | A, B, D | 1 | 330THRFKFc | … | … | … | … |
| orf19.940 | Bud2 | B | 1 | 716LRKGKSc | … | … | … | … |
a Letter designations refer to descriptors found in the following sources: A, “cell wall” in the protein description obtained from the Candida Genome Database (as of January 2011); B, “cell wall” in the Gene Ontology annotation (as of January 2011) obtained from the Candida Genome Database; C, “cell wall proteins” in the review by Alberti-Segui et al [12]; and D, “cell wall proteins” in the review by Chaffin [11].
b Weintraub motif.
c Cardin motif.
No Effects of Heparin on C. albicans Growth and Morphology
Heparin at 100 U/mL—the concentration recommended to prevent clotting of central venous catheters [2]—had no effect on doubling times of planktonic yeast cells grown in YPD, RPMI-HEPES, RPMI-MOPS, or Complete Supplemental Medium (CSM). There were no differences between heparin-treated and untreated organisms in formation of hyphae, integrity of membrane sterols, or location of septin rings (Supplementary Figure).
Binding of Heparin by C. albicans In Vitro
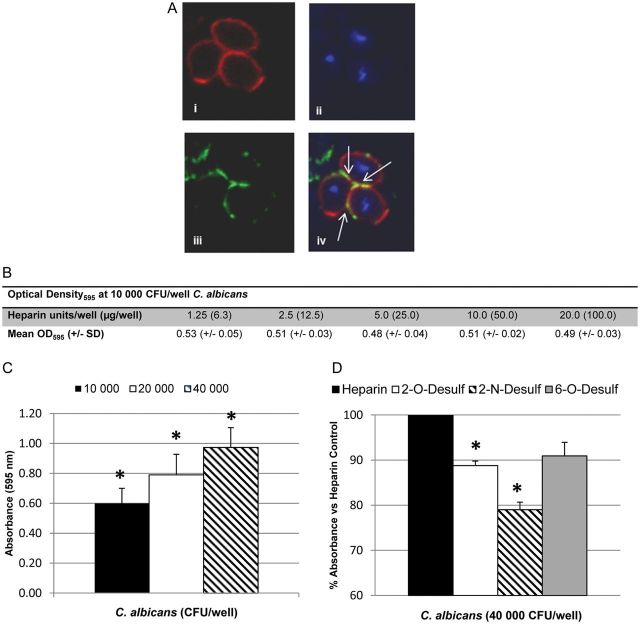
Deposition of soluble heparin–Alexa Fluor 488 was seen at the cell surface (Figure 1A iii) and at the interface of adjoining yeast cells (Figure 1A iv). In an ELISA, binding of C. albicans to solid-phase heparin remained stable from 1.25 U (6.3 µg) to 20 U (100 µg) per well (Figure 1B). Absorbance increased linearly as the input of C. albicans increased from 1 × 104 to 4 × 104 CFU per well (Figure 1C).
Figure 1.
Candida albicans binds heparin in vitro. A, Confocal microscopy of C. albicans after staining with PKH26 (top left), 4′,6-diamidino-2-phenylindole dihydrochloride (top right), or heparin–Alexa Fluor 488 (bottom left). Arrows (bottom right) show colocalization of heparin with C. albicans cell surface. B, Binding of 10 000 colony-forming units (CFU) of C. albicans (optical density at 595 nm [OD595]) to increasing concentrations of heparin immobilized on an allylamine-coated 96-well microtiter plate. Values represent means±standard deviations (SDs) of duplicate wells. C, Heparin-binding enzyme-linked immunosorbent assay with 2.5 U of heparin per well and increasing C. albicans input. Graph represents means ± SDs of 4 experiments; *P < .007 for all inputs. D, Binding of C. albicans (40 000 CFU per well) to equimolar amounts of heparin analogs desulfated at the 2-O, 2-N, or 6-O positions versus heparin control (normalized to 100%). Graph represents means ± SDs of 3 experiments, performed in triplicate *P ≤ .003 vs heparin control.
Heparin's extremely high anionic charge is contributed by sulfated residues at position 2-O of iduronic acid and the 2-N and 6-O positions of glucosamine. With equimolar amounts of desulfated heparins, binding of 4 × 104 C. albicans was decreased by 11% when heparin was desulfated at the 2-O position (P = .003), and by 21% with desulfation at the 2-N position (P = .002; Figure 1D). Desulfation at the 6-O position did not significantly reduce binding. The 2-O and 2-N sulfate residues are adjacent in the helical wheel structure of heparin, potentially indicating a preferential “face” for heparin binding [27].
Int1, the surface protein that encodes the largest number of putative HBMs (Table 1), is a serodominant antigen involved in adhesion, filamentation, and virulence [28, 29]. To understand whether predicted HBMs in Int1 were functional, overlapping His-tagged polypeptides encompassing amino acids 51–385, 385–659, 656–1193, 1188–1551, and 1548–1711 of Int1 were expressed in Saccharomyces cerevisiae. Polypeptides spanning amino acids 656–1193 and amino acids 1548–1711 bound to a heparin-Sepharose column and were eluted with 0.5–1 mol/L NaCl. Polypeptides spanning amino acids 51–385, 385–659, and 1188–1551 failed to bind.
Amino acids 656–1193 encompass one potential heparin-binding site, 804QKKHQIHK (basic residues underlined; motif 1 in Table 1). Amino acids 1548–1711 encompass a canonical Weintraub motif 1593FKKRFFKL (motif 4 in Table 1) and a canonical Cardin motif 1612SHKTRA (motif 5). The 3 lysine residues and single arginine residue in motif 4 are located on the rim of a positively charged pocket that might facilitate binding via electrostatic interaction to a strong anion, such as heparin. Motif 5 did not share this conformation.
Standard PCR-mediated mutagenesis [17] was used to derive a set of isogenic mutants with alanine substitutions in basic residues in HBMs 1 and 4. In addition to the wild-type strain BWP17wt and the ▵int1 double disruptant, 4 reintegrants were made by reinsertion of a single copy of the INT1 gene into the double disruptant. The reintegrant contained 1 wild-type copy of INT1. In the remaining single-copy reintegrants, alanine substitutions were made in 2 basic residues in motif 1 (804QAAHQIHK), motif 4 (1593FKAAFFKL), or both.
Influence of HBMs on Biofilm Production in Central Venous Catheters
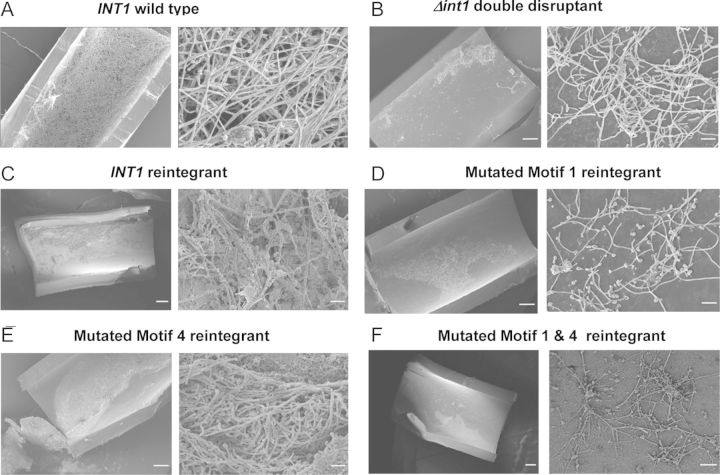
In a rat model of biofilm formation in heparinized jugular venous catheters [25], the wild-type strain BWP17wt generated a profuse biofilm (Figure 2A). Biofilm formation by the ▵int1 double disruptant was much reduced (Figure 2B). Reintegration of 1 wild-type copy of INT1 restored a profuse biofilm (Figure 2C). However, alanine substitution of lysines K805/K806 in motif 1 greatly impaired biofilm formation (Figure 2D). Although alanine substitution of lysine K1595 and arginine K1596 in motif 4 did not reduce biofilm formation, the motif 1&4 mutant again produced sparse biofilm (Figure 2E and 2F). Preincubation of C. albicans with an affinity-purified IgG antibody recognizing motif 1 also markedly inhibited biofilm formation, although preimmune IgG had no effect (Figure 3A and 3B). Postimmune IgG antibody recognizing motif 1 bound 10-fold more effectively to C. albicans compared to preimmune IgG, as assessed by flow cytometry (Figure 3C). These results show that lysines K805/K806 in motif 1 are essential for biofilm formation in vivo and that an antibody against motif 1 effectively inhibits biofilm formation.
Figure 2.
Lysine residues K805/K806 in motif 1 are essential for biofilm formation in vivo. Intraluminal biofilm formation was assessed with scanning electron microscopy in heparinized central venous catheters in rats after injection of wild type (A), INT1 double disruptant (B), INT1 reintegrant (C), motif 1 mutant (D), motif 4 mutant (E), or motif 1&4 mutant (F). In each pair, scanning electron microscopy shows the intraluminal biofilm at ×100 (left panel) and ×2000 (right panel). Micrographs are representative of 4 animals infected with each strain.
Figure 3.
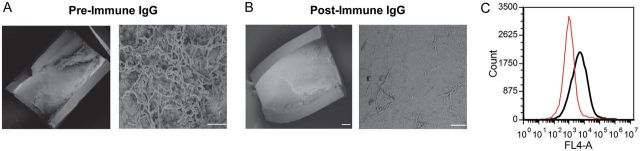
Immunoglobulin G (IgG) antibody recognizing motif 1 abolishes biofilm. Intraluminal biofilm after preincubation of Candida albicans with preimmune IgG (A) or postimmune IgG recognizing the peptide 799HKQEKQKKHQIHKV812 (B), which encompasses motif 1 (underlined). Scanning electron microscopy at ×100 (left panel) and ×2000 (right panel). Micrographs are representative of observations in 3 animals. C, Histograms representing binding of preimmune (red curve) and postimmune (black curve) IgG.
Elution of C. albicans Surface Proteins
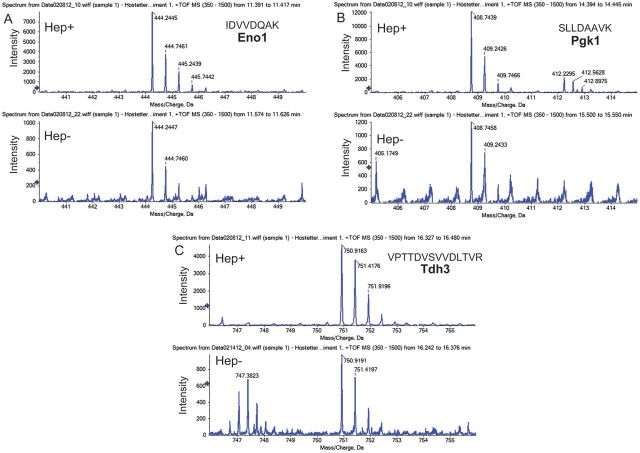
In other systems, heparin has been reported to cleave surface proteins [30] and change protein conformation [31]. After incubation with 250 U/mL heparin at 37°C for 1 hour, soluble peptides from >50 C. albicans surface proteins were detected in the supernatant, as determined by mass spectrometry. Peptides representing 12 proteins were found in supernatants of untreated organisms. Heparin treatment led to a 7-fold increase in intensity of IDVVDQAK (Eno1), a 10-fold increase in intensity of SLLDAAVK (Pgk1), and a 5-fold increase in VPTTDVSVVDLTVR (Tdh3) peptides (Figure 4A–C), compared with untreated organisms. The 10 proteins whose peptides were found in highest concentration in the supernatant (Table 2) are known to be localized to the cell surface. Six of 10—Eno1, Tdh3, Pgk1, Ssb1, and Ssa1/2—contain putative HBMs (Table 1) and are considered important antigens for innate and adaptive immune responses against C. albicans [32–35]. Whether the presence of these antigens in the supernatant represents protein secretion, cleavage, or another form of removal is not known; however, their displacement from the candidal cell surface may impair host recognition. Because soluble Int1 peptides were not identified after heparin treatment, Int1 remains a viable surface-expressed target for antibody therapy.
Figure 4.
Elution of candidal peptides after heparin treatment. Semiquantitative analysis of soluble peptide intensity after incubation of Candida albicans with (Hep+) or without (Hep−) 100 U/mL heparin. A, Eno1 peptide. B, Pgk1 peptide. C, Tdh3 peptide. Abbreviation: TOF MS, time-of-flight mass spectrometry.
Table 2.
Proteins With the Most Peptides Recovered From Supernatant After Heparin Treatment of Candida albicans
| ID | Protein Score | Protein Mass (Da) | Peptide Matches, No. |
|---|---|---|---|
| Eno1a,b | 1751 | 47 202 | 53 |
| Tdh3a,b | 1154 | 35 925 | 57 |
| Pgk1a,b | 886 | 45 266 | 30 |
| Ssb1a,b | 849 | 66 610 | 25 |
| Met6b | 830 | 85 763 | 25 |
| Ssa2a,b | 827 | 70 199 | 24 |
| Ssa1a,b | 772 | 70 452 | 20 |
| Atp2b | 704 | 54 035 | 20 |
| Pdc1b | 594 | 62 744 | 17 |
| Hsp90b | 581 | 80 773 | 13 |
a Proteins with putative linear heparin-binding motifs.
bSurface-associated and cell wall proteins according to Gene Ontology annotations (Candida Genome Database, January 2011).
DISCUSSION
One of a plethora of virulence mechanisms, biofilm formation in C. albicans involves numerous genes that affect adhesion, hyphal production, and secretion of extracellular matrix [36, 37]. Many of these genes are controlled by a network of 6 master transcriptional regulators [26]. In evaluating a possible relationship between C. albicans, heparin, and biofilm formation, attention to heparin concentrations used in humans is critical. Although in one study heparin concentrations (2000 to 10 000 U/mL) inhibited C. albicans growth and biofilm formation in vitro [38], these concentrations of heparin are 2- to 100-fold greater than those routinely prescribed for use in central venous catheters [2]. Moreover, because in vitro assays of biofilm do not necessarily predict in vivo phenotypes [39], definitive testing is best performed in vivo.
Our results show that C. albicans binds both soluble and plate-bound heparin. Sulfation at the 2-O residue of iduronic acid and the 2-N residue of glucosamine correlates positively with binding. In vivo, lysines K805/K806 of motif 1 in Int1 are essential for biofilm production in central venous catheters. An IgG antibody directed against motif 1 inhibits biofilm formation. As shown by the flow cytometry experiment (Figure 3C), the antibody to motif 1 does not lyse the organisms. Although we do not yet know the mechanism of biofilm formation that is targeted by the antibody, there are at least possibilities. First, heparin may serve as a bridge between HBMs on individual Candida cells, thereby facilitating cell-to-cell adhesion, an important early step in biofilm formation [36]. Alternatively, heparin may also link Candida cells and heparin-binding proteins of the endothelial matrix, such as fibronectin, and thereby serve to tether the Candida cells to the endothelialized surface of the catheter.
These results have important clinical implications because of the use of heparin in central venous catheters, in which setting Candida spp. are the fourth most common cause of infections [40]. Indeed, the presence of central venous catheters is considered a major risk factor in patients at highest risk of candidemia [41]. In a comparison of chlorhexidine- and heparin-coated catheters, all candidal colonization and candidemias were found in patients who received heparin-coated catheters [42]. From this standpoint, monthly infusion of a humanized antibody directed against motif 1 might be an effective preventative for patients requiring long-term central venous catheters.
Putative linear HBMs are also present in Staphylococcus epidermidis and Staphylococcus aureus (Jason Long Lu and Margaret Hostetter, unpublished data), 2 organisms that are even more prominent causes of catheter-associated infection [40]. Both clinical and laboratory studies have implicated heparin in biofilm formation by S. aureus [43, 44]. The presence of putative linear HBMs in several organisms that figure prominently in central line-associated bloodstream infections suggests that these motifs may serve as biomarkers for organisms able to form biofilms in the presence of heparin or may, as in the case of C. albicans, directly mediate this complication. In characterizing the mechanisms underlying the interaction between C. albicans surface proteins and heparin, the studies reported here define both a new mechanism of biofilm formation and a novel strategy for inhibiting catheter-associated biofilms.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. The authors gratefully acknowledge the assistance of Matt Kofron, PhD, Confocal Microscopy Core, and Amanda White, MBA, Flow Cytometry Core at Cincinnati Children's Research Foundation.
Financial support. This work was supported by Cincinnati Children's Hospital Medical Center (Innovator's Award to M. K. H.) and the Center for Clinical and Translational Science & Training, University of Cincinnati (Just-in-Time Core pilot grant 1171).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Capila I, Linhardt RJ. Heparin-protein interactions. Angew Chem Int Ed Engl. 2002;41:391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 2.Hamilton RA, Plis JM, Clay C, Sylvan L. Heparin sodium versus 0.9% sodium chloride injection for maintaining patency of indwelling intermittent infusion devices. Clin Pharm. 1988;7:439–43. [PubMed] [Google Scholar]
- 3.Harrop HA, Coombe DR, Rider CC. Heparin specifically inhibits binding of v3 loop antibodies to HIV-1 gp120, an effect potentiated by CD4 binding. AIDS. 1994;8:183–92. doi: 10.1097/00002030-199402000-00005. [DOI] [PubMed] [Google Scholar]
- 4.Rusnati M, Tulipano G, Spillmann D, et al. Multiple interactions of HIV-1 Tat protein with size-defined heparin oligosaccharides. J Biol Chem. 1999;274:28198–205. doi: 10.1074/jbc.274.40.28198. [DOI] [PubMed] [Google Scholar]
- 5.Kim S, Wolyniak MJ, Staab JF, Sundstrom P. A 368-base-pair cis-acting HWP1 promoter region, HCR, of Candida albicans confers hypha-specific gene regulation and binds architectural transcription factors Nhp6 and Gcf1p. Eukaryot Cell. 2007;6:693–709. doi: 10.1128/EC.00341-06. PMCID: 1865660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Margalit H, Fischer N, Ben-Sasson SA. Comparative analysis of structurally defined heparin binding sequences reveals a distinct spatial distribution of basic residues. J Biol Chem. 1993;268:19228–31. [PubMed] [Google Scholar]
- 7.Torrent M, Nogues MV, Andreu D, Boix E. The "CPCclip motif": A conserved structural signature for heparin-binding proteins. PLoS One. 2012;7:e42692. doi: 10.1371/journal.pone.0042692. PMCID: 3412806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cardin AD, Weintraub HJ. Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis. 1989;9:21–32. doi: 10.1161/01.atv.9.1.21. [DOI] [PubMed] [Google Scholar]
- 9.Sobel M, Soler DF, Kermode JC, Harris RB. Localization and characterization of a heparin binding domain peptide of human von Willebrand factor. J Biol Chem. 1992;267:8857–62. [PubMed] [Google Scholar]
- 10.Farshi P, Ohlig S, Pickhinke U, et al. Dual roles of the Cardin-Weintraub motif in multimeric Sonic Hedgehog. J Biol Chem. 2011;286:23608–19. doi: 10.1074/jbc.M110.206474. PMCID: 3123124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaffin WL. Candida albicans cell wall proteins. Microbiol Mol Biol Rev. 2008;72:495–544. doi: 10.1128/MMBR.00032-07. PMCID: 2546859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alberti-Segui C, Morales AJ, Xing H, et al. Identification of potential cell-surface proteins in Candida albicans and investigation of the role of a putative cell-surface glycosidase in adhesion and virulence. Yeast. 2004;21:285–302. doi: 10.1002/yea.1061. [DOI] [PubMed] [Google Scholar]
- 13.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: Signalp 3.0. J Mol Biol. 2004;340:783–95. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 14.Fankhauser N, Maser P. Identification of GPI anchor attachment signals by a kohonen self-organizing map. Bioinformatics. 2005;21:1846–52. doi: 10.1093/bioinformatics/bti299. [DOI] [PubMed] [Google Scholar]
- 15.Pierleoni A, Martelli PL, Casadio R. Predgpi: A GPI-anchor predictor. BMC Bioinformatics. 2008;9:392. doi: 10.1186/1471-2105-9-392. PMCID: 2571997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mao Y, Zhang Z, Gast C, Wong B. C-terminal signals regulate targeting of glycosylphosphatidylinositol-anchored proteins to the cell wall or plasma membrane in Candida albicans. Eukaryot Cell. 2008;7:1906–15. doi: 10.1128/EC.00148-08. PMCID: 2583546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson RB, Davis D, Mitchell AP. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–74. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osmond RI, Kett WC, Skett SE, Coombe DR. Protein-heparin interactions measured by Biacore 2000 are affected by the method of heparin immobilization. Anal Biochem. 2002;310:199–207. doi: 10.1016/s0003-2697(02)00396-2. [DOI] [PubMed] [Google Scholar]
- 19.Marson A, Robinson DE, Brookes PN, et al. Development of a microtiter plate-based glycosaminoglycan array for the investigation of glycosaminoglycan-protein interactions. Glycobiology. 2009;19:1537–46. doi: 10.1093/glycob/cwp132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–72. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch FE, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. New York, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 22.Devore-Carter D, Kar S, Vellucci V, Bhattacherjee V, Domanski P, Hostetter MK. Superantigen-like effects of a Candida albicans polypeptide. J Infect Dis. 2008;197:981–9. doi: 10.1086/529203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cormack B. Directed Mutagenesis Using the Polymerase Chain Reaction. Current Protocols in Molecular Biology. 2001;37:8.5.1–8.5.10. [Google Scholar]
- 24.Eismann T, Huber N, Shin T, et al. Peroxiredoxin-6 protects against mitochondrial dysfunction and liver injury during ischemia-reperfusion in mice. Am J Physiol Gastrointest Liver Physiol. 2009;296:G266–74. doi: 10.1152/ajpgi.90583.2008. PMCID: 2643922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andes D, Nett J, Oschel P, Albrecht R, Marchillo K, Pitula A. Development and characterization of an in vivo central venous catheter Candida albicans biofilm model. Infect Immun. 2004;72:6023–31. doi: 10.1128/IAI.72.10.6023-6031.2004. PMCID: 517581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nobile CJ, Fox EP, Nett JE, et al. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 2012;148:126–38. doi: 10.1016/j.cell.2011.10.048. PMCID: 3266547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raman R, Sasisekharan V, Sasisekharan R. Structural insights into biological roles of protein-glycosaminoglycan interactions. Chem Biol. 2005;12:267–77. doi: 10.1016/j.chembiol.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 28.Gale CA, Bendel CM, McClellan M, et al. Linkage of adhesion, filamentous growth, and virulence in Candida albicans to a single gene, INT1. Science. 1998;279:1355–8. doi: 10.1126/science.279.5355.1355. [DOI] [PubMed] [Google Scholar]
- 29.Mochon AB, Jin Y, Kayala MA, et al. Serological profiling of a Candida albicans protein microarray reveals permanent host-pathogen interplay and stage-specific responses during candidemia. PLoS Pathogens. 2010;6:e1000827. doi: 10.1371/journal.ppat.1000827. PMCID: 2845659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao K, Shenoy SK, Nobles K, Lefkowitz RJ. Activation-dependent conformational changes in β-arrestin 2. J Biol Chem. 2004;279:55744–53. doi: 10.1074/jbc.M409785200. [DOI] [PubMed] [Google Scholar]
- 31.Lortat-Jacob H, Grimaud JA. Interferon-gamma c-terminal function: new working hypothesis—heparan sulfate and heparin, new targets for IFN-gamma, protect, relax the cytokine and regulate its activity. Cell Mol Biol. 1991;37:253–60. [PubMed] [Google Scholar]
- 32.Sundstrom P, Aliaga GR. Molecular cloning of cDNA and analysis of protein secondary structure of Candida albicans enolase, an abundant, immunodominant glycolytic enzyme. J Bacteriol. 1992;174:6789–99. doi: 10.1128/jb.174.21.6789-6799.1992. PMCID: 207354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pitarch A, Diez-Orejas R, Molero G, et al. Analysis of the serologic response to systemic Candida albicans infection in a murine model. Proteomics. 2001;1:550–9. doi: 10.1002/1615-9861(200104)1:4<550::AID-PROT550>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 34.Pitarch A, Abian J, Carrascal M, Sanchez M, Nombela C, Gil C. Proteomics-based identification of novel Candida albicans antigens for diagnosis of systemic candidiasis in patients with underlying hematological malignancies. Proteomics. 2004;4:3084–106. doi: 10.1002/pmic.200400903. [DOI] [PubMed] [Google Scholar]
- 35.Li XS, Reddy MS, Baev D, Edgerton M. Candida albicans Ssa1/2p is the cell envelope binding protein for human salivary histatin 5. J Biol Chem. 2003;278:28553–61. doi: 10.1074/jbc.M300680200. [DOI] [PubMed] [Google Scholar]
- 36.Nobile CJ, Mitchell AP. Genetics and genomics of Candida albicans biofilm formation. Cell Microbiol. 2006;8:1382–91. doi: 10.1111/j.1462-5822.2006.00761.x. [DOI] [PubMed] [Google Scholar]
- 37.Mayer FL, Wilson D, Hube B. Candida albicans pathogenicity mechanisms. Virulence. 2013;4:119–28. doi: 10.4161/viru.22913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miceli MH, Bernardo SM, Ku TS, Walraven C, Lee SA. In vitro analyses of the effects of heparin and parabens on Candida albicans biofilms and planktonic cells. Antimicrob Agents Chemother. 2012;56:148–53. doi: 10.1128/AAC.05061-11. PMCID: 3256088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nobile CJ, Andes DR, Nett JE, et al. Critical role of BCR1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathogens. 2006;2:e63. doi: 10.1371/journal.ppat.0020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mermel LA, Allon M, Bouza E, et al. Clinical practice guidelines for the diagnosis and management of intravascular catheter-related infection: 2009 update by the Infectious Diseases Society of America. Clin Infect Dis. 2009;49:1–45. doi: 10.1086/599376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beck-Sague C, Jarvis WR. Secular trends in the epidemiology of nosocomial fungal infections in the United States, 1980–1990: national nosocomial infections surveillance system. J Infect Dis. 1993;167:1247–51. doi: 10.1093/infdis/167.5.1247. [DOI] [PubMed] [Google Scholar]
- 42.Carrasco MN, Bueno A, de las Cuevas C, et al. Evaluation of a triple-lumen central venous heparin-coated catheter versus a catheter coated with chlorhexidine and silver sulfadiazine in critically ill patients. Intensive Care Med. 2004;30:633–8. doi: 10.1007/s00134-003-2093-4. [DOI] [PubMed] [Google Scholar]
- 43.Diskin CJ, Stokes TJ, Dansby LM, Radcliff L, Carter TB. Is systemic heparin a risk factor for catheter-related sepsis in dialysis patients? an evaluation of various biofilm and traditional risk factors. Nephron Clinical Practice. 2007;107:c128–32. doi: 10.1159/000110032. [DOI] [PubMed] [Google Scholar]
- 44.Shanks RM, Donegan NP, Graber ML, et al. Heparin stimulates Staphylococcus aureus biofilm formation. Infect Immun. 2005;73:4596–606. doi: 10.1128/IAI.73.8.4596-4606.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.