Abstract
Disruption of the high-affinity nitrate transporter NRT2.1 activates the priming defense against Pseudomonas syringae, resulting in enhanced resistance. In this study, it is demonstrated that the high-affinity ammonium transporter AMT1.1 is a negative regulator of Arabidopsis defense responses. The T-DNA knockout mutant amt1.1 displays enhanced resistance against Plectosphaerella cucumerina and reduced susceptibility to P. syringae. The impairment of AMT1.1 induces significant metabolic changes in the absence of challenge, suggesting that amt1.1 retains constitutive defense responses. Interestingly, amt1.1 combats pathogens differently depending on the lifestyle of the pathogen. In addition, N starvation enhances the susceptibility of wild type plants and the mutant amt1.1 to P. syringae whereas it has no effect on P. cucumerina resistance. The metabolic changes of amt1.1 against P. syringae are subtler and are restricted to the phenylpropanoid pathway, which correlates with its reduced susceptibility. By contrast, the amt1.1 mutant responds by activating higher levels of camalexin and callose against P. cucumerina. In addition, amt1.1 shows altered levels of aliphatic and indolic glucosinolates and other Trp-related compounds following infection by the necrotroph. These observations indicate that AMT1.1 may play additional roles that affect N uptake and plant immune responses.
Keywords: AMT1.1, basal resistance, metabolomics, transceptor, NRT2.1
Introduction
Arabidopsis defense responses against P. syringae and P. cucumerina have been widely studied. These pathogens have very different life styles, in that P. syringae is a hemibiotrophic bacterial pathogen that penetrates through natural openings, whereas P. cucumerina is a necrotrophic fungus that can overcome cell barriers and penetrate into the cytoplasm (Berrocal-Lobo et al., 2002). Salicylic acid-dependent defenses have emerged as the most effective resistance mechanism against the bacterium (Brooks et al., 2005; Glazebrook, 2005). The effector secretion machinery of P. syringae is currently under study because of its effectiveness in suppressing Arabidopsis defenses. Although the direct link remains unknown, Camañes et al. (2012b) demonstrated that coronatine, a P. syringae effector, targets NRT2.1 and subsequently causes a H2O2 burst. The nrt2 mutant remains insensitive to this manipulation and the increased JA-signaling that is responsive to coronatine (Melotto et al., 2006). P. cucumerina is more complex. Arabidopsis defenses against this pathogen are horizontal and involve several defense signaling pathways, such as SA, JA, IAA, and ABA-mediated callose deposition (Lorenzo and Solano, 2005; García-Andrade et al., 2011; Sánchez-Vallet et al., 2012; Gamir et al., 2014). N fertilization influences plant-pathogen interactions; therefore, the following two factors affect plant resistance: the total amount of N and the source of N. Over-fertilization increases the severity of mildew in several grapevine cultivars and many crop plants (Keller et al., 2003; Marschner, 2012). The level of N in fertilization programs also affects plant resistance based on the pathogen lifestyle. For example, elevated levels of N promote susceptibility against biotrophs, such as powdery mildew, whereas it results in reduced disease development of Botrytis cinerea (Walters and Bingham, 2007). However, the source of N appears to have a greater influence in plant resistance (Gupta et al., 2013). Fertilizing tobacco plants with NO−3 accelerates the hypersensitive response following P. syringae pv. Phaseolicola infections and provides enhanced resistance. This resistance is related to NO−3 reduction and NO production that triggers SA synthesis (Durner and Klessig, 1999). Plants fed NO−3 display increased PR1 expression and higher levels of SA. By contrast, tobacco plants fed with NH+4 are more susceptible to the bacterium. The bypass of nitrate reduction avoids NO production and triggers 4-aminobutyric accumulation, which is a nutrient for the pathogen (Solomon and Oliver, 2001), increasing the susceptibility of the plant.
Previously, first in animal sciences and then in plant sciences, the term transceptor is applied to membrane proteins that perform a dual transport and signaling function. Gojon et al. (2011) revised the transceptor role of the nitrate transporter family member NRT1. In addition to binding to low affinity nitrate, this protein can sense nitrate in the root environment. Additional studies demonstrated that both NRT1.1 and the high-affinity transporter NRT2.1 are transceptors. The sensing network regulated by NRT1.1 has not yet been elucidated, although several of its components have been recently described. One target of NRT1.1 is the transcription of NRT2.1. The dual role of the induction of NRT2.1 during early time-points of nitrate supply and repression during later time-points both directly and indirectly involve NRT1.1. Of the N metabolism components, mutants impaired in NRT1.1, such as chl1 are also affected in NIA1, NiR (encoding nitrate reductase and nitrite reductase, respectively) (Wang et al., 2009) and CIPK8, which has a regulatory role on NRT2.1 (Hu et al., 2009). Glutamic acid (Glu) is important in N flow and also regulates root architecture by promoting root branching. This effect is antagonized by excess NO−3, and this antagonistic action is also regulated by NRT1.1-dependent signaling (Walch-Liu and Forde, 2008). Root development and branching is also conditioned to auxin signaling, and it appears that NRT1.1 couples NO−3 sensing and auxin signaling.
There is recent evidence for other NO−3 sensors coordinating signaling events in Arabidopsis. NRT2.1 displays NRT1.1-independent roles and is also unrelated to NO−3 uptake activity. This gene is a repressor of lateral root initiation under high sucrose and NO−3 supply, but surprisingly, this gene seems to sense abiotic stress and concomitantly suppresses biotic stress responses. We have demonstrated that the mutant nrt2 is affected in bacterial effector manipulation (Camañes et al., 2012b). Mutation of NRT2.1 has significant consequences in the transcriptome of Arabidopsis upon P. syringae infection, suggesting a complex regulatory function for the NRT2.1 gene and also the NRT2.1 protein (Camañes et al., 2012b). Therefore, we hypothesize that NRT2.1 should be designated a transceptor as previously proposed by Gojon et al. (2011). In Arabidopsis, the link between the immune system and nitrate transport is not fully understood. However, recent advances indicate two different relevant events. First, low nitrate treatment induces a rapid burst of ethylene production and upregulates CTR1, EIN3, and EIL1 (Zheng et al., 2013). Second, we demonstrated that a low N treatment enhances P. syringae susceptibility (Camañes et al., 2012b), which may be related to the increase of ET-dependent signaling that is antagonistic to the SA-signaling that is recognized as an efficient defense against biotrophic pathogens (Pieterse et al., 2009).
Interactions with the plant immune system are not restricted to NRT2.1. Other NRT family members, such as NRT2.6 show enhanced expression upon Erwinia amylovora infection (Dechorgnat et al., 2012). Despite high NO−3 levels that trigger NRT2.6 expression, no nitrate-related phenotype has been associated with the nrt2.6 mutant. Furthermore, nrt2.6 is less tolerant to E. amylovora because of the reduced ability to produce H2O2 upon infection.
Some members of the AMT1 family are high-affinity ammonium transporters. Of these, AMT1.1 has the highest affinity for NH+4 (Shelden et al., 2001). The removal of N increases AMT1.1 expression, and this correlates with an increase in NH+4 uptake. This function is affected by 30–40% in the insertional T-DNA mutant amt1.1. Interestingly, amt1.1 plants are indistinguishable from wild type under optimal fertilization and growth conditions. Furthermore, amt1.1 displays wild type levels of total N in the presence of N or after 4 days of N starvation (Kaiser et al., 2002). Although the ammonium supply may affect the plant responses to pathogens, to the best of our knowledge, no NH+4 transporter is involved in the plant immune system. NRT2.1 and AMT1.1 are the primary high-affinity transport systems (HATS) involved in NO−3 and NH+4 uptake, respectively (Cerezo et al., 2001; Kaiser et al., 2002), and reciprocal regulation between AtNRT2.1 and AtAMT1.1 expression has been observed (Camañes et al., 2012a). High-affinity ammonium and nitrate transporters act as sensors of low NH+4 and NO−3 in the root environment; therefore, thus the gene expression of AMT1.1 and NRT2.1 is enhanced upon low fertilization. Interestingly, its role in HATS under normal N supply conditions (>1 mM NO−3 or NH+4) is negligible (Gansel et al., 2001). Using an experimental system with optimal fertilization, we focused on the role of the high-affinity ammonium transporter gene in the Arabidopsis immune responses against the hemibiotrophic bacterium P. syringae and the necrotrophic fungus P. cucumerina. Metabolomic and genetic studies revealed an interplay between AMT1.1 and plant resistance.
Materials and methods
Plant material and growth conditions
Seeds of the Arabidopsis accession Col-0 were obtained from the SALK institute (Alonso et al., 2003), the EMS lin1 line (in Col-0 background) was generously supplied by Jocelyn Malamy (University of Chicago, EEUU, Little et al., 2005) and the line nrt3.1 (salk_043672) in the Col-0 background was obtained from SALK institute (Alonso et al., 2003). The background Col3-gl1 and the mutant amt1.1 (in Col3-gl1 background) were provided by Alain Gojon (INRA, Montpelier, France).
For the bioassays of bacterial resistance, 1 week after germination, the seedlings were transferred to 33-ml soil pots. The plants were cultivated at 20°C day/18°C night with 8.5 h of light (105 μE m−2 s−1) per 24 h and 60% relative humidity.
All plant genotypes were germinated in soil, and, 1 week after germination, seedlings were individually transferred to 33 ml pots containing commercial potting soil (TKS1, Floragard GmbH; http://www.floragard.de). Plants were cultivated at 20°C day/18°C night temperatures with 8.5 h of light (105 μE m−2 s−1) per 24 h and 60% relative humidity.
For fungal resistance assays. About 50 seeds were germinated in 33-ml soil pots. Plants were grown in the same conditions as described for bacterial experiments.
Pathogen strains and resistance assays
The bacterial strain Pseudomonas syringae pv. tomato DC3000 was grown overnight at room temperature in King's B solid medium with appropriate antibiotics and diluted to the desired concentration with 10 mM MgSO4 for plant inoculation. The bacteria were used to infect 5-week-old (or otherwise mentioned) Arabidopsis plants by dipping in a suspension of 2.5 × 105 colony-forming units (cfu)/ml using 0.02% Silwet L-77 (Tornero and Dangl, 2001), control plants were treated with Silwet accordingly. Three days after challenge inoculation, the disease level was determined by harvesting the plants a plating a range of dilutions in agar-KB medium containing rifampicin. After incubation at 28°C for 3 days, the number of rifampicin-resistant colony forming units per gram of infected leaf tissue was determined, and bacterial proliferation over the 3-day time interval was calculated.
The adapted strain of P. cucumerina BMM was kindly provided by Brigitte Mauch-Mani (Universitè de Neuchâtel, Switzerland). Two week old plants were sprayed with 1 × 103 spores/mL and maintained at 100% relative humidity. Two days after inoculation leaves were sampled by freezing in liquid N2 and stored at −80°C until analysis. Disease rate was determined by microscopical analysis after tripan blue stainings of infected leaves (Flors et al., 2008). Leaves were classified in a disease ranking as explained in the figure legend.
Growth conditions for starvation experiments
Arabidopsis thaliana was grown hydroponically as described in Lejay et al. (1999). The seeds were germinated directly on top of modified Eppendorf tubes filled with pre-wet sand. Plants were grown until the age of 5 weeks on a 1 mM NH4NO3 nutrient solution (repressed plants), which prevented any growth difference between the two genotypes (Lejay et al., 1999). Before inoculation experiments, plants were transferred for 48 h to N-free solution (de-repressed plants).
HPLC-ESI full scan mass spectrometry (Q-TOF instrument)
Metabolome analysis was performed using an Acquity UPLC system (Waters, Mildford, MA, USA) interfaced to hybrid quadrupole time-of-flight (QTOF Premier). The LC separation was performed by HPLC SunFire C18 analytical column, 5 μm particle size, 2.1 × 100 mm (Waters). Analytes were eluted with a gradient of methanol and water containing 0.01% HCOOH. Chromatographic conditions and QTOF MS parameters were followed as described in Gamir et al. (2014).
HPLC-ESI tandem mass spectrometry (triple quadrupole instrument). hormonal analysis
An Acquity ultra-performance liquid chromatography system (UPLC) (Waters, Mildford, MA, USA) was interfaced to a triple quadrupole mass spectrometer (TQD, Waters, Manchester, UK). The chromatographic separation conditions were closely related to those described previously. Chromatograpic conditions and TQD parameters were followed as described in Flors et al. (2008) and Gamir et al. (2012). Masslynx v 4.1 (Waters, Manchester, UK) software was used to process the quantitative data obtained from calibration standards and samples.
Full scan data analysis
Raw data obtained from Masslynx software was transformed to.CDF using Databrigde provide by Masslynx package. The.CDF data was process with R for statistical computing using XCMS package for relative quantification (Smith et al., 2006). Principal Component Analysis (PCA) were used as explained in http://www.numericaldynamics.com/ as a tool to define major changes in the metabolome of the plant under priming condition during fungal infections.
For heatmap construction and clustering of metabolites it was used the software MarVis Filter and MarVis cluster (http://marvis.gobics.de/; Kaever et al., 2012).
Kinetics of 15NH+4 influx
The kinetics of 15NH+4 influx as a function of external 15NH+4 concentrations ([15NH+4]0) was measured with [15NH+4]0 ranging from 0.02 to 0.8 mM. To kinetics studies control plants and de-repressed plants of three genotypes (Col-0 and nrt3.1) were used. Influx of 15NH+4 into the roots were assayed as described by Gazzarrini et al. (1999). Col-0 and nrt3.1 plants with a normal fertilization or N depletion were sequentially transferred to 0.1 mM CaSO4 for 1 min and to complete nutrient solution (pH 6.0) containing 15NH+4 (98% atom excess 15N) for 5 min, at the concentrations indicated in figures. At the end of the 15N labeling, roots were washed for 1 min in 0.1 mM CaSO4 and were separated from shoots. The roots were dried at 70°C for 48 h, weighed, crushed in a hammer-mill and analyzed for total 15N content using an integrated system for continuous flow isotope ratio mass spectrometry (Euro-EA elemental analyser, EuroVector S.P.A.; and Isoprime mass spectrometer; GV Instruments). Root influx is expressed in μmol 15NH+4 (g root DW)−1 h−1. To kinetics studies data-transformation method based on the Michaelis-Menten formalism was used. The experiment was repeated three times.
RNA extraction and real-time PCR analysis
Gene expression by quantitative real-time RT-PCR was performed using RNA samples extracted from root tissue using the RNA kit (Omega Bio-Tek Inc, Doraville, GA, USA) according to the manufacturer instructions. To avoid contaminating DNA, the samples were treated with DNAse I. A total of 1 μg of total RNA was annealed to oligo-dT and reverse transcribed using Omniscript Reverse Transcription kit (QIAGEN) to obtain cDNA. The sequences of the gene-specific oligonucleotides designed and used for real-time PCR are the following: AMT1.1 forward: 5′acactgtggccagttaggcg3′ and reverse: 5′ccgtggggatgtctttgaga3′, Tubuline (TUB) forward: 5′cgattccgttctcgatgttgt3′ and reverse: 5′aatgagtgacacacttggaatcctt3′ and EF1α forward: 5′gtcgattctggaaagtcgacc3′ and reverse: 5′aatgtcaatggtgataccacgc3′. Real-time PCR was conducted using the QuantiTect™ SYBR Green PCR Kit (QIAGEN) and the SmartCycler II instrument (Cepheid). The experiment was repeated three times.
Results
Alteration in nitrate and ammonium transporters enhances basal resistance
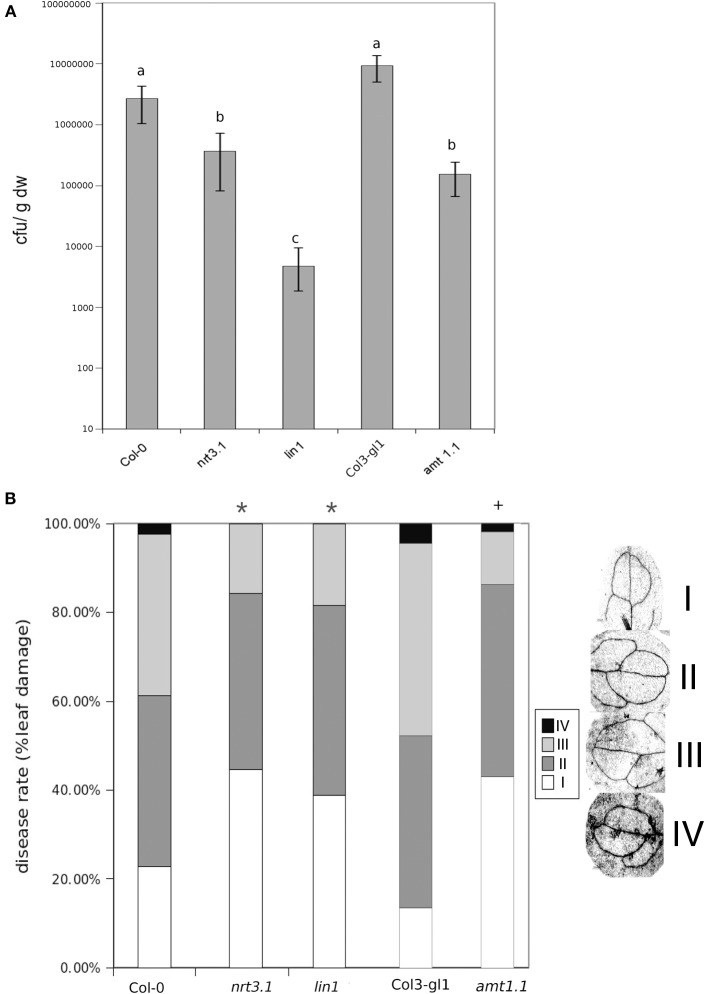
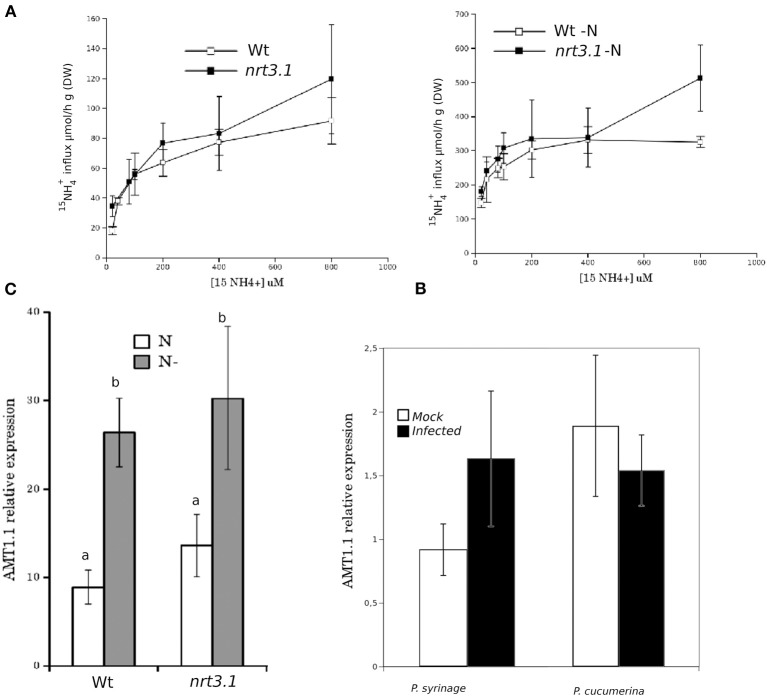
It has been reported that deletions in NRT2.1 and NRT2.2 cause the reduced susceptibility of Arabidopsis to P. syringae (Camañes et al., 2012b). NRT2.1 has been proposed as a transceptor that delivers and coordinates distal signaling that affects primary metabolism and defense signaling (Gojon et al., 2011; Camañes et al., 2012b). The function of NRT2.1 in transporting nitrate is also influenced by the membrane protein NRT3.1, which is not a transporter itself, but its mutation significantly affects the HATS of nitrate (Okamoto et al., 2006; Orsel et al., 2006). To determine whether deficiencies in NRT3.1 and the ammonium transporter gene AMT1.1 have consequences in the basal resistance, we characterized the responses of the mutant lines nrt3.1 and amt1.1 toward P. syringae and P. cucumerina. The enhanced resistance mutant lin1 (blocked in NRT2.1) was used as a control. All three mutants showed enhanced resistance against both P. syringae and P. cucumerina compared to their respective control wild type plants (Figures 1A,B). The mutant lin1 was more resistant to the bacterium, whereas nrt3.1 and amt1.1 showed significantly reduced susceptibility, although lower than lin1. Because NRT3.1 influences nitrate transport, we tested whether it also affects ammonium transport. The mutant nrt3.1 showed wild type 15NH+4 uptake kinetics both with N and after 48 h of N depletion (Figure 2A). In addition, we also determined whether the AMT1.1 gene is upregulated in the absence of N under the same experimental conditions. N depletion induces (de-represses) AMT1.1 expression in wild type plants (Gazzarrini et al., 1999), whereas nrt3.1 shows wild type AMT1.1 expression (Figure 2B). Both results suggest that NRT3.1 does not modulate ammonium kinetics uptake and that it does not alters AMT1.1 de-repression.
Figure 1.
Bacterial proliferation and disease rate in Col-0, lin1, nrt3.1, and Col3-gl/1 and amt1.1 plants infected with P. syringae and P. cucumerina respectivelly. (A) Five week old plants were challenge-inoculated by dipping in a bacterial suspension of P. syringae at 2 × 105 c.f.u. mL−1. The values presented are means (±SD) of the log of the proliferation values. Data represent the average of three independent experiments (n = 3). Different letters mean significant statistical differences (ANOVA, LSD test; p < 0.05). (B) Two week old plants were inoculated by spraying with 1 × 103 spores × mL−1 with PcBMM. Disease symptoms were recorded by trypan-blue staining at 5 days post inoculation. Disease rate was ranked according to the infected leaf surface: level I no infection, level II less than 25% of infected leaf surface, level III between 25 and 50% of infected leaf surface, level IV more than 50% of infected leaf surface. The figure shows a representative experiment that was repeated three times with the same results. Data presented are the means of the percentage of diseased leaves per plant. Asterisk indicate statistically significant differences compared with non-induced control plants (t-test; *p < 0.05; +p < 0.05 with their respective controls; n~50 leaves).
Figure 2.
(A) Kinetics of 15NH+4 influx in Col-0 and nrt3.1 roots in the low 15NH+4 concentration range. Plants were grown hydroponically with N supplied as 1 mM NH4NO3 during 6 weeks. After that one group of plants were transferred to 1 mM NH4NO3, other group of plants were transferred to N-free nutrient solution (-N) during 48 h. 15NH+4 influx was measured at different concentrations of external 15NH+4. Each data is the mean of 30 replicates ±SE. (B) Real-time PCR analysis of the expression of AMT1.1 in Col-0 and nrt3.1 plants fertilized normally along 5 weeks and exposed to N starvation 2 days (-N). The AMT1.1 transcript levels were normalized to the expression of TUB measured in the same samples. The experiment was repeated using EF1α with similar results. Each bar represents average data with standard error bars from two technical replicates three independent experiments (n = 6). (C) Real-time PCR analysis of the expression of AMT1.1 in mock and P. syringae or P. cucumerina wild type infected plants. The AMT1.1 transcript levels were normalized to the expression of TUB measured in the same samples. The experiment was repeated using EF1α with similar results. Each bar represents average data with standard error bars from two technical replicates three independent experiments (n = 6).
Finally, we tested whether infection with P. syringae or P. cucumerina affects AMT1.1 expression in a manner that may act as targets for bacterial or fungal effectors. Following infection, AMT1.1 expression was not significantly modified by any of the pathogens (Figure 2C).
Nitrogen starvation enhances P. syringae susceptibility but does not affect P. cucumerina resistance
Previously, we determined that nitrate starvation for 48 h increased the susceptibility of Arabidopsis toward P. syringae and that nrt2.1 was also impaired (Camañes et al., 2012b). To determine whether N depletion affects the basal resistance against both pathogens and to determine the contribution of the ammonium transporter AMT1.1 to this interaction, we performed a hydroponic experiment by inducing N depletion for 48 h. According to our previous study (Camañes et al., 2012b), wild type starved plants were more susceptible to P. syringae (Figure 3), whereas they were not affected in their resistance to the fungus (Figure 3). Notably, amt1.1 starved plants also showed increased susceptibility against P. syringae, whereas their resistance to the fungus was not affected.
Figure 3.
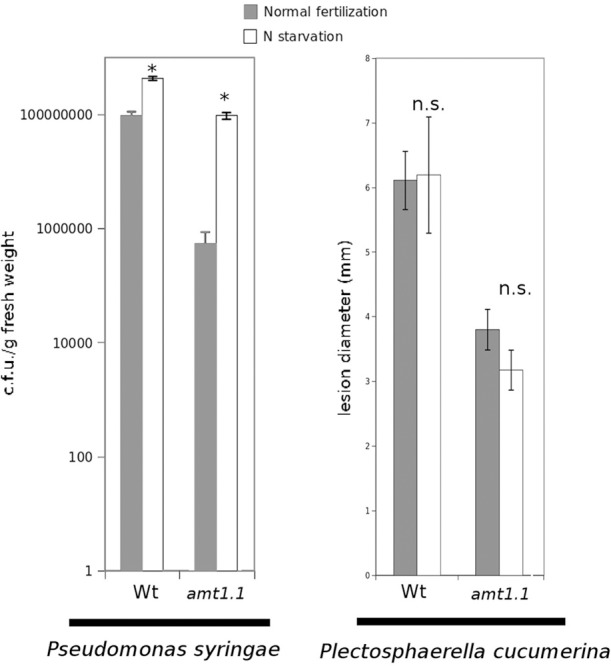
P. syringae proliferation and lesion diameter cause by P. cucumerina in Col3-gl1 and amt1.1 plants fertilized normally (Normal fertilization) along 5 weeks and exposed to N starvation 2 days before inoculation (N starvation). Hydroponically growing plants were challenge-inoculated either with a bacterial suspension of P. syringae at 2.5 × 105 c.f.u./ml or with a drop of 1 × 105 spores × mL−1 of PcBMM. Data represent the average of three independent experiments (n = 3). The values presented are means of infected plants (±SD). Asterisk indicates statistically significant differences (LSD test; p < 0.05).
amt1.1 displays altered hormonal defense responses against P. syringae and P. cucumerina
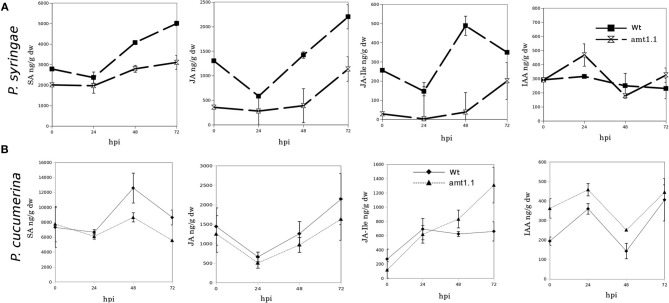
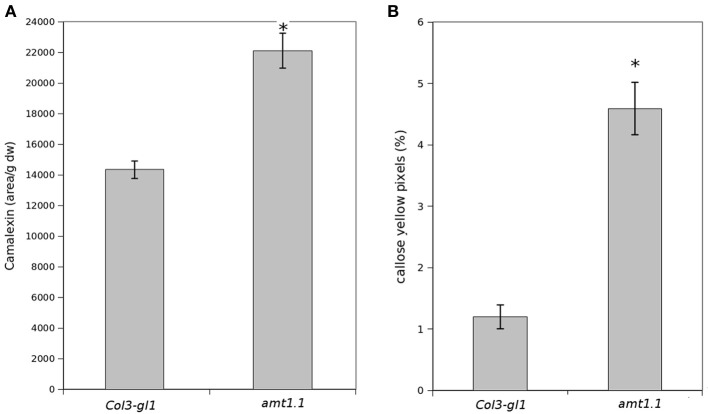
Because amt1.1 showed reduced susceptibility and enhanced resistance toward the bacterium and the fungus, respectively, compared to the wild type plants, we analyzed the main hormones involved in defense signaling. Surprisingly, the SA, JA, and JA-Ile levels remained lower in the amt1.1 plants infected with P. syringae for the entire experiment compared to the wild type plants (Figure 4A). Indoleacetic acid (IAA), however, did not significantly change in the amt1.1 plants. A reduction of JA and JA-Ile in amt1.1 plants suggests that this mutation may counteract bacterial effectors by enhancing resistance. However, this hypothesis is unlikely because the SA levels remained lower in the mutant. Therefore, the hormonal analysis did not clearly explain the reduced susceptibility of amt1.1 to the bacterium. By contrast, the hormonal responses of the mutant against the fungus may contribute to resistance. JA-Ile and IAA following infection are increased compared to the wild type plants; however, SA at 48 and 72 hpi is reduced in the mutant infected with P. cucumerina (Figure 4B). We also tested other common defense responses that effective against P. cucumerina, such as the camalexin levels and callose accumulation (Ton and Mauch-Mani, 2004; Gamir et al., 2014). Both defensive responses were strongly enhanced in the mutant response to the pathogen. Camalexin and callose in amt1.1 remained significantly higher at 48 hpi (Figure 5).
Figure 4.
SA, JA, JA-Ile, and IAA profiling upon P. syringae and P. cucumerina infection. Plants were challenged as described in Figure 1. Both mock and pathogen infected plants were harvested at different time-points. Freeze dried material was processed for a targeted quantification analysis by TQD-MS. The concentration of the hormones was determined in all the samples by normalizing the chromatographic area for each compound with the dry weight of the corresponding sample. Leaf material from 15 individual plants for P. syringae (A) resistance assays and 150 plants for P. cucumerina (B) resistance assays were pooled together for each treatment × genotype combination. Data represent average three independent experiments ±SD; n = 3.
Figure 5.
Camalexin and callose levels upon P. syringae and P. cucumerina infection. Plants were challenged as described in Figure 1. Either mock or pathogen infected plants were harvested at 48 hpi. (A) Freeze dried material was processed for a targeted quantification analysis of camalexin by TQD-MS. The relative concentration was determined in all the samples by normalizing the chromatographic area for each compound with the dry weight of the corresponding sample. Leaf material from 150 individual plants were pooled together for each treatment × genotype combination. Data represent average three independent experiments. (B) Callose was visualized by aniline blue staining and epifluorescence microscopy (UV). Quantification was performed by determining the number of yellow pixels per million pixels corresponding to pathogen-induced callose on digital photographs of infected leaf areas. Asterisk indicates statistically significant differences (LSD test; p < 0.05). Data shown are means (±SD; n = 20) of the relative number of yellow pixels per photograph.
Metabolomic profiling of amt1.1 in response to pathogen attack
The hormonal profile of amt1.1 in response to bacterial infection did not explain its enhanced resistance; therefore, we performed full metabolomic profiling to understand the metabolic changes that may contribute to the resistance of this mutant. For P. cucumerina, the classical defenses against necrotrophs, such as JA-Ile, callose, and camalexin, over-accumulated in amt1.1 plants infected with the fungus. We also performed metabolic profiling following infection with the fungus to determine the extent to which amt1.1 affects the responses to fungal infection. We used untargeted reverse-phase liquid chromatography-mass spectrometry (HPLC-QTOF-MS) for the profiling. Col3-gl1 and amt1.1 plants were either mock or P. syringae or P. cucumerina infected. Metabolomic analysis and data reporting were performed as described by Pitzschke and Hirt (2010), Fernie et al. (2011), and Kaever et al. (2012).
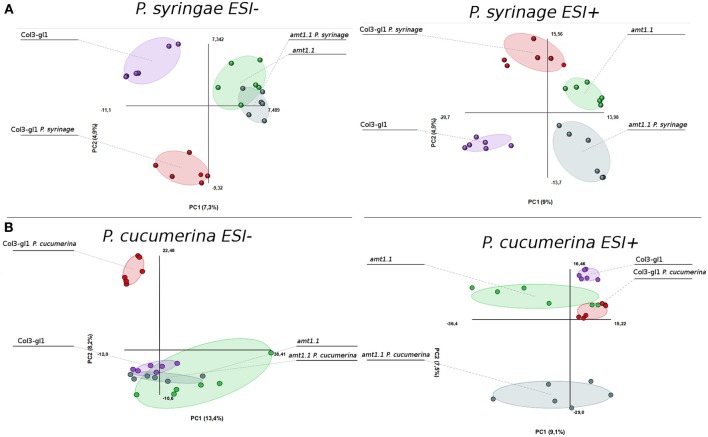
The acquired raw data were transformed into.cdf files using Databridge from the Masslynx 4.1 software (Masslynx 4.1, Waters). These data were subsequently subjected to analysis using the free software R for statistical purposes. The signals from the positive and negative electro-spray analysis (ESI+; ESI−) were processed separately, these ionization modes assure that all compounds that tend to form either cations or anions will be detected by the mass spectrometer. We performed a PCA of all signals obtained in ESI+ and ESI−. The infection with P. syringae and P. cucumerina induced strong changes in the metabolome of the wild type plants and amt1.1 plants that differed for the positively and negatively ionized compounds (Figure 6). The impact of the infection in ESI+ appeared stronger in amt1.1 upon P. cucumerina infection compared with P. syringae; however the changes in ESI− are more subtle because there is an overlap in the variability of amt1.1 with or without infection independently of the pathogen.
Figure 6.
Non-supervised Principal Component Analysis. (PCA) analysis representation of major sources of variability of ESI+ and ESI− signals obtained from a non-targeted analysis by HPLC-QTOFMS to monitor metabolomic changes during bacterial (A) and fungal invasion (B). (A) Five week old plants were dip inoculated with P. syringae with 2.5 × 10E5 c.f.u/ml. 48 hpi Leaf material from 15 individual plants were pooled together for each treatment × genotype combination. (B) Two week old plants were sprayed inoculated with 10E3 spores/ml of P. cucumerina and samples for analysis were collected 48 hpi. Leaf material from 150 individual plants were pooled together for each treatment × genotype combination. Data points represent two technical replicates from three independent experiments (biological replicates; n = 6) injected randomly into the HPLC-QTOFMS. The signals corresponding to different treatments were compared using the non-parametric Kruskal-Wallis test, and only data with a P-value lower than 0.01 between groups was used for subsequent processing.
Notably, in the absence of infection, the basal metabolomes of Col3-gl1 and amt1.1 do not overlap in any of the PCA with the exception of ESI− in the seedlings. This suggests that many metabolites are already altered in amt1.1 in the absence of infection, and these changes may contribute to its enhanced resistance, discarding a general priming phenomenon to explain the phenotypes observed against both pathogens. Additionally, the wild type plants and amt1.1 plants show differences in the basal responses between both experiments without inoculation, which may be due to a difference in the age of the plants (5 and 2 week-old plants for the P. syringae and P. cucumerina experiments, respectively) but also to the different mock treatments (dip inoculated Silwet for P. syringae and sprayed 10 mM MgSO4 for P. cucumerina). A heatmap analysis was performed to identify metabolites involved in the resistance phenotypes of amt1.1, and the clusters of metabolites that over-accumulated in amt1.1 either in the absence or presence of the infection were selected (Figure S1). Clusters from the heatmaps that showed overaccumulated compounds in the mutant (red color) were selected for a detailed metabolic study and pathway analysis.
Prior to identifying the signals in the selected clusters, we performed a detailed analysis of the amino acids in the metabolome. For such purpose, we constructed a library of amino acid standards for the identity assignation using the exact mass provided by the Q-TOF analyser and the retention time parameter (Gamir et al., 2014).
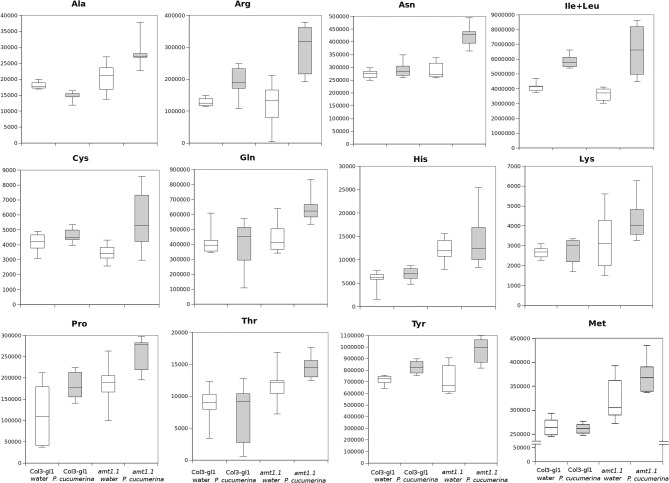
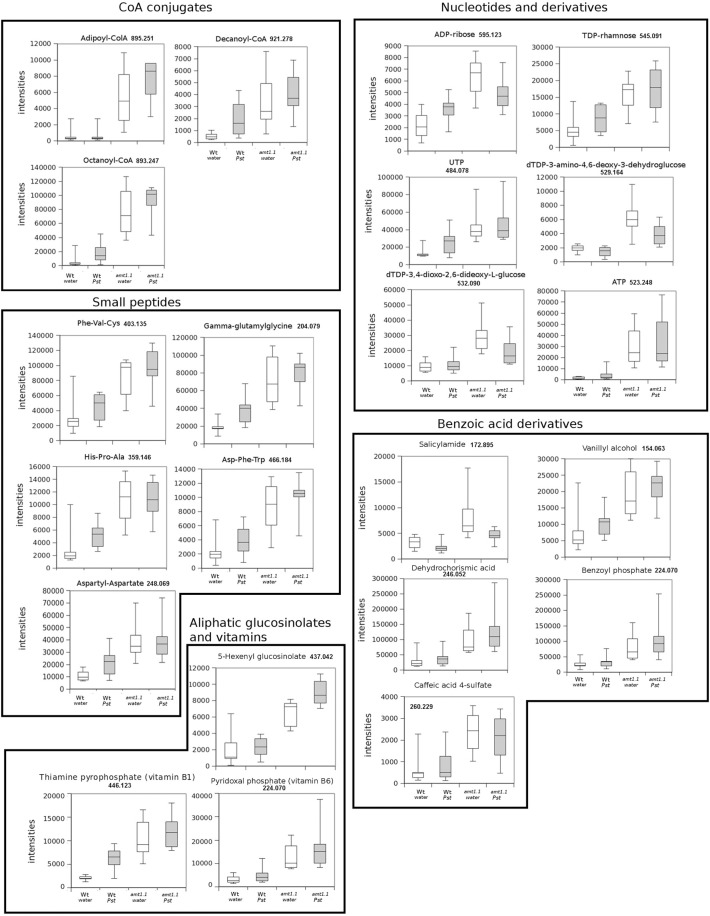
In response to P. syringae, all the amino acids, together with azelaic acid and pipecolic, were not increased in the amt1.1 mutant in response to the bacterium (data not shown). However, a different profile was observed following P. cucumerina infection. The majority of amino acids were increased in amt1.1 plants with or without infection (Figure 7). Pro, Thr, His, and Met were previously elevated in the absence of infection and this increase occurred until 48 hpi. Ala, Arg, Asn, Ile+Leu (not separated in our chromatographic analysis), Cys, Gln, Lys, and Tyr showed a primed profile because they remained at the same levels in the wild-type plants before the inoculation; however, after infection, amt1.1 displayed elevated levels compared to Col3-gl1. Only Asp, Glu, Phe, Trp, and Val in the mutant remained at the same levels as wild type infected plants (data not shown). Next, we performed a full comparative analysis of the metabolome. To identify the compounds, we used a library of standards with both the exact mass provided by the Q-TOF detector and the retention time. For the compounds with no available standards, we used the exact mass and fragmentation spectrum, when available, from the Massbank and Metlin databases. For such compounds, we assigned a tentative identification. After the signals were either exactly or tentatively identified, the compounds were searched against the Aracyc (http://pathway.gramene.org/gramene/aracyc.shtml) and Kegg (http://www.genome.jp/kegg/) databases to assign a putative metabolic pathway and a biological function. The analysis of the hormones or amino acids in amt1.1 yielded a plausible explanation for its reduced susceptibility to P. syringae. However, the selection of the metabolite clusters from the heatmap that over-accumulated in amt1.1 compared to the wild type plants following infection provides valuable information. Following the tentative identification of metabolites and their classification into metabolic pathways of Arabidopsis, we observed that fatty acid-CoA conjugates were increased in amt1.1 either in the absence or presence of P. syringae (Figure 8). Nucleotides, such as ATP or UTP were also increased in amt1.1. Interestingly, three chemical species derived from TDP were also found for the amt1.1 over-accumulated compounds. Dipeptides and tripeptides were another group of interest, which was surprising because the free amino acids in amt1.1 are not different from the wild type plants. Although SA was not increased in the mutant, several benzoyl compounds over-accumulated in amt1.1, both with and without infection. The metabolic pathways with a higher number of hits in the cluster of over-accumulated compounds in amt1.1 were the flavonoid and phenylpropanoid phytoalexins (Table 1). Although infection induced a strong increase in these compounds in Col3-gl1, amt1.1 showed higher levels in the absence of infection. Finally, an aliphatic glucosinolate and vitamins B1 and B6, all of which are involved in defense, were also increased in the mutant (Figure 8). For the interaction with P. cucumerina, the identified compounds matched four secondary metabolism pathways of Arabidopsis, indolic glucosinolates, aliphatic glucosinolates, the Trp pathway and phenylpropanoids (with a total of 19 compounds) (Figure 9). Notably, many secondary metabolites in these groups accumulated in amt1.1 without infection, and all were then either increased or maintained in response to P. cucumerina in amt1.1 plants compared to wild type plants. In addition to these pathways, although there were fewer hits, tentative identification of the metabolites showed that amt1.1 responded to infection by increasing three mevalonic acid derivatives, four nucleotide intermediaries, three shikimic acid metabolites, two polyamines, and several hormones, such as GA3, GA-A58, and ABA (Figure S2).
Figure 7.
Aminoacid profiling upon P. cucumerina infection. Two week old Col3-gl/1 and amt1.1 plants either mock or P. cucumerina inoculated were processed for relative quantification analysis by HPLC-QTOFMS data. The concentration of the metabolites was determined in all the samples by normalizing the chromatographic area for each compound with the dry weight of the corresponding sample. White bars are mock inoculated and filled bars are P. cucumerina infected plants. Leaf material from 150 individual plants were pooled together for each treatment × genotype combination. Boxplots represent average three independent experiments with two technical replicates (n = 6).
Figure 8.
Profiling of the main hits in the overaccumulated compounds upon P. syringae (Pst) infection in amt1.1 compared with Col3-gl1. Five week old Col3-gl1 and amt1.1 were infected as described in Figure 1. After 48 hpi plants were processed for relative quantification analysis by HPLC-QTOFMS data. The concentration of the metabolites was determined in all the samples by normalizing the chromatographic area for each compound with the dry weight of the corresponding sample. The compounds were tentatively identified using the exact mass criteria using the METLIN and Massbank databases. The compounds were grouped by metabolic pathways according to KEGG and AraCyc databases. White bars are mock inoculated and filled bars are P. syringae infected plants. Leaf material from 15 individual plants were pooled together for each treatment × genotype combination. Boxplots represent average three independent experiments with two technical replicates. Only data showing a p-value below 0.05 after a Kuskal Wallys test were used for pathway assignation (n = 6).
Table 1.
Phenylpropanoid profiling upon P. syringae infection.
| Neutral mass | Col3-gl1-Mock | Col3-gl1-P. syringae | amt1.1-Mock | amt1.1-P. syringae | Tentative identification |
|---|---|---|---|---|---|
| ESI− | |||||
| 868.141 | 5717.574 ± 1339.009 | 7903.077 ± 1171.697 | 15069.108 ± 3900.508 | 10608.243 ± 1332.910 | Quercetagetin 7-methyl ether 3-(2‴-caffeoylglucosyl)-(1→2)-glucuronide |
| 784.183 | 1993.974 ± 416.230 | 5584.543 ± 1443.404 | 6600.123 ± 1439.933 | 5310.700 ± 908.973 | 6-Hydroxyluteolin 7-[6″-(3-hydroxy-3-methylglutaryl)glucoside]-3-glucuronide |
| 920.272 | 1741.940 ± 548.621 | 4257.727 ± 1356.214 | 9965.213 ± 1679.580 | 8535.086 ± 1714.162 | Quercetin 3-(2‴-caffeylsambubioside)-7-glucoside |
| 546.072 | 2872.295 ± 527.433 | 5379.807 ± 1033.293 | 10884.914 ± 2827.895 | 20974.750 ± 3776.673 | 4,6-Dideoxy-4-oxo-dTDP-D-glucose |
| 982.276 | 1986.003 ± 537.055 | 6063.620 ± 1171.669 | 11885.748 ± 1351.898 | 15605.830 ± 897.168 | Malvidin-3-(p-coumaroyl)-rutinoside-5-glucoside |
| 356.221 | 3751.581 ± 770.196 | 2451.929 ± 1357.613 | 11677.959 ± 1955.930 | 11193.410 ± 1678.297 | 4,8,11,14-Eicosatetraenoic acid, 6-hydroxy-, (E,Z,Z,Z)- |
| 402.132 | 97604.298 ± 15455.088 | 190757.113 ± 33780.888 | 343773.018 ± 49529.885 | 404922.286 ± 53046.318 | 7-Hydroxyflavanone beta-D-glucopyranoside |
| 892.242 | 16249.517 ± 9694.477 | 39546.429 ± 13717.218 | 164163.569 ± 34206.807 | 186117.251 ± 20320.571 | Palargonidin 3-(6″-ferulylglucoside)-5-(6‴-malonylglucoside) |
| 528.126 | 834.255 ± 209.720 | 1438.870 ± 373.901 | 6122.663 ± 1052.023 | 6546.308 ± 1472.074 | 4,2′-Dihydroxy-3,4′,6′-trimethoxychalcone 4-glucoside |
| 344.123 | 6814.904 ± 2501.614 | 9237.129 ± 1791.413 | 17129.393 ± 2994.036 | 18527.512 ± 1827.166 | 5,6,7,4′-Tetramethoxyflavanone |
| 460.129 | 25299.855 ± 13806.666 | 40022.020 ± 15639.629 | 158482.998 ± 18759.482 | 169688.892 ± 18730.315 | 7,8,3′,4′-Tetramethoxy-6″,6″-dimethylpyrano[2″,3″:5,6]flavone |
| 894.252 | 2544.518 ± 1377.062 | 5227.236 ± 2004.211 | 25842.316 ± 5738.193 | 29016.835 ± 3447.355 | Genistein 7,4′-bis(O-glucosylapioside) |
| 622.151 | 2809.060 ± 709.473 | 7451.498 ± 1212.428 | 12048.485 ± 1529.533 | 12526.350 ± 1257.233 | Kaempferol 3-[2′”-acetyl-alpha-L-arabinopyranosyl-(1→6)-galactoside] |
| ESI+ | |||||
| 292.042 | 1186.017 ± 392.336 | 1807.786 ± 476.655 | 4104.516 ± 661.600 | 4953.756 ± 1425.747 | 5,7,3′-Trihydroxyisoflavone |
| 360.181 | 853.215 ± 287.879 | 1370.833 ± 540.537 | 6396.268 ± 2111.103 | 4320.185 ± 1595.289 | 5,2′,3′-Trihydroxy-3,7,8-trimethoxyflavone |
| 306.072 | 5958.788 ± 1705.843 | 7086.008 ± 2216.577 | 23547.293 ± 5398.893 | 19686.483 ± 5614.565 | Pelargonidin |
| 400.116 | 30044.197 ± 4429.060 | 48455.063 ± 8051.467 | 89620.850 ± 16202.253 | 69018.310 ± 10018.920 | Flavonol 3-O-D-galactoside |
| 544.609 | 1029.246 ± 505.676 | 2338.742 ± 704.251 | 5440.121 ± 1619.737 | 5301.904 ± 2113.984 | Quercetin 3-glucoside-3′-sulfate |
| 498.147 | 65764.676 ± 21821.696 | 45436.512 ± 10350.166 | 202317.111 ± 40047.133 | 192242.002 ± 55136.009 | Quercetin 7-methyl ether 3,3′-disulfate |
| 460.138 | 12122.235 ± 2423.017 | 16230.111 ± 3295.099 | 27447.715 ± 4518.375 | 24785.374 ± 4133.228 | Luteolin 4′-methyl ether 7,3′-disulfate |
| 636.387 | 137818.486 ± 17769.746 | 86637.426 ± 7725.291 | 71514.777 ± 4284.190 | 63127.380 ± 9084.052 | flavonoid Isoscutellarein 4′-methyl ether 8-(2″,4″-disulfatoglucuronide) |
Five week old plants either mock Col3-gl/1 and amt1.1 or P. syringae inoculated (Col3-gl/1 Pc and amt1.1 Pc) plants were processed for relative quantification analysis by HPLC-QTOFMS data. Tentative identification of signals was performed by contrasting the exact mass in the METLIN and Massbank databases. The concentration of selected signals within the phenylpropanoid metabolism was determined in all the samples by normalizing the chromatographic area for each compound with the dry weight of the corresponding sample. Data show average values of three independent biological replicates containing pools of 15 plants in each replicate are shown with their standard deviation.
Figure 9.
Profile of the main hits in the overaccumulated compounds upon P. cucumerina infection in amt1.1 compared with Col3-gl1. Two week old Col3-gl1 and amt1.1 were infected as described in Figure 1. After 48 hpi plants were processed for relative quantification analysis by HPLC-QTOFMS data. The concentration of the metabolites was determined in all the samples by normalizing the chromatographic area for each compound with the dry weight of the corresponding sample. The compounds were tentatively identified using the exact mass criteria using the METLIN and Massbank databases. Those compounds tentatively identified have the exact mass indicated. Te compounds fully identified have no reference to the mass. The compounds were grouped by metabolic pathways according to KEGG and AraCyc databases. White bars are mock inoculated and filled bars are P. cucmerina infected plants. Leaf material from 150 individual plants were pooled together for each treatment × genotype combination. Boxplots represent average three independent experiments with two technical replicates. + indicates outliers. Only data showing a p-value below 0.01 after a Kuskal wallys test were used for pathway assignation (n = 6).
Discussion
We previously reported (Camañes et al., 2012b) that under optimal nutritional conditions, the total contents of N in nrt2.1 and amt1.1 plants are identical, which indicates that the N content is not responsible for altering the basal resistance in the mutants. Based on our previous study, we performed all resistance experiments, except otherwise noted, under the optimal fertilization conditions. First, we demonstrated that mutations in several genes related to N transport resulted in an increased resistance against two pathogens with different lifestyles. The HATS of nitrate uses a membrane complex in which the NRT3.1 protein modulates the function of NRT2.1, and mutation of NRT3.1 results in the most significant losses of HATS function compared to NRT2.1 mutations (Orsel et al., 2006). Therefore, we tested two different hypotheses. First, we tested whether this gene may also regulate ammonium transport, and second, we tested whether NRT3.1 is involved in the plant defense response. The uptake experiments with 15NH+4 showed that the absorption of ammonium in nrt3.1 does not differ from Col-0 plants, which suggests that it is not involved in ammonium HATS. We also demonstrated that AMT1.1 gene expression with or without N was similar for Col-0 and nrt3.1 plants. These observations confirm that this gene is not involved in AMT1.1 modulation. Because nrt3.1 plants show resistance against P. cucumerina and P. syringae, this suggests that it may have overlapping mechanisms with NRT2.1 to stimulate defense; however, this hypothesis must be confirmed.
We ascertained the influence of AMT1.1 on plant resistance because our experiments suggest that AMT1.1 acts as a transceptor to coordinate plant tolerance against N depletion and defense against pathogens as suggested previously for NRT1 and NRT2.1 (Gojon et al., 2011; Camañes et al., 2012b). The mutant amt1.1 showed reduced susceptibility against P. syringae, and surprisingly, amt1.1 showed strong resistance toward P. cucumerina infection compared to wild type plants. Previously, we demonstrated that environmental factors contribute to the enhanced expression of NRT2.1. For example, N depletion affects the resistance of Arabidopsis to P. syringae. Although the depletion of N induces AMT1.1 gene expression (Figure 2), it also increased the susceptibility in amt1.1 plants, suggesting that it is the absence and not the overexpression of AMT1.1 that alters resistance mechanisms. Notably, neither infection with P. syringae or P. cucumerina significantly altered the AMT1.1 transcripts. We tested whether this also applies to resistance against necrotrophs. The enhanced resistance of amt1.1 against P. cucumerina was not altered by N depletion; therefore, the absence of these genes impacts Arabidopsis defense against this pathogen but not their overexpression, suggesting posttranscriptional or posttranslational regulation. This is not so surprising because NRT1.1 and NRT2.1 have a complex protein regulation mechanism mediated by proteins, such as CIPK23 and NAR2.1 (Okamoto et al., 2006; Ho et al., 2009).
To identify the molecular mechanisms for the amt1.1 phenotypes, we performed several targeted and non-targeted metabolomic studies. For the targeted studies, hormones regulate the plant immune system (Pieterse et al., 2009). Active SA-dependent responses against P. syringae promote resistance in Arabidopsis. Additionally, because of the existing negative crosstalk between the SA and JA signaling pathways, resistance against biotrophs is associated with the concomitant downregulation of JA and JA-Ile levels (Pieterse et al., 2009). This has been observed in amt1.1 plants upon P. syringae infection. In fact, SA, JA, and JA-Ile remain lower compared to wild type plants during a time-course experiment. Therefore, the reduced susceptibility of amt1.1 to the bacterium cannot be caused by changes in the hormonal balance. Except for IAA, the hormones tested remained at lower levels in the absence of infection. This suggests that disruption of AMT1.1 affects the basal levels of SA, JA, and JA-Ile, producing several constitutive changes. Interestingly, these basal changes are not visible in 2-week old plants; therefore, some alterations affected by the mutation are age dependent. For non-host resistance, such as for P. cucumerina, the defense responses are horizontal, multigenic and not controlled by single hormonal pathway, because other hormones in addition to SA, JA, ABA, and ET influence Arabidopsis responses against this necrotroph (Sanchez-Vallet et al., 2010). These compounds include the β-subunit of the heterotrimeric G-protein (Delgado-Cerezo et al., 2012) and glucosinolates and other Trp derivatives (Sánchez-Vallet et al., 2012; Gamir et al., 2014). In our first approach, we showed that amt1.1 influences the SA, JA-Ile, and IAA profile during infection. Both JA-Ile and IAA over-accumulate, whereas SA is at lower levels compared to Col3-gl1. This may partially explain the observed phenotype because P. cucumerina has a necrotrophic lifestyle and is sensitive to JA-dependent signaling (Thomma et al., 2000). Recent research has suggested complex interplay between hormones in addition to the negative crosstalk between SA and JA (Pieterse et al., 2009), IAA also has antagonistic effects with SA, and both may explain the lower amounts of SA observed in amt1.1, although the final link between the ammonium transporter and hormonal signaling remains to be elucidated. Arabidopsis can also resist P. cucumerina through two other major mechanisms, callose deposition and camalexin (Ton and Mauch-Mani, 2004; García-Andrade et al., 2011; Sánchez-Vallet et al., 2012). Both mechanisms are directly or indirectly influenced by AMT1.1. Early callose deposition is enhanced in the mutant, and the camalexin levels remain higher at 48 hpi compared to Col3-gl1.
In our Arabidopsis-P. syringae system, we observed that amt1.1 does not influence the basal amino acid accumulation in response to the bacterium. By contrast, there is a significant change in the amino acid profile for P. cucumerina. The mutant amt1.1 over-accumulates several amino acids in the absence of infection, but these changes are even more robust after infection. Up to 11 amino acids over-accumulate in amt1.1 infected plants compared to Col3-gl1. Surprisingly, Trp remains at the same level as the wild type plants with or without P. cucumerina. This may be explained by the over-accumulation of other Trp derivatives, such as indolic glucosinolates, IAA, camalexin, and other indole conjugates (Figure 9). These compounds are metabolic sinks that may trap the putative overproduction of Trp.
According to the PCA, the distance between Col3-gl1 and amt1.1 upon infection by both pathogens is rather marked; however, the diversity of the metabolic responses against P. cucumerina is much wider compared with the bacterium. In fact, of all of the compounds analyzed by targeted and untargeted chromatography that were found in clusters of over-accumulated metabolites in amt1.1 upon P. syringae infection, only five pathways were identified, and of 45 signals, 24 were tentatively assigned to the phenylpropanoids (Table 1) and 5 to benzoic acid derivatives. This suggests that shikimic acid derivatives may compete with SA accumulation, which is a likely explanation for the reduced susceptibility of amt1.1 and the reduced levels of free SA. Phenylpropanoids have been associated with plant resistance against a wide range of pathogens (Shadle et al., 2003). Two other compounds over-accumulated in amt1.1 plants are vitamins B1 and B6. Both have been linked to disease resistance (Denslow et al., 2005; Ahn et al., 2007). Exogenous treatment with vitamin B1 can induce defense priming in Arabidopsis against P. syringae mediated by H2O2 accumulation, callose deposition, and NPR1. Vitamin B6 is involved in superoxide quenching and stress responses, and during P. syringae infection in Nicotiana, vitamin B6 acts as an antioxidant and modulator of active oxygen species (Denslow et al., 2005). The relevance of the other metabolites, such as small peptides, nucleotides and CoA conjugates in the defense against pathogens must be clarified. Of the signals over-accumulated in amt1.1 infected with P. cucumerina, 51 were tentatively or fully identified. These compounds were mainly involved in the indolic and aliphatic glucosinolates pathways, Trp pathway, flavonoids, nucleotides, and fatty acids. Notably, glucosinolates have been directly linked to resistance against P. cucumerina Sánchez-Vallet et al., 2012; Gamir et al., 2014, and also callose deposition in response to PAMPs (Clay et al., 2009). Although no direct evidence for AMT1.1 and glucosinolates has been previously reported, this may be worth investigating. The NRT/PTR transporters have been associated with glucosinolate translocation in Arabidopsis, mainly aliphatic (Nour-Eldin et al., 2012). Additionally, Met is a precursor of aliphatic glucosinolates (Kraker and Gershenzon, 2011). This amino acid highly accumulates in amt1.1 plants infected with P. cucumerina. Furthermore, three precursors of aliphatic glucosinolates are also elevated in amt1.1 (Figure 9). These findings suggest a repressive influence of AMT1.1 on plant defenses against necrotrophic infection that result in changes in amino acid metabolism and/or peptide transport that causes increased defenses, most likely mediated by aliphatic and indolic glucosinolates. Other relevant metabolic contributions from different pathways may also be involved, such as chlorogenic acid, ABA, spermidine, several fatty acids, and other unclassified compounds that are also over-accumulated in amt1.1 plants upon infection.
We previously demonstrated that the member of the high-affinity transport system of nitrate, the NRT2.1 gene, in addition to transporting nitrate, is involved in the coordination of plant defense responses (Camañes et al., 2012b). This gene is de-regulated upon low nitrate concentrations in the roots, subsequently, the HATS is activated, providing tolerance mechanism against abiotic stresses (Gansel et al., 2001). Despite these functions, NRT2.1 represses plant responses against bacterial and fungal pathogens. The metabolic interplay between nitrate uptake and resistance against pathogens is being elucidated. Because this resistance mechanism is multicomponent and non-pathogen-specific, the disruption of NRT2.1 results in enhanced resistance against P. syringae and P. cucumerina through different defense mechanisms (Camañes et al., 2012b; Gamir et al., 2014).
Here, we demonstrate that AMT1.1 acts as a negative regulator of Arabidopsis defense. Its disruption has a moderate impact on metabolomic changes upon P. syringae infection, which is correlated with a reduced susceptibility to the bacterium. By contrast, the metabolomic changes of amt1.1 upon P. cucumerina challenge are significant, targeting Trp-derivatives and indolic glucosinolates. These changes may be responsible for the enhanced resistance of amt1.1 against the fungus. This study and previous findings from our laboratory indicate that N transporters are transceptors that impact the transcriptome (Camañes et al., 2012b) and metabolome of Arabidopsis (Gamir et al., 2012, 2014) to coordinate abiotic stress tolerance and the negative regulation of biotic stress responses. The manner in which NRT2.1 and AMT1.1 regulate stress signaling remains to be identified; however, it is clear that the plant has mechanisms to distinguish and regulate different sources of stress. Transceptors appear to play a key role in integrating the environmental signals and plant defense responses.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the SCIC of the Universitat Jaume I and particularly to Cristian Barrera, Jorge Collado and Carla Ordiñana for their technical support. This work was financially supported by the Plan de Promoción de la Investigación-Universitat Jaume I P1·1B2010-06 and P1-1B2013-68 and the Spanish ministry MINECO AGL2012-39923-C02.
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00231/abstract
Heat map analysis performed with Marvis (Filter and Cluster packages). Five and two week old plants either mock (Col3-gl/1 and amt1.1) or either P. syringae Col3-gl/1 P. syringae and amt1.1 P. syringae or P. cucumerina inoculated (Col3-gl/1 Pc and amt1.1 Pc) plants were processed for relative quantification analysis by HPLC-QTOFMS data. The concentration of the metabolites was determined in all the samples by normalizing the chromatographic area for each compound with the dry weight of the corresponding sample. Heatmaps are generated by using Mar-Vis Filter and Cluster following a Kruskal-Wallys test (p < 0.01). Clusters overepresented amt1.1 mock or infected (intense red colors) compared with wild type plants were selected for subsequent data analysis.
Profile of additional hits in the overaccumulated compounds upon P. cucumerina infection in amt1.1 compared with Col3-gl1. For experimental details see Figures 1, 9. The concentration of the metabolites was determined in all the samples by normalizing the chromatographic area for each compound with the dry weight of the corresponding sample. The compounds were tentatively identified using the exact mass criteria using the METLIN and Massbank databases. Those compounds tentatively identified have the exact mass indicated. Te compounds fully identified have no reference to the mass. The compounds were grouped by metabolic pathways according to KEGG and AraCyc databases. White bars are mock inoculated and filled bars are P. cucmerina infected plants. Leaf material from 150 individual plants were pooled together for each treatment × genotype combination. Boxplots represent average three independent experiments with two technical replicates. Only data showing a p-value below 0.01 after a Kuskal wallys test were used for pathway assignation (n = 6).
References
- Ahn I.-P., Kim S., Lee Y.-H., Suh S.-C. (2007). Vitamin B1-induced priming is dependent on hydrogen peroxide and the NPR1 gene in arabidopsis Plant Physiol. 143, 838–848 10.1104/pp.106.092627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H., Shinn P., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657 10.1126/science.1086391 [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M., Segura A., Moreno M., López G., García-Olmedo F., Molina A. (2002). Snakin-2, an antimicrobial peptide from potato whose gene is locally induced by wounding and responds to pathogen infection1. Plant Physiol. 128, 951–961 10.1104/pp.010685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks D. M., Bender C. L., Kunkel B. N. (2005). The Pseudomonas Syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defences is Arabidopsis thaliana. Mol. Plant Pathol. 6, 629–639 10.1111/j.1364-3703.2005.00311.x [DOI] [PubMed] [Google Scholar]
- Camañes G., Bellmunt E., García-Andrade J., García-Agustín P., Cerezo M. (2012a). Reciprocal regulation between AtNRT2.1 and AtAMT1.1 expression and the kinetics of NH+4 and NO−3 influxes. J. Plant Physiol. 169, 268–274 10.1016/j.jplph.2011.10.011 [DOI] [PubMed] [Google Scholar]
- Camañes G., Pastor V., Cerezo M., García-Andrade J., Vicedo B., García-Agustín P., et al. (2012b). A deletion in NRT2.1 attenuates pseudomonas syringae-induced hormonal perturbation, resulting in primed plant defenses. Plant Physiol. 158, 1054–1066 10.1104/pp.111.184424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerezo M., Tillard P., Filleur S., Munos S., Daniel-Vedele F., Gojon A. (2001). Major alterations of the regulation of root NO−3 uptake are associated with the mutation of Nrt2.1 and Nrt2.2 genes in arabidopsis Arabidopsis. Plant Physiol. 127, 262–271 10.1104/pp.127.1.262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay N. K., Adio A. M., Denoux C., Jander G., Ausubel F. M. (2009). Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323, 95–101 10.1126/science.1164627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechorgnat J., Patrit O., Krapp A., Fagard M., Daniel-Vedele F. (2012). Characterization of the Nrt2.6 gene in Arabidopsis thaliana: a link with plant response to biotic and abiotic stress. PLoS ONE 7:e42491 10.1371/journal.pone.0042491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Cerezo M., Sánchez-Rodríguez C., Escudero V., Miedes E., Fernández P. V., Jordá L., et al. (2012). Arabidopsis heterotrimeric G-protein regulates cell wall defense and resistance to necrotrophic fungi. Mol. Plant. 5, 98–114 10.1093/mp/ssr082 [DOI] [PubMed] [Google Scholar]
- Denslow S. A., Walls A. A., Daub M. E. (2005). Regulation of biosynthetic genes and antioxidant properties of vitamin B6 vitamers during plant defense responses. Physiol. Mol. Plant Pathol. 66, 244–255 10.1016/j.pmpp.2005.09.004 [DOI] [Google Scholar]
- Durner J., Klessig D. F. (1999). Nitric oxide as a signal in plants. Curr. Opin. Plant Biol. 2, 369–374 10.1016/S1369-5266(99)00007-2 [DOI] [PubMed] [Google Scholar]
- Fernie A. R., Aharoni A., Willmitzer L., Stitt M., Tohge T., Kopka J., et al. (2011). Recommendations for reporting metabolite data. Plant Cell. 23, 2477–2482 10.1105/tpc.111.086272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flors V., Ton J., van Doorn R., Jakab G., García-Agustín P., Mauch-Mani B. (2008). Interplay between JA, SA and ABA signalling during basal and induced resistance against Pseudomonas syringae and Alternaria brassicicola. Plant J. 54, 81–92 10.1111/j.1365-313X.2007.03397.x [DOI] [PubMed] [Google Scholar]
- Gamir J., Pastor V., Cerezo M., Flors V. (2012). Identification of indole-3-carboxylic acid as mediator of priming against Plectosphaerella cucumerina. Plant Physiol. Biochem. 61, 169–179 10.1016/j.plaphy.2012.10.004 [DOI] [PubMed] [Google Scholar]
- Gamir J., Pastor V., Kaever A., Cerezo M., Flors V. (2014). Targeting novel chemical and constitutive primed metabolites against Plectosphaerella cucumerina. Plant J. 78, 227–240 10.1111/tpj.12465 [DOI] [PubMed] [Google Scholar]
- Gansel X., Munos S., Tillard P., Gojon A. (2001). Differential regulation of the NO3- and NH4+ transporter genes AtNrt2.1 and AtAmt1.1 in Arabidopsis: relation with long-distance and local controls by N status of the plant. Plant J. 26, 143–155 10.1046/j.1365-313x.2001.01016.x [DOI] [PubMed] [Google Scholar]
- García-Andrade J., Ramírez V., Flors V., Vera P. (2011). Arabidopsis ocp3 mutant reveals a mechanism linking ABA and JA to pathogen-induced callose deposition. Plant J. 67, 783–794 10.1111/j.1365-313X.2011.04633.x [DOI] [PubMed] [Google Scholar]
- Gazzarrini S., Lejay T., Gojon A., Ninnemann O., Frommer W. B., von Wirén N. (1999). Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into Arabidopsis roots. Plant Cell 11, 937–947 10.1105/tpc.11.5.937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Ann. Rev. Phytopathol. 43, 205–227 10.1146/annurev.phyto.43.040204.135923 [DOI] [PubMed] [Google Scholar]
- Gojon A., Krouk G., Perrine-Walker F., Laugier E. (2011). Nitrate transceptor(s) in plants. J. Exp. Bot. 62, 2299–2308 10.1093/jxb/erq419 [DOI] [PubMed] [Google Scholar]
- Gupta K. J., Brotman Y., Segu S., Zeier T., Zeier J., Persijn S. T., et al. (2013). The form of nitrogen nutrition affects resistance against Pseudomonas syringae pv. phaseolicola in tobacco. J. Exp. Bot. 64, 553–568 10.1093/jxb/ers348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C. H., Lin S. H., Hu H. C., Tsay Y. F. (2009). CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194 10.1016/j.cell.2009.07.004 [DOI] [PubMed] [Google Scholar]
- Hu H. C., Wang Y. Y., Tsay Y. F. (2009). AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J. 57, 264–278 10.1111/j.1365-313X.2008.03685.x [DOI] [PubMed] [Google Scholar]
- Kaever A., Landesfeind M., Possienke M., Feussner K., Feussner I., Meinicke P. (2012). MarVis-filter: ranking, filtering, adduct and isotope correction of mass spectrometry data. J. Biomed. Biotechol. 2012:263910 10.1155/2012/263910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser B. N., Rawat S. R., Siddiqi M. Y., Masle J., Glass A. D. M. (2002). Functional analysis of an Arabidopsis T-DNA Knockout of the high-affinity NH+4 transporter AtAMT1.1. Plant Physiol. 130, 1263–1275 10.1104/pp.102.010843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller M., Rogiers S., Schultz H. (2003). Nitrogen and ultraviolet radiation modify grapevines' susceptibility to powdery mildew. Vitis 42, 87–94 [Google Scholar]
- Kraker J.-W., Gershenzon J. (2011). From amino acid to glucosinolate biosynthesis: protein sequence changes in the evolution of methylthioalkylmalate synthase in Arabidopsis. Plant Cell 23, 38–53 10.1105/tpc.110.079269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejay L., Tillard P., Lepetit M., Olive F. D., Filleur S., Daniel-Vedele F., et al. (1999). Molecular and functional regulation of two NO−3 uptake systems by N- and C- status of Arabidopsis plants. Plant J. 18, 509–519 10.1046/j.1365-313X.1999.00480.x [DOI] [PubMed] [Google Scholar]
- Little D. Y., Rao H., Oliva S., Daniel-Vedele F., Krapp A., Malamy J. E. (2005). The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc. Natl. Acad. Sci. U.S.A. 102, 13693–13698 10.1073/pnas.0504219102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Solano R. (2005). Molecular players regulating the jasmonate signaling network. Curr. Opin. Plant Biol. 8, 532–540 10.1016/j.pbi.2005.07.003 [DOI] [PubMed] [Google Scholar]
- Marschner P. (2012). Marschner's Mineral Nutrition of Higher Plants. 3rd Edn. Amsterdam; Boston: Elsevier/Academic Press [Google Scholar]
- Melotto M., Underwood W., Koczan J., Nomura K., He S. Y. (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126, 969–980 10.1016/j.cell.2006.06.054 [DOI] [PubMed] [Google Scholar]
- Nour-Eldin H. H., Andersen T. G., Burow M., Madsen S. R., Jørgensen M. E., Olsen C. E., et al. (2012). NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature 488, 531–534 10.1038/nature11285 [DOI] [PubMed] [Google Scholar]
- Okamoto M., Kumar A., Li W., Wang Y., Siddiqi M. Y., Crawford N. M., et al. (2006). High-affinity nitrate transport in roots of Arabidopsis depends on expression of the NAR2-like gene AtNRT3.1. Plant Physiol. 140, 1036–1046 10.1104/pp.105.074385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsel M., Chopin F., Leleu O., Smith S. J., Krapp A., Daniel-Vedele F., et al. (2006). Characterization of a two-component high-affinity nitrate uptake system in Arabidopsis. Physiology and protein–protein interaction. Plant Physiol. 142, 1304–1317 10.1104/pp.106.085209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse C. M. J., Leon-Reyes A., Van der Ent S., Van Wees S. C. M. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316 10.1038/nchembio.164 [DOI] [PubMed] [Google Scholar]
- Pitzschke A., Hirt H. (2010). Bioinformatic and systems biology tools to generate testable models of signaling pathways and their targets. Plant Physiol. 152, 460–469 10.1104/pp.109.149583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Vallet A., López G., Ramon B., Delgado-Cerezo M., Riviere M. P., Llorente F., et al. (2012). Disruption of abscisic acid signaling constitutively activates Arabidopsis resistance to the necrotrophyic fungus Plectosphaerella cucumerina. Plant Phys. 160, 2109–2124 10.1104/pp.112.200154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Vallet A., Ramos B., Bednarek P., López G., Piślewska-Bednarek M., Schulze-Lefert P., et al. (2010). Tryptophan-derived secondary metabolites in Arabidopsis thaliana confer non-host resistance to necrotrophic Plectosphaerella cucumerina fungi. Plant J. 63, 115–127 10.1111/j.1365-313X.2010.04224.x [DOI] [PubMed] [Google Scholar]
- Shadle G. L., Wesley S. V., Korth K. L., Chen F., Lamb C., Dixon R. A. (2003). Phenylpropanoid compounds and disease resistance in transgenic tobacco with altered expression of L-phenylalanine ammonia-lyase. Phytochemistry 64, 153–161 10.1016/S0031-9422(03)00151-1 [DOI] [PubMed] [Google Scholar]
- Shelden M. C., Dong B., de Bruxelles G. L., Trevaskis B., Whelan J., Ryan P. R., et al. (2001). Arabidopsis ammonium transporters, AtAMT1;1 and AtAMT1;2, have different biochemical properties and functional roles. Plant Soil 231, 151–160 10.1023/A:1010303813181 [DOI] [Google Scholar]
- Smith C. A., Want E. J., O'Maille G., Abagyan R., Siuzdak G. (2006). XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 78, 779–787 10.1021/ac051437y [DOI] [PubMed] [Google Scholar]
- Solomon P. S., Oliver R. P. (2001). The nitrogen content of the tomato leaf apoplast increases during infection by Cladosporium fulvum. Planta 213, 241–249 10.1007/s004250000500 [DOI] [PubMed] [Google Scholar]
- Thomma B. P. H. J., Eggermont K., Broekaert W. F., Cammue B. P. A. (2000). Disease development of several fungi on Arabidopsis can be reduced by treatment with methyl jasmonate. Plant Physiol. Biochem. 38, 421–427 10.1016/S0981-9428(00)00756-7 [DOI] [Google Scholar]
- Ton J., Mauch-Mani B. (2004). β-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J. 38, 119–130 10.1111/j.1365-313X.2004.02028.x [DOI] [PubMed] [Google Scholar]
- Tornero P., Dangl J. L. (2001). A high throughput method for quantifying growth of phytopathogenic bacteria in Arabidopsis thaliana. Plant J. 28, 475–481 10.1046/j.1365-313X.2001.01136.x [DOI] [PubMed] [Google Scholar]
- Walch-Liu P., Forde B. G. (2008). Nitrate signaling mediated by the NRT1.1 nitrate transporter antagonizes L-glutamate-induced changes in root architecture. Plant J. 54, 820–828 10.1111/j.1365-313X.2008.03443.x [DOI] [PubMed] [Google Scholar]
- Walters D., Bingham I. (2007). Influence of nutritions on disease development caused by fungal pathogens: implications for plant disease control. Ann. Appl. Biol. 85, 901–909 10.1111/j.1744-7348.2007.00176.x [DOI] [Google Scholar]
- Wang R., Xing X., Wang Y., Tran A., Crawford N. M. (2009). A genetic screen for nitrate regulatory mutants captures the nitrate transporter gene NRT1.1. Plant Physiol. 151, 472–478 10.1104/pp.109.140434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D., Han X., An Y., Guo H., Xia X., Yin W. (2013). The nitrate transporter NRT2.1 functions in the ethylene response to nitrate deficiency in Arabidopsis. Plant Cell Environ. 36, 1328–13337 10.1111/pce.12062 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Heat map analysis performed with Marvis (Filter and Cluster packages). Five and two week old plants either mock (Col3-gl/1 and amt1.1) or either P. syringae Col3-gl/1 P. syringae and amt1.1 P. syringae or P. cucumerina inoculated (Col3-gl/1 Pc and amt1.1 Pc) plants were processed for relative quantification analysis by HPLC-QTOFMS data. The concentration of the metabolites was determined in all the samples by normalizing the chromatographic area for each compound with the dry weight of the corresponding sample. Heatmaps are generated by using Mar-Vis Filter and Cluster following a Kruskal-Wallys test (p < 0.01). Clusters overepresented amt1.1 mock or infected (intense red colors) compared with wild type plants were selected for subsequent data analysis.
Profile of additional hits in the overaccumulated compounds upon P. cucumerina infection in amt1.1 compared with Col3-gl1. For experimental details see Figures 1, 9. The concentration of the metabolites was determined in all the samples by normalizing the chromatographic area for each compound with the dry weight of the corresponding sample. The compounds were tentatively identified using the exact mass criteria using the METLIN and Massbank databases. Those compounds tentatively identified have the exact mass indicated. Te compounds fully identified have no reference to the mass. The compounds were grouped by metabolic pathways according to KEGG and AraCyc databases. White bars are mock inoculated and filled bars are P. cucmerina infected plants. Leaf material from 150 individual plants were pooled together for each treatment × genotype combination. Boxplots represent average three independent experiments with two technical replicates. Only data showing a p-value below 0.01 after a Kuskal wallys test were used for pathway assignation (n = 6).