Abstract
The presence of an abnormal vaginal microflora in early pregnancy is a risk factor for preterm delivery. There is no investigation on vaginal flora dominated by lactic acid bacteria and possible association with preterm delivery. We assessed the dominant vaginal Lactobacillus species in healthy pregnant women in early pregnancy in relation to pregnancy outcome. We observed 111 low risk pregnant women with a normal vaginal microflora 11 + 0 to 14 + 0 weeks of pregnancy without subjective complaints. Vaginal smears were taken for the identification of lactobacilli using denaturing gradient gel electrophoresis (DGGE). Pregnancy outcome was recorded as term or preterm delivery (limit 36 + 6 weeks of gestation). The diversity of Lactobacillus species in term vs. preterm was the main outcome measure. L. iners alone was detected in 11 from 13 (85%) women who delivered preterm. By contrast, L. iners alone was detected in only 16 from 98 (16%) women who delivered at term (p < 0.001). Fifty six percent women that delivered at term and 8% women that delivered preterm had two or more vaginal Lactobacillus spp. at the same time. This study suggests that dominating L. iners alone detected in vaginal smears of healthy women in early pregnancy might be associated with preterm delivery.
The healthy human vagina is dominated by a variety of Lactobacillus species which play an essential role in protecting women from genital infection. A deficiency in lactobacilli can upset the microbial balance in the vagina, frequently resulting in the syndrome of bacterial vaginosis (BV)1,2, which is associated with a quantitative and qualitative shift from normally occurring lactobacilli to a mixed microflora dominated by anaerobic bacteria3.
According to Nugent et al.4, bacterial vaginosis is characterized by a complete loss of lactobacilli and a concomitant increase in Gram-variable and Gram-negative rods, primary among them Gardnerella vaginalis as well as Bacteroides, Prevotella, and Mobiluncus species1,2. The presence of an abnormal vaginal microbiota in early pregnancy is a recognized risk factor for preterm delivery (PTD) and low birth weight5,6,7,8,9,10.
Women with a normal vaginal microbiota in the first trimester have been found to have a 75% lower risk of delivery before 35 weeks of pregnancy than women with an abnormal vaginal microflora10. By contrast, the absence of lactobacilli and the presence of BV or aerobic vaginitis in the first trimester were associated with increased risks of PTD between 25 and 35 weeks10. Because vaginal infection is an important mechanism responsible for preterm birth5, maintaining the natural, healthy balance of the Lactobacillus microbiota in the vagina is particularly important during pregnancy11. The protective role of lactobacilli against genital pathogens includes production of lactic acid, hydrogen peroxide, and bacteriocins such as acidolin, lactacin B, or lactocin 16012,13,14.
For some time, the vaginal microbiota of healthy women of child-bearing age was believed to be dominated by L. acidophilus and L. fermentum, followed by L. brevis, L. jensenii, L. casei, and other species15. More recently, molecular methods have shown L. crispatus, L. jensenii, and L. iners to be the most common isolates16,17. In our recent study, L. crispatus and L. gasseri, followed by L. jensenii and L. rhamnosus, were the most frequently occurring species in the vaginas of pregnant women, with the vaginal microflora dominated by either a single or a combination of two Lactobacillus species18. These results were in agreement with those by Vásquez et al.19 and Antonio et al.20 who had also found L. jensenii, L. crispatus, and L. gasseri to be the predominant Lactobacillus species in the vagina.
To gain a better understanding of the potential protective role specific species of lactobacilli in pregnancy, the present study was undertaken to characterise the dominant Lactobacillus species colonizing the vagina of healthy pregnant women in early pregnancy. Further on this observation intended to describe if different diversity of vaginal lactobacilli in first trimester of pregnancy could have an influence on pregnancy outcome.
Results
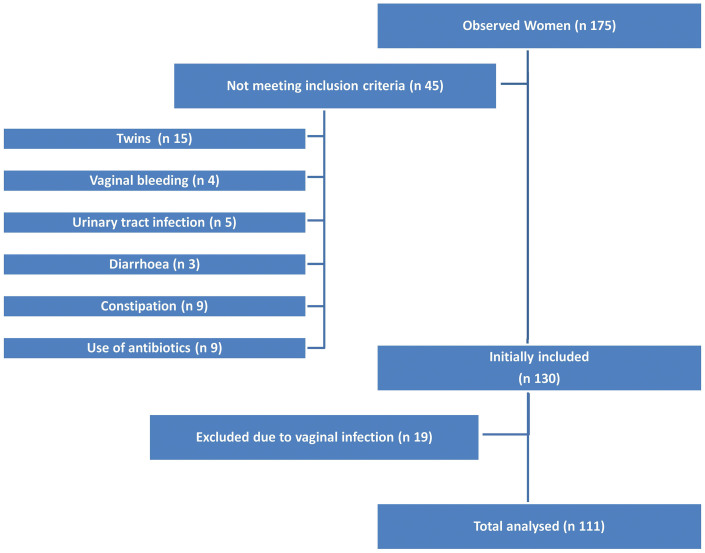
The patient selection diagram is presented in Figure 1. Overall, 175 patients were considered for inclusion into the study. One hundert thirty women fulfilled the inclusion criteria. Of those, 19 patients were excluded because of vaginal infection, so that a total of 111 low-risk pregnant women of white European, Middle Eastern, or Asiatic origin were included in the final analysis.
Figure 1. Exclusion criteria and patient selection diagram.
The demographic data are shown in Table 1. Thus, 98 women (88%) were delivered after 37 + 0 weeks of pregnancy (mean 40 ± 1 week), and 13 (12%) were delivered at less than 36 + 6 weeks (mean 35 ± 1 week). Of these, one was delivered in week 33, two each in weeks 34 and 35, and 8 women in week 36. Mean birth weight was 3455 ± 351 g in term deliveries and 2316 ± 205 g in preterm deliveries. There were two cases of premature preterm rupture of membranes (pPROM) in the preterm delivery group. Cases of preeclampsia and foetal growth restriction were observed only in deliveries after gestational week 37.
Table 1. Demographic characteristics and general pregnancy data.
| Term delivery | Preterm delivery | |
|---|---|---|
| Number of patients, n = 111 (%) | 98 (88) | 13 (12) |
| Age (years), mean ± SD | 35 ± 4.61 | 36 ± 4.31 |
| Gestational age at birth, mean ± SD | 39.6 ± 0.98 | 35.1 ± 0.85 |
| Birth weight, mean ± SD | 3455 ± 351 g | 2316 ± 205 g |
| Obstetrical history, n (%) | 98 (100) | 13 (100) |
| Preterm delivery < 33 weeks, n (%) | 9 (9) | 1 (8) |
| Preterm delivery 33–36 weeks, n (%) | 11 (11) | 1 (8) |
| Parity, n (%) | ||
| Nulliparous | 5 (5) | 2 (15) |
| Primiparous | 37 (38) | 4 (31) |
| Multiparous | 56 (57) | 7 (54) |
SD, standard deviation.
Overall, 44% of women delivered at term and 92% women delivered preterm had only one Lactobacillus spp. detectable by PCR denaturing gradient gel electrophoresis (DGGE) and sequencing in their vaginal specimens. Around 56% women who delivered at term, and 8% who delivered preterm had a combination of 2, or more Lactobacillus spp. This statisticaly significant difference (p < 0.0009) is presented in table 2. Comparing mean number of Lactobacillus species detected from pregnant women we observed statistical significant difference between term and preterm delivery group, too (p < 0.004), (1.8 ± 0.9 vs. 1.2 ± 0.8).
Table 2. Lactobacillus species or combinations of species in women with term and preterm delivery (n = 111) as determined by denaturing gradient gel electrophoresis (DGGE).
| Number of vaginal Lactobacillus species | Term delivery (n = 98) | Preterm delivery (n = 13) | ||
|---|---|---|---|---|
| n | % | n | % | |
| 0 | 0 | 0 | 0 | 0 |
| 1 | 43 | 44 | 12 | 92 |
| 2 | 38 | 39 | 0 | 0 |
| 3 | 11 | 11 | 0 | 0 |
| 4 | 4 | 4 | 1 | 8 |
| 5 | 2 | 2 | 0 | 0 |
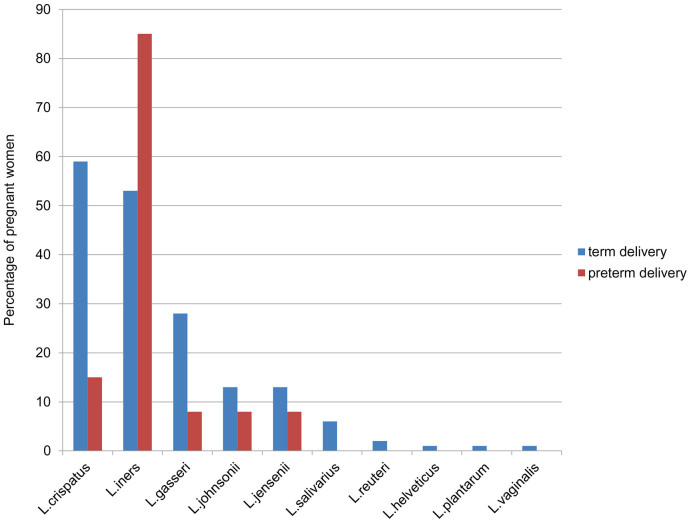
We verified by DGGE 190 Lactobacillus-specific DGGE bands belonging to 10 species of this genus that dominate the vaginal Lactobacillus microbiota of pregnant women. In women delivered at term 174 Lactobacillus spp.- specific bands belonging to 10 species of this genus were detected in vaginal smears. Contrastingly, five Lactobacillus species were detected among the 16 DGGE bands from women who delivered preterm.
The Lactobacillus species detected in women delivered at term were mostly L. crispatus, L. iners, L. gasseri, followed by L. johnsonii, L. jensenii, and L. salivarius. Further on L. reuteri, L. helveticus, L. plantarum and L. vaginalis were detected in only few women with term deliveries. In women delivered preterm, L. iners was the predominant vaginal Lactobacillus spp. In the 16 DGGE bands from women delivered preterm, L. crispatus was detected twice and L. gasseri, L. johnsonii and L. jensenii were detected once only (Figure 2).
Figure 2. Percentage of the most frequent vaginal Lactobacillus species in women with term (TD) and preterm delivery (PTD) as determined by denaturing gradient gel electrophoresis (DGGE).
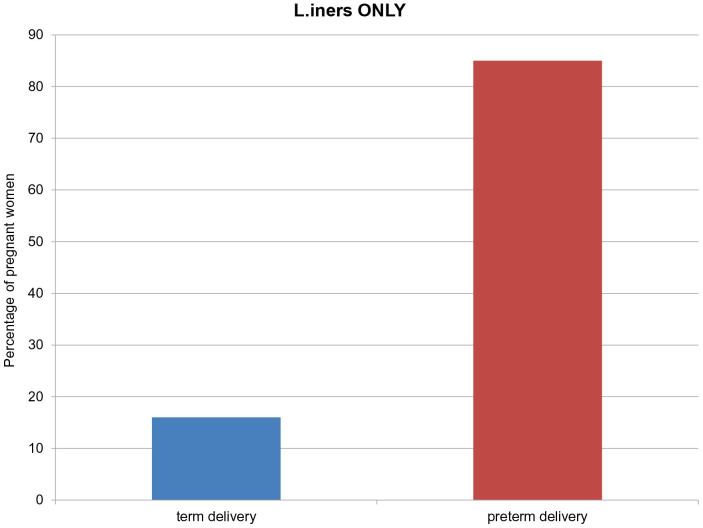
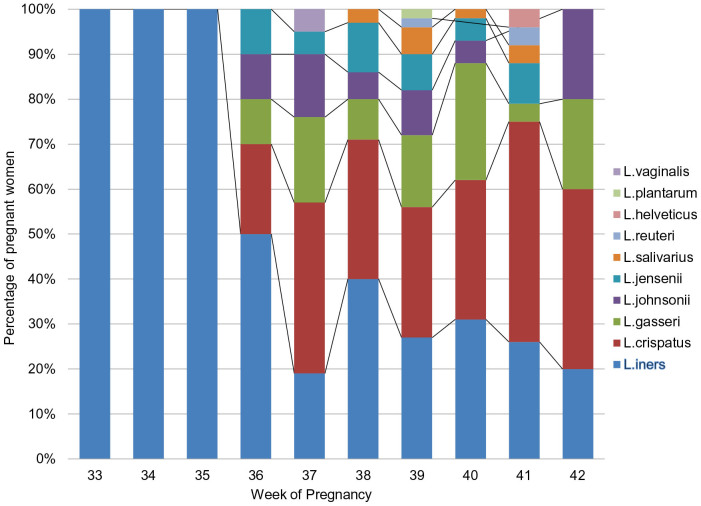
The presence of L. iners alone was observed in 16/98 women (16%) delivered at term and in 11/13 women (85%) delivered preterm (p < 0.001) (Figure 3). The distribution of Lactobacillus species detected at the beginning of the study, i.e., between 11 + 0 and 14 + 0 weeks of pregnancy, by corresponding gestational age of birth is presented in Figure 4. L. iners alone was detected in women delivered until 35 + 0 weeks of pregnancy. Further on we observed that the raising diversity of lactobacillar species has a beneficial influence on pregnancy duration.
Figure 3. Percentage of L. iners as only single Lactobacillus species in women with term (TD) and preterm delivery (PTD) as determined by denaturing gradient gel electrophoresis (DGGE).
Figure 4. Lactobacilli status obtained at the beginning of the study (i.e., between 11 + 0 and 14 + 0 weeks of gestation) compared to gestational age at birth.
Discussion
The purpose of this study was to characterize dominant and possible combination of vaginal lactobacilli among healthy pregnant women and to compare the results between women delivered at term and those delivered preterm. We observed an association between the vaginal presence of a single vaginal Lactobacillus species in late first trimester of pregnancy, mostly L. iners, and preterm delivery.
Different species of Lactobacillus have been reported to populate the vagina to varying degrees. Evidence suggests that there is considerable ethnical and geographical variation in the composition of the normal Lactobacillus microbiota in the vagina15,16,17,19,20,21,22, although these varied results may be due to the heterogeneity of the populations investigated. To minimize the influence of age and health status, we included only healthy women in the late first trimester of low-risk pregnancies into our cohort. Other reasons for the differences in reported results between studies characterizing the vaginal Lactobacillus microbiota include differences in the methodology used and a lack of reliable identification methods. To avoid the diagnostic limitations of Lactobacillus spp. isolation, we used DGGE fingerprinting and sequencing, a highly sensitive culture-independent method for the detection and identification of different Lactobacillus species, which even allows for the detection of species that are otherwise difficult to detect, such as L. iners23,24,25.
The protective potential of lactobacilli in pregnancy is based on their ability to inhibit the growth of vaginal microorganisms known to cause vaginal infection10,11,26. Displacement of lactobacilli from the vagina frequently leads to an abnormal vaginal microflora, including BV, which, if present in early pregnancy, is a risk factor for PTD and low birth weight5,6,7,8,9,10.
According to a Cochrane database27 analysis and recent reviews on the management of vaginal infection28,29, the screening for BV alone is still a matter of discussion. We therefore excluded all patients with abnormal flora and infection in the first trimester and focused on the possible association between a normal vaginal microflora in early pregnancy and pregnancy outcome.
Patients with diarrhoea and constipation were excluded from this investigation because of the possible influence of the rectal on the vaginal microbiota30,31,32,33. Previous studies have shown that the vagina may be colonised by a combination of Lactobacillus species18,26. In our study, 56% of women who delivered at term had colonisation with two or more Lactobacillus species, with L. crispatus, L. iners, L. gasseri, L. johnsonii, and L. jensenii the leading vaginal species. The remaining 44% of women with term pregnancies had only one vaginal Lactobacillus species. By contrast, 92% of the women who delivered preterm had only one Lactobacillus species. Considering all observed participants included in this study it seems that higher diversity in lactobacillar species is more health-contributing and protective due to pregnancy duration (p < 0.0009). Even a diference in mean number of species between term and preterm group was statistically significant (p < 0,004). Analysing data on single strain colonisation we surprisingly observed that L. iners alone was identified in only 16% of patients that delivered at term, and in 85% of women that delivered preterm, with statistically significant difference between those two groups (p < 0.001).
Due to avaliable data on L. iners, there is a an upcoming discussion necessary on the influence of this lactic acid bacterium in the pregnancy. L. iners is the smallest Lactobacillus discovered to date34,35 and although it is a frequently detected bacterial species in the vagina that demands special nutrient requierements, little is known about its characteristics16,34,35,36. Due to recent report by Macklaim et al, L. iners can respond to different changes in vaginal environments by ability to regulate its genomic function37. L. iners is a dominant part of the vaginal microbiota in the transitional phase between normal and abnormal that may predispose to the development of an abnormal vaginal microflora38,39. In a BV environment L. iners increases expression of a cholesterol-dependent cytolysin, mucin, glycerol transport and related metabolic enzymes. Genes belonging to a CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) system were greatly up regulated suggesting that bacteriophage influence the community. Reflective of L. iners, the bacterial community as a whole demonstrated a preference for glycogen and glycerol as carbon sources under BV conditions37. Furthermore, recolonisation of the vagina with L. iners after BV may be a risk factor for a recurrence of BV40. Due to our data we can not report that L. iners alone is subsequently responsible for infections in pregnancy but considering the possible lack of protection of L. iners against pathogens in the vaginal microbiota and according to the results of our study, we can assume that the presence of L. iners alone may accelerate some mechanisms potentially associated with preterm delivery. The presence of vaginal infection in the first trimester has been associated with an increased risk of PTD between 25 and 35 weeks10. Because none of our women were delivered before week 33 and the mean gestational age among women with preterm birth was 35 weeks, we assume that a mechanism other than first-trimester vaginal infection—one involving L. iners as a single Lactobacillus species—may have played a role in these preterm deliveries. Absence of other lactobacilli than L. iners and very low diversity in lactobacillar species with protective role against genital pathogens12,13,14 in first-trimester could be a potential risk for preterm delivery.
By contrast, L. iners in combination with other Lactobacilli strains had no influence on the vaginal microbiota and did not impair the protective role of lactic acid bacteria.
This study has several limitations, first among them the sample size. However, although over one hundred pregnant women fulfilled our stringent inclusion criteria and were included in final data evaluation, we found a statistically significant difference between women with term and preterm deliveries. Second, because all included patients had a normal bacterial microflora at the beginning of the study, we did not evaluate further the presence of lactobacilli or vaginal infections in the second or third trimester of pregnancy.
We found that the presence of L. iners as a single Lactobacillus species in the vagina of pregnant women in early pregnancy may be involved in PTD through a mechanism still to be elucidated. To this end, future studies investigating the presence of different lactobacilli in relation to the complete vaginal microflora and providing a longitudinal picture of the vaginal microbiota throughout pregnancy are needed.
Methods
This study was performed in 2012 with the approval of the ethics committee of the Medical University of Vienna (EK Nr. 139/2008) in accordance with the Declaration of Helsinki and the guidelines of Good Clinical Practice and supported by the head of the institute. Written informed consent was obtained from all study participants prior to enrolment. Our institution is in mainly specialized for prenatal care of high risk pregnancies. To obtain the representative group of pregnant women for this study we chose the study population consisted of women aged 18–40 years with low-risk singleton pregnancies between 11 + 0 and 14 + 0 weeks of gestation scheduled to give birth at our department.
This study regarding to our previous investigation on Lactobacillus strains in pregnant women published in 200718 was performed to investigate the lactobacillary microflora in the first trimester of pregnancy in women with low-risk singleton pregnancies and without vaginal infection. We included all women committed to undergo prenatal care at our tertiary care institution, including scheduled birth at our department. To be eligible, women had to be free of subjective complaints, vaginal bleeding, and clinical signs of vaginal infection and abnormal vaginal flora. Patients with diarrhoea and constipation, assuming disturbed potential role of the rectum as reservoir for lactobacilli, were not eligible for enrolment into the study. Women with urinary tract infection, women having received antibiotic therapy in the previous 4 weeks, and pregnancies with major foetal morphological abnormalities or twin pregnancies were also excluded.
After enrolment into the study, one vaginal smear lateral vaginal walls and the posterior fornix was taken from each participant, transferred to a microscopy slide, Gram-stained, and evaluated at an experienced central laboratory at the Department of Obstetrics and Gynecology, Medical University of Vienna, using the Nugent scoring system4. Only women with a normal vaginal microbiota, i.e., women with Nugent scores 0–3, were eligible for inclusion in the investigational part of the study. These women underwent vaginal smear examination for molecular profiling of lactobacilli using PCR denaturing gradient gel electrophoresis (DGGE) and sequencing. The smears were transferred to transport medium (Copan innovation Italy) to obtain the stability of present vaginal microflora and sent to the University of Natural Resources and Life Sciences, Vienna, for further processing.
All women were followed up until delivery. Pregnancy outcome was assessed on the basis of gestational age at delivery and recorded as term delivery at or later than 37 weeks of gestation and as preterm delivery at or less than 36 + 6 weeks of gestation. Mean birth weight was also recorded, with low birth weight defined as a weight of less than 2500 g.
Examination of vaginal swabs
The swabs were agitated in 0.5 ml sterile TE buffer (pH 8.0) in 2 ml reaction tubes for 1 minute. The suspension was then stored at −80°C.
Bacterial strains and growth conditions
A set of Lactobacillus strains, mainly type-strains (T) of the species, were applied as ladder strains in the DGGE analysis: L. gasseri (LMG 9203T), L. johnsonii (LMG 9436T), L. crispatus (LMG 9479T), L. plantarum (LMG 6907T), L. jensenii (LMG 6414T), L. salivarius (LMG 9477T), L. paracasei (LMG 13087T), L. vaginalis (LMG 12891T), and L. iners (LMG 18913). The lactobacilli except the L. iners strain were cultured anaerobically (80% N2, 10% CO2, 10% H2) in MRS broth (Merck, Darmstadt, Germany) at 37°C. L. iners was grown on Columbia Agar with 5% sheep blood (PB5039A, Oxoid Deutschland GmbH, Wesel, Germany) at 37°C and microaerophilic conditions (85% N2, 10% CO2, 5% O2) for 3 days.
DNA extraction
DNA was extracted from 0.5 ml of Lactobacillus cultures and the swab suspension by using the PeqGOLD Bacterial DNA Kit (PEQLAB Biotechnologie GMBH, Erlangen, Germany) according to the protocol for Gram-positive bacteria.
Denaturing gradient gel electrophoresis (DGGE)
The extracted bacterial DNA was amplified applying the primer set Lac1-f (5′-AGCAGTAGGGAATCTTCCA-3′) and Lac2r (5′-ATT(CT)CACCGCTACACATG-3′)21. For DGGE separation a GC-clamp (5′-CGC CCG GGG CGC GCC CCG GGC GGC CCG GGG GCA CCG GGG G-3′) was attached to the 5′-end of the reverse primer21. The PCR was carried out in a Mastercycler (Eppendorf, Hamburg, Germany). Each PCR reaction was composed of 5 μl 10× PCR buffer (Finnzymes, Espoo, Finland), 37 μl sterile distilled water, 2 μl of each primer (10 pmol/μl, Eurofins MWG Operon, Ebersberg, Germany), 1 μl dNTPs (10 mM, Carl Roth, Karlsruhe, Germany), 1 μl Dynazyme DNA polymerase (2 U/μl; Finnzymes, Espoo, Finland) and 2 μl DNA template. The PCR was performed at 95°C for 4 min, 35 cycles of 95°C for 30 s, 61°C for 40 s, 72°C for 60 s, and a final elongation step at 72°C for 5 min22. The DGGE was conducted with the DCode™ Universal Mutation Detection System (BioRad, Munich, Germany) according to the manufacturer's instructions: In detail, an 8% polyacrylamide gel (37.5:1 acrylamide-bisacrylamide, Serva, Heidelberg, Germany) with a denaturing gradient of 35%–55% formed by urea and formamide was applied. The gels were run in 1× TAE buffer at 60°C and 70 V for 16 hours. The DGGE derived bands were visualized by ethidium bromide staining, documented by digital photography (GelDoc XR+ Imager, Bio-Rad), and identified by comparing their position in the gel to the ladder consisting of bands originating from pure Lactobacillus strains of different species23. DGGE bands with gel positions different than the DGGE ladder were excised from the DGGE gel using a clean scalpel and incubated overnight in 1× PCR buffer at 4°C. Re-amplification was carried out using the primer pair Lac1-f and Lac-2r without GC-clamp. The PCR products were purified applying the PCRExtract Mini Kit (5Prime, Hamburg, Germany) and sequenced (Eurofins MWG Operon, Ebersberg, Germany). The received sequences were analyzed with the BLASTn tool (http://blast.ncbi.nlm.nih.gov), and a minimum sequence identity of 98% was chosen as criterion for species identification.
Statistical analysis
Sample size calculations were approximately based on our previous investigation on Lactobacillus strains in pregnant women published in 200718. We included all patients matching the inclusion criteria for our observation. The demographic and background information was summarized and displayed using descriptive statistics. The proportion of women with lactobacilli was compared between women with term vs. preterm delivery using a Fisher's exact test. Lactobacillary combination and single lactobacilli strain detection as well as mean lactobacillary number was compared between women with term vs. preterm delivery using Kruskal-Wallis-Test. A p-value < 0.05 (two-sided) was considered statistically significant.
Author Contributions
All authors fulfilled all conditions required for authorship. L.P. as a first and corresponding author was responsible for organisation of the research and writing the paper. K.J.D. was microbiology researcher and writing the paper. F.J.N. provided clinical support acquisition of data. M.J.S. provided clinical support acquisition of data. M.F. provided microbiology research. I.K. provided microbiology research. P.H. was senior study supervisor and responsible for writing the paper. W.K. was senior microbiology research supervisor and responsible for writing the paper. H.K. was senior author, responsible for co-organisation of the research and writing the paper.
Acknowledgments
The authors thank Gabriele Berghammer, the text clinic, for medical writing services.
References
- Spiegel C. A. et al. Anaerobic bacteria in nonspecific vaginitis. N. Engl. J. Med. 303, 601–607 (1980). [DOI] [PubMed] [Google Scholar]
- Spiegel C. A. Bacterial vaginosis. Clin. Microbiol. Rev. 4, 485–502 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsum U. et al. Bacterial vaginosis - a microbiological and immunological enigma. APMIS. 113, 81–90 (2005). [DOI] [PubMed] [Google Scholar]
- Nugent R. P., Krohn M. A. & Hillier S. L. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of gram stain interpretation. J. Clin. Microbiol. 29, 297–301 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenberg R. L., Hauth J. C. & Andrews W. W. Intrauterine infection and preterm delivery. N. Engl. J. Med. 342, 1500–1507 (2000). [DOI] [PubMed] [Google Scholar]
- Gravett M. G. et al. Independent associations of bacterial vaginosis and Chlamydia trachomatis infection with adverse pregnancy outcome. JAMA. 256, 1899–1903 (1986). [PubMed] [Google Scholar]
- Hay P. E. et al. Abnormal bacterial colonisation of the genital tract and subsequent preterm delivery and late miscarriage. BMJ. 308, 295–298 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krohn M. A., Hillier S. L., Lee M. L., Rabe L. K. & Eschenbach D. A. Vaginal Bacteroides species are associated with an increased rate of preterm delivery among women in preterm labor. J Infect Dis. 164, 88–93 (1991). [DOI] [PubMed] [Google Scholar]
- McGregor J. A. et al. Bacterial vaginosis is associated with prematurity and vaginal fluid mucinase and sialidase: results of a controlled trial of topical clindamycin cream. Am J Obstet Gynecol. 170, 1048–1059 (1994). [DOI] [PubMed] [Google Scholar]
- Donders G. et al. Predictive value for preterm birth of abnormal vaginal flora, bacterial vaginosis and aerobic vaginitis during the first trimester of pregnancy. BJOG. 116, 1315–1324 (2009). [DOI] [PubMed] [Google Scholar]
- Reid G. & Bocking A. The potential for probiotics to prevent bacterial vaginosis and preterm labor. Am J Obstet Gynecol. 189, 1202–1208 (2003). [DOI] [PubMed] [Google Scholar]
- Aroutcheva A. A. et al. Defense factors of vaginal lactobacilli. Am J Obstet Gynecol. 185, 375–379 (2001). [DOI] [PubMed] [Google Scholar]
- Turovskiy Y., Ludescher R. D., Aroutcheva A. A., Faro S. & Chikindas M. L. Lactocin 160, a Bacteriocin Produced by Vaginal Lactobacillus rhamnosus, Targets Cytoplasmic Membranes of the Vaginal Pathogen, Gardnerella vaginalis. Probiotics Antimicrob Proteins. 20, 67–74 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover S. E., Aroutcheva A. A., Faro S. & Chikindas M. L. Natural antimicrobials and their role in vaginal health: a short review. Int J Probiotics Prebiotics. 3, 219–230 (2008). [PMC free article] [PubMed] [Google Scholar]
- Kandler O., Weiss N. [Genus Lactobacillus.]. Bergey's Manual of Systematic Bacteriology vol. 2, 9th edition. [Sneath, P.H. A.,Mair, N. S., Sharpe, M. E.,Holt, J. G. (eds.)] [1063–1065] (Williams and Wilkins, Baltimore,1986). [Google Scholar]
- Burton J. P., Cadieux P. A. & Reid G. Improved understanding of the bacterial vaginal microbiota of women before and after probiotic instillation. Appl. Environ. Microbiol. 69, 97–101 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid G., McGroarty J. A., Tomeczek L. & Bruce A. W. Identification and plasmide profiles of Lactobacillus species from the vagina of 100 healthy women. FEMS Immunol. Med. Microbiol. 15, 23–26 (1996). [DOI] [PubMed] [Google Scholar]
- Kiss H. et al. Vaginal Lactobacillus microbiota of healthy women in the late first trimester of pregnancy. BJOG. 114, 1402–1407 (2007). [DOI] [PubMed] [Google Scholar]
- Vasquez A., Jakobsson T., Ahrne S., Forsum U. & Molin G. Vaginal Lactobacillus flora of healthy Swedish women. J. Clin Microbiol. 40, 2746–2749 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonio M. A. D., Hawes S. E. & Hillier S. L. The Identification of Vaginal Lactobacillus Species and the Demographic and Microbiologic Characteristics of Women Colonized by These Species. J. Infect Dis. 180, 1950–1956 (1999). [DOI] [PubMed] [Google Scholar]
- Ravel J. et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA. 108, 4680–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespers V. et al. Quantification of bacterial species of the vaginal microbiome in different groups of women, using nucleic acid amplification tests. BMC Microbiol. 12, 83 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J. et al. Detection of Lactobacillus, Pediococcus, Leuconostoc and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl Environ Microbiol. 67, 2578–85 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan P. D., Shanahan F., O'Mahony C. & Marchesi J. R. Culture-independent analyses of temporal variation of the dominant fecal microbiota and targeted bacterial subgroups in Crohn's Disease. J Clin Microbiol. 44, 3980–3988 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y. L. et al. Rapid identification of 11 human intestinal Lactobacillus species by multiplex PCR assays using group- and species-specific primers derived from the 16S-23S rRNA intergenic spacer region and its flanking 23S rRNA. FEMS Microbiol Lett. 187, 167–173 (2000). [DOI] [PubMed] [Google Scholar]
- Verstraelen H. et al. Longitudinal analysis of the vaginal microflora in pregnancy suggests that L. crispatus promotes the stability of the normal vaginal microflora and that L. gasseri and/or L. iners are more conducive to the occurrence of abnormal vaginal microflora. BMC Microbiol. 2, 116 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald H. M., Brocklehurst P. & Gordon A. Antibiotics for treating bacterial vaginosis in pregnancy. Cochrane Database Syst Rev. 24, CD000262 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donders G. Diagnosis and management of bacterial vaginosis and other types of abnormal vaginal bacterial flora: a review. Obstet Gynecol Surv. 65, 462–73 (2010). [DOI] [PubMed] [Google Scholar]
- Riggs M. A. & Klebanoff M. A. Treatment of vaginal infections to prevent preterm birth: a meta-analysis. Clin Obstet Gynecol. 47, 796–807 (2004). [DOI] [PubMed] [Google Scholar]
- Antonio M. A., Rabe L. K. & Hillier S. L. Colonization of the rectum by Lactobacillus species and decreased risk of bacterial vaginosis. J Infect Dis. 192, 394–398 (2005). [DOI] [PubMed] [Google Scholar]
- Reid G. et al. Oral probiotics can resolve urogenital infections. FEMS Immunol Med Microbiol. 30, 49–52 (2001). [DOI] [PubMed] [Google Scholar]
- El Aila N. A. et al. Identification and genotyping of bacteria from paired vaginal and rectal samples from pregnant women indicates similarity between vaginal and rectal microflora. BMC Infect Dis. 14, 167 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricevic L. et al. Characterisation of the oral, vaginal and rectal Lactobacillus flora in healthy pregnant and postmenopausal women. Eur J Obstet Gynecol Reprod Biol. 160, 93–99 (2012). [DOI] [PubMed] [Google Scholar]
- Falsen E., Pascual C., Sjödén B., Ohlén M. & Collins M. D. Phenotypic and phylogenetic characterization of a novel Lactobacillus species from human sources: description of Lactobacillus iners sp. nov. Int J Syst Bacteriol. 49, 217–221 (1999). [DOI] [PubMed] [Google Scholar]
- Ventura M. et al. Genome-scale analyses of health-promoting bacteria: probiogenomics. Nat Rev Microbiol. 7, 61–71 (2009). [DOI] [PubMed] [Google Scholar]
- Jakobsson T. & Forsum U. Lactobacillus iners: a marker of changes in the vaginal flora? J Clin Microbiol. 45, 3145 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macklaim J. M. et al. Comparative meta-RNA-seq of the vaginal microbiota and differential expression by Lactobacillus iners in health and dysbiosis. Microbiome. 1, 12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macklaim J. M., Gloor G. B., Anukam K. C., Cribby S. & Reid G. At the crossroads of vaginal health and disease, the genome sequence of Lactobacillus iners AB-1. Proc Natl Acad Sci U S A. 15, 4688–4695 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago G. L. et al. Longitudinal qPCR study of the dynamics of L. crispatus, L. iners, A. vaginae, (sialidase positive) G. vaginalis, and P. bivia in the vagina. PLoS One. 7, 45281 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra A., Palcu C. T., Sobel J. D. & Akins R. A. Bacterial vaginosis: culture- and PCR-based characterizations of a complex polymicrobial disease's pathobiology. Curr Infect Dis Rep. 9, 485–500 (2007). [DOI] [PubMed] [Google Scholar]