Abstract
INK4 locus at chromosome 9p21 has been reported to be associated with primary open angle glaucoma (POAG) and its subtypes along with the associated optic disc parameters across the populations of European, Japanese and African ancestries. The locus encodes three tumor suppressor genes namely CDKN2A, ARF, CDKN2B and a long non-coding RNA CDKN2B-AS1 (also known as ANRIL). Here, we report association study of 34 SNPs from INK4 locus with POAG in a population of Indo-European ancestry from the eastern part of India (350 patients and 354 controls). With 81% power to detect genetic association we observed only nominal association of rs1011970 (uncorrected p = 0.048) with POAG and rs10120688 (uncorrected p = 0.048) in patients without a high intra-ocular pressure (IOP<21 mm of Hg) compared to controls. This study, in contrast to the previous reports, suggests lack of significant genetic association of INK4 locus with POAG in East Indian population which needs to be replicated in larger studies in diverse world populations.
Glaucoma is a group of optic neuropathies which is the foremost cause of irreversible blindness. The recent estimates from population-based surveys indicate that 60 million people are affected by glaucoma worldwide which is estimated to reach 79.6 million by 2020 and India will surpass Europe to have the second largest population of glaucoma patients1. In India, the prevalence of blindness is 1% and glaucoma causes 8.3% of the total blindness2. Primary open angle glaucoma (POAG) is the most common subtype, which is a multifactorial complex disease. According to the latest review of the field, family based linkage analyses have established 17 loci for POAG out of which only five underlying genes have been identified3. In parallel, case-control associations in candidate gene regions as well as genome wide association studies have suggested additional loci to be associated with POAG3.
The INK4 locus at chromosome 9p21 has been found to be associated with POAG and its subtypes along with the associated traits like vertical cup-disc ratio (summarised in supplementary Table S1). The association was observed in populations of Caucasian, Japanese and some populations of African ancestry.
The locus encodes three tumor suppressor genes namely CDKN2A, ARF, CDKN2B and a long non-coding RNA CDKN2B-AS1 (also known as ANRIL). Recent reviews suggest that CDKN2A and CDKN2B function as cell growth regulator controlling cell cycle G1 progression by preventing the activation of cyclin dependent kinases4. It has been reported that ANRIL is involved in the epigenetic regulation of genes at the 9p21 locus4 as well as the other genes in trans5.
Given the apparent importance of this locus in POAG, we tested genetic association of SNPs at this locus in a cohort of POAG cases and controls from the state of West Bengal in the eastern part of India.
Results
A total of 1392 samples were enrolled for the study. Of these 704 samples (350 cases and 354 controls) were used to analyse genotype data for the entire region (Discovery phase) and 688 samples (368 cases and 320 controls) were further used to validate candidate tag SNPs from the region. The mean age at onset of the patients recruited in the discovery phase was observed to be 48.99 ± 16.6 years and the mean age of controls was 49.75 ± 11.36 years. The average IOP of patients was 22.94 ± 7.76 mm Hg.
9p21 locus SNPs do not show allelic association in Indian POAG patients
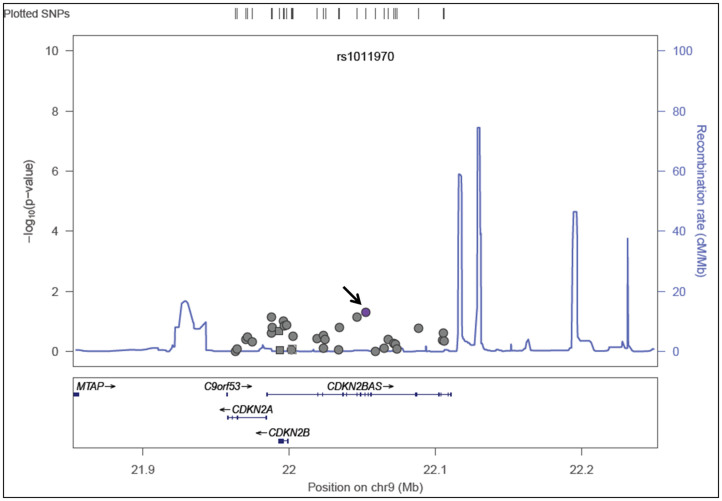
Chi-Square analysis did not reveal allelic association of any of the 34 SNPs with POAG as represented in regional association plot (Fig. 1) except the marginal association of rs1011970. The minor allele frequencies and respective p-values are given in Table 1. In addition, a marginal association of GG genotype [p = 0.029, OR = 2.1 (CI = 1.01–4.4)] of rs1412832 was observed as a risk towards POAG albeit with a low frequency (0.074 cases/0.036 controls). Our study had >80% power to detect an allelic association for this region. The details of the power analysis along with comparison with other studies reporting positive association is summarized in Table 2. To further confirm the negative association of 9p21 locus in Indian POAG patients we selected three tag SNPs from this region and genotyped them in an independent cohort of 368 cases and 320 controls from the same population background. We did not observe any allelic association (Table 3). To rule out any possible error in the analysis we also analysed publically available GLAUGEN data for this locus which reported positive association and could reproduce association in 12 SNPs from this locus (data not shown).
Figure 1. Regional association plot of INK4 locus in East Indian population.
This plot shows association results of studied SNPs from 9p21 region with POAG in East Indian cohort. The circles represent the genotyped SNPs. The horizontal axis is the genomic context of the region studied (along with the genes). The left vertical axis represents negative logarithm of p-value and the right vertical axis is the recombination frequency of the region. Purple circle (marked by arrow) represents rs1011970 which has minimum p-value. Overall the data suggests minimal association (if any) of this locus with POAG.
Table 1. Details of INK4 locus SNPs at 9p21 in East Indian cohort.
| Controls (n = 708) | POAG (n = 700) | HTG** (n = 464) | Non-HTG** (n = 208) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Minor allele | MAF | MAF | p-value | OR(95%CI) | MAF | p-value | OR(95%CI) | MAF | p-value | OR(95%CI) |
| rs2811708 | A | 0.17 | 0.17 | 0.96 | 0.99(0.75-1.31) | 0.16 | 0.90 | 0.98(0.72-1.34) | 0.16 | 0.93 | 0.98(0.65-1.48) |
| rs3731239 | G | 0.23 | 0.22 | 0.79 | 0.97(0.75-1.24) | 0.22 | 0.99 | 1(0.76-1.33) | 0.19 | 0.38 | 0.84(0.57-1.24) |
| rs2811709 | A | 0.05 | 0.04 | 0.38 | 0.79(0.47-1.33) | 0.034 | 0.31 | 0.73(0.4-1.34) | 0.028 | 0.26 | 0.61(0.25-1.47) |
| rs4074785 | A | 0.17 | 0.19 | 0.32 | 1.15(0.88-1.51) | 0.18 | 0.43 | 1.13(0.83-1.53) | 0.19 | 0.45 | 1.17(0.78-1.74) |
| rs3731217 | C | 0.11 | 0.13 | 0.46 | 1.13(0.82-1.55) | 0.12 | 0.66 | 1.08(0.76-1.56) | 0.13 | 0.33 | 1.25(0.8-1.98) |
| rs3218020* | A | 0.37 | 0.34 | 0.07 | 1.22(0.98-1.51) | 0.41 | 0.11 | 1.21(0.95-1.54) | 0.43 | 0.08 | 1.32(0.97-1.81) |
| rs2811712 | G | 0.15 | 0.13 | 0.23 | 0.83(0.62-1.12) | 0.13 | 0.24 | 0.82(0.58-1.15) | 0.12 | 0.3 | 0.79(0.5-1.24) |
| rs3218018 | C | 0.06 | 0.04 | 0.16 | 0.7(0.43-1.15) | 0.038 | 0.14 | 0.66(0.37-1.16) | 0.033 | 0.17 | 0.57(0.25-1.28) |
| rs3217992 | A | 0.39 | 0.42 | 0.21 | 1.15(0.93-1.42) | 0.42 | 0.28 | 1.14(0.9-1.45) | 0.44 | 0.16 | 1.25(0.91-1.71) |
| rs1063192* | G | 0.25 | 0.24 | 0.9 | 0.98(0.77-1.26) | 0.24 | 0.94 | 1.01(0.77-1.33) | 0.22 | 0.56 | 0.9(0.62-1.29) |
| rs2069426 | A | 0.06 | 0.04 | 0.09 | 0.66(0.4-1.08) | 0.036 | 0.08 | 0.6(0.34-1.07) | 0.033 | 0.15 | 0.55(0.24-1.25) |
| rs3217980 | A | 0.01 | 0 | 0.13 | 0.38(0.1-1.43) | 0.004 | 0.20 | 0.38(0.08-1.79) | 0.004 | 0.4 | 0.42(0.05-3.4) |
| rs2069422 | C | 0.06 | 0.04 | 0.12 | 0.69(0.42-1.11) | 0.038 | 0.12 | 0.64(0.36-1.13) | 0.033 | 0.15 | 0.55(0.24-1.25) |
| rs573687* | A | 0.23 | 0.23 | 0.86 | 1.02(0.8-1.31) | 0.23 | 0.79 | 1.04(0.79-1.37) | 0.22 | 0.78 | 0.95(0.66-1.38) |
| rs13298881 | G | 0.02 | 0.02 | 0.83 | 1.08(0.53-2.2) | 0.02 | 0.97 | 1.02(0.45-2.29) | 0.028 | 0.52 | 1.37(0.53-3.58) |
| rs545226 | G | 0.39 | 0.42 | 0.31 | 1.12(0.9-1.38) | 0.42 | 0.36 | 1.12(0.88-1.42) | 0.43 | 0.25 | 1.2(0.88-1.64) |
| rs7049105* | A | 0.4 | 0.37 | 0.35 | 0.9(0.73-1.12) | 0.37 | 0.41 | 0.9(0.71-1.15) | 0.35 | 0.28 | 0.84(0.61-1.16) |
| rs2157719* | G | 0.25 | 0.24 | 0.76 | 0.96(0.75-1.23) | 0.24 | 0.84 | 0.97(0.74-1.28) | 0.23 | 0.6 | 0.91(0.63-1.31) |
| rs1759417 | A | 0.03 | 0.02 | 0.28 | 0.67(0.32-1.4) | 0.017 | 0.35 | 0.67(0.29-1.56) | 0.019 | 0.6 | 0.75(0.25-2.23) |
| rs2151280* | G | 0.4 | 0.38 | 0.39 | 0.91(0.73-1.13) | 0.37 | 0.48 | 0.92(0.72-1.17) | 0.35 | 0.25 | 0.83(0.6-1.14) |
| rs1412829* | G | 0.24 | 0.23 | 0.85 | 0.98(0.76-1.25) | 0.23 | 0.93 | 0.99(0.75-1.3) | 0.22 | 0.63 | 0.91(0.63-1.32) |
| rs1333034 | G | 0.06 | 0.04 | 0.15 | 0.7(0.43-1.15) | 0.038 | 0.14 | 0.66(0.37-1.16) | 0.033 | 0.17 | 0.57(0.25-1.28) |
| rs10120688* | G | 0.39 | 0.35 | 0.07 | 0.82(0.66-1.02) | 0.35 | 0.20 | 0.85(0.67-1.09) | 0.31 | 0.048 | 0.72(0.52-1) |
| rs1011970 | A | 0.27 | 0.23 | 0.048 | 0.78(0.62-1) | 0.23 | 0.11 | 0.8(0.61-1.05) | 0.21 | 0.08 | 0.72(0.49-1.04) |
| rs4977756* | G | 0.24 | 0.24 | 0.95 | 0.99(0.78-1.27) | 0.24 | 0.72 | 1.05(0.8-1.38) | 0.21 | 0.41 | 0.86(0.59-1.25) |
| rs7855162 | G | 0.09 | 0.09 | 0.76 | 0.94(0.65-1.36) | 0.086 | 0.81 | 0.95(0.63-1.44) | 0.086 | 0.86 | 0.95(0.55-1.65) |
| rs1412832* | G | 0.22 | 0.24 | 0.38 | 1.12(0.87-1.43) | 0.24 | 0.50 | 1.1(0.83-1.45) | 0.24 | 0.63 | 1.09(0.76-1.57) |
| rs10116277* | C | 0.38 | 0.36 | 0.51 | 0.93(0.75-1.15) | 0.37 | 0.81 | 0.97(0.76-1.24) | 0.33 | 0.25 | 0.83(0.6-1.15) |
| rs1547705 | C | 0.03 | 0.03 | 0.56 | 0.83(0.45-1.54) | 0.028 | 0.66 | 0.86(0.43-1.71) | 0.028 | 0.79 | 0.88(0.36-2.2) |
| rs1333040 | G | 0.36 | 0.35 | 0.77 | 0.97(0.78-1.21) | 0.35 | 0.95 | 0.99(0.78-1.27) | 0.33 | 0.5 | 0.89(0.64-1.24) |
| rs4977574 | A | 0.48 | 0.45 | 0.16 | 0.86(0.7-1.06) | 0.45 | 0.34 | 0.89(0.71-1.13) | 0.42 | 0.16 | 0.8(0.59-1.09) |
| rs10965235 | A | 0.08 | 0.07 | 0.41 | 0.85(0.58-1.25) | 0.073 | 0.48 | 0.85(0.55-1.32) | 0.067 | 0.42 | 0.78(0.43-1.43) |
| rs944797 | A | 0.46 | 0.43 | 0.23 | 0.88(0.71-1.09) | 0.44 | 0.47 | 0.92(0.72-1.16) | 0.41 | 0.2 | 0.82(0.6-1.12) |
| rs2383207* | A | 0.38 | 0.36 | 0.44 | 0.92(0.74-1.14) | 0.36 | 0.73 | 0.96(0.75-1.22) | 0.34 | 0.4 | 0.87(0.63-1.2) |
*SNPs which have been found to be associated with POAG in the previous studies,
**HTG and Non-HTG cohorts are the subtypes of POAG cohort.
Note: p-values in bold represents marginal association of SNPs. Italicized rs-IDs represent 20 tag SNPs chosen for haplotype association test shown in supplementary Table S2; Control data is same for POAG, HTG and Non-HTG groups.
Abbreviations- n: Number of chromosomes, HTG: High Tension Glaucoma, Non-HTG: Non-High Tension Glaucoma, MAF: Minor allele frequency, OR: Odds ratio, CI: Confidence interval.
Table 2. Power calculations to detect Genetic association.
| Wiggs et al. 2012(3146 cases, 3487 controls) | Fan B.J. et al 2011(536 cases, 336 controls) | East Indian population(350 cases, 354 controls) | |||||
|---|---|---|---|---|---|---|---|
| SNP | Allele | POWER (%) | Allele frequency | POWER (%) | Allele frequency | POWER (%) | Allele frequency |
| rs1063192 | A | 100 | 0.57 | 90.32 | 0.58 | 81.21 | 0.75 |
Note: Power calculations were done for ‘A' allele by keeping relative risk as 1.8 (Homozygous genotype) and 1.2 (Heterozygous genotype) calculated from genotype data given by Fan B.J. et al, 2011and allele frequency were taken from respective studies (Supplementary Table S1).
Table 3. Details of association of three replicated tag SNPs representing three different LD blocks.
| Discovery phase350 cases, 354 controls | Replication phase368 cases, 320 controls | Combined718 cases, 674 controls | |||||
|---|---|---|---|---|---|---|---|
| SNP | Minor allele | MAF(Cases/Controls) | p-value | MAF(Cases/Controls) | p-value | MAF(Cases/Controls) | p-value |
| rs3217992 | A | 0.42/0.39 | 0.20 | 0.39/0.4 | 0.74 | 0.40/0.39 | 0.51 |
| rs4977756 | G | 0.24/0.24 | 0.94 | 0.27/0.25 | 0.38 | 0.25/0.24 | 0.53 |
| rs10116277 | C | 0.36/0.38 | 0.50 | 0.41/0.39 | 0.39 | 0.39/0.38 | 0.84 |
Genotype association analysis with cup-disc ratio
Among the markers examined at this locus, rs1063192 has been shown to be associated with reduced cup-disc ratio in Caucasian and Japanese population backgrounds (supplementary Table S1). We tested association of cup-disc ratio with the alleles and genotypes of this SNP in 215 POAG cases by linear regression analysis but did not find any association (data not shown).
Selection of tag SNPs and haplotype association
Further, for haplotype analysis of this locus, 20 SNPs were chosen as tag SNPs on the basis of their pairwise r2 values in our dataset (r2>0.8) (Supplementary Fig. 1). Thirteen haplotypes were generated among which only one showed significant frequency difference between cases and controls although the overall frequency of occurrence was low in both groups (0.005 cases/0.017 controls) (Supplementary Table S2).
Association analysis in HTG and Non-HTG subjects
Many studies have reported association of this locus with POAG patients who do not suffer from high IOP (Supplementary Table S1). In accordance to these studies, we divided our patients into two subgroups: (a) high tension glaucoma (HTG) cases having IOP>21 mm of Hg in at least one eye (n = 232), and (b) non-HTG cases with IOP<21 mm of Hg in both eyes (n = 104) and compared the allele frequencies of SNPs separately against 354 controls. The average IOP estimate of HTG subgroup was 25.63 ± 7.79 mm Hg and of non-HTG subgroup was 16.98 ± 2.71 mm Hg. In non-HTG subgroup analysis, rs10120688 was observed to have a marginal association (p<0.048); but none were found to be associated with the HTG subgroup (Table 1).
Discussion
Our study suggests a negative association of the INK4 locus with POAG in Indian sub-continent. There are very marginal/weak associations of (a) rs1011970 with POAG, and (b) rs10120688 with non-HTG subgroup of the disease which will not sustain statistical correction for multiple testing. The association of rs10120688 with normal-tension glaucoma was found in the meta-analysis of GLAUGEN and NEIGHBOR study populations by Wiggs et al (Supplementary table S1) and also in African-Americans, however it did not pass multiple-testing correction6. Burdon et al also found association of this SNP with IOP (Supplementary table S1). The genetic association might be found positive if a larger study is undertaken with exclusively normal tension glaucoma patients.
It is worthwhile to mention here that the 9p21 locus has been demonstrated to have remarkable variability in its genetic association and potential role in POAG pathogenesis. For example, some GWA studies identified this locus to be significantly associated (Supplementary Table S1), while other studies carried on similar population did not support the association7. Further, a study on Afro-Caribbean ancestry claimed association of this locus8 which is supported by similar observation made in African-Americans though with marginal association6. However, in the same study the association did not sustain when investigated in populations from Ghana, West Africa6. Similarly, a study in Brazilian population on one SNP (rs4977756) from this locus also reported lack of any association with POAG9.
It is possible that the effect of this locus on the disease might be exerted by, yet unknown, genetic or environmental factor(s) that are variable in different population groups and not being considered in any of the current studies undertaken to address POAG pathogenesis.
Recently, Janssen et al found that among the reported 65 POAG candidate genes, which includes 5 familial genes, 33 highly likely candidate genes and 27 less-likely candidate genes for POAG belong to only four functional molecular networks3. Genes at 9p21 locus are part of their molecular network ‘4' constructed by Ingenuity knowledge database, which consisted of 12 candidate POAG genes where main functionalities were ophthalmic disease, cell cycle, connective tissue development and function. It might be possible that in the populations where this locus has not been found to be associated, other molecular networks play major roles but it will require more extensive genetic analysis supported by gene expression assays and functional characterization.
Our finding of lack of genetic association of INK4 locus in a modest cohort of East Indian POAG patients indicates biological complexity of this region. This requires further validation by undertaking a much larger study population; but the underlying variable observations made in different populations across the globe remains a greater challenge to resolve.
Methods
Selection criteria of study individuals
Diagnosis of patients has been done in Dristi Pradip eye clinic, Jodhpur Park, Kolkata. The patients have been diagnosed through clinical ocular and systemic examinations. The inclusion and exclusion criteria for samples were the same as reported earlier10. Briefly, the patients were recruited if they were positive for 2 out of the 3 criteria, namely, Intra-ocular pressure (IOP)>21 mm of Hg, glaucomatous field damage and significant cupping of the optic disc. Individuals with ocular hypertension and with any history of inflammation or ocular trauma (past & present) were excluded from this study.
Controls were selected from the general population without any history of ocular disease and who tested negative for POAG by means of routine eye examination for glaucoma as described above. The study protocol adhered to the tenets of the Declaration of Helsinki and was approved by the Institutional Review Board.
Collection of blood samples and genomic DNA preparation
Peripheral blood was collected with EDTA from the POAG patients and the individuals without POAG with their written consent. Genomic DNA was prepared from fresh whole blood using the PAX gene blood DNA isolation kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. The DNA was dissolved in TE (10 mM Tris-HCl, 1 mM EDTA, pH 8.0).
Genotyping
The genotype data of 38 SNPs were extracted from a genome-wide genotyping dataset for 350 POAG cases and 354 controls which was generated using Illumina Human660W-Quad chip (Illumina, Inc., San Diego, CA, USA). Four SNPs were removed in QC for minor allele frequency (MAF<0.01) and Hardy-Weinberg equilibrium (HWE p<0.01). Chi-square association was performed on 34 SNPs spanning 142.53 kb in reference human genome (hg18). The SNP and the sample call rates were >95% and >98%, respectively. The selected tag SNPs for validation were genotyped using the GoldenGate genotyping assay (Illumina GGGT assay).
Statistical analysis
Power calculations were done for ‘A' allele of rs1063192 which is the most replicated SNP in different reported studies by Genetic power calculator software (http://pngu.mgh.harvard.edu/~purcell/gpc/)11.The relative risk was kept constant as 1.8 (for homozygous genotype) and 1.2 (for heterozygous genotype) as reported by Fan B.J. et al (Supplementary Table S1) and allele frequency of respective populations were taken as parameters. Chi-square test of association was performed using PLINK software12. The regional plots were generated by an online available Locus Zoom tool (http://csg.sph.umich.edu/locuszoom/)13. LD calculations and haplotype association were performed using haploview software version 414.
Author Contributions
M.V. has done sample collection, genotyping, data analysis and written the manuscript which was critically revised by A.M. and K.R.; A.S. has helped in data analysis; L.K. has helped in DNA sample preparation and genotyping; S.C. has helped in collection of samples with necessary clinical information; J.R. has provided a subset of control samples; A.S. has clinically characterized the patients whose biological samples have been included in the study and played lead role as the clinical collaborator; A.M. and K.R. has conceptualized and led this project including arranging for the required funds. All authors reviewed the manuscript.
Supplementary Material
Supplementary information
Acknowledgments
The authors are thankful to the donors who participated in this study and Dr. Keya Sen for helping with clinical evaluation of patients along with Dr. A. Sen. We acknowledge Ms. Subhashree Nayak for her technical help needed for the study. This work was supported by the Council of Scientific & Industrial Research (CSIR), India (grant number MLP-0016 and BSC-0123). MV and Anchal Sharma were supported by fellowships from Indian Council of Medical Research (ICMR) and Department of Science and Technology (DST), India, respectively. LK and SC acknowledge CSIR for their fellowships. Funding support for the GLAUGEN study was provided through the NIH Genes, Environment and Health Initiative [GEI] (U01HG004728). The GLAUGEN study is one of the genome-wide association studies funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Funding support for genotyping, which was performed at the Broad Institute of MIT and Harvard, was provided by the NIH GEI (U01 HG04424).
References
- Quigley H. A. & Broman A. T. The number of people with glaucoma worldwide in 2010 and 2020. Br. J. Ophthalmol. 90, 262–267 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian D. Molecular and cellular approaches to understand and treat some disease of the eye. Current Science 82, 948–957 (2002). [Google Scholar]
- Janssen S. F. et al. The vast complexity of primary open angle glaucoma: Disease genes, risks, molecular mechanisms and pathobiology. Prog. Ret. Eye Research 37, 31–67 (2011). [DOI] [PubMed] [Google Scholar]
- Congrains A., Kamide K., Ohishi M. & Rakugi H. ANRIL: Molecular Mechanisms and Implications in Human Health. Int J Mol Sci 14, 1278–92 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdt L. M. et al. Alu Elements in ANRIL Non-Coding RNA at Chromosome 9p21 Modulate Atherogenic Cell Functions through Trans-Regulation of Gene Networks. PLoS Genet 9, e1003588 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. et al. Investigation of Known Genetic Risk Factors for Primary Open Angle Glaucoma in Two Populations of African Ancestry Invest. Ophthalmol. Vis. Sci. 17, 6248–6254 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorleifsson G. et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma. Nat. Genet. 42, 906–909 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D. et al. CDKN2B polymorphism is associated with primary open-angle glaucoma (POAG) in the Afro-Caribbean population of Barbados, West Indies. PLoS One 7, e39278 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes H. F., Oliveira M. B., Vasconcellos J. P. C., Costa V. P. & de Melo M. B. Investigation of rs4236601 and rs4977756 SNPs in a primary open-angle glaucoma Brazilian population; (Abstract no. 2291, Page no. 217).Presented at the 62nd Annual Meeting of The American Society of Human Genetics, November 8, 2012, San Francisco, California (http://abstracts.ashg.org/cgi-bin/swish-ashgw).
- Banerjee D. et al. Mitochondrial genome analysis of primary open angle glaucoma patients. PLoS One 8, e70760 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Cherny S. S. & Sham P. C. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 19, 149–150 (2003). [DOI] [PubMed] [Google Scholar]
- Purcell S. et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim R. J. et al. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics 15, 2336–2337 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J. C., Fry B., Maller J. & Daly M. J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information