Abstract
The synthesis and subsequent genomic integration of DNA that is complementary to the genomes of non-retroviral RNA viruses are rarely observed. However, upon infection of various human cell lines and primary fibroblasts with the vesicular stomatitis virus (VSV), we detected DNA complementary to the VSV RNA. The VSV DNA was detected in the cytoplasm as single-stranded DNA fully complementary to the viral mRNA from the poly(A) region to the 7-methyl guanosine cap. The formation of this DNA was cell-dependent. Experimentally, we found that the transduction of cells that do not produce VSV DNA with the long interspersed nuclear element 1 and their infection with VSV could lead to the formation of VSV DNA. Viral DNA complementary to other RNA viruses was also detected in the respective infected human cells. Thus, the genetic information of the non-retroviral RNA virus genome can flow into the DNA of mammalian cells expressing LINE-1-like elements.
There are few reports on the synthesis of deoxyribonucleic acid (DNA) corresponding to non-retroviral ribonucleic acid (RNA) viruses, and the biological and clinical implications of this phenomenon are unclear1,2,3,4,5,6,7. Zhdanov reported that DNA complementary to some non-retroviral RNA viruses, including the measles virus, could be detected in vitro and in vivo; Zhdanov discussed the possible implications of this viral DNA in autoimmune diseases1. However, the existence of such viral DNAs was not confirmed until recently2,4. Complementary DNA (cDNA) to the lymphocytic choriomeningitis virus (LCMV), a murine arenavirus, was detected after retrovirus-producing but not retrovirus-nonproducing mouse cells were infected with LCMV3. In a more recent study, the same group reported that parts of the LCMV DNA are integrated into the genome of infected cultured mouse cells and recombined with a long terminal repeat (LTR) retrotransposon, i.e., an intracisternal A-type particle (IAP)5. Although the biological effects of the LCMV DNA in mice are unclear, the integrated viral genome could persistently produce viral antigens in vivo3.
Bornavirus6 and filovirus7 sequences are present in the genome sequences of many animal species, suggesting that cDNAs were synthesized from viral RNAs in germline cells of some ancient animals and these cDNAs were integrated into chromosomes of the germline cells. The mechanism of reverse transcription that produces cDNA from these RNA viruses is unclear; however, IAP and long interspersed nuclear elements (LINEs) have been implicated in the reverse transcription of LCMV5 and bornavirus6, respectively. LINE-1 is a non-LTR retrotransposon containing two open reading frames (ORFs), ORF1 and ORF2, which encode a nucleic acid binding protein and an endonuclease/reverse transcriptase, respectively8. The reverse transcription step is initiated at a nick on the nuclear genomic DNA through target-primed reverse transcription (TPRT)9.
The vesicular stomatitis virus (VSV), a member of the Rhabdoviridae family, is a negative-sense single-stranded RNA virus. The VSV genome consists of five genes that encode the following proteins: nucleocapsid (N), polymerase co-factor (P), polymerase (L), peripheral matrix protein (M), and surface glycoprotein (G). The mRNA expressed from each gene consists of a 7-methyl guanosine (m7G) cap, the coding sequence (N, P, M, G, or L), and a polyA tail. There are leader and trailer sequences at the 5′ and 3′ ends of the VSV antigenome, respectively. The VSV-encoded mRNAs are generated from the antigenome RNA and are transcribed from the start site of the gene, and the termination of mRNA transcription likely occurs by polyadenylation. Two hours after inoculation, viral protein production from the VSV mRNA can be detected. VSV G has been widely used for the pseudotyping of retroviral and lentiviral vectors10,11, and VSV was proposed to be a useful oncolytic virus for the selective killing of tumour cells12. Echovirus type 30 and respiratory syncytial virus (RSV) are the member of the positive-sense single-stranded RNA virus family Picornaviridae and the negative-sense single-stranded RNA virus family Paramyxoviridae, respectively.
We describe the synthesis of viral DNA that is complementary to the non-retroviral RNA viruses VSV, echovirus type 30, and RSV in retrovirus-free human cell lines or primary human fibroblasts after acute infection. The VSV DNA was detected in the cytoplasm in a non-integrated form and was fully complementary to the VSV mRNA. The VSV DNA structure was unique and potentially reverse transcribed from the viral mRNA of the polyA tail to the m7G cap. Furthermore, our data suggest that LINE-1-related retrotransposons may be responsible for VSV DNA synthesis.
Results
Generation of VSV cDNA in human 293T cells after infection
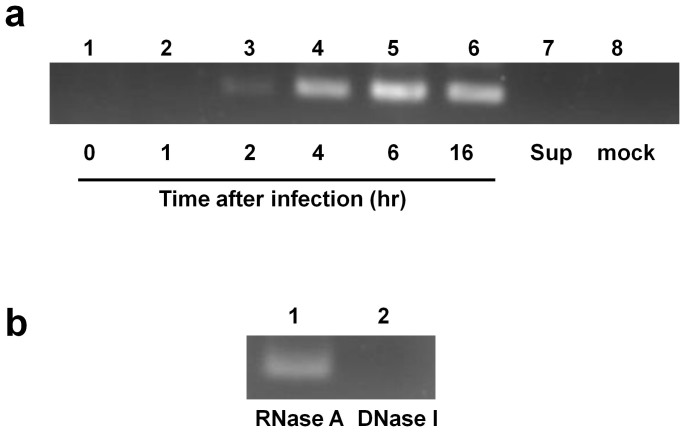
We detected cDNA to the VSV RNA genome in lysates of 293T cells that were incubated overnight after VSV infection. We examined the synthesis of viral cDNA at various post-infection time points via polymerase chain reaction (PCR). For PCR amplification, the VSV N gene was detected using the N478-F/N681-R primer pair (all PCR primers used are listed in Supplementary Table S1). The viral DNA was initially detected 2 hr after the infection of 293T cells with VSV at an MOI of 0.1, and the level of viral DNA increased over time (Fig. 1a); however, PCR bands were not detected earlier than 2 hr. The resulting PCR products were confirmed to be of VSV origin (Indiana strain) via DNA sequencing. To confirm that the VSV DNA was present in the cell lysate, the nucleic acids extracted after 16 hr were treated with RNase A or DNase I and used for PCR. The PCR-generated band disappeared when the templates were treated with DNase I but not after treatment with RNase A (Fig. 1b). These data indicate that the viral DNA was generated after VSV infection. When lysates of 293T cells infected with VSV were treated with S1 nuclease before PCR for the VSV N gene and β globin, a PCR band for the N gene but β globin gene was disappeared as shown in supplementary Figure S1. This result suggests that there is a single-stranded region in VSV DNA for the N gene.
Figure 1. Detection of VSV DNA in 293T human cells.
(a) DNA was extracted from 293T cells at 0, 1, 2, 4, 6, or 16 hr after infection with VSV at an MOI of 0.1 and subjected to PCR amplification for the VSV N gene using the N478-F/N681-R primer pair. The VSV inoculum (Sup) (lane 7) and mock-infected samples (lane 8) were included as controls. The samples prepared 1 hr after inoculation (lane 2) or VSV Sup did not produce VSV DNA. The DNA sequence of the PCR product obtained from the cells harvested at 16 hr post-infection (lane 6) was verified to be the expected VSV N amplicon. (b) DNA samples were harvested at 16 hr post-infection and were treated with RNase A or DNase I and subsequently used for PCR. The VSV DNA was not detected after DNase I treatment (lane 2). All samples used in Figures 1a and 1b were prepared in a single experiment, and cropped gel images are shown.
Localisation of VSV DNA
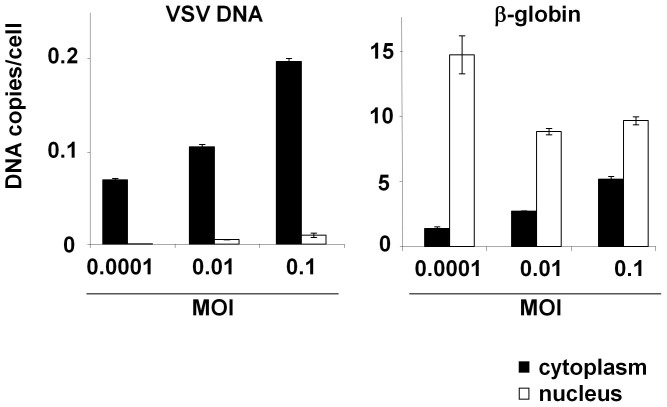
To determine the localisation of the viral DNA, cell fractionation experiments were performed after the infection of 293T cells with VSV. Different MOI (0.1, 0.01, and 0.0001) were used to optimise the cell fractionation experiments (Fig. 2). The cells were harvested 16 hr after infection and fractionated; the cytoplasmic and nuclear lysates were subjected to real-time PCR to amplify the VSV N gene using the primer pair described above. The PC03/PC04 primer pair was used to amplify the cellular β-globin gene. At an MOI of 0.01 or 0.0001, the VSV DNA was only detected in the cytoplasmic fraction, whereas the β-globin DNA was detected primarily in the nuclear fraction and weakly in the cytoplasmic fraction, indicating the accurate separation of the cytoplasmic and nuclear fractions. At an MOI of 0.1, the β-globin DNA was also detected in the cytoplasmic fraction, likely due to the cytopathic effects of VSV infection (Fig. 2). These data suggest that the VSV DNA is localised to the cytoplasm and may be generated there.
Figure 2. Fractionation of VSV-infected 293T cells.
293T cells were infected with VSV at different MOI (0.0001 to 0.1) and incubated for 16 hr. The nuclear and cytoplasmic fractions were separated and subjected to real-time PCR using the VSV-N (left panel) and β-globin primers (right panel). The DNA copy number per cell was estimated using the VSV N gene or β-globin gene in the control plasmid and is indicated on the axis. The means ± SE of two independent experiments performed in duplicate are shown.
Structure of the VSV DNA
To characterise the structure of the VSV DNA, PCR primers targeting the coding (N and L) and non-coding regions (leader and trailer) of the VSV genome were used. The accumulation of viral proteins synthesised from primary mRNA transcripts leads to the replication of the genome, which involves the synthesis of an antigenome. The VSV N and L mRNA lack the non-coding regions (leader and trailer) but contain m7G and polyA at the 5′ and 3′ ends, respectively. The antigenome contains the viral RNA sequences from the leader to the trailer RNA (Fig. 3a).
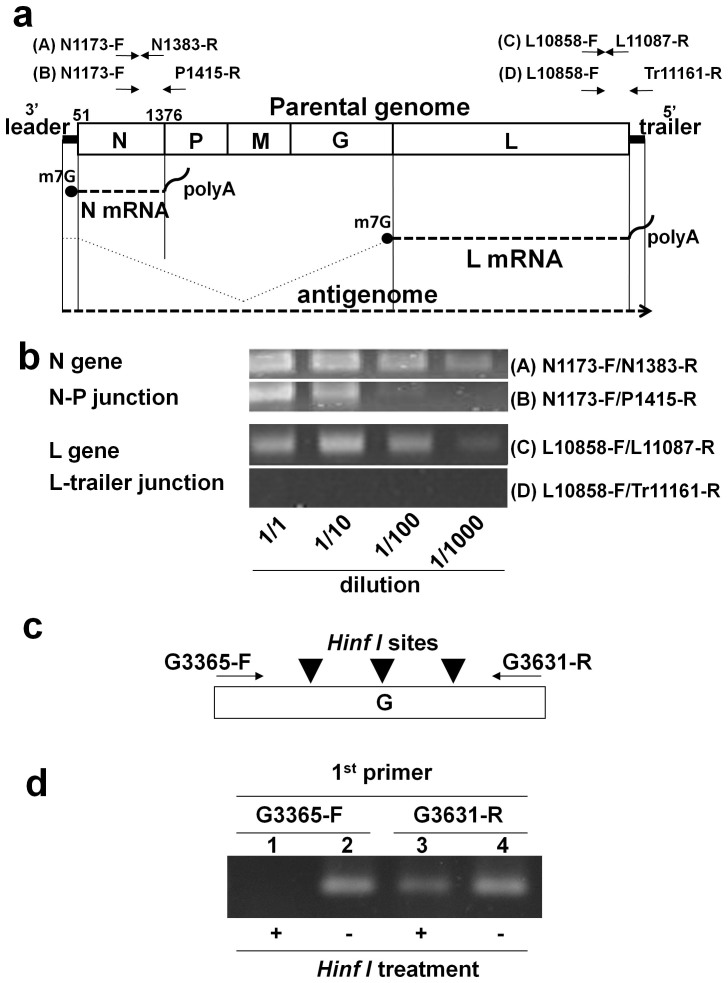
Figure 3. Structure and strand polarity of the VSV DNA and viral mRNAs.
(a) The PCR primers (A), (B), (C), and (D) are indicated above the diagram showing the VSV structure. The PCR primer pairs targeted (A) a region within the N gene (N1173-F/N1383-R), (B) a region spanning the N-P gene junction (N1173-F/P1415-R), (C) a region within the L gene (L10858-F/L11087-R), and (D) a region spanning the L gene-trailer junction (L10858-F/Tr11161-R). The N and L mRNA are shown schematically. (b) The DNA was extracted from 293T cells at 16 hr after infection with VSV at an MOI of 0.1 and subjected to PCR amplification. The cell lysates were diluted 1- to 1000-fold prior to PCR, and the PCR products were visualised using gel electrophoresis. (c) Unidirectional primer extension was performed using the positive-strand G3365-F primer or negative-strand G3631-R primer. After treatment with HinfI, the samples were subjected to PCR using the G3365-F/G3631-R primer pair. The three arrowheads indicate the positions of the HinfI restriction sites between the G3365-F and G3631-R primers. (d) The first and second lanes show the PCR results using G3365-F as the extension primer with and without HinfI treatment, respectively. The third and fourth lanes show the PCR results with and without HinfI treatment, respectively, using G3631-R as the extension primer. Note that cropped gel images, (b) and (d), are shown, and the gels shown in (b) were run under identical conditions.
The DNA was extracted from the 293T cells infected with VSV as described above. Using a primer pair targeting the N gene (N1173-F/N1383-R), the viral DNA was detected in the lysates of the infected cells even at a 1/1000 dilution of the lysates. However, the primer pair N1173-F/P1415-R, which amplifies a DNA fragment spanning the N and P genes, yielded discrete PCR bands at dilutions of 1/1 and 1/10 but not at higher dilutions (Fig. 3a and b). Similarly, the L gene was amplified using the L10858-F/L11087-R primer pair even at a 1/1000 dilution of the lysates, whereas the fragment spanning the L gene and trailer region could not be amplified with the L10858-F/Tr11161-R primer pair even at a 1/1 dilution of the lysates (Fig. 3a and b). These data suggest that the majority of VSV DNA molecules were fragmented and that each viral DNA corresponded to a single viral gene. This fragmentation pattern suggests that these molecules may be cDNAs that were reverse transcribed from viral mRNAs produced after VSV infection.
Strand polarity of the VSV DNA
To further investigate the above possibility, we determined the strand polarity of the VSV DNA. Unidirectional primer extension was performed with either the positive strand (coding sequence polarity) primer G3365-F or the negative strand primer G3631-R, and the resulting reaction mixtures were treated with the HinfI restriction enzyme. There are three HinfI sites in this PCR fragment. PCR was subsequently performed using the G3365-F/G3631-R primer pair (Fig. 3c). If the first unidirectional primer extension produced double-stranded DNA, no PCR band was expected after HinfI treatment. When the G3365-F primer was used for unidirectional primer extension, PCR products were not detected after HinfI treatment (Fig. 3d). This result demonstrates that the VSV DNA is single-stranded and has the opposite polarity relative to the coding sequence. However, we observed that the viral RNA with the same polarity as the genome was not reverse transcribed, although this RNA is expected to be present abundantly in infected cells (Fig. 3a).
VSV DNA synthesis in human cells
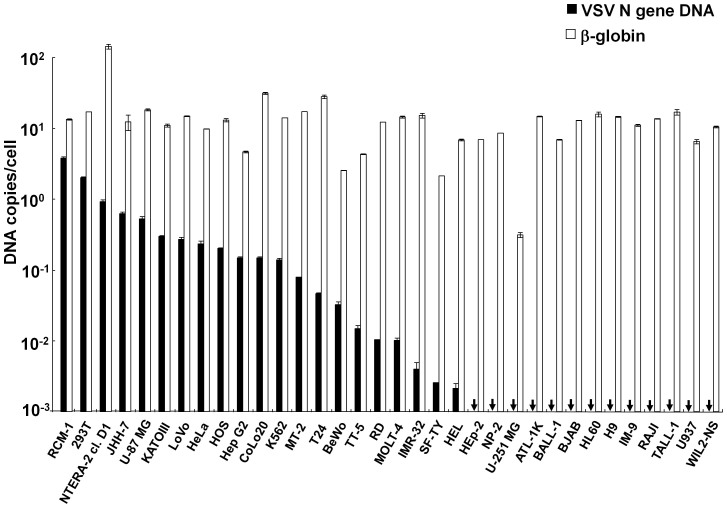
We tested the ability of various human cells to produce the VSV DNA upon acute VSV infection. The total DNA was extracted from 32 human cell lines and two primary cells, SF-TY and TT-5 primary skin fibroblasts (Table S2) that had been infected with VSV and incubated for 16 hr; the extracted DNA was used for real-time PCR to amplify the VSV N gene and cellular β-globin gene (Fig. 4). Many of the tested human cell lines contained the VSV DNA, but a third of the cell lines contained no detectable VSV DNA (<1 × 10−3 DNA copies/cell). Even among the positive cell lines, the VSV DNA levels varied across a three-log range (2.2 × 10−3–4.2 × 100 copies/cell): RCM, 293T or NTERA-2 -cl. D1 cells contained high levels of VSV DNA. There was no clear dependence of VSV DNA content on cell lineage. Many suspension cells or hematopoietic cells, such as ATL-1K, BALL-1, BJAB, HL60, H9, IM-9, RAJI, TALL-1, U937, and WIL2-NS, did not contain the VSV DNA, whereas the K562 or HEL erythroleukaemia cell line and the HTLV-I-positive MT-2 T-cell line contained the VSV DNA. Notably, even the adherent NP-2 and U-251 MG glial cell lines but U87-MG cell line did not contain VSV DNA. Both the SF-TY and TT-5 primary skin fibroblasts contained the VSV DNA, indicating that primary cells can generate VSV DNA. Therefore, most adherent cell lines, some primary cells, and some hematopoietic cell lines of human origin can produce VSV DNA.
Figure 4. Cell-based differences in VSV DNA production.
Various human cells were infected with VSV at an MOI of 0.1. The DNA was extracted 16 hr later and subjected to PCR amplification. Real-time PCR was performed using the N478-F/N681-R or β-globin primer pair. The copy numbers of the VSV N and β-globin genes in each cell line are shown in closed and open bars, respectively. The arrows indicate that the copy number per cell was below the detection limit. The positive cells are ordered according to the VSV N DNA level, followed by the VSV DNA-negative adherent cells. Subsequently, the VSV DNA-negative suspension cells are arranged in alphabetical order. The means ± SE of two independent experiments performed in duplicate are shown.
Effects of LINE-1 (L1.2) transduction on VSV DNA formation
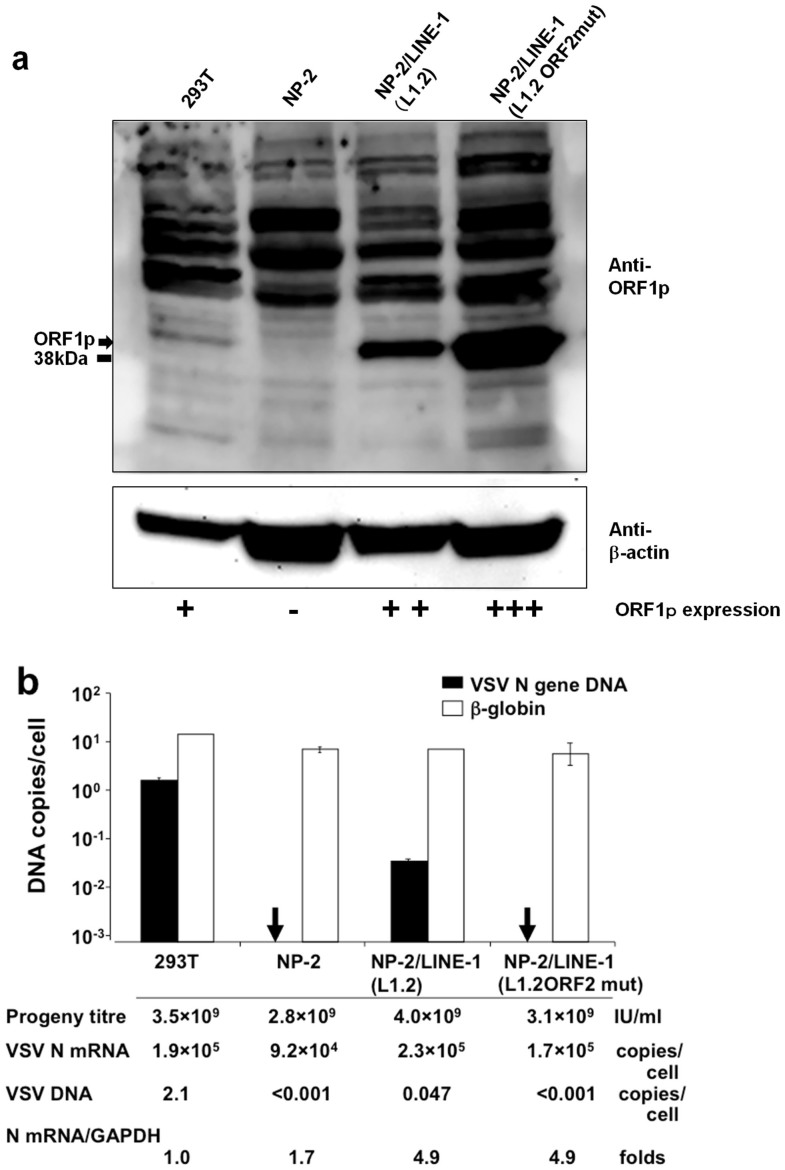
We postulated that LINE-1 produced the VSV DNA. LINE-1 (L1.2) has a moderate retrotransposition activity, whereas LINE-1 (L1.2 ORF2mut) lacks this activity13. Therefore, we introduced VSV DNA via transduction into non-producing NP-2 cells with either wild-type LINE-1 (L1.2) or a LINE-1 mutant to generate NP-2/LINE-1 (L1.2) and NP-2/LINE-1 (L1.2 ORF2mut) cells (Fig. 5a).
Figure 5. LINE-1 transduction of the NP-2 cell line and its effect on VSV DNA formation.
(a) LINE-1 transduction of the NP-2 cell line. The LINE-1 ORF1p and β-actin levels in the 293T, NP-2, NP-2/LINE-1 (L1.2), and NP-2/LINE-1 (L1.2 ORF2mut) cells were examined via Western blot using the anti-ORF1p IgY antibody. The ORF1p protein was observed at a molecular weight of ~41 kDa. The membrane was re-probed with an anti-β-actin antibody. The size marker at 38 kDa is indicated. Note that cropped western blots are shown and that full-length images are presented in the supplementary information. (b) The effect of LINE-1 transduction on VSV DNA production. The cells described above were infected with VSV, and the cell lysates were prepared. Real-time PCR was performed with either the N478-F/N681-R or β-globin primer pair. In addition, the VSV progeny produced by these cells were titrated and the VSV N mRNA expression levels were compared among these cells.
NP-2/LINE-1 (L1.2) and NP-2/LINE-1 (L1.2 ORF2mut) cells and the parental NP-2 cells and 293T cells were infected with VSV at an MOI of 0.1, incubated for 16 hr, and tested for the formation of VSV DNA (Fig. 5b). The time course of VSV cytopathic effects was similar among these cells; all cells produced similar levels of progeny virus (2.8 × 109–4.0 × 109 IU/ml) and VSV mRNA (9.2 × 104–2.3 × 105 copies/cell) (Fig. 5b). The VSV DNA was detected in the lysate of 293T cells as described above (Fig. 4). No VSV DNA was detected in the parental NP-2 cells or the NP-2/LINE-1 (L1.2 ORF2mut) cells (<1 × 10−3 copies/cell), whereas the NP-2/LINE-1 (L1.2) cells produced an amount of VSV DNA (4.7 × 10−2 copies/cell) two logs higher than the detection limit (Fig. 5b). Subsequent inverse PCR analyses and the sequencing of PCR products also confirmed that the VSV N gene cDNA was produced in the NP-2/LINE-1 (L1.2) cells, as shown in Figures 6a and 6b. Furthermore, the transduction of U-251 MG cells (which did not produce VSV DNA (Fig. 4)) with LINE-1 (L1.2) also led to the generation of VSV DNA (data not shown). These data indicate that the LINE-1 reverse transcriptase can generate VSV DNA in human cells.
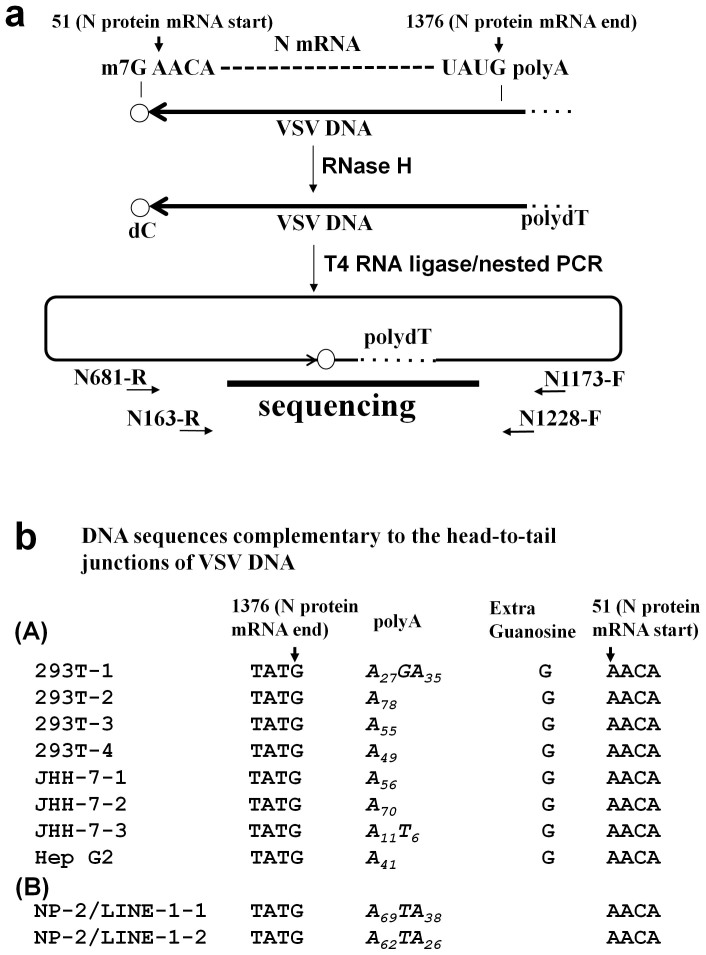
Figure 6. Sequence analysis of the 5′ and 3′ ends of the VSV DNA.
(a) Schematic diagram of the inverse PCR method used to amplify the 5′ and 3′ ends of the VSV DNA. According to the results shown in Fig. 3, we proposed that the VSV mRNA was reverse transcribed to DNA. The start (nucleotide 51) and end (nucleotide 1376) sites of the VSV N mRNA are indicated (GenBank: J02428.1). The cell lysates were treated with RNase H to digest the VSV RNA and then incubated with T4 RNA ligase to induce intra-molecular DNA end joining. The DNA was then amplified across the junction via nested PCR and sequenced. (b) The VSV DNA sequences of the head-to-tail junctions are shown. (A) VSV DNAs from four clones of 293T cells, three clones of JHH-7 cells, and one clone of Hep G2 cells are shown. All clones contained sequences corresponding to the 5′ end of the N mRNA (nt position 1376), followed by polyA tails consisting of 11 to 78 adenylate residues. (B) No additional G residue was found in two clones from the NP-2/LINE-1 (L1.2) cells infected with VSV.
Although the intensity of the Western blot band detected in 293T cells using the anti-ORF1p IgY antibody against LINE-1 ORF1p was significantly weaker than that of NP-2/LINE-1 (L1.2) (Fig. 5a), the amount of VSV DNA formed in the VSV-infected 293T cells was nearly 2 logs higher than that of NP-2/LINE-1 (L1.2). This result suggests that LINE-1 (L1.2) is not a major factor responsible for VSV DNA formation in 293T cells, although LINE-1 (L1.2) can produce VSV DNA.
Structures of the 5′ and 3′ ends of viral DNA
To resolve the hypothesis that the VSV mRNA rather than the VSV genomic RNA or antigenome RNA was used as the template for VSV DNA synthesis (Fig. 3b and d) and to determine the 5′ and 3′ end structures of the VSV DNA, the DNA fragments that were expected to contain both ends of the VSV DNA were cloned using inverse PCR following single-stranded head-to-tail ligation (Fig. 6a). Any extra DNA sequences at the 5′ or 3′ end of the VSV DNA would be detected using this assay. The lysates of the 293T, JHH-7, and Hep G2 cells prepared 16 hr after VSV infection were treated with RNase H to digest the template VSV RNA; T4 RNA ligase was then added to produce circular VSV DNA (Fig. 6a). The DNA was then subjected to PCR using primers that directed extension from the N gene to either the P gene or leader sequence (Fig. 6a; also see Fig. 3a). Nested PCR products (approximately 300 bp each) were cloned and sequenced. DNA sequences complementary to the VSV DNA were detected (Fig. 6b).
All clones contained the sequences corresponding to the 5′ end of the N mRNA (nt position 1376), followed by polyA tails consisting of 11 to 78 adenylate residues. In seven of the eight clones obtained from the VSV-infected 293T, JHH-7, or Hep G2 cells, the 5′ end of the N mRNA (nt position 51) was ligated to the 3′ polyA tail with an additional G residue inserted between them. The seventh clone had six additional T residues. The origin of the additional G residue was unclear, but a C residue could have been incorporated into the VSV cDNA by pairing with the m7G 5′ mRNA cap. However, no additional G residue was found in the two clones from the NP-2/LINE-1 (L1.2) cells infected with VSV.
Effects of reverse transcriptase inhibitors on VSV DNA formation
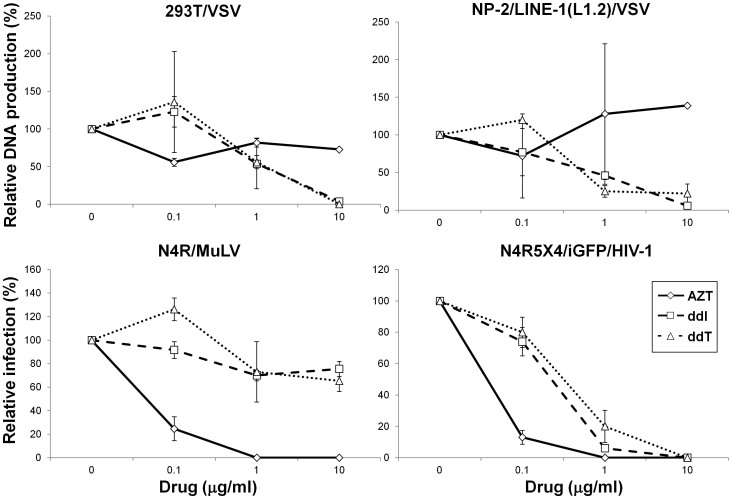
We examined whether the synthesis of VSV DNA was sensitive to reverse transcriptase inhibitors. The 293T and NP-2/LINE-1 (L1.2) cells were treated with 2′,3′-dideoxyinosine (ddI), 2′,3′-dideoxythymidine (ddT), or 3′-azido-3′-deoxythymidine (AZT) for 1 hr and then infected with VSV at an MOI of 0.1. After a 16-hr incubation with these drugs, the DNA was extracted and examined for the presence of the VSV N DNA. These drugs were also used to test cells infected with HIV-1 (IIIB strain) and a murine leukaemia virus (MuLV) pseudovirus; the synthesis of VSV DNA in the 293T and NP-2/LINE-1 (L1.2) cells was similarly and markedly inhibited by ddI and ddT but not AZT (Fig. 7). In conclusion, these results suggest that LINE-1 may generate the VSV DNA.
Figure 7. Effects of reverse transcriptase inhibitors on VSV DNA production.
The synthesis of VSV DNA in the 293T and NP-2/LINE-1 (L1.2) cells in the presence of AZT, ddI, or ddT was examined via real-time PCR using the VSV N primers. We also tested the effects of these drugs on infection with recombinant MuLV pseudotype virus or HIV-1 strain IIIB. The means ± SE of two independent experiments performed in duplicate are shown.
Synthesis of DNA complementary to other single-stranded RNA viruses in human cells
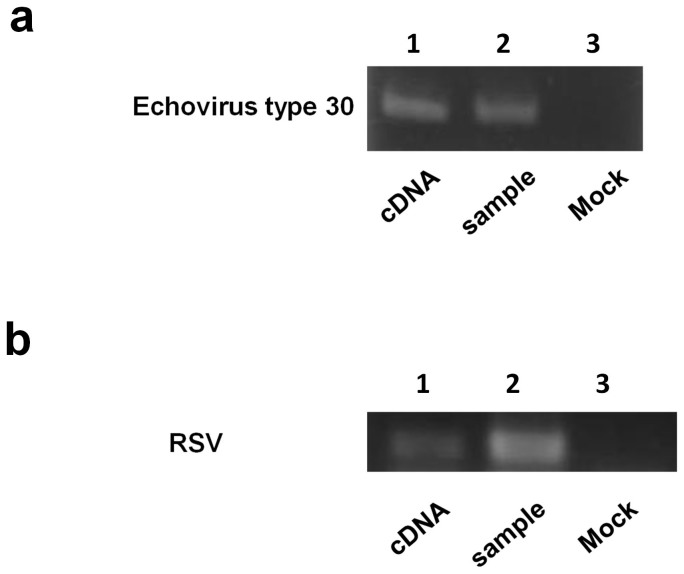
We examined whether the DNA that was complementary to other cytopathic RNA viruses, specifically a positive-sense single-stranded RNA virus (echovirus type 30) and negative-sense single-stranded RNA virus (RSV), was produced in human cells that were permissive to these viruses (Fig. 8). Only a few cell lines are susceptible to these viruses. DNA complementary to the echovirus type 30 and RSV were detected in the lysates of the RD and HEp-2 cells, respectively. This finding suggests that the infection of human cells with other non-retroviral RNA viruses could lead to the production of viral cDNA.
Figure 8. Synthesis of complementary DNA to RNA of echovirus type 30 and RSV in human cells.
(a) The DNA was extracted from RD cells 48 hr after infection with echovirus type 30 and subjected to PCR with the echovirus type 30-specific primers. (b) The RSV DNA was detected in HEp-2 cell lysates prepared five days post-infection with RSV. The DNA sequences of these bands were determined after cloning and found to be identical to the sequences of the viral strains used (GenBank: echovirus type 30, AF311938; RSV, AY911262). Viral RNAs were extracted from echovirus type 30 and RSV propagated in RD and Hep-2 cells, respectively. cDNAs to these RNAs were made, purified and used as templates for PCR (lane 1). RD and HEp-2 cells were infected with echovirus type 30 and RSV, incubated and lysed as described in the Methods section. The cell lysates were used for PCR (lane2). The lysates of mock-infected RD and Hep-2 cells were used as controls (lane 3). Note that cropped gel images are shown and full-length gel images are presented in the supplementary information.
Discussion
There are few reports on the cDNA formation of RNA viruses1,14. However, illicit viral DNA formation has not been confirmed2,4, with the exception of recent studies on LCMV and bornavirus3,5,6,15. Viral DNA complementary to the LCMV RNA is produced in retrovirus-infected mouse cells but not in retrovirus-uninfected mouse cells or human cells3. This study suggested the involvement of a retrovirus in LCMV DNA formation. However, we showed that the VSV DNA was formed in various human cell lines and primary human fibroblasts.
In this report, we clarified the structure of the cytoplasmic VSV DNA and showed that a retrotransposon-related reverse transcriptase can generate the VSV DNA. Dhellin et al. showed that when cells are transfected with expression plasmids for LINE-1 and unspliced RNA, spliced RNA is made abundantly and DNA complementary to the spliced RNA is formed. This finding suggests that cellular mRNA as well as mRNA to the transduced gene can be reverse transcribed when LINE-1 is expressed. We showed that VSV DNA is formed in primary fibroblastic cells infected with VSV and that DNA complementary to echovirus type 30 or RSV was detected in human cells infected with them. It is probable that the reverse transcription of human cellular RNA can occur under natural conditions.
In HEp-2 cells, DNA complementary to RSV RNA but not to VSV RNA was detected after infection (Figs. 4 and 8). It was even difficult to detect cDNA to RSV RNA despite repeated attempts to detect it using several PCR primer pairs under different PCR conditions as described. Fortunately, we could detect it using one PCR primer pair. Regarding the detection of VSV DNA, more than 30 different types of cells (Fig. 4) were examined using only one PCR primer pair under similar assay conditions. It is probable that the viral DNA produced in the HEp-2 cells infected with VSV or RSV was of a small amount, which produced contradictory results.
The polarity of the VSV DNA was complementary to viral mRNA (Fig. 3d); the mRNA of the VSV N gene was reverse transcribed from the m7G cap to the polyA tail (Fig. 6b). The VSV DNA was initially detected as early as 2 hr after inoculation (Fig. 1a), which was consistent with the fact that the first viral mRNA transcription in the replication cycle occurs 2 hr after inoculation16. These data suggest that the VSV DNA is generated in the infected cells when the mRNA becomes available for reverse transcription.
The inverse PCR and subsequent sequencing revealed that additional G residues, which correspond to m7G in Figure 6b, were present before the initiation site of the N gene mRNA. Interestingly, the extra G residue was not observed in the NP-2 cells overexpressing L1.2 (Fig. 6b). Similar additional G residues were detected in retrotransposition experiments by Gilbert et al17. Volloch et al. also showed that reverse transcriptase of avian myeloblastosis virus (AMV) can reverse transcribe m7G cap18. In that study, HeLa cells were transduced with replication-competent LINE-1s (RC-L1s), highly potent L1.3 and lowly potent L1.2A and LRE-2, and L1 retrotransposition events were characterised17. Two of the six full-length L1s obtained after the transduction of the highly potent L1.3 contained an additional G residue next to the start of the transposed LINE-1 sequence, whereas the L1s obtained after the transduction of the lower potency elements L1.2A and LRE-2 did not contain an additional G. This study suggests that highly potent LINE-1 can reverse transcribe full length LINE-1 mRNA, including m7G, while lowly potent LINE-1 will reverse transcribe mRNA before m7G. We have made similar observations. That is, the full-length viral mRNA of the N gene from the polyA to the m7G cap was reverse transcribed into VSV DNA in the 293T, JHH-7, and Hep G2 cells, whereas the NP-2 cells transduced with a lowly potent LINE-1, L1.2, NP-2/LINE-1 (L1.2) produced VSV DNA that lacked a residue complementary to the m7G cap. Additionally, the NP-2 cells transduced with LINE-1 (L1.2) did not produce as much VSV DNA as the 293T cells after VSV infection (Fig. 5a and b). Thus, 293T, JHH-7, and Hep G2 cells may express a highly potent LINE-1 or another LINE-1-related reverse transcriptase.
We detected the expression of LINE-1 via Western blot using a chicken antibody against recombinant ORF1p expressed in Escherichia coli. The use of a specific antibody against LINE-1 reverse transcriptase ORF2p is recommended if such an antibody is available. Using the ORF1p antibody, we confirmed that the ORF1p protein was abundantly expressed in the NP-2 cells transfected with the L1.2 expression plasmids (Fig. 5a). However, the 293T cells that produced VSV DNA in amounts that were two logs higher than the NP-2/LINE-1 (L1.2) cells had a weak signal (Fig. 5b). These results suggest that the Western blot signal using the ORF1p antibody may not fully reflect the activity of LINE-1. The expression of LINE-1 in human cells may be strongly suppressed; LINE-1 transduction experiments have suggested that there are various types of LINE-1 with different activities. Our findings (Figure 4 and 5) suggest that different types of LINE-1 are expressed in human cells. Whether the expression of LINE-1 in human cells can be monitored via the detection of DNA complementary to the VSV RNA remains to be determined.
Primers are required for the reverse transcription of VSV RNA by the reverse transcriptase encoded by LINE-1. The inverse PCR analyses indicated that special DNA primers may not be required for VSV DNA formation because no extra DNA sequences were detected using these assays (Fig. 6b). The mechanism of production and accumulation of VSV DNA in the cytoplasm remains unclear. VSV RNA is present in the cytoplasm, and we showed that VSV DNA was detected in the cytoplasm. It still remains to be determined how VSV DNA formation is initiated in cells infected with VSV. OligoU or oligoT in VSV-infected cells can be used as a primer. Furthermore, RNA molecules with an oligoU stretch at their 3′ ends, such as uridine-rich snRNAs, are good candidate primers for VSV DNA synthesis. In particular, U6 RNA sequences are frequently mobilised by LINE-1, and U6 often fuses with the 3′ end of LINE-119. The U6 RNA may associate with the LINE-1 complex20. The identity of the primer used for VSV DNA synthesis remains unclear.
For formation of double-stranded DNA, a specific primer or specific mechanism will be required to start transcription of second DNA strands. In the case of retrovirus infection, DNA-RNA hybrid is made and polypurine tracts in viral genome RNA play a role to start the transcription of the second DNA strand. Similar sequences are not present in VSV RNA. In the case of double-stranded DNA formation by LINE-1, cellular DNA nicks and repair system in the nucleus are used to make second DNA strand. Such DNA repair system will not operate for VSV DNA.
The biological relevance of the formation of cDNA to non-retroviral RNA virus genomes is unclear. The viral genes integrated into a germline cell genome may confer resistance to the infections of hosts by related exogenous viruses7. Although LCMV DNA can be integrated into the host genome and contributes to persistent viral gene expression5,15, the VSV DNA was primarily found in the cytoplasm of cells dying due to the cytopathic effects of VSV (Fig. 2). Therefore, the VSV DNA may be recognised using DNA sensors21, e.g., DAI (DLM-1/ZBP1)22, inflammasome23, or Toll-like receptor 924. Stetson et al. reported that the nuclease Trex1 prevents the accumulation of single-stranded DNA derived from endogenous retro-elements and inhibits the autoimmunity triggered by DNA sensors of the interferon-stimulatory DNA (ISD) pathway25. Jones et al. showed that HIV-1 infection results in the accumulation of L1 DNA in primary CD4+ cells and that the majority of the L1 DNA is extra-chromosomal. They discussed the implications of the innate immune response to HIV-1 infection by an unrecognised interaction between HIV-1 and the endogenous retrotransposable elements. We showed that the genetic information of the non-retroviral RNA virus genome can flow into DNA in mammalian cells. The biological significance of the cytoplasmic VSV DNA should be investigated further.
Methods
Cell lines and plasmids
The NP-2 cells were derived from human glioma cells. The N4R (NP-2/CD4/ecoR) cells were NP-2 cells transduced with CD4 and an ecotropic murine leukaemia virus (MuLV) receptor26. The N4R5X4/iGFP cells, which were used for HIV-1 titration, were N4R cells that were transduced with CXCR4, CCR5, and an HIV-LTR-driven GFP. The N4R5X4/iGFP cells clearly expressed GFP when HIV-1 infection was established. The NP-2/LINE-1 cells were NP-2 cells that were transfected with a LINE-1 (L1.2) expression plasmid with a pCEP4 backbone13. The NP-2/LINE-1 (L1.2 ORF2mut) cells were transfected with a LINE-1 (L1.2) expression plasmid with an ORF2 mutation (D702Y)13. The other human cells used in this study are presented in Table S2. All cells were maintained at 37°C in a humidified atmosphere containing 5% CO2. N4R and N4R5X4/iGFP were used for the infection assays with MuLV and HIV-1, respectively.
VSV preparation and titration
To prepare the VSV samples, 4.0 × 105 HOS cells were seeded onto 60 mm plates. After 24 hr, the cells were infected with VSV (Indiana strain) at an MOI of 0.5. After washing with phosphate buffered saline (PBS), the cells were incubated at 37°C for 16 hr. The cells and debris were removed by low-speed centrifugation, and the supernatant was harvested and stored in small aliquots at −80°C until use. The viral titres were determined via standard plaque assays using feline kidney 8C cells27. The 8C cells were seeded into 35 mm plastic culture plates at a density of 1.2 × 106 cells/plate. The following day, the cells were infected with 300 μl of diluted VSV at 37°C for 1 hr. The cells were then washed once with culture medium, and agar-containing medium was placed over the plates. The following day, the cells were stained with neutral red, and the number of plaques was counted. The VSV stocks showed infectious titres of 1.0 × 108 pfu/ml. Therefore, to obtain an MOI of 0.1, the VSV stocks were usually diluted 100-fold. The echovirus type 30 and RSV were propagated in RD and HEp-2 cells, respectively; their culture supernatants were harvested after cultivation for 48 hr and five days, respectively. Because these viral titres were very low, the cells were infected with undiluted virus stocks and incubated for significantly longer periods compared with VSV infection before the cytopathic effects became evident.
VSV infection and DNA purification
A total of 1.0 × 106 293T cells were infected with VSV at an MOI of 0.1 for 1 hr. The cells were washed once with PBS and incubated for 16 hr. The cell lysates were prepared using the Wizard® Genomic DNA Purification Kit (Promega, Madison, WI, USA) according to the manufacturer's instructions. The extracted DNA was dissolved in 100 μl of TE buffer. For the experiments examining the differences in VSV DNA production between various cell lines, the cells were lysed in 160 μl of buffer containing 10 mM Tris-HCl (pH 8.0), 1 mM ethylenediaminetetraacetic acid (EDTA), 0.45% Nonidet P-40 (Sigma, St. Louis, MO, USA), 0.45% Tween 20 (Sigma), and 20 mg/ml proteinase K (Sigma), incubated at 52°C for 2 hr, and heated to 96°C for 10 min to inactivate the proteinase K.
PCR amplification
To detect the viral DNA via PCR, 15 μl of a reaction mixture containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 2 mM of each dNTP, 60 ng of the sense and antisense PCR primers, and Taq DNA polymerase (Roche, Branchburg, NJ, USA) was added to 5 μl of the cell lysate. PCR was performed in a thermal cycler (Perkin-Elmer Cetus, Norwalk, CT, USA). Thirty-seven cycles of PCR were performed under the following conditions: denaturation at 94°C for 1 min, annealing at 55°C for 45 sec, and extension at 72°C for 1 min. The resulting PCR products were confirmed via DNA sequencing to be of VSV origin.
Real-time PCR amplification
Cell lysate aliquots (2.5 μl) were subjected to real-time PCR using 25 μl of Brilliant SYBR® Green QPCR Master Mix (Agilent Technologies, La Jolla, CA, USA) with 0.4 μM of each primer. The amount of VSV DNA in each sample was normalised using the β-globin gene as a control. The assay was performed in duplicate using the Mx3000P QPCR system (Agilent Technologies). The thermo-cycling conditions were as follows: denaturation at 95°C for 30 sec, annealing at 55°C for 1 min, and extension at 72°C for 30 sec. The copy number of each product was estimated using standard curves that were obtained using serial dilutions of the plasmids containing the VSV N and β-globin genes (pGEM®-T Easy vector) (Promega).
PCR primers
The oligonucleotide primers were synthesised by Hokkaido System Science Co., Ltd. (Sapporo, Japan). PC03/PC04 and HA031578-F/HA031578-R were ready-made primer pairs designed for the amplification of β-globin and glyceraldehyde 3-phosphate dehydrogenase (GAPDH), respectively (TAKARA, Otsu, Shiga, Japan). A list of PCR primers is provided in Table S1. The primer pairs used to determine the presence of VSV DNA were N1173-F and N1383-R (targeting a region within the N gene), N1173-F and P1415-R (targeting a region spanning the N and P genes), L10858-F and L11087-R (targeting a region within the L gene), and L10858-F and Tr11161-R (targeting a region spanning the L gene and trailer region). The efficiencies of these PCR primer pairs were assessed using cloned VSV cDNA fragments as the control template. The primers were shown to have similar efficiencies (data not shown).
Time course of VSV DNA synthesis and the treatment of cell lysates with RNase A or DNase I
The time course of VSV DNA synthesis was determined as follows: 1.0 × 106 293T cells were seeded in 60 mm plates and infected with 1.0 × 105 infectious units (IUs) of VSV (MOI of 0.1). After 1, 2, 4, 6, and 16 hr, the cellular DNA was extracted and dissolved in 100 μl of TE buffer. A 1-μl aliquot of this sample was then subjected to PCR, which enabled the detection of DNA resulting from the inoculation of 1.0 × 103 IUs of VSV. The virus lysate derived from 4.0 × 104 IUs of VSV was used as a supernatant (sup) control (Fig. 1a) to detect VSV DNA.
At 16 hr post-infection, 1 μl of the cell lysate was treated with 1 μl of RNase A (Wako Pure Chemical Industries, Osaka, Japan) or DNase I (Promega) and incubated at 37°C for 1 hr. The cell lysates were extracted with phenol/chloroform, and the DNA was precipitated in ethanol. The DNA pellet was dissolved in 10 μl of distilled water. One microliter of dissolved DNA was subjected to PCR using the N478-F/N681-R primer pair.
Cell fractionation
293T cells were infected with VSV as described above and incubated for 16 hr. The cells were then lysed with 100 μl of buffer containing 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, 0.65% Nonidet P40, and 150 mM NaCl, and then vortexed for 10 sec and centrifuged at 5000 rpm for 5 min to separate the supernatant from the precipitate. The supernatant was treated with 20 mg/ml proteinase K, incubated at 52°C for 2 hr, and then heated to 96°C for 10 min to inactivate the proteinase K. The precipitate was washed with buffer, and 100 μl of buffer containing 20 mg/ml proteinase K was added. The samples were then incubated at 52°C for 2 hr and heated again to 96°C for 10 min. The VSV N gene and β-globin gene in the cell lysates were detected via real-time PCR (Brilliant SYBR® Green QPCR Master Mix) using the N478-F/N681-R and β-globin primer pairs, respectively. The copy numbers of the VSV N gene and β-globin gene in each cell were estimated with standard curves obtained using serial dilutions of plasmids (pGEMT®-easy vector) containing the VSV N and β-globin genes. The β-globin DNA was detected to examine whether the nuclear fractions were separated from the cytoplasm fractions.
Determination of strand polarity of the VSV DNA
One cycle of unidirectional primer extension was performed at 72°C for 1 min using either the positive-strand primer G3365-F or negative-strand primer G3631-R. The product was treated with HinfI at 37°C for 1 hr. After the phenol/chloroform extraction and ethanol precipitation, the sample was subjected to PCR using the G3365-F/G3631-R primer pair. There are three HinfI restriction sites between G3365-F and G3631-R (Fig. 3c). If double-stranded DNA was synthesised during the first extension, this DNA would be digested by HinfI, leading to the disappearance of the PCR bands.
Inverse PCR to determine the end structures of VSV DNA
Inverse PCR was performed to clone the 5′ and 3′ ends of the VSV DNA (Fig. 6a). Human cells were infected with VSV and lysed 16 hr later. The cell lysates were treated with RNase H to digest the template VSV RNA. The lysates were then incubated with T4 RNA ligase (TAKARA) overnight at 37°C28 to induce intra-molecular DNA joining and produce circular VSV DNA (Fig. 6a). The ligated DNA was purified using phenol/chloroform and subjected to PCR with the N1173-F/N681-R primer pair. Nested PCR was performed using the N1228-F/N163-R primer pair, and the nested PCR products were cloned into the pGEM®-T Easy vector and sequenced using the SP6 and T7 promoter primers.
Western blot analysis to detect LINE-1 proteins
The cells were lysed in a solution containing 25 mM Tris-HCl (pH 7.4), 137 mM NaCl, 2.7 mM KCl, 1% Nonidet-P40, and a proteinase inhibitor cocktail (Roche). The lysate of 3 × 105 cells was centrifuged to pellet the debris, and the supernatant was harvested and boiled in a reducing sodium dodecyl sulphate (SDS) sample buffer prior to the immunoblot analyses using an anti-ORF1p IgY antibody and control anti-β-actin antibody (A4700, Sigma) according to the manufacturer's protocols. Antibodies against LINE-1 ORF1p were generated by immunising chickens with purified human L1 ORF1p, which was overexpressed in E. coli and purified from yolks29.
Effects of LINE-1 transduction on the synthesis of VSV DNA
The 293T, NP-2, NP-2/LINE-1, and NP-2/LINE-1(ORF2mut) cells were seeded at a density of 1 × 106 cells/plate in 60 mm plates. The following day, the cells were infected with VSV at an MOI of 0.1 and incubated at 37°C for 16 hr. The whole cell extracts were subjected to real-time PCR using the N478-F/N681-R and β-globin primer pairs. The VSV progeny produced by these cells were titrated as described above. To compare the VSV N mRNA expression levels in these cell lines, real-time RT-PCR was performed. The total cellular RNA was isolated using ISOGEN® according to the manufacturer's protocol and treated with DNase I to digest any contaminating cellular DNA. The cDNA was synthesised using the SuperScript® II First-Strand Synthesis System with an oligo(dT) primer. The cDNA was then subjected to real-time PCR using the N478-N/N681-R primer pair, and the results were normalised to GAPDH mRNA expression using the HA031578-F/HA031578-R primer pair (Table S1). The PCR conditions were the same as those described above.
Effects of reverse transcriptase inhibitors on the synthesis of VSV DNA
We seeded 5.0 × 105 293T and NP-2/LINE-1(L1.2) cells in six-well plates. The following day, the cells were treated with ddI, ddT, or AZT for 1 hr at concentrations ranging from 0.1 to 10 μg/ml and then infected with VSV at an MOI of 0.1. After a 16-hr incubation period in the presence of the drugs, the cells were lysed and subjected to real-time PCR using the N478-F/N681-R primer pair. No apparent cell damage was observed in the drug-treated, mock-infected cells.
A recombinant MuLV pseudotype virus and HIV-1 (IIIB strain) were also tested for sensitivity to AZT, ddI, and ddT. For the MuLV pseudotype, Phoenix-E cells were transfected with the pCX bsr-GFP plasmid, which is a MuLV-based retroviral vector (pCX bsr)30 containing the GFP gene, using FuGENE6 (Roche). The viral supernatants were harvested and stored at −80°C until further use. The N4R cells were seeded by adding 125 μl of medium (5.0 × 104 cells/ml) to the wells of a 96-well plate. The following day, the cells were treated for 1 hr with the reverse transcriptase inhibitors at concentrations of 0.1–10 μg/ml and then infected with the MuLV pseudotype virus. After three days of culture in the presence of the drug, the fluorescent cells were counted. For HIV-1 infection, the N4R5X4/iGFP cells were seeded into 96-well plates at a concentration of 1.0 × 104 cells/ml. The following day, the cells were treated with AZT, ddI, or ddT for 3 hr at concentrations of 0.1–10 μg/ml and then infected with HIV-1 (strain IIIB). The number of fluorescent cells was recorded after two days of culture in the presence of the drug.
Synthesis of the cDNA of other non-retroviral RNA viruses
For the infection with echovirus type 30, 4.0 × 105 RD cells were seeded into six-well plates. The following day, the cells were infected with the virus at an MOI of 0.01 for 1 hr and washed once with PBS. Two days later, a cytopathic effect was observed, and the cell lysates were prepared using cell lysis buffer as described for VSV experiments. A PCR was performed using the ECHO6462-F/ECHO6668-R primer pair targeting the polyprotein (Table S1). We performed 40 cycles of PCR using the following conditions: denaturation at 94°C for 30 sec, annealing at 60°C for 30 sec, and extension at 72°C for 30 sec. The amplified reaction mixture was then separated via electrophoresis on agarose gels that were subsequently stained with ethidium bromide.
For infection with RSV, 4.0 × 105 HEp-2 cells were seeded into six-well plates. The following day, the cells were infected with the virus at an MOI of 0.04 for 1 hr and washed once with PBS. Five days later, a cytopathic effect was observed, and the cell lysates were prepared as described. A PCR was then performed using the RSV3334-F/RSV3518-R primer pair targeting the M gene (Table S1). We performed 40 cycles of PCR using the following conditions: denaturation at 98°C for 10 sec, annealing at 59°C for 20 sec, and extension at 72°C for 20 sec. The amplified reaction mixture was then separated via electrophoresis on agarose gels that were subsequently stained with ethidium bromide. The RSV and echovirus type 30 synthesised via RT-PCR were used as positive controls. We used several different primer pairs to detect echovirus type 30 and RSV; however, only the primer pairs described above yielded positive bands. The DNA samples extracted from these bands were sequenced and confirmed to be of echovirus type 30 and RSV origins.
Author Contributions
A.S., O.I. and H.H. designed the project. A.S., Y.T. and H.H. performed the experiments and wrote the paper. A.S., Y.N. and T.N. performed the experiments. J.B. provided the anti-ORF1p IgY antibody. All authors contributed to the data interpretation.
Supplementary Material
Supplementary information
Acknowledgments
We thank Haig H Kazazian Jr. for kindly providing the LINE-1 expression vectors (pJM101 (L1.2) and pJM105 (L1.2)). The RSV and echovirus type 30 were kindly provided by Hirokazu Kimura. We thank Takahisa Mori for generating the data on the HIV-1 antiviral drug responses, and we thank Atsushi Tanaka, Nobuaki Shimizu, Atsushi Jinno-Oue, Hiroki Isomura, and Tomoyasu Hattori for their helpful comments. We are also grateful to Robin A. Weiss for a critical reading of the manuscript, helpful comments, and encouragement. This work was supported in part by the 21st Century COE Program in addition to Grants-in-Aid from the Japanese Society for the Promotion of Science and the Japan Health Sciences Foundation.
References
- Zhdanov V. M. Integration of viral genomes. Nature 256, 471–473 (1975). [DOI] [PubMed] [Google Scholar]
- Viola M. V., Gann K., Scott C. & Rothfield N. Absence of measles proviral DNA in systemic lupus erythematosus. Nature 275, 667–669 (1978). [DOI] [PubMed] [Google Scholar]
- Klenerman P., Hengartner H. & Zinkernagel R. M. A non-retroviral RNA virus persists in DNA form. Nature 390, 298–301 (1997). [DOI] [PubMed] [Google Scholar]
- Weiss R. A. & Kellam P. Illicit viral DNA. Nature 390, 235–236 (1997). [DOI] [PubMed] [Google Scholar]
- Geuking M. B. et al. Recombination of retrotransposon and exogenous RNA virus results in nonretroviral cDNA integration. Science 323, 393–396 (2009). [DOI] [PubMed] [Google Scholar]
- Horie M. et al. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature 463, 84–87 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. J., Leach R. W. & Bruenn J. Filoviruses are ancient and integrated into mammalian genomes. BMC Evol Biol 10, 193 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier J. L. & Kazazian H. H. Jr Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 135, 23–35 (2008). [DOI] [PubMed] [Google Scholar]
- Luan D. D., Korman M. H., Jakubczak J. L. & Eickbush T. H. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72, 595–605 (1993). [DOI] [PubMed] [Google Scholar]
- Hoshino H., Shimoyama M., Miwa M. & Sugimura T. Detection of lymphocytes producing a human retrovirus associated with adult T-cell leukemia by syncytia induction assay. Proc Natl Acad Sci U S A 80, 7337–7341 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapham P., Nagy K. & Weiss R. A. Pseudotypes of human T-cell leukemia virus types 1 and 2: neutralization by patients' sera. Proc Natl Acad Sci U S A 81, 2886–2889 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojdl D. F. et al. Exploiting tumor-specific defects in the interferon pathway with a previously unknown oncolytic virus. Nat Med 6, 821–825 (2000). [DOI] [PubMed] [Google Scholar]
- Moran J. V. et al. High frequency retrotransposition in cultured mammalian cells. Cell 87, 917–927 (1996). [DOI] [PubMed] [Google Scholar]
- Simpson R. W. & Iinuma M. Recovery of infectious proviral DNA from mammalian cells infected with respiratory syncytial virus. Proc Natl Acad Sci U S A 72, 3230–3234 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciurea A. et al. Persistence of lymphocytic choriomeningitis virus at very low levels in immune mice. Proc Natl Acad Sci U S A 96, 11964–11969 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields B. N., Knipe D. M. & Howley P. M. Fields virology, (Wolters Kluwer/Lippincott Williams & Wilkins Health, Philadelphia, 2013). [Google Scholar]
- Gilbert N., Lutz S., Morrish T. A. & Moran J. V. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol Cell Biol 25, 7780–7795 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volloch V. Z., Schweitzer B. & Rits S. Transcription of the 5′-terminal cap nucleotide by RNA-dependent DNA polymerase: possible involvement in retroviral reverse transcription. DNA Cell Biol 14, 991–996 (1995). [DOI] [PubMed] [Google Scholar]
- Buzdin A. et al. A new family of chimeric retrotranscripts formed by a full copy of U6 small nuclear RNA fused to the 3′ terminus of l1. Genomics 80, 402–406 (2002). [DOI] [PubMed] [Google Scholar]
- Garcia-Perez J. L., Doucet A. J., Bucheton A., Moran J. V. & Gilbert N. Distinct mechanisms for trans-mediated mobilization of cellular RNAs by the LINE-1 reverse transcriptase. Genome Res 17, 602–611 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai H., Savitsky D., Tamura T. & Taniguchi T. Regulation of the cytosolic DNA-sensing system in innate immunity: a current view. Curr Opin Immunol 21, 17–22 (2009). [DOI] [PubMed] [Google Scholar]
- Takaoka A. et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448, 501–505 (2007). [DOI] [PubMed] [Google Scholar]
- Muruve D. A. et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature 452, 103–107 (2008). [DOI] [PubMed] [Google Scholar]
- Hemmi H. et al. A Toll-like receptor recognizes bacterial DNA. Nature 408, 740–745 (2000). [DOI] [PubMed] [Google Scholar]
- Stetson D. B., Ko J. S., Heidmann T. & Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134, 587–598 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soda Y. et al. Establishment of a new system for determination of coreceptor usages of HIV based on the human glioma NP-2 cell line. Biochem Biophys Res Commun 258, 313–321 (1999). [DOI] [PubMed] [Google Scholar]
- Fischinger P. J., Peebles P. T., Nomura S. & Haapala D. K. Isolation of RD-114-like oncornavirus from a cat cell line. J Virol 11, 978–985 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C. A., Manthey A. E. & Gumport R. I. Using T4 RNA ligase with DNA substrates. Methods Enzymol 100, 38–52 (1983). [DOI] [PubMed] [Google Scholar]
- An W. et al. Characterization of a synthetic human LINE-1 retrotransposon ORFeus-Hs. Mob DNA 2, 2 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi T., Shishido T., Murata K. & Hanafusa H. v-Crk activates the phosphoinositide 3-kinase/AKT pathway in transformation. Proc Natl Acad Sci U S A 97, 7290–7295 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information