ABSTRACT
Proteins associated with primary cilia and basal bodies mediate numerous signaling pathways, but little is known about their role in Notch signaling. Here, we report that loss of the Bardet-Biedl syndrome proteins BBS1 or BBS4 produces increased Notch-directed transcription in a zebrafish reporter line and in human cell lines. Pathway overactivation is accompanied by reduced localization of Notch receptor at both the plasma membrane and the cilium. In Drosophila mutants, overactivation of Notch can result from receptor accumulation in endosomes, and recent studies implicate ciliary proteins in endosomal trafficking, suggesting a possible mechanism by which overactivation occurs in BBS mutants. Consistent with this, we observe genetic interaction of BBS1 and BBS4 with the endosomal sorting complexes required for transport (ESCRT) gene TSG101 and accumulation of receptor in late endosomes, reduced endosomal recycling and reduced receptor degradation in lysosomes. We observe similar defects with disruption of BBS3. Loss of another basal body protein, ALMS1, also enhances Notch activation and the accumulation of receptor in late endosomes, but does not disrupt recycling. These findings suggest a role for these proteins in the regulation of Notch through endosomal trafficking of the receptor.
KEY WORDS: Alstrom Syndrome, Bardet-Biedl Syndrome, Notch, Basal body, Cilia, Endosomal sorting
INTRODUCTION
Proteins associated with primary cilia and their anchoring basal bodies play a variety of intracellular roles, including trafficking of ciliary cargo, maintenance of microtubule dynamics and control of cell cycle (Cardenas-Rodriguez and Badano, 2009). Additionally, an increasing body of evidence implicates these proteins in the intracellular regulation of developmental signaling pathways, such as the Shh and Wnt pathways (Berbari et al., 2009; Cardenas-Rodriguez and Badano, 2009; Gerdes et al., 2009; Goetz and Anderson, 2010). Experimental insights into these pathways have revealed varying mechanisms of regulation, including the localization of signaling components to the cilium, as is the case with Shh (Rohatgi et al., 2007), and regulation of downstream effectors, such as the proteasomal degradation of β-catenin in Wnt signaling (Gerdes et al., 2007). Recently, Notch signaling was identified as another major developmental pathway that is potentially associated with cilia (Ezratty et al., 2011). Little is known, however, about the mechanistic link underlying this regulation.
Notch signaling is mediated by transmembrane receptors at the cell surface, which are activated by Delta and Serrate/Jagged (DSL) ligands on the surface of neighboring cells. Upon activation, the Notch receptor is cleaved to produce the Notch intracellular domain (NICD), which translocates to the nucleus to direct the transcription of downstream targets, such as members of the HES gene family (Guruharsha et al., 2012). Prior to ligand binding, the Notch receptor undergoes several processing events, including endosomal sorting, which results either in trafficking through late endosomes or multi-vesicular bodies (MVBs) to lysosomes for degradation or recycling back to the plasma membrane for ligand binding and activation (Le Borgne, 2006). Altering this processing, either by disruption of recycling or disruption of MVB sorting, deregulates receptor signaling. For example, disruption of sorting in Drosophila mutants for the endosomal sorting complex required for transport (ESCRT) proteins results in overactivation of the Notch pathway as a result of the accumulation of receptor in late endosomes from where it can actively signal in a ligand-independent manner (Moberg et al., 2005; Thompson et al., 2005; Vaccari and Bilder, 2005; Vaccari et al., 2008). Similarly, the Notch antagonist Numb impedes pathway activation by inhibiting recycling of the receptor to the plasma membrane (Nilsson et al., 2008; Nilsson et al., 2011; Smith et al., 2004) and sorting of the receptor through late endosomes for lysosomal degradation (McGill et al., 2009).
Recent evidence links basal body and ciliary proteins to endosomal recycling. The ciliary pocket is a site of clathrin-dependent endocytosis, indicating a possible spatial relationship (Clement et al., 2013). The core complex of Bardet-Biedl syndrome (BBS) proteins, known as the BBSome, is also functionally associated with the recycling endosome factor, Rab11 (Westlake et al., 2011). Further, the basal body protein ALMS1 interacts directly with endosome components (Collin et al., 2012). Similarly, ARL13B is required for endocytic recycling (Barral et al., 2012), pointing towards the importance of ciliary proteins in mediating the recycling and trafficking of endosomal cargo. Given that endosomal sorting and recycling is integral to proper Notch transduction, we hypothesized that disruption of this process by the ablation of basal body proteins would directly perturb signaling. Here, we report that the loss of BBS proteins results in upregulation of Notch signaling both in a transgenic zebrafish Notch reporter line and in cultured human cells. We provide evidence to suggest that loss of BBS proteins results in aberrant trafficking of the receptor through late endosomes to lysosomes and impaired recycling of the Notch receptor. We also present evidence that loss of another basal body protein, ALMS1, results in similar defects in Notch receptor trafficking through late endosomes and subsequent overactivation of the pathway, though it does not inhibit ciliary localization of the receptor. These findings support a model whereby basal body proteins mediate the endosomal recycling and sorting of Notch receptor, thereby mediating proper signal transduction.
RESULTS
Loss of BBS proteins results in upregulation of Notch signaling
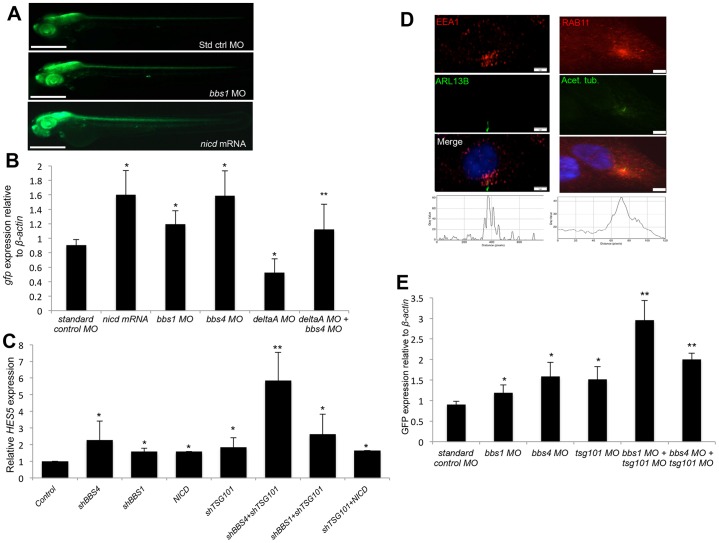
To investigate the role of BBS proteins in Notch signaling, we took advantage of a transgenic zebrafish Notch reporter line (Tp1bglob:eGFP) expressing eGFP under the control of an element containing NICD-responsive RBP-Jκ-binding regions (Parsons et al., 2009). We injected two-cell stage transgenic embryos with morpholinos that suppress translation of either of two BBS proteins, Bbs1 or Bbs4, as verified by western blotting (Zaghloul et al., 2010; supplementary material Table S1; Fig. S1A). We assessed the expression of the GFP reporter by fluorescence microscopy and by quantitative (q)RT-PCR. By 48 hours post-fertilization (hpf), a visible enhancement of fluorescence in morphant embryos could be observed in comparison with those injected with a standard control morpholino (Fig. 1A). This correlated with a 1.2-fold (bbs1) or 1.6-fold (bbs4) increase in relative GFP mRNA expression in 24 hpf embryos (Fig. 1B), the latter being comparable to the increase observed with injection of mRNA encoding the NICD fragment of zebrafish notch3 (nicd; supplementary material Table S1; Fig. 1A,B). To validate the specificity of this effect, we co-injected the corresponding mRNA for each gene. For each of the bbs morpholinos tested, GFP expression was either fully or partially rescued by addition of the corresponding mRNA (supplementary material Fig. S1B). To better characterize the observed Notch upregulation, we asked whether it is dependent on ligand binding. Upon injection of a morpholino against deltaA (Latimer et al., 2002; supplementary material Table S1), we observed a 43% decrease in GFP expression compared with controls (Fig. 1B). Co-injection of bbs4 morpholino, however, overcame this effect to increase GFP expression more than twofold (Fig. 1B), indicating ligand-independent activation of the pathway.
Fig. 1.
Loss of BBS proteins and genetic interaction with endosomal components enhances Notch pathway activation. (A) GFP fluorescence in representative Tp1blgob:eGFP transgenic embryos at 48 hours post fertilization (hpf) in bbs1 morphant (MO) embryos and nicd-mRNA-injected embryos. Scale bar: 0.5 mm (B) qRT-PCR quantification of GFP mRNA levels relative to β-actin in 24 hpf Tp1blgob:eGFP embryos. *P≤0.01 compared with standard control morpholino; **P≤0.005 compared with deltaA morpholino (Student's t-test). (C) qRT-PCR analysis of relative HES5 expression in HEK293 cells expressed as the fold change relative to control cells. Data represent the mean±s.d. (seven separate experiments). *P≤0.01 compared with control; **P≤0.01 compared with either short hairpin alone (Student's t-test). (D) Double immunofluorescent labeling of either early endosomes (EEA1) or recycling endosomes (RAB11) and primary cilia (ARL13B or acetylated tubulin) in hTERT-RPE1 cells. Histograms represent the intensity of red fluorescence across the image. Scale bars: 5 µm. (E) qRT-PCR analysis showing relative GFP expression in Tp1bglob:eGFP zebrafish embryos at 24 hpf. Data show the mean±s.d. *P≤0.005 compared with standard control morpholino; **P≤0.005 compared with morpholinos alone (Student's t-test).
To explore the possibility of perturbed Notch signaling in ciliated human cells, we investigated the transcription of Notch targets in the ciliated human cell line HEK293, which endogenously expresses Notch pathway components (Gerdes et al., 2007; Hansson et al., 2010). We obtained shRNA constructs that, after 48 hours of transient transfection, specifically decrease mRNA and protein expression of BBS1 or BBS4 (supplementary material Fig. S1C–E). Upon loss of BBS1 or BBS4 we observed a 1.6- and 2.3-fold increase in the expression of HES5, respectively (Fig. 1C), compared with cells transfected with no DNA, a control that was not significantly different in Notch target expression compared with cells transfected with empty vector (supplementary material Fig. S2A). The observed increase in HES5 expression was comparable to that observed with transfection of a plasmid construct expressing the NOTCH1 NICD fragment (Fig. 1C) and is consistent with our observations in zebrafish embryos. This corresponded to an increase in activation of the Notch receptor, detected by increased nuclear localization of the C-terminal portion of the Notch receptor – from which the activated NICD is produced – and by western blot analysis of cleaved NICD (supplementary material Fig. S2B). To further confirm the observed Notch misregulation, we assayed for expression of another transcriptional target, HES1. Similar to HES5, HES1 expression was significantly increased in shBBS4-treated cells (supplementary material Fig. S2C). Additionally, to preclude the possibility of a cell-type-specific defect, we replicated these observations in another ciliated cell line, hTERT-RPE1 cells (supplementary material Fig. S2D).
BBS genes interact genetically with the ESCRT gene TSG101 to mediate Notch signaling
The ligand-independent upregulation of Notch signaling with suppression of Bbs4 is consistent with the possibility that the loss of BBS proteins results in endosomal trafficking defects of Notch receptor and signaling from endosomes. To test this hypothesis, we first asked whether there might be a spatial relationship between BBS proteins and endosomal trafficking, by investigating the intracellular localization of endosomal activity using double immunofluorescent staining of either early endosomes (anti-EEA1) or recycling endosomes (anti-RAB11), along with markers of primary cilia (anti-ARL13B or anti-acetylated-tubulin) in hTERT-RPE1 cells (Fig. 1D; supplementary material Table S1). Consistent with previous reports, we observed an enrichment of both early and recycling endosomes near the base of the primary cilium, as indicated by analysis of fluorescence intensity (Fig. 1D, histograms; Knödler et al., 2010). To further examine this relationship, we investigated the possibility of a genetic interaction between BBS1 or BBS4 and the ESCRT gene TSG101 (supplementary material Table S1). Mutations in the Drosophila homolog of TSG101 are characterized by Notch overactivation due to the accumulation and activation of the Notch receptor in late endosomes (Moberg et al., 2005; Vaccari et al., 2008). Concordantly, transfection of HEK293 or hTERT-RPE1 cells with shTSG101 alone resulted in a significant increase in HES5 expression (Fig. 1C; supplementary material Fig. S2D) as well as HES1 (supplementary material Fig. S2C,D). Importantly, however, co-transfection of either shBBS1 or shBBS4 produced a threefold or sixfold increase, respectively, in HES5 expression and a threefold increase in HES1 expression, indicating a greater-than-additive enhancement of Notch target transcription (Fig. 1C; supplementary material Fig. S2C,D). To determine whether this interaction is specific to BBS genes, we co-transfected HEK293 cells with shTSG101 and NICD. Although either construct alone increased the transcription of HES5 1.8-fold and 1.6-fold, respectively, the transfection of both did not produce a significantly greater fold change difference in expression by comparison (Fig. 1C), supporting the idea of a genetic interaction between TSG101 and BBS1 or BBS4. Likewise, expression of HES1 was not more significantly enhanced (supplementary material Fig. S2C).
To verify this interaction in vivo, we designed a splice-blocking morpholino targeting the exon4-intron4 splice junction of the tsg101 transcript. Morpholino efficacy was validated by RT-PCR using primers flanking the included intron and by western blotting (supplementary material Fig. S2E,F). As expected, injection of the tsg101 morpholino alone into Tp1bglob:eGFP embryos resulted in increased GFP expression in 24 hpf embryos (Fig. 1E). Combined injection, however, of tsg101 morpholino with either bbs1 or bbs4 morpholino increased relative GFP expression in a greater-than-additive manner (Fig. 1E), corroborating our observations in cultured cells.
Loss of BBS proteins results in a shift in Notch receptor localization away from the plasma membrane
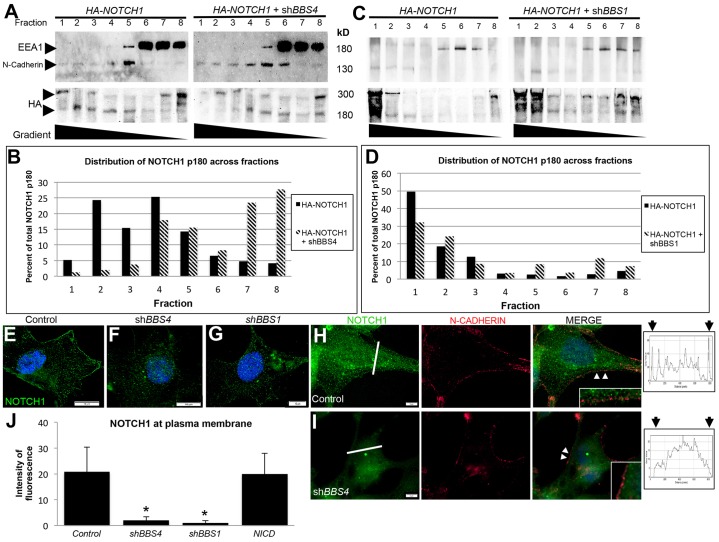
A genetic interaction with endosomal sorting components that leads to enhanced Notch activation compounded with a spatial relationship between the base of the cilium and endosomes suggests the possibility that endosomal trafficking of the Notch receptor might be implicated in the misactivation of the pathway that is observed with loss of BBS1 or BBS4. If recycling of the receptor is compromised, we would expect to see loss of receptor localization at the plasma membrane. Furthermore, if sorting in MVBs is disrupted, we would expect to see accumulation in endosomes. To determine whether this was the case, we first performed a step-wise (50%, 30% and 10%) sucrose gradient separation of shBBS4-transfected cell homogenates. Using N-cadherin as a marker of the plasma membrane (supplementary material Table S1) and EEA1 as a marker of endosomes, we observed localization of N-cadherin to the more dense fractions (fractions 1–5), whereas EEA1 localized to the least dense fractions (fractions 5–8) in cells transfected with and without shBBS4 (Fig. 2A). To assess the localization of the Notch receptor, we transfected cells with an N-terminal HA-tagged full-length NOTCH1 construct (Hansson et al., 2010). Upon transfection into either HEK293 or hTERT-RPE1 cells, HA could be detected as two bands: the full-length p300 protein and the p180 post-site1-cleavage N-terminal portion of the heterodimeric receptor, which is found at the cell membrane and recycled through endosomes (supplementary material Fig. S3A; McGill et al., 2009). As expected, HA antibody detection of the p180 protein in control cells was more robust in membrane fractions, with some localization in endosomal fractions in both cell types (Fig. 2A,B; supplementary material Fig. S3B). Cells co-transfected with shBBS4, however, exhibited a marked increase in the amount of HA localization to the EEA1-labeled fractions (Fig. 2A,B). Quantification of the amount of p180 fragment in each fraction taken as a proportion of the total across all fractions confirmed this shift (Fig. 2B), which was also observed in hTERT-RPE1 cells (supplementary material Fig. S3B). Likewise, shBBS1 treatment of cells produced a decrease in the proportion of p180 in the plasma membrane fractions and an increase in the endosomal fractions relative to that of control cells (Fig. 2C,D). No shift could be observed for the C-terminal p120 portion of the receptor heterodimer, likely as a result of the detection of the activated NICD fragment in addition to detection of the pre-activated receptor (supplementary material Fig. S3C). The full-length receptor protein (p300) also did not exhibit a consistent change in localization with loss of BBS4 or BBS1 (supplementary material Fig. S3D,E), suggesting that the shift is specific to the heterodimeric form of the receptor.
Fig. 2.
Notch receptor localization shifts away from plasma membrane in BBS-depleted cells. (A) Sucrose-gradient fractionation of HEK293 cells transfected with HA-NOTCH1 alone or HA-NOTCH1+shBBS4 probed for HA, N-cadherin (plasma membrane marker), and EEA1 (endosome marker). (B) Quantification of the p180 band in A displayed as the proportion in each fraction relative to the total across all fractions. (C) Sucrose-gradient fractionation of HEK293 cells transfected with HA-NOTCH1 alone or HA-NOTCH1+shBBS1 probed for HA, N-cadherin and EEA1. (D) Quantification of p180 from C displayed as the proportion in each fraction relative to the total across all fractions. (E–G) hTERT-RPE1 cells immunostained with antibody against endogenous NOTCH1 (green). Scale bars: 10 µm. (H,I) hTERT-RPE1 cells coimmunostained for endogenous NOTCH1 (green) and plasma membrane marker N-cadherin (red) showing colocalization (arrowheads, inset). Histograms represent the intensity of green fluorescence across the white line. Arrows above histograms represent regions of plasma membrane fluorescence. Scale bars: 5 µm (J) Quantification of all intensity values within individual membrane peaks. Data represent the mean±s.d. (≥50 cells per treatment, two cross-sections per cell). *P≤0.001 compared with control (Student's t-test).
To better characterize the intracellular localization of the receptor, we visualized endogenous Notch1 receptor in hTERT-RPE1 cells by using an antibody targeted against it. Consistent with previous reports and with our sucrose gradient data, endogenous NOTCH1 localized strongly to the plasma membrane of control cells (Fig. 2E). Cells with reduced BBS4 or BBS1 expression, however, exhibited a visible reduction in the intensity of membrane localization of the receptor (Fig. 2F,G). Co-labeling of cells with antibodies against NOTCH1 and the plasma membrane marker N-cadherin confirmed these observations (Fig. 2H,I). To quantify the extent of Notch receptor localization to the plasma membrane, we analyzed the intensity of NOTCH1 fluorescence in cross-sections spanning cells from membrane to membrane (Fig. 2H,I, white lines). The fluorescence intensity at the membrane was determined by the values at the edges of the histograms, representing the membrane edges of each cross-section (Fig. 2H,I, arrowheads). We then plotted the average of all intensity values under individual membrane peaks (Fig. 2J). Loss of BBS4 resulted in a 94% decrease in NOTCH1 average fluorescence intensity at the plasma membrane and loss of BBS1 decreased membrane expression by 74% (Fig. 2J). This change was not observed upon NICD overexpression (Fig. 2J), indicating that it is not secondary to pathway activation.
Notch receptor accumulates in late endosomes in BBS4- or BBS1-depleted cells
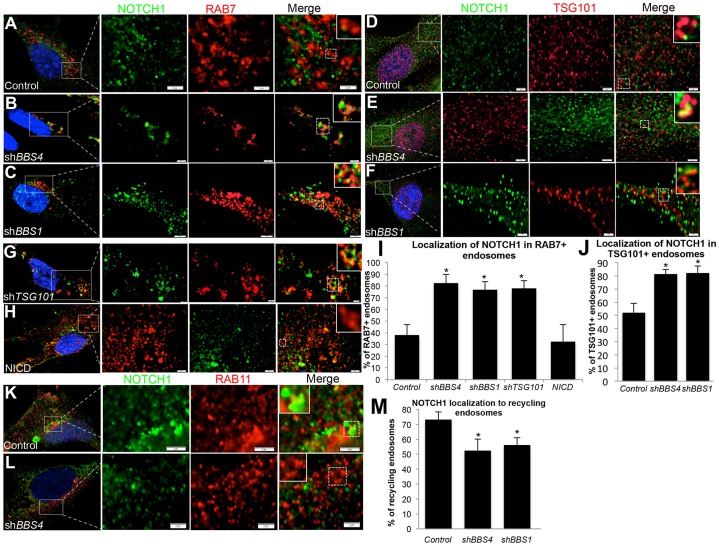
The shift in receptor localization from plasma membrane to endosomal fractions suggests that the activation of Notch resulting from the loss of BBS proteins might be caused by the accumulation of Notch receptor in endosomes, similar to that observed in Drosophila ESCRT mutants. Given that the accumulation of Notch receptor in endosomal mutants occurs in late endosomes and MVBs, we examined receptor localization by co-labeling cells with markers of both of these organelles (supplementary material Table S1) and quantified the proportion of marker-labeled vesicles that also contained NOTCH1. Compared with control cells, where Notch receptor could be detected in 38% of RAB7-labeled late endosomes and 51% of TSG101-labeled MVBs, we observed colocalization in 83% of late endosomes and 81% of MVBs in shBBS4-treated cells (Fig. 3A,B,D,E,I,J). Reduction in BBS1 expression also resulted in the accumulation of Notch receptor in both RAB7- (77%) and TSG101-labeled (82%) endosomes (Fig. 3C,F,I,J). This is consistent with the increased Notch receptor localization in RAB7-labeled late endosomes of shTSG101-treated cells (78%; Fig. 3G,I). To assess whether perturbation in receptor localization could be a secondary result of pathway upregulation, we evaluated NOTCH1 localization in cells transfected with NICD. We did not observe aberrant accumulation of NOTCH1 in RAB7-labeled endosomes (32%; Fig. 3H,I).
Fig. 3.
NOTCH1 accumulates in late endosomes but is decreased in recycling endosomes. (A–H) hTERT-RPE1 cells immunostained with antibodies against endogenous NOTCH1 (green) and markers of late endosomes (RAB7, red) or multi-vesicular bodies (TSG101, red). Scale bars: 2 µm. (I,J) Quantification of the proportion of either RAB7-positive or extra-nuclear TSG101-positive endosomes that are also positive for NOTCH1 staining shown as a percentage of total RAB7- or TSG101-positive endosomes. Data show the mean±s.d. (K,L) hTERT-RPE1 cells coimmunostained for endogenous NOTCH1 (green) and recycling endosomes (RAB11, red). All immunofluorescence images represent a single plane of focus at the denoted endosomes and are at 100× magnification, with enlarged regions outlined by white boxes and enlarged individual vesicles outlined by dashed boxes and shown as insets. Scale bars: 2 µm (M) Quantification of the proportion of RAB11-positive endosomes that are also positive for NOTCH1 shown as a percentage of total RAB11-positive endosomes. Data show the mean±s.d. *P≤0.001 compared with control (Student's t-test).
In light of the loss of receptor localization to the plasma membrane, we hypothesized that the defective trafficking of the receptor might be related to a defect in recycling. To assess this possibility, we co-labeled control and short-hairpin-treated cells with antibodies against NOTCH1 and RAB11 (Fig. 3K,L). Upon quantification, we observed a significant decrease in the proportion of RAB11-positive endosomes that also contained NOTCH1 in shBBS4- or shBBS1-treated cells (28% and 23% decrease, respectively; Fig. 3M).
Lysosomal degradation of the Notch receptor is impaired with loss of BBS proteins
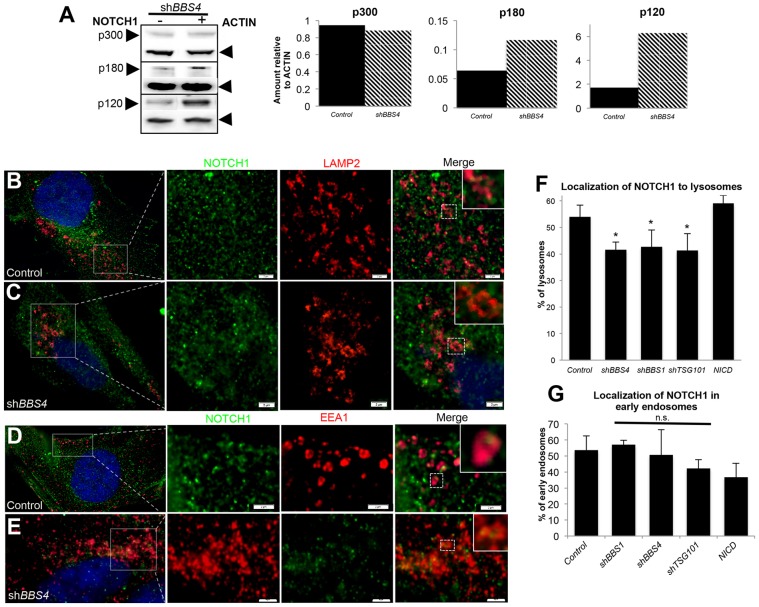
If increased Notch signaling is the result of endosomal accumulation, we would expect a decrease in receptor degradation in lysosomes (Chastagner et al., 2008). To determine whether this was the case, we first assessed by western blotting whether endogenous NOTCH1 in HEK293 cells was degraded properly. Using antibodies that detect either the N-terminal p180 region of the heterodimeric receptor or the p120 C-terminal fragment, which are targeted for degradation in lysosomes (Tien et al., 2009), we assessed the amount of either portion as well as the full-length (p300) unprocessed receptor protein (Fig. 4A). Upon transfection with shBBS4, we observed a threefold increase in the amount of either the p120 or the p180 protein relative to β-actin, an increase that was neither due to increased production of the full-length receptor protein, as demonstrated by a lack of change in the p300 band (Fig. 4A), nor due to increased transcription of the gene encoding it, as detected by qRT-PCR (supplementary material Fig. S3F). Similar accumulations of the p180 and p120 fragments were observed in hTERT-RPE1 cells (supplementary material Fig. S3G).
Fig. 4.
Loss of NOTCH1 degradation and lysosomal localization in BBS-depleted cells. (A) Detection of full-length (p300) and post-site1-cleaved N-terminal (p180) NOTCH1 by HA detection in HA–NOTCH1-transfected cells, and detection of C-terminal domains (p120) of endogenous NOTCH1 in whole-cell lysates of HEK293 cells transfected with or without shBBS4. Graphs show the quantification of band intensity for each, measured using ImageJ software, relative to β-actin. (B,C) Double immunostaining of hTERT-RPE1 cells for NOTCH1 (green) and the lysosomal marker, LAMP2 (red) (D,E) Double immunostaining of hTERT-RPE1 cells for NOTCH1 (green) and the early endosomal marker, EEA1 (red). Immunofluorescence images are at 100× magnification, with enlarged regions denoted by white boxes and enlarged individual vesicles denoted by dashed boxes and shown as insets. Scale bars: 2 µm. (F) Quantification of the percentage of LAMP2-positive lysosomes that also contain NOTCH1. *P≤0.001 compared with control (Student's t-test). (G) Quantification of EEA1-positive early endosomes containing NOTCH1. n.s., non-significant. Data shown the mean±s.d.
To determine whether this reduction in degradation is a result of impaired trafficking to lysosomes, we co-labeled transfected hTERT-RPE1 cells with antibodies against the lysosome marker LAMP2 and endogenous NOTCH1. In control cells, we found that 54% of LAMP2-positive lysosomes also contained NOTCH1. This colocalization decreased to 41% and 42% in cells with suppressed BBS4 or BBS1 expression, respectively, decreases similar to those observed in shTSG101-treated cells (41%; Fig. 4B,C,F; supplementary material Fig. S3H). This change was not seen in NICD-transfected cells (Fig. 4F). To verify that this defect is not due to a general loss of endocytosis, we labeled early endosomes with an antibody against EEA-1. The amount of localization of NOTCH1 in EEA1-positive endosomes was not significantly different in BBS1- or BBS4-depleted cells compared with control cells (Fig. 4D,E,G; supplementary material Fig. S3I).
Disruption of BBS3 or ALMS1 perturbs endosomal trafficking of the Notch receptor
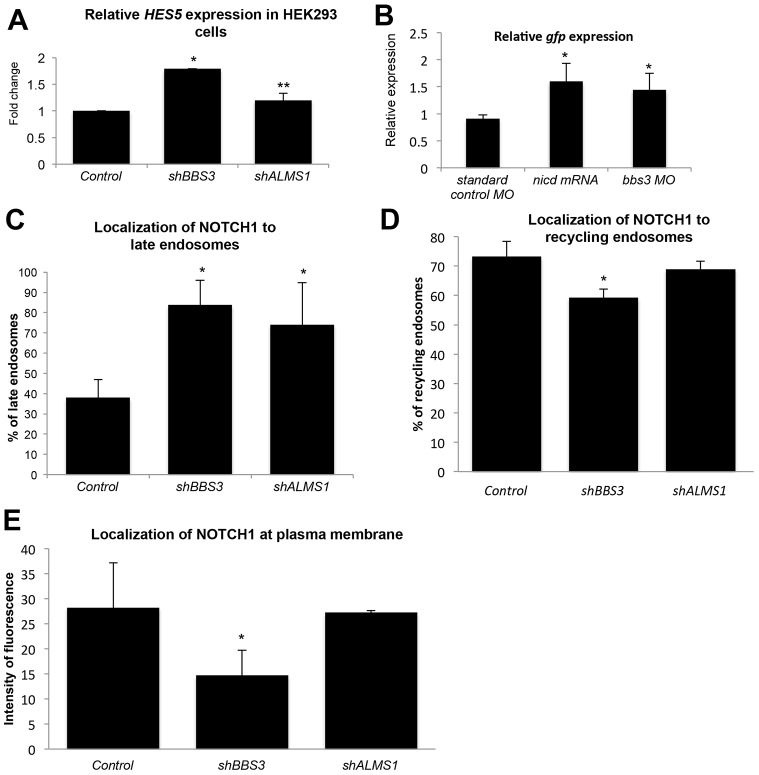
To determine whether our observations extended to other basal body proteins, we investigated the role of BBS3, a BBS protein that is not a core member of the BBSome but that associates with it (Jin et al., 2010), and ALMS1, a basal body ciliopathy protein that is not related to the BBSome but that is important for regulation of developmental signaling (Hearn et al., 2005; Jagger et al., 2011). Suppression of either protein by short hairpin treatment in HEK293 cells increased HES5 expression, as detected by qRT-PCR (1.8-fold and 1.2-fold, respectively; Fig. 5A). Similarly, injection of a morpholino designed to block the translation of bbs3 (Zaghloul et al., 2010) in Tp1bglob:eGFP transgenic zebrafish embryos resulted in a significant increase in GFP expression (Fig. 5B). To investigate possible endosomal mechanisms for this upregulation, we examined the localization of the NOTCH1 receptor in BBS3- and ALMS1-depleted cells by immunofluorescence and quantified the proportion of RAB7-labeled endosomes that colocalized with NOTCH1. Similar to BBS1 and BBS4, loss of either BBS3 or ALMS1 resulted in a significant increase in RAB7 colocalization with NOTCH1 (Fig. 5C). The effect on recycling and plasma membrane localization, however, was discrepant between the two proteins. Loss of BBS3 resulted in a modest but significant decrease in the localization of NOTCH1 to RAB11-positive recycling endosomes and a 50% decrease in the intensity of plasma membrane localization (Fig. 5D,E). Depletion of ALMS1, however, did not significantly alter either (Fig. 5D,E), suggesting that it is neither necessary for recycling of the Notch receptor nor for subsequent localization to the plasma membrane.
Fig. 5.
Enhancement of Notch signaling and accumulation of receptor in late endosomes with loss of BBS3 or ALMS1. (A) qRT-PCR analysis of relative HES5 expression in HEK293 cells shown as the fold change relative to control cells. *P≤0.001, **P≤0.05 compared with control (Student's t-test). (B) qRT-PCR analysis of expression of GFP mRNA relative to that of β-actin in Tp1bglob:eGFP zebrafish embryos at 24 hpf. *P≤0.01 compared with standard control morpholino (MO) (Student's t-test). (C) Quantification of double immunostaining experiments for RAB7 and NOTCH1 in hTERT-RPE1 cells. The graph represents the percentage of RAB7-positive endosomes that also contain NOTCH1. *P≤0.001 compared with control (Student's t-test). (D) Quantification of double immunostaining experiments for RAB11 and NOTCH1 in hTERT-RPE1 cells. The graph represents the percentage of RAB11-positive endosomes that also contain NOTCH1. *P≤0.005 compared with control (Student's t-test). (E) Quantification of plasma membrane staining intensity as measured by intensity of fluorescence. The graph represents the average across ≥5 cells of the average of intensity values for each membrane peak, measured by ImageJ analysis. *P≤0.001 compared with control (Student's t-test). All data show the mean±s.d.
Loss of BBS1 or BBS4 perturbs ciliary localization of Notch receptor
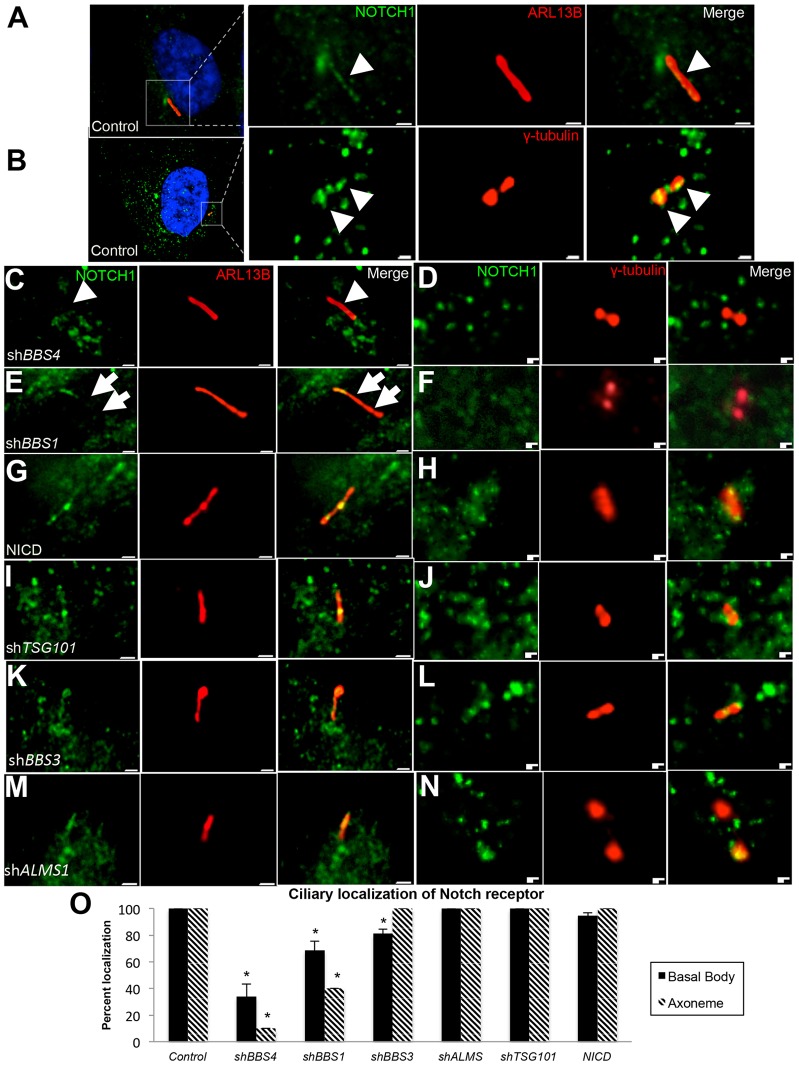
In addition to plasma membrane localization, at least one Notch receptor, NOTCH3, localizes to cilia in mouse skin epithelium (Ezratty et al., 2011), suggesting that trafficking to and away from the cilium might be related to proper receptor localization and function. Given that the BBSome mediates trafficking of cargo to the cilium (Jin et al., 2010), and that our data suggest the importance of BBS4 and BBS1 in regulating intracellular trafficking of the receptor through endosomes, loss of these proteins might also result in loss of ciliary localization of the Notch receptor. To examine this possibility, we first assessed whether NOTCH1 localizes to primary cilia by co-labeling control hTERT-RPE1 cells with antibodies against the NOTCH1 receptor and either an axonemal (anti-ARL13B) or a basal body marker (anti-γ-tubulin; supplementary material Table S1). We found that, in addition to its plasma membrane and intracellular localization, the receptor consistently localized to axonemes and to basal bodies in 100% of control cells (Fig. 6A,B,O). When BBS4 expression was suppressed, however, axonemal localization was diminished in 90% of cells, and less than half (42%) of cells exhibited basal body localization (Fig. 6C,D,O). Loss of BBS1 also produced a significant (32%) decrease in the basal body localization of Notch receptor, and axonemal expression could not be detected beyond the most proximal region in 60% of cells (Fig. 6E,F,O). Depletion of BBS3 had discrepant effects on Notch receptor localization. Of shBBS3-treated cells, 81% exhibited basal body localization of the receptor, a significant reduction, but no change in axonemal localization could be observed (Fig. 6K,L,O). Interestingly, and in contrast to BBS proteins, loss of ALMS1 did not alter either basal body or axonemal localization of Notch receptor (Fig. 6M–O). Neither NICD expression nor shTSG101 treatment significantly perturbed the ciliary localization of the receptor (Fig. 6G–J,O), indicating that pathway upregulation or defective endosomal sorting alone might not be sufficient for disruption of ciliary localization. Loss of ciliary localization in BBS1- or BBS4-deficient cells might indicate that the cilium itself is important for proper localization of NOTCH1. To determine whether this is the case, we assessed NOTCH1 localization in ciliated hTERT-RPE1 cells or subconfluent cells that are not ciliated (Pitaval et al., 2010). The plasma membrane localization pattern in both control and shBBS4 was identical under both conditions (supplementary material Fig. S4A), indicating that Notch receptor localization is not dependent on the cilium and that the mislocalization in shBBS4-treated cells is likely due to loss of the protein.
Fig. 6.
Ciliary localization of the NOTCH1 receptor is disrupted with loss of BBS proteins. (A) Control hTERT-RPE1 cells immunostained for NOTCH1 (green, arrowheads) and ARL13B (red). (B) Control hTERT-RPE1 cells immunostained for NOTCH1 (green) and γ-tubulin (red). Enlarged regions are outlined by white boxes. Scale bars: 0.5 µm. (C–N) hTERT-RPE1 cells transfected with the indicated constructs and coimmunostained for NOTCH1 (green) and either ARL13B (red; C,E,G,I,K,M) or γ-tubulin (D,F,H,J,L,N). Arrowheads, regions of co-localization. Arrows, lack of axonemal localization. Scale bars: 0.5 µm. (O) Quantification of NOTCH1 localization at basal bodies and axonemes. The graph represents the average proportion of basal bodies or axonemes for which NOTCH1 colocalization could be observed. Axonemal localization reflects localization throughout the axoneme. Data show the mean±s.d. *P≤0.001 compared with control (Chi-squared test).
Markers of Notch-inhibited cell types are suppressed in bbs4 morphant embryos
The increased Notch signaling observed in BBS-deficient zebrafish embryos suggests that development of tissues and cell types associated with this pathway might also be perturbed. To investigate this possibility, we examined markers of two tissues associated with BBS phenotypes, the expression of which is inhibited by Notch signaling. We first assayed expression of neurogenin1, a marker of neurogenesis that is repressed by active Notch signaling (Cornell and Eisen, 2002; Takke et al., 1999). Consistent with our observed activation of Notch, we observed a reduction in the domain of ngn1 expression by whole-mount in situ hybridization in 24 hpf embryos injected with bbs4 morpholino (supplementary material Fig. S4B). Similarly, tsg101 morpholino suppressed ngn1 expression at this stage, suggesting that increased Notch signaling through endosomal defects produces similar effects.
BBS is also often characterized by renal defects similar to those seen in other ciliopathies (Zaghloul and Katsanis, 2009). Evidence suggests that ciliopathic renal defects in zebrafish might be attributable to loss of pronephric multi-ciliated cells (MCCs) due to increased Notch signaling (Liu et al., 2007). To determine whether this might be true in bbs4-deficient embryos, we examined the expression of a marker specific to pronephric MCCs, shippo1 (Liu et al., 2007). At 24 hpf, we observed a dramatic decrease in shippo1 expression in the pronephros in both bbs4 morphants and tsg101 morpholino-injected embryos (supplementary material Fig. S4C), indicating that these animals are deficient in these Notch-inhibited cells.
DISCUSSION
In this study, we provide evidence that BBS1, BBS3, BBS4 and ALMS1 are necessary for the regulation of Notch signaling. Loss of expression of these genes in zebrafish or cultured cells resulted in enhanced activation of the pathway. Our assessment of Notch receptor localization revealed that it accumulated in late endosomes and is likely not properly degraded when BBSome proteins are depleted, leading to increased receptor activation and target transcription. In addition, loss of BBSome proteins reduced the localization of Notch receptor to the plasma membrane and cilium, and reduced its localization to recycling endosomes, implicating the BBSome in proper recycling of Notch. Interestingly, although loss of ALMS1 also significantly increased receptor accumulation in late endosomes, it altered neither membrane nor ciliary localization and did not reduce recycling of the receptor, suggesting a trafficking defect that is independent of recycling.
BBS protein regulation of endosomal recycling and sorting
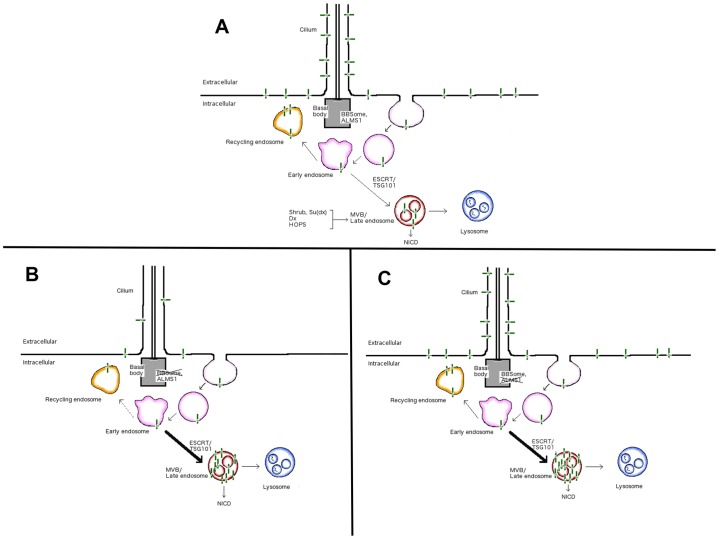
Our findings offer a mechanistic link between BBS proteins and Notch signaling through endosomal recycling and sorting. This extends previous studies directly linking ciliary and basal body proteins to proteins involved in endosomal trafficking (Collin et al., 2012; Knödler et al., 2010; Westlake et al., 2011) and now implicates the BBSome proteins in proper recycling of endosomal cargo. In control cells, the Notch receptor is endocytosed away from the plasma membrane. From early endosomes, the receptor is either trafficked back to the membrane by recycling endosomes or it is directed to lysosomal degradation by late endosomes (Fig. 7A). The loss of plasma membrane localization seen with disruption of BBS1, BBS3 or BBS4 compounded with the significant decrease in localization of receptor to recycling endosomes suggests that BBSome-associated proteins are necessary for endosomal recycling. When this is lost, Notch receptor is potentially redirected to the late endosomes (Fig. 7B). The result is a significant increase in the amount of receptor in the late endosomes, overwhelming the lysosomal degradation pathway and resulting in activation of the receptor from endosomes. Loss of lysosomal localization is modest but, compounded with significantly increased late endosome localization and the accumulation of the heterodimeric receptor in whole cells (Fig. 4A; supplementary material Fig. S3G), this suggests that degradation is at least perturbed. This might be a result of defective sorting of cargo through late endosomes, a process regulated by ESCRT proteins, including TSG101. This latter step is likely disrupted in ALMS1-deficient cells, as these cells do not exhibit recycling defects but do accumulate Notch receptor in the late endosomes similar to BBS-deficient cells (Fig. 7C).
Fig. 7.
Model of Notch receptor trafficking in endosomes of cells deficient in basal body proteins. (A) In control cells, the Notch receptor (green lines) localizes to the cilium and plasma membrane, from where it is endocytosed into early endosomes (pink). The receptor is either recycled back to the membrane by recycling endosomes (yellow) or is sorted to the late endosomes and multi-vesicular bodies (MVBs, red) for degradation in the lysosomes (blue), a process that is mediated in part by proteins such as Shrub, Deltex (Dx), Suppressor of Deltex [Su(Dx)] and HOPS (also known as TMUB1), which are involved in endosomal trafficking and degradation of the receptor. (B) Loss of BBSome proteins does not disrupt endocytosis, but results in reduced membrane and ciliary localization as a result of reduced recycling (dashed arrow). The disruption in recycling results in increased trafficking to the late endosomes (heavy black arrow), from where active NICD can be produced. (C) Loss of ALMS1 does not disrupt recycling or membrane or ciliary localization, but increases the accumulation of receptor in late endosomes (heavy black arrow) and increases signaling.
Our observations suggest that the loss of Notch receptor localization at the cilium correlates with the loss of plasma membrane localization. A possible exception to this is the normal axonemal localization of Notch receptor in BBS3-depleted cells. Although this might be consistent with the more modest recycling defect in shBBS3 cells, it also suggests that receptor localization to the ciliary membrane might be subject to other mechanisms. The BBSome and BBS3 are implicated in the transport of proteins to the cilium (Jin et al., 2010) but their role in trafficking to other parts of the cell membrane is unclear. It is possible that defects in endosomal recycling affect trafficking to both ciliary and plasma membranes through similar mechanisms, but our data do not definitively establish this link. Our observations do, however, suggest that proper localization of Notch receptor to the plasma membrane is not dependent on the presence of a cilium (supplementary material Fig. S4A). Loss of membrane localization in the absence of BBS4 occurs irrespective of the presence of a cilium. Therefore, the trafficking role of BBS and Alstrom proteins might be independent of cilia. Trafficking studies that can continuously monitor individual cargo will be needed to clarify the link between membrane localization, ciliary localization, endosomal trafficking and the role of these proteins.
It is possible that the observed disruption in Notch localization is due to an inability of the receptor to be transported initially to either the cilium or the plasma membrane from the Golgi. Indeed, Golgi-to-membrane transport can occur through recycling endosomes (Ang et al., 2004), raising the possibility that the reduction in recycling endosome localization is due to an inability of the Notch receptor to properly exit the Golgi. However, this alone is not sufficient to explain our observations. If the Notch receptor is trapped in either the Golgi or in post-Golgi transport vesicles, we would not expect to see an increase in Notch signaling, nor would an accumulation of receptor in the late endosomes be likely. Therefore, although we cannot preclude the possibility of a Golgi-to-membrane or Golgi-to-cilia transport defect, the evidence suggests that increased Notch receptor activation is more likely due to a defect in trafficking after it has been endocytosed.
These findings also raise the question of the molecular mechanism responsible for basal body protein-dependent trafficking of Notch receptor through endosomes. Interestingly, an important molecular cue that targets Notch receptor in endosomes for lysosomal degradation is ubiquitylation. Evidence suggests that proteins, including Shrub and Deltex (Fig. 7A), that drive Notch receptor towards lysosomal degradation do so through polyubiquitylation (Chastagner et al., 2008). Defective sorting, resulting in increased endosomal activation of the receptor, is associated with a change in ubiquitylation state, likely to be a shift from polyubiquitylation to monoubiquitylation (Hori et al., 2011). Importantly, the aberrant activation is ligand-independent, similar to our observations with BBS proteins. At least one BBS protein, BBS11, is an E3 ubiquitin ligase (Chiang et al., 2006). It is possible that BBS proteins are involved in ubiquitylation of Notch receptor in early or sorting endosomes, which localize to the base of the cilium where BBS proteins are found (Fig. 1D; Ansley et al., 2003). It remains to be seen whether the ubiquitylation of the Notch receptor is altered in BBS-deficient cells but, if this is the case, it might provide a mechanistic explanation for why the sorting of endosomal cargo is disrupted.
An alternative explanation relates to the importance of microtubule dynamics in endosomal trafficking. It has long been recognized that the transport of endosomes and trafficking to lysosomes is dependent on microtubules and microtubule-based motors (Matteoni and Kreis, 1987; Oda et al., 1995). Furthermore, disruption of BBS proteins results in defects in microtubule architecture and nucleation (Kim et al., 2004). In addition, ALMS1 interacts directly with RILPL1 (Collin et al., 2012), which is related to RILP, a Rab7-interacting lysosomal protein that mediates endolysosomal trafficking through microtubule transport (Jordens et al., 2001; Progida et al., 2007). These reports suggest that BBS and Alstrom proteins might be necessary for proper trafficking of endosomal cargo by regulation of microtubule dynamics.
BBS and Alstrom proteins are necessary for regulation of Notch signaling
This study is the first demonstration of a role for BBS and Alstrom proteins in the regulation of Notch. In light of the role of these proteins in disease, our observation of enhanced Notch signaling might implicate a previously unexplored mechanism for disease phenotypes associated with BBS or Alstrom Syndrome. Notch is a crucial regulator of the differentiation of various cell types, and misregulation might have ramifications for a number of tissues. Notch has been studied extensively in neurogenesis, for example (Pierfelice et al., 2011), and neurological defects have been reported across the ciliopathies, including BBS (Lee and Gleeson, 2010). Impaired Notch regulation might therefore suggest a mechanism for these phenotypes. Consistent with this possibility, we observed decreased expression of ngn1, a marker of neurogenesis that is inhibited by Notch (Cornell and Eisen, 2002; Takke et al., 1999). Likewise, defects in Notch signaling contribute to renal defects, which are prevalent in the ciliopathies and in BBS (Arts and Knoers, 2013; Sirin and Susztak, 2012; Zaghloul and Katsanis, 2009). Maintenance of Notch activation suppresses the production of MCCs in the zebrafish pronephros, producing renal cysts, and suppression of the pathway can ameliorate this phenotype (Liu et al., 2007). We observed impaired production of MCCs in bbs4 morphants, supporting a potential role for increased Notch signaling in a major ciliopathy phenotype. Although the extent to which this pathway plays a role in BBS, Alstrom and other ciliopathies remains to be seen, our findings indicate its potential importance in pathogenesis.
Our observation of pathway amplification is in contrast to a recent report of reduced Notch signaling in the developing epidermis of conditional Ift88-, Ift74- and Kif3a-knockout mice (Ezratty et al., 2011), suggesting potential mechanistic discrepancies. First, the observation of muted Notch signaling in conditional Ift mutants suggests the possibility of protein-specific mediation of the pathway or a dependence on intraflagellar transport (Ezratty et al., 2011). Second, the finding that Notch signaling is muted in the epidermis suggests the possibility of tissue-specific effects. Although we did not focus our study on one particular tissue, we observed, for example, that enhancement of GFP in bbs1-morpholino-injected Tp1bglob:eGFP zebrafish embryos was more pronounced in some tissues relative to others. Finally, because Notch signaling is important in differentiation, timing is crucial. Indeed, at a later stage of development (E18.5) some cells of Ift88 conditional knockout epidermis exhibited ectopic Hes1 expression (Ezratty et al., 2011). To investigate this discrepancy, it will be necessary to assay signaling in ciliary mutants at various developmental stages.
Our findings underscore the importance of intracellular trafficking in the context of ciliopathy proteins. This study represents the first report of a role for these proteins in trafficking and the regulation of Notch, providing new mechanistic insight into the mediation of signaling by basal body proteins. This link raises the possibility of other signaling pathways mediated in this manner. Endosomal sorting is important in sequestering GSK-3β, a central molecule in canonical Wnt signaling (Taelman et al., 2010). Given the role of BBS proteins in mediating Wnt (Gerdes et al., 2007), these findings suggest an additional mechanism by which this regulation might occur. Likewise, components of other pathways, such as Shh, accumulate in endosomes and therefore might be similarly regulated (Incardona et al., 2002). In light of these possible links, it will be necessary to investigate the link between ciliopathy proteins and endosomal regulation of signaling to clarify the cellular mechanisms of signaling disruption in ciliopathies.
MATERIALS AND METHODS
Cell culture and transfection
hTERT-RPE1 and HEK293 cells (ATCC) were cultured in DMEM∶F-12 with 10% FBS and 0.1% hygromycin or DMEM with 10% FBS and 1% penicillin-streptomycin, respectively. For transfection, cells in growth medium without antibiotics were plated onto coverslips or in culture dishes and transfected at 90–95% confluency using Lipofectamine™ 2000 (Life Technologies) according to the manufacturer's protocol with the following constructs at 4 µg in 2 ml: shBBS4 in pSUPER (Gerdes et al., 2007); shBBS1, shBBS3, shALMS or shTSG in pLKO.1-puro (Sigma-Aldrich MISSION® collection). Transfection reagent alone (No DNA) or with empty vector (pSUPER or pLKO.1-puro) was used as a control. Transfected cells were harvested at 48 hours without change of culture medium after confirmation of 85–95% transfection efficiency (by GFP expression vector).
Immunofluorescence
Cells plated onto coverslips were fixed at 48 hours post-transfection in 4% paraformaldehyde (PFA) or ice-cold methanol for 10 minutes, followed by a 10-minute permeabilization with 0.1% Triton X-100 and two 1× PBS washes. Double immunostaining was carried out by first blocking for 1 hour in PBS-Tween plus 10% serum and 1% bovine serum albumin (BSA), followed by overnight incubation with primary antibody at 4°C, a 1-hour incubation with secondary antibody at room temperature, a second block, a second overnight incubation with primary antibody and a second one-hour secondary antibody incubation. Primary antibodies against the following proteins were used: NOTCH1 (C-20, Santa Cruz Biotechnology, 1∶50), ARL13B (Proteintech, 1∶1000), acetylated tubulin (Sigma, 1∶1000), γ-tubulin (Sigma, 1∶1000), EEA1 (BD Transduction Laboratories, 1∶250), RAB7 (Cell Signaling Technology, 1∶100), TSG101 (4A10, Abcam, 1∶500), LAMP2 (H4B4, Abcam, 1∶100) and RAB11 (Cell Signaling Technology, 1∶100). Species-specific secondary antibodies (Alexa Fluor®, Invitrogen) were used at 1∶1000. Following primary and secondary antibody incubations, coverslips were washed in 1× PBS, incubated in DAPI (0.2 µg/ml in PBS), mounted with Prolong Gold Antifade (Invitrogen) and imaged at 100× magnification using an Olympus IX50 or an Olympus IX81 with cellSens imaging software using deconvolution and z-stacking capability.
qRT-PCR
RNA was extracted from cells or zebrafish embryos using TRIzol reagent (Life Technologies) according to the manufacturer's protocol and was purified using the RNeasy Kit (Qiagen). cDNA was transcribed using the Fermentas First Strand cDNA Transcription Kit (Thermo Scientific) according to the manufacturer's protocol, diluted 1∶9 (for all zebrafish genes and all human genes except reference) or 1∶90 (reference human gene only) and added to a reaction including target-specific primers (sequences available upon request) and LightCycler 480 SybrGreen (Roche) and run on a LightCycler 480 (Roche) for 5 minutes at 95°C, followed by 40 cycles of 95°C (10 seconds), 58°C (15 seconds), 72°C (10 seconds) then 5 minutes at 72°C. A reverse-transcriptase-free sample was used as a negative control. All samples were run in duplicate with the CT value normalized to GAPDH (cells) or β-actin (zebrafish) to calculate relative expression for each gene in each sample. Biological replicates were repeated a minimum of three times and up to seven times per treatment. Fold change was calculated relative to control samples.
Western blotting and sucrose gradient
HEK293 or hTERT-RPE1 cells were plated in six-well or 10-cm culture dishes for western blotting of whole-cell lysates or sucrose-gradient fractionation, respectively. For the former, cells were washed twice in ice-cold PBS, incubated for 30 minutes on ice in Tris/NaCl/NP-40 buffer containing 1% protease inhibitor (Sigma) and centrifuged at 10,000 g for 10 minutes at 4°C. Supernatant was collected and assayed by western blotting according to standard protocols using Life Technologies reagents and instrumentation. The absorbance of each protein band was measured using ImageJ software and quantified as the area under the curve. For sucrose gradients, cells were washed in cold HEPES/MgCl2, collected in the same solution plus protease inhibitor and spun at 1000 g for 2 minutes at 4°C. The resulting pellet was resuspended in Tris/MgCl2/protease inhibitor with 8% sucrose and homogenized on ice for 1 minute using a Dounce homogenizer. The suspension was centrifuged for 2 minutes at 600 g at 4°C, the supernatant was added to a sucrose gradient of 50% (500 µl), 30% (750 µl) and 10% (500 µl) and centrifuged for 1 hour at 40,000 g using a Beckman Ultracentrifuge. Gradient fractions were collected by gravity drip in ∼250 µl aliquots and run using a standard western blotting protocol. Total protein for each fraction was calculated by using the bicinchoninic acid (BCA) assay kit (Pierce) according to the manufacturer's protocol and a Perkin Elmer Wallac Victor plate reader. For each sucrose gradient western blot, 2 µg of total protein was run for fractions 1–5 and 6 µg of total protein was run for fractions 6–8, owing to the reduced concentration and reduced sensitivity of detecting Notch receptor in the endosomal fractions. Total protein for whole-cell lysates was calculated by using the BCA assay kit to ensure equal protein amounts (30 µg) were run in each lane. Primary antibodies against the following proteins were used for whole-cell lysate and/or sucrose gradient fractions: NOTCH1 (Abcam, 1∶1000), HA-tag (Sigma, 1∶10,000), N-cadherin (BD Transduction Laboratories, 1∶1000), EEA1 (BD Transduction Laboratories, 1∶5000), β-actin (Cell Signaling Technology, 1∶1000), BBS1 (Santa Cruz Biotechnologies, 1∶1000), BBS4 (Sigma, 1∶1000), BBS3 (Proteintech, 1∶1000) and TSG101 (Abcam, 1∶1000). Horseradish peroxidase (HRP)-conjugated anti-mouse-IgG (Cell Signaling Technology) and anti-rabbit-IgG Poly-HRP (Pierce) secondary antibodies were used at 1∶2000.
Zebrafish lines, morpholino injection and in situ hybridization
Adult Tp1bglob:eGFP transgenic zebrafish were housed and naturally mated according to standard protocols. Embryos were collected for morpholino injection at the two- to four-cell stage in embryo medium and cultured at 28.5°C. Translation blocking morpholinos were injected against bbs1, bbs4, bbs3 (Zaghloul et al., 2010) and deltaA (Latimer et al., 2002), as well as a splice-blocking morpholino against tsg101 (5′-TGGGTGTTGGGAGACATACATGTTT-3′). Overexpression was carried out by injection of mRNA (transcribed from pC3+MT vector using the mMessage mMachine Kit, Ambion) encoding the active intracellular domain fragment of notch3 (nicd) at 50 pg. Transgenic embryos were harvested for RNA at 24 hpf in groups of 20 or observed for GFP expression at 48 hpf using a SteREO Zeiss Lumar.V12 dissecting microscope and imaged at 30× magnification with an Axiocam MRc 5 and Axiovision software. In situ hybridization was carried out as per standard protocols (Thisse and Thisse, 2008) using previously described riboprobes against either neurogenin1 or shippo1 (Geling et al., 2004; Liu et al., 2007).
Quantification of NOTCH1 localization by immunofluorescence
Immunostained cells were imaged as described above. Exposure and camera settings were optimized for each experiment using control cells. For endosomal and lysosomal localization, the number of marker-labeled vesicles that were also labeled with NOTCH1 was counted as a proportion of the total number of marker-labeled vesicles for a given focal plane. The average percentage of co-labeled vesicles containing NOTCH1 was calculated for a minimum of 30 cells per treatment. Each cell was distinguished from neighboring cells by nuclear DAPI staining. The focal plane was adjusted to focus only on the structure being assessed (i.e. endosomes, basal body, axoneme). For TSG101-labeled MVBs, we assessed only vesicles that were definitively outside of the nucleus to preclude the possibility of counting intranuclear NICD localization. For endosomal accumulation at the cilium, the focal plane at the base of the cilium was used. For membrane staining intensity, fluorescence was observed and quantified across multiple focal planes obtained as z-stacks for each cell. A minimum of two regions spanning the width of each cell were assessed by fluorescence intensity analyses using ImageJ software. The average of all intensity values within each intensity peak was calculated and the average for all peaks measured per treatment was plotted for a minimum of 50 cells.
Supplementary Material
Acknowledgments
We thank Michael Parsons (Johns Hopkins University, Baltimore, MD) for Tp1bglob:eGFP transgenic zebrafish, Masahako Hibi (Nagoya University, Japan) for the notch3-ICD expression vector, Nicholas Katsanis (Duke University, Durham, NC) for the shBBS4/pSUPER construct, Pier Paolo Di Fiore (IFOM, Milan, Italy) for the HA-NOTCH1 construct, Laure Bally-Cuiff (CNRS, France) for the ngn1 construct and Iain Drummond (Massachusetts General Hospital/Harvard University, Cambridge, MA) for the shippo1 construct.
Footnotes
Competing interests
The authors declare no competing interests.
Author contributions
C.C.L., S.L., V.P.E. carried out all experiments and analyzed data. S.L. created the model. J.L.B. and N.A.Z. conceived of experiments and analyzed data. N.A.Z. wrote the paper.
Funding
This work was funded by a grant from the National Institutes of Health (National Institute of Diabetes and Digestive and Kidney Diseases) [grant number K01DK092402 to N.A.Z.]; a grant from the Programa de Desarrollo de las Ciencias Básicas to J.L.B.; and by a grant from the Agencia Nacional de Investigación e Innovación, Uruguay to J.L.B. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.130344/-/DC1
References
- Ang A. L., Taguchi T., Francis S., Fölsch H., Murrells L. J., Pypaert M., Warren G., Mellman I. (2004). Recycling endosomes can serve as intermediates during transport from the Golgi to the plasma membrane of MDCK cells. J. Cell Biol. 167, 531–543 10.1083/jcb.200408165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansley S. J., Badano J. L., Blacque O. E., Hill J., Hoskins B. E., Leitch C. C., Kim J. C., Ross A. J., Eichers E. R., Teslovich T. M. et al. (2003). Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature 425, 628–633 10.1038/nature02030 [DOI] [PubMed] [Google Scholar]
- Arts H. H., Knoers N. V. (2013). Current insights into renal ciliopathies: what can genetics teach us? Pediatr. Nephrol. 28, 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral D. C., Garg S., Casalou C., Watts G. F., Sandoval J. L., Ramalho J. S., Hsu V. W., Brenner M. B. (2012). Arl13b regulates endocytic recycling traffic. Proc. Natl. Acad. Sci. USA 109, 21354–21359 10.1073/pnas.1218272110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari N. F., O'Connor A. K., Haycraft C. J., Yoder B. K. (2009). The primary cilium as a complex signaling center. Curr. Biol. 19, R526–R535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas-Rodriguez M., Badano J. L. (2009). Ciliary biology: understanding the cellular and genetic basis of human ciliopathies. Am. J. Med. Genet. 151C, 263–280 10.1002/ajmg.c.30227 [DOI] [PubMed] [Google Scholar]
- Chastagner P., Israël A., Brou C. (2008). AIP4/Itch regulates Notch receptor degradation in the absence of ligand. PLoS ONE 3, e2735 10.1371/journal.pone.0002735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang A. P., Beck J. S., Yen H. J., Tayeh M. K., Scheetz T. E., Swiderski R. E., Nishimura D. Y., Braun T. A., Kim K. Y., Huang J. et al. (2006). Homozygosity mapping with SNP arrays identifies TRIM32, an E3 ubiquitin ligase, as a Bardet-Biedl syndrome gene (BBS11). Proc. Natl. Acad. Sci. USA 103, 6287–6292 10.1073/pnas.0600158103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement C. A., Ajbro K. D., Koefoed K., Vestergaard M. L., Veland I. R., Henriques de Jesus M. P., Pedersen L. B., Benmerah A., Andersen C. Y., Larsen L. A. et al. (2013). TGF-beta signaling is associated with endocytosis at the pocket region of the primary cilium. Cell Rep. 3, 1806–1814 [DOI] [PubMed] [Google Scholar]
- Collin G. B., Marshall J. D., King B. L., Milan G., Maffei P., Jagger D. J., Naggert J. K. (2012). The Alström syndrome protein, ALMS1, interacts with α-actinin and components of the endosome recycling pathway. PLoS ONE 7, e37925 10.1371/journal.pone.0037925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell R. A., Eisen J. S. (2002). Delta/Notch signaling promotes formation of zebrafish neural crest by repressing Neurogenin 1 function. Development 129, 2639–2648 [DOI] [PubMed] [Google Scholar]
- Ezratty E. J., Stokes N., Chai S., Shah A. S., Williams S. E., Fuchs E. (2011). A role for the primary cilium in Notch signaling and epidermal differentiation during skin development. Cell 145, 1129–1141 10.1016/j.cell.2011.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geling A., Plessy C., Rastegar S., Strähle U., Bally-Cuif L. (2004). Her5 acts as a prepattern factor that blocks neurogenin1 and coe2 expression upstream of Notch to inhibit neurogenesis at the midbrain-hindbrain boundary. Development 131, 1993–2006 10.1242/dev.01093 [DOI] [PubMed] [Google Scholar]
- Gerdes J. M., Liu Y., Zaghloul N. A., Leitch C. C., Lawson S. S., Kato M., Beachy P. A., Beales P. L., DeMartino G. N., Fisher S. et al. (2007). Disruption of the basal body compromises proteasomal function and perturbs intracellular Wnt response. Nat. Genet. 39, 1350–1360 10.1038/ng.2007.12 [DOI] [PubMed] [Google Scholar]
- Gerdes J. M., Davis E. E., Katsanis N. (2009). The vertebrate primary cilium in development, homeostasis, and disease. Cell 137, 32–45 10.1016/j.cell.2009.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz S. C., Anderson K. V. (2010). The primary cilium: a signalling centre during vertebrate development. Nat. Rev. Genet. 11, 331–344 10.1038/nrg2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruharsha K. G., Kankel M. W., Artavanis-Tsakonas S. (2012). The Notch signalling system: recent insights into the complexity of a conserved pathway. Nat. Rev. Genet. 13, 654–666 10.1038/nrg3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson E. M., Lanner F., Das D., Mutvei A., Marklund U., Ericson J., Farnebo F., Stumm G., Stenmark H., Andersson E. R. et al. (2010). Control of Notch-ligand endocytosis by ligand-receptor interaction. J. Cell Sci. 123, 2931–2942 10.1242/jcs.073239 [DOI] [PubMed] [Google Scholar]
- Hearn T., Spalluto C., Phillips V. J., Renforth G. L., Copin N., Hanley N. A., Wilson D. I. (2005). Subcellular localization of ALMS1 supports involvement of centrosome and basal body dysfunction in the pathogenesis of obesity, insulin resistance, and type 2 diabetes. Diabetes 54, 1581–1587 10.2337/diabetes.54.5.1581 [DOI] [PubMed] [Google Scholar]
- Hori K., Sen A., Kirchhausen T., Artavanis-Tsakonas S. (2011). Synergy between the ESCRT-III complex and Deltex defines a ligand-independent Notch signal. J. Cell Biol. 195, 1005–1015 10.1083/jcb.201104146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incardona J. P., Gruenberg J., Roelink H. (2002). Sonic hedgehog induces the segregation of patched and smoothened in endosomes. Curr. Biol. 12, 983–995 [DOI] [PubMed] [Google Scholar]
- Jagger D., Collin G., Kelly J., Towers E., Nevill G., Longo-Guess C., Benson J., Halsey K., Dolan D., Marshall J. et al. (2011). Alström Syndrome protein ALMS1 localizes to basal bodies of cochlear hair cells and regulates cilium-dependent planar cell polarity. Hum. Mol. Genet. 20, 466–481 10.1093/hmg/ddq493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., White S. R., Shida T., Schulz S., Aguiar M., Gygi S. P., Bazan J. F., Nachury M. V. (2010). The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell 141, 1208–1219 10.1016/j.cell.2010.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordens I., Fernandez-Borja M., Marsman M., Dusseljee S., Janssen L., Calafat J., Janssen H., Wubbolts R., Neefjes J. (2001). The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr. Biol. 11, 1680–1685 [DOI] [PubMed] [Google Scholar]
- Kim J. C., Badano J. L., Sibold S., Esmail M. A., Hill J., Hoskins B. E., Leitch C. C., Venner K., Ansley S. J., Ross A. J. et al. (2004). The Bardet-Biedl protein BBS4 targets cargo to the pericentriolar region and is required for microtubule anchoring and cell cycle progression. Nat. Genet. 36, 462–470 10.1038/ng1352 [DOI] [PubMed] [Google Scholar]
- Knödler A., Feng S., Zhang J., Zhang X., Das A., Peränen J., Guo W. (2010). Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc. Natl. Acad. Sci. USA 107, 6346–6351 10.1073/pnas.1002401107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latimer A. J., Dong X., Markov Y., Appel B. (2002). Delta-Notch signaling induces hypochord development in zebrafish. Development 129, 2555–2563 [DOI] [PubMed] [Google Scholar]
- Le Borgne R. (2006). Regulation of Notch signalling by endocytosis and endosomal sorting. Curr. Opin. Cell Biol. 18, 213–222 10.1016/j.ceb.2006.02.011 [DOI] [PubMed] [Google Scholar]
- Lee J. H., Gleeson J. G. (2010). The role of primary cilia in neuronal function. Neurobiol. Dis. 38, 167–172 10.1016/j.nbd.2009.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Pathak N., Kramer-Zucker A., Drummond I. A. (2007). Notch signaling controls the differentiation of transporting epithelia and multiciliated cells in the zebrafish pronephros. Development 134, 1111–1122 10.1242/dev.02806 [DOI] [PubMed] [Google Scholar]
- Matteoni R., Kreis T. E. (1987). Translocation and clustering of endosomes and lysosomes depends on microtubules. J. Cell Biol. 105, 1253–1265 10.1083/jcb.105.3.1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill M. A., Dho S. E., Weinmaster G., McGlade C. J. (2009). Numb regulates post-endocytic trafficking and degradation of Notch1. J. Biol. Chem. 284, 26427–26438 10.1074/jbc.M109.014845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moberg K. H., Schelble S., Burdick S. K., Hariharan I. K. (2005). Mutations in erupted, the Drosophila ortholog of mammalian tumor susceptibility gene 101, elicit non-cell-autonomous overgrowth. Dev. Cell 9, 699–710 10.1016/j.devcel.2005.09.018 [DOI] [PubMed] [Google Scholar]
- Nilsson L., Conradt B., Ruaud A. F., Chen C. C., Hatzold J., Bessereau J. L., Grant B. D., Tuck S. (2008). Caenorhabditis elegans num-1 negatively regulates endocytic recycling. Genetics 179, 375–387 10.1534/genetics.108.087247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson L., Jonsson E., Tuck S. (2011). Caenorhabditis elegans numb inhibits endocytic recycling by binding TAT-1 aminophospholipid translocase. Traffic 12, 1839–1849 10.1111/j.1600--0854.2011.01271.x [DOI] [PubMed] [Google Scholar]
- Oda H., Stockert R. J., Collins C., Wang H., Novikoff P. M., Satir P., Wolkoff A. W. (1995). Interaction of the microtubule cytoskeleton with endocytic vesicles and cytoplasmic dynein in cultured rat hepatocytes. J. Biol. Chem. 270, 15242–15249 10.1074/jbc.270.25.15242 [DOI] [PubMed] [Google Scholar]
- Parsons M. J., Pisharath H., Yusuff S., Moore J. C., Siekmann A. F., Lawson N., Leach S. D. (2009). Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mech. Dev. 126, 898–912 10.1016/j.mod.2009.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierfelice T., Alberi L., Gaiano N. (2011). Notch in the vertebrate nervous system: an old dog with new tricks. Neuron 69, 840–855 10.1016/j.neuron.2011.02.031 [DOI] [PubMed] [Google Scholar]
- Pitaval A., Tseng Q., Bornens M., Théry M. (2010). Cell shape and contractility regulate ciliogenesis in cell cycle-arrested cells. J. Cell Biol. 191, 303–312 10.1083/jcb.201004003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Progida C., Malerød L., Stuffers S., Brech A., Bucci C., Stenmark H. (2007). RILP is required for the proper morphology and function of late endosomes. J. Cell Sci. 120, 3729–3737 10.1242/jcs.017301 [DOI] [PubMed] [Google Scholar]
- Rohatgi R., Milenkovic L., Scott M. P. (2007). Patched1 regulates hedgehog signaling at the primary cilium. Science 317, 372–376 10.1126/science.1139740 [DOI] [PubMed] [Google Scholar]
- Sirin Y., Susztak K. (2012). Notch in the kidney: development and disease. J. Pathol. 226, 394–403 10.1002/path.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. A., Dho S. E., Donaldson J., Tepass U., McGlade C. J. (2004). The cell fate determinant numb interacts with EHD/Rme-1 family proteins and has a role in endocytic recycling. Mol. Biol. Cell 15, 3698–3708 10.1091/mbc.E04--01--0026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taelman V. F., Dobrowolski R., Plouhinec J. L., Fuentealba L. C., Vorwald P. P., Gumper I., Sabatini D. D., De Robertis E. M. (2010). Wnt signaling requires sequestration of glycogen synthase kinase 3 inside multivesicular endosomes. Cell 143, 1136–1148 10.1016/j.cell.2010.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takke C., Dornseifer P., v Weizsäcker E., Campos-Ortega J. A. (1999). her4, a zebrafish homologue of the Drosophila neurogenic gene E(spl), is a target of NOTCH signalling. Development 126, 1811–1821 [DOI] [PubMed] [Google Scholar]
- Thisse C., Thisse B. (2008). High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69 10.1038/nprot.2007.514 [DOI] [PubMed] [Google Scholar]
- Thompson B. J., Mathieu J., Sung H. H., Loeser E., Rørth P., Cohen S. M. (2005). Tumor suppressor properties of the ESCRT-II complex component Vps25 in Drosophila. Dev. Cell 9, 711–720 10.1016/j.devcel.2005.09.020 [DOI] [PubMed] [Google Scholar]
- Tien A. C., Rajan A., Bellen H. J. (2009). A Notch updated. J. Cell Biol. 184, 621–629 10.1083/jcb.200811141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccari T., Bilder D. (2005). The Drosophila tumor suppressor vps25 prevents nonautonomous overproliferation by regulating notch trafficking. Dev. Cell 9, 687–698 10.1083/jcb.200811141 [DOI] [PubMed] [Google Scholar]
- Vaccari T., Lu H., Kanwar R., Fortini M. E., Bilder D. (2008). Endosomal entry regulates Notch receptor activation in Drosophila melanogaster. J. Cell Biol. 180, 755–762 10.1083/jcb.200708127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westlake C. J., Baye L. M., Nachury M. V., Wright K. J., Ervin K. E., Phu L., Chalouni C., Beck J. S., Kirkpatrick D. S., Slusarski D. C. et al. (2011). Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc. Natl. Acad. Sci. USA 108, 2759–2764 10.1073/pnas.1018823108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaghloul N. A., Katsanis N. (2009). Mechanistic insights into Bardet-Biedl syndrome, a model ciliopathy. J. Clin. Invest. 119, 428–437 10.1172/JCI37041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaghloul N. A., Liu Y., Gerdes J. M., Gascue C., Oh E. C., Leitch C. C., Bromberg Y., Binkley J., Leibel R. L., Sidow A. et al. (2010). Functional analyses of variants reveal a significant role for dominant negative and common alleles in oligogenic Bardet-Biedl syndrome. Proc. Natl. Acad. Sci. USA 107, 10602–10607 10.1073/pnas.1000219107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.