Abstract
Summary
Therapeutic upregulation of macroautophagy in cancer cells provides an alternative mechanism for cell death. Prolactin (PRL) and its receptor (PRLR) are considered attractive therapeutic targets because of their roles as growth factors in tumor growth and progression. We utilized a novel antagonist peptide of PRL, G129R, to block activity of the tumoral PRL/PRLR axis, which resulted in inhibition of tumor growth in orthotopic models of human ovarian cancer. Prolonged treatment with G129R induced accumulation of redundant autolysosomes in three-dimensional cancer spheroids, leading to a type II programmed cell death. This inducible autophagy was a non-canonical beclin-1–independent pathway and was sustained by an astrocytic phosphoprotein (PEA-15) and protein kinase C zeta interactome. Lower levels of tumoral PRL/PRLR in clinical samples were associated with longer patient survival. Our findings provide a new understanding of the mechanisms of tumor growth inhibition through targeting PRL/PRLR and may have clinical implications.
Introduction
The multifunctional hormone, prolactin (PRL), is not only essential for normal reproduction and maintenance of pregnancy but also contributes to pathogenesis of gynecologic malignances, including ovarian and endometrial cancers (Levina et al., 2009; Mor et al., 2005; Tan et al., 2011). Human PRL has pro-proliferative effects on ovarian and endometrial cancer cells (Asai-Sato et al., 2005). Recent studies support a robust role for PRL in ovarian cancer cell survival and invasion, which implicates it as a therapeutic target (Tan et al., 2011). PRL binding to its membrane-associated prolactin receptor (PRLR) is followed by activation of oncogenic signaling pathways such as JAK2 and STAT3, stimulating proliferation of cancer cells and tumor growth (Rui et al., 1994; Xie et al., 2002). Despite the importance of the PRL/PRLR signaling complex in tumor growth, the underlying mechanisms are not well understood, and the ability to target this pathway is limited by incomplete knowledge of its activity. G129R, a variant of normal human PRL that differs by a single amino acid substitution mutation, inhibited PRL-induced oncogenic signaling responsible for cancer cell proliferation (Llovera et al., 2000).
Autophagy is a lysosome-dependent cellular degradation pathway that can be triggered by many stimuli, including metabolic stress, hypoxia, or treatment with chemotherapy agents or radiation (Rubinsztein et al., 2007). Key proteins regulate the formation and expansion of vesicular structures such as autophagosomes, which then fuse with lysosomes to form autolysosomes. Under normal conditions, basal levels of autophagy in proliferative cells function as a survival mechanism (Mathew et al., 2007). Prolonged exposure to therapeutic agents, however, can lead to progression of destructive autophagy and eventual programmed cell death (Dalby et al., 2010; Kamat et al., 2009; White et al., 2010). Targeted molecular therapies that can induce sustained autophagy offer new therapeutic opportunities (Shimizu et al., 2004), particularly in breast, prostate, and ovarian cancers that are known for high rates of loss of tumor-suppressor gene BECN1 (Kubes et al., 1998; Liang et al., 1999).
Here we describe that prolonged treatment with G129R antagonized the activities of the tumoral PRL/PRLR axis and inhibited tumor growth through induction of destructive autophagy. Our results indicate that inhibition of the tumoral PRL/PRLR axis may have implications for anticancer therapy through promotion of autophagy related cell death.
Results
In vivo antitumor efficacy of PRLR antagonist G129R
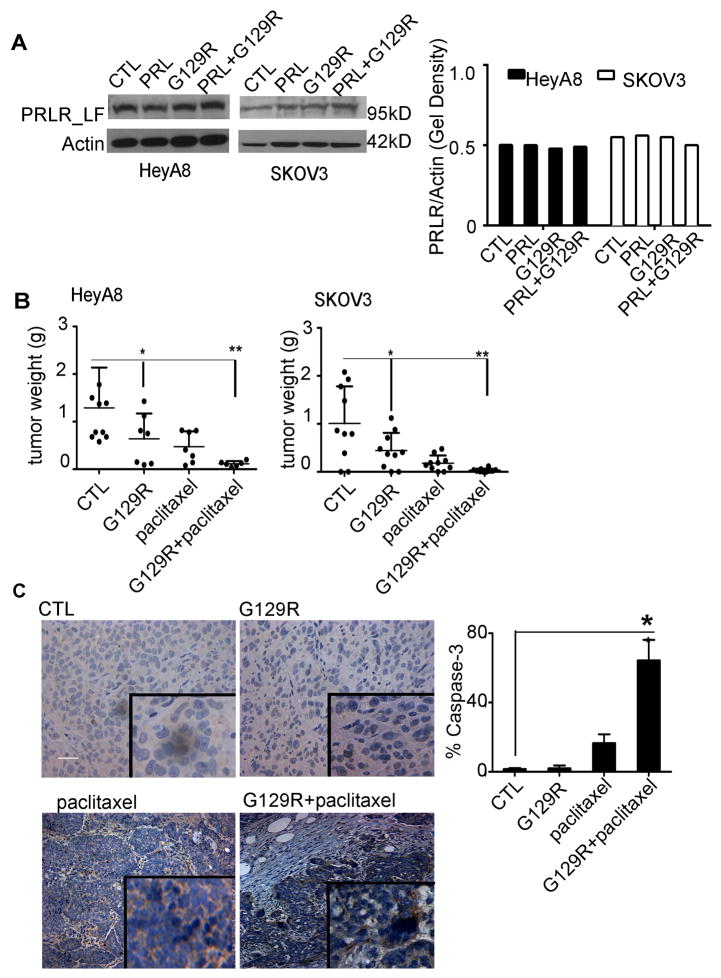
To gain insights into the effects of blockade of PRL/PRLR activities on tumor growth, we evaluated the effects of G129R in orthotopic mouse models of human ovarian cancers that express PRLR. Expression of the long form of PRLR (PRLR-LF, 95 kDa) was detected in both HeyA8 and SKOV3 ovarian cancer cells (Fig. 1A). An in vivo dose-finding experiment showed that increasing doses of G129R (100, 200, or 400 μg daily) were inversely related to tumor growth without affecting body weight (Fig. S1A & S1B). Therefore, we chose the dose of 100 μg/day and used mannitol as the control since it is the principal excipient in the G129R formulation (Wang, 2000). After 28 days of G129R monotherapy, tumor weights were 50% lower in both HeyA8 and SKOV3 models than in controls (Fig. 1B). Given that taxane-based chemotherapy is used frequently for ovarian cancer, we tested a combination of G129R and paclitaxel, which resulted in >90% lower tumor weights than in controls (Fig. 1B). The number of tumor nodules was significantly lower in G129R and G129R+paclitaxel treatment groups than in controls in both models (Fig. S1C & S1D, p<0.05). We did not observe any obvious toxic effects of G129R, as shown by stability of mouse body weights (Fig. S1E & S1F) and of complete blood counts and by histological evaluation of organs, including liver, kidney, and spleen, from mice treated with G129R at 100 μg/day for 2 weeks (Fig. S1G & S1H). We also examined the HeyA8 tumors for level of cleaved caspase-3, which was significantly higher in the G129R+paclitaxel group than in the paclitaxel-only group (Fig. 1C).
Figure 1.
G129R inhibited tumor growth in ovarian cancer models.
(A) (Left panel) PRLR-LF was detected in HeyA8 and SKOV3 cells treated with PRL (0.1 μg/mL), G129R (10 μg/mL), a combination (PRL+G129R), or control (CTL). (Right panel) The fold gel density was normalized to that of β-actin.
(B) Effects of G129R and paclitaxel on tumor growth in HeyA8 and SKOV3 orthotopic mouse models. Weight of the tumor from each mouse is shown; (Left panel) HeyA8: *p < 0.05 (CTL vs. G129R); **p < 0.001 (CTL vs. G129R+paclitaxel). (Right panel) SKOV3: *p< 0.05 (CTL vs. G129R); **p < 0.001 (CTL vs. G129R+paclitaxel).
(C) (Left panel) Representative images of cleaved caspase-3 in HeyA8 tumor tissues (inset shown at 200× magnification); scale bar = 100 μm. Results were confirmed with triplicate experiments. (Right panel) Percentage of tumor with cleaved caspase-3 staining is shown graphically (error bar = 95% confidence interval); differences between groups were compared by unpaired two-tailed t-test: n=5, *p = 0.0008 (paclitaxel vs. G129R+paclitaxel).
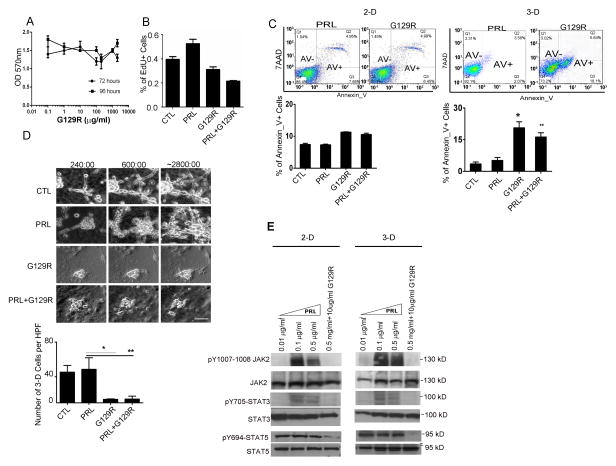
Inhibition of tumor cell growth by G129R in 3-D but not 2-D conditions
To identify the mechanism of G129R’s inhibition of tumor growth, we examined the effects of G129R in vitro in ovarian cancer cells lines. We observed no significant effects on cell viability or proliferative property of SKOV3 cells under 2-dimensional (2-D) conditions (Fig. 2A &2B, p>0.05). Moreover, G129R had no significant effect on migration or invasive properties of HeyA8, SKOV3, or IG10 cells cultured under 2-D monolayer conditions (Fig. S2A, 2B, & 2C).
Figure 2.
G129R inhibited growth of tumor cells in 3-D but not 2-D conditions.
(A) Effects of G129R on viability of 2-D cultured SKOV3 cells. ANOVA analysis was performed to compare the viability of cells treated with G129R at various doses (0.01 to 1000 μg/mL) and for various time periods (72 and 96 hours; p>0.05).
(B) Effects of G129R on cell proliferation (as measured by 5-ethynyl-2′-deoxyuridine [EdU] incorporation assay) of 2-D SKOV3 cells. n=6, p=0.09 (control [CTL] vs. G129R); p=0.08 (CTL vs. PRL+G129R).
(C) G129R significantly increased the AnnexinV–positive (AnV+) proportion of SKOV3 cells cultured in 3-D but not in 2-D conditions. Representative FACS plots are shown, and corresponding AnV+ percentages in 3-D cultured SKOV3 cells were, by treatment: PRL (6.74%), G129R (23.8%), and PRL+G129R (18.3%); n=6, *p < 0.001 (CTL vs. G129R); **p < 0.001 (CTL vs. PRL+G129R).
(D) G129R inhibited the growth of 3-D HeyA8 spheroids. (Upper panel) Representative photomicrographs indicate the approximate time elapsed: (a) 240, (b) 600, and (c) 2800 minutes (see Movies 1–4 in Supplemental Materials). (Lower panel) 3-D structure scoring was based on the number of 3-D cells per high-power field (HPF) after 48 hours of treatment. Over 20 HPFs were scored, and average numbers of cells were graphed. n=20, **p < 0.01 (PRL vs. G129R); *p < 0.05 (PRL vs. PRL+G129R).
(E) G129R antagonized PRL activation of JAK2, STAT3, and STAT5 in HeyA8 3-D spheroids. The effects of PRL at various doses on phosphorylation of JAK2 and STAT3/5 were detected after 1 hour of treatment. The antagonistic effect of G129R against PRL was captured by adding a combination of PRL (0.5 μg/mL) and G129R (10 μg/mL).
Given the substantial difference between the effects of G129R in in vivo and 2-D conditions, we reasoned that the inhibitory mechanism of G129R may be recapitulated by 3-D cultured cancer spheroids (Debnath and Brugge, 2005). G129R did not significantly increase the percentage of AnnexinV-positive SKOV3 cells in 2-D culture but significantly increased the percentage of AnnexinV-positive 3-D SKOV3 spheroids (Fig. 2C). Similar results were noted with the 3-D cultured HeyA8 cells (Fig. S2D) but not IG10 cells, which have very low levels of endogenous PRLR-LF, suggesting that the 3-D growth inhibition induced by G129R depends on PRLR-LF expression.
We next utilized time-lapse video microscopy to monitor the longitudinal effects of G129R in 3-D cancer spheroids. The growth of 3-D spheroids was significantly faster in the control or PRL-treated groups (Fig. 2D and Movies 1 & 2) than in the groups treated with G129R or PRL+G129R (Fig. 2D and Movies 3 & 4). The number of 3-D cultured cells in spheroids per high-power field for each group was statistically analyzed at the end of each video. The numbers of G129R-treated or PRL+G129R-treated spheroids were significantly lower than the numbers of PRL-treated spheroids (Fig. 2D). Moreover, G129R or PRL+G129R treatments blocked activation of JAK2, STAT3, and STAT5 in 3-D cultured HeyA8 cells (Fig. 2E), which showed that the inhibition of oncogenic JAK2-STATs pathways by G129R is mediated through antagonism of the PRL/PRLR axis (Lin et al., 2011).
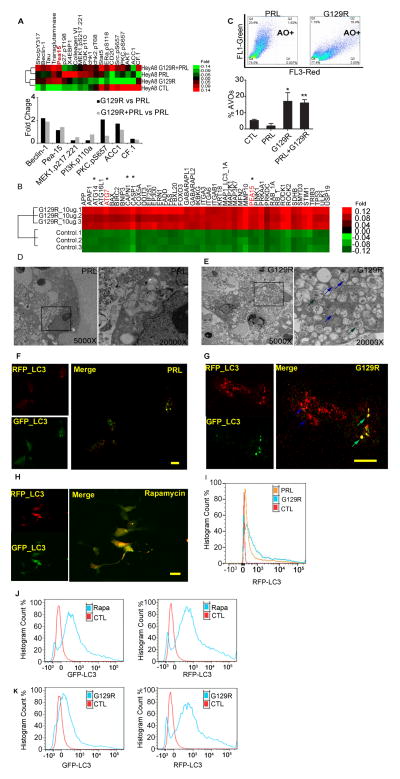
Sustained autophagy following G129R treatment
To gain insight into mechanisms of growth inhibition by G129R, we analyzed the proteomic profile of HeyA8 3-D spheroids using reverse-phase protein array analysis (RPPA) and found that a number of autophagic factors were upregulated by treatment with G129R or PRL+G129R, including beclin-1 and phosphoprotein enriched in astrocytes (PEA-15) (Fig. 3A). We also analyzed the genomic profile of HeyA8 3-D spheroids regulated by G129R. Multiple genes involved in autophagy progression and cell death, including ATG14, ATG16L1, ATG7, BCL2 (Akar et al., 2008; Levine et al., 2008), BIRC2 (Meng et al., 2010), BNIP3 (Quinsay et al., 2010), CAPN1 (Demarchi et al., 2007), CASP1, CASP2, MAP1_LC3_1A, MMP10, PEA-15 (Bartholomeusz et al., 2008), PINK1, and others, were upregulated by G129R treatment. Among the upregulated genes, ATG7 is known to be involved in triggering autophagic cell death (Yu et al., 2004); others have known activity in autophagy (marked by asterisks in Fig. 3B).
Figure 3.
Autophagy in 3-D cancer spheroids was induced by G129R.
(A) (Upper panel) Heat map clustered from RPPA analysis revealed an array of autophagic factors in HeyA8 3-D spheroids regulated by G129R or PRL+G129R. (Lower panel) Graph of normalized fold changes indicated that PEA-15 was increased by G129R or PRL+G129R as compared to PRL.
(B) Heat map clustered from cDNA microarray analysis identified genes involved in autophagy that were upregulated by G129R in HeyA8 3-D spheroids (one-way ANOVA, n=3, p < 0.001). Genes denoted with asterisks are those with roles in autophagy.
(C) Representative FACS plot showed the percentage of acidic vesicular organelles (AVOs, as detected by AO staining) in HeyA8 3-D spheroids treated with PRL or PRL+G129R; corresponding AVO percentages were, by treatment: PRL (1.62%), G129R (23.4%), PRL+G129R (18.4%). n=6, *p < 0.05 (control [CTL] vs. G129R), **p < 0.001 (CTL vs. PRL+G129R).
(D,E) Representative images from transmission electron microscopy showed autophagic vesicles formed in HeyA8 3-D spheroids treated with PRL (D) or G129R (E). Substantial amounts of autophagosomes (green arrows) and autolysosomes (blue arrows) were identified in the cytoplasm of the G129R-treated spheroids, while no such structure was observed in PRL-treated spheroids. Substructure was observed under 5000× and 20,000× magnification. Ultra-thin sections in representative areas from 15 fields are shown. For higher resolution images, please refer to Supplemental Figure S2E.
(F,G,H) Tandem LC3 microscopy showed autophagic flux in pGFP-RFP-LC3–expressing HeyA8 3-D spheroids treated with PRL (F) or G129R (G) for 72 hours or 50 nM rapamycin (H) for 24 hours. Vesicles containing both GFP+ and RFP+LC3 represent early stage autophagosomes (green arrow), while vesicles containing RFP+LC3 only represent late-stage autolysosomes (blue arrow). Scale bar (F,H) = 10 μm; Scale bar (G) = 20 μm.
(I) FACS analysis indicated a significant shift of the RFP+ population for HeyA8 3-D spheroids treated with G129R in comparison with PRL or CTL.
(J,K) FACS analysis for GFP+LC3 or RFP+LC3 population of HeyA8 spheroids treated with rapamycin (50 nM; J) or G129R (10 μg/ml; K) indicated the differential effects of rapamycin and G129R in autophagy.
We next used acridine orange (AO) to stain the acidic vesicular organelles (AVOs) in G129R-treated cancer cell spheroids and analyzed them by FACS. There were significantly more AO-positive cells (i.e., AVOs) in HeyA8 3-D spheroids treated with G129R (18.8%) or PRL+G129R (18.4%) than in controls (6.2%) or PRL-treated 3-D spheroids (1.62%) (Fig. 3C). Similar results were not observed in 2-D cultured HeyA8 cells (Fig. S2E). In addition, we observed no significant effects of G129R on IG10 cells with low-level PRLR-LF (Fig. S2E).
Next, we employed transmission election microscopy to identify the subcellular organelles formed in PRL-treated (Fig. 3D) and G129R-treated (Fig. 3E) 3-D spheroids. G129R treatment induced substantial amounts of autophagosomes and late autophagic vacuoles, or autolysosomes. Furthermore, the G129R-treated HeyA8 spheroids had intact nuclear membranes and no apoptosomes. We did not observe the same amounts of autophagosomes or autolysosomes in PRL-treated 3-D spheroids (Fig. 3D; high-resolution images are shown in Fig. S2F).
We then performed monomeric pGFP-RFP-LC3 microscopy to monitor autophagic flux in 3-D cancer cell spheroids (Klionsky et al., 2012). A pEGFP-RFP–tagged microtubule-associated protein 1 light chain 3 (LC3) plasmid (Kimura et al., 2007) was stably transfected into HeyA8 cells prior to 3-D culture. The mGFP protein is acid sensitive and is degraded following fusion of autophagosomes with lysosomes, while the mRFP is relatively stable. PRL-treated HeyA8 3-D cancer spheroids expressed little GFP or RFP signal, which indicated that PRL did not induce formation of autophagosomes (Fig. 3F). However, we observed autophagic puncta with mGFP/mRFP-merged signals in cytoplasm of G129R-treated HeyA8 3-D spheroids, which indicated the formation of autophagosomes (RFP+andGFP+LC3, Fig. 3G). More importantly, as a result of autophagic flux and quenching of acid-sensitive fluorescent signal of GFP due to low pH in the lysosomes, presence of red puncta by RFP+onlyLC3 (RFP+GFP−) became highly visible under the treatment of G129R, representing the formation of autolysosomes (Fig. 3G). The autophagic vesicles expressing RFP+andGFP+LC3 were validated in HeyA8 3-D cancer spheroids treated with rapamycin (50 nM) for 24 hours (Fig. 3H).
To quantitatively measure the autophagic flux in a high-throughput manner, we used fluorescence-activated cell sorting (FACS) to count the late-stage autolysosomes (RFP+). Treatment with G129R induced a significant shift toward RFP+LC3 in comparison with control or PRL treatment (Fig. 3I). These findings indicate that different with rapamycin, G129R induces substantial autophagy in 3-D cancer spheroids by forming abundant autophagic vacuoles, particularly autolysosomes. To distinguish the autophagy induced by G129R, we further compared the GFP+LC3 and RFP+LC3 populations of 3-D HeyA8 cells following treatment with rapamycin or G129R. In comparison with control, rapamycin (50 nM) increased both GFP+ and RFP+ populations, while G129R increased the population of RFP+LC3 but not GFP+ (Fig. 3J & 3K). Using time-lapse video microscopy, we monitored the conversion of 3-D cancer spheroids from GFP+andRFP+LC3 to RFP+onlyLC3 following prolonged treatment with G129R; the 3-D spheroids treated with rapamycin remained GFP+andRFP+LC3 (Fig. S3A, and Movies 5 & 6). Thus, our results indicate that rapamycin induced autophagosomes containing GFP+andRFP+ LC3, while G129R induced enrichment of late-stage autolysosomes containing RFP+onlyLC3.
To determine the effects of G129R-induced autophagy in cell survival, we measured the viability of 3-D cultured cancer spheroids. Under 2-D conditions, neither G129R nor PRL+G129R changed the viability of HeyA8 or SKOV3 cells. However, G129R significantly decreased the viability of cells cultured under 3-D conditions (Fig. S3B). The inhibitory effect of G129R under 3-D conditions was reversed by addition of autophagy inhibitors bafilomycin A1 (Yamamoto et al., 1998) or hydroxychloroquine (Amaravadi et al., 2011). The cell death was further validated by FACS analysis with SYTOX green (Fig. S3C). We confirmed this finding by examining the conversion of LC3 from isoform I to II (Klionsky et al., 2012) under these conditions and found the same results: the autophagy inhibitors reversed effects of G129R on LC3 I/II conversion (Fig. S3D).
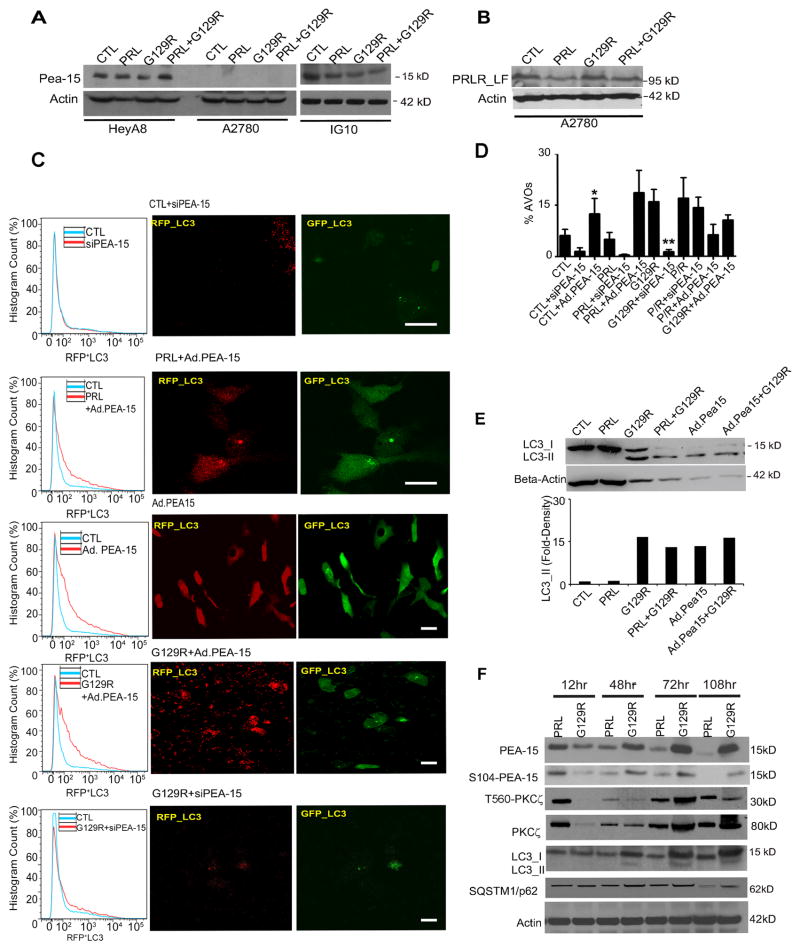
G129R-induced autophagy is mediated by PEA-15
To explore the mechanism of autophagy induced by G129R, we examined an array of autophagic factors identified in Fig 3. We used a short-hairpin (shRNA) complex against BECN1 to knockdown beclin-1 in HeyA8 cells and discovered that knockdown of beclin-1 did not decrease the autophagic flux in HeyA8 3-D spheroids treated with G129R, as shown by FACS analysis of AO staining and LC3-I/II conversion (Fig. S4A & S4B). The proportions of autolysosomes containing RFP+LC3 in 3-D HeyA8 spheroids treated with G129R alone were not significantly different than those in HeyA8-shBECN1 spheroids treated with G129R (Fig. S4C & S4D). However, when we targeted ATG7 with shRNA in HeyA8 spheroids, the G129R-induced LC3-I/II conversion and enrichment of RFP+LC3 containing autolysosomes were reversed by ATG7 knockdown (Fig. S4E & S4F). Additionally, G129R significantly decreased the viability of HeyA8-shBECN1 spheroids, but not HeyA8-shATG7 spheroids (Fig. S4G). FACS analysis with SYTOX-green staining showed that shATG7 sufficiently reversed the cell death induced by G129R, however, shBECN1 did not have such effects (Fig. S4H). In addition, we determined the effect of ATG5 knockdown on cell viability and cell death in 3-D cancer spheroids treated with G129R, but did not observe significant effects from either analysis (Data not shown). These results indicate that G129R-induced autophagy is beclin-1 independent but executed by ATG7.
Next, we investigated the role of PEA-15 during G129R-induced autophagy through siRNA knockdown or adenoviral expression. During the initial 12 hours of treatment with PRL or G129R, PEA-15 was expressed in HeyA8 and IG10 cells (Fig. 4A) but not in A2780 cells, which express PRLR-LF (Fig. 4B). We manipulated the levels of PEA-15 in pGFP-RFP-LC3-HeyA8 cells with siRNA (siPEA-15) or with an adenoviral construct (Ad.PEA-15) and treated these cells with PRL or G129R. The formation of autophagic vesicles was assessed by FACS analysis of RFP+ (left panels, Fig. 4C) or by confocal microscopy (right panels, Fig. 4C). Knockdown of PEA-15 did not induce accumulation of autophagic vacuoles. Overexpression of PEA-15 enriched autophagosomes containing EGFP+/RFP+LC3 (Fig. 4C, and high-resolution images in Fig. S4I), as previously reported (Bartholomeusz et al., 2008). Interestingly, G129R alone or in combination with Ad.PEA-15 significantly increased autolysosomes containing RFP+LC3 only (Fig. 4C, also shown in Fig. S4I). This finding indicates that Ad.PEA-15 was able to initiate autophagy, while G129R substantially enriched the late-stage autolysosomes (formed through fusion of autophagosomes and lysosomes containing RFP+LC3 only). Statistical analysis of RFP-integrated densities from 15 independent images demonstrated significantly greater density in cells treated with G129R+Ad.PEA-15 than in cells treated with Ad.PEA-15 (Fig. S4J). Given G129R significantly decreased the viability of cancer cell spheroids (Fig. S3), these findings of abundant autolysosomes enriched by G129R point to an alternative death pathway through sustained autophagy.
Figure 4.
G129R-induced autophagy is mediated by PEA-15.
(A) Expression of PEA-15 in HeyA8, A2780, and IG10 cells.
(B) Expression of PRLR-LF in A2780 cells. Only HeyA8 cells expressed both PRLR-LF and PEA-15.
(C) Role of PEA-15 in G129R-induced autophagy assessed by tandem tagged LC3 microscopy and FACS. The pGFP-RFP-LC3–expressing HeyA8 3-D spheroids were treated with control (CTL)+siPEA-15, PRL+Ad.PEA-15, Ad.PEA-15, Ad.PEA-15+G129R, or G129R+siPEA-15.
(Left panel) Histograms show results of FACS analysis indicating percentage of RFP+LC3 cells under each treatment in comparison with non-treated controls. Each analysis represents 10,000 cells, and three individual replicates per condition were performed. (Right panel) Representative images of cells with GFP+ and RFP+LC3 are shown; n=20. For higher-resolution images of GFP+andRFP+LC3 in Ad.PEA-15 and RFP+LC3 only in G129R+Ad.PEA-15, please see Supplemental Figure S4A.
(D) Percentage of AVOs in 3-D spheroids measured under manipulation of PEA-15. Knockdown of PEA-15 with siRNA abolished the G129R-induced AVOs. n=3, *p < 0.01 (CTL vs. CTL+Ad.PEA-15); **p < 0.001 (G129R vs. G129R+siPEA-15).
(E) Measurement of autophagic flux by LC3-I to -II conversion. (Upper panel) Expression of LC3-II was increased in HeyA8 3-D spheroids by G129R, PRL+G129R, or Ad.PEA-15. (Lower panel) Gel density of LC3-II in each condition was normalized with β-actin and measured by ImageJ 3.0.
(F) Time dynamics of PEA-15 activation in HeyA8 3-D spheroids. Phosphorylation of PEA-15 and PKC-ζ was detected by pS104 and pT560, respectively. Phosphorylation of PEA-15 and PKC-ζ was decreased in spheroids treated with PRL but increased in those treated with G129R, reaching its peak at 72 hours of treatment.
We further measured the percentage of AVOs in HeyA8 3-D spheroids. Overexpression of PEA-15 significantly increased the percentage of AVOs, while knockdown of PEA-15 lowered the percentage of AVOs (Fig. 4D). Exogenous PEA-15 increased the percentage of AVOs independent of G129R. We did not observe the increased AVOs after G129R treatment in either A2780 or IG10 cells (data not shown), which largely lack endogenous PEA-15 or PRLR (A2780, Fig. 4A; IG10, Fig. 5H).
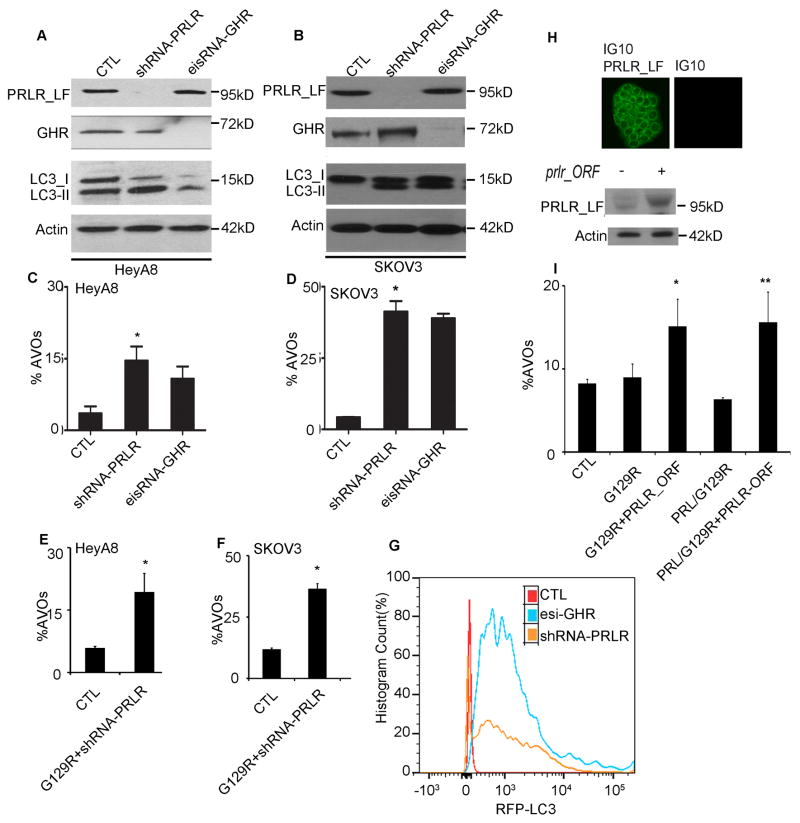
Figure 5.
PRLR silencing induced autophagic flux in 3-D cancer spheroids.
(A,B) LC3-I to -II conversion in HeyA8 (A) and SKOV3 (B) 3-D spheroids stably transfected with shRNA-prlr or esiRNA-GHR, respectively.
(C,D) Knockdown of PRLR or GHR prominently increased percentages of AVOs in HeyA8 (C) and SKOV3 (D) 3-D spheroids. n=3, *p < 0.0001 (CTL vs. shRNA-prlr, HeyA8; C); *p < 0.005 (CTL vs. shRNA-prlr, SKOV3; D).
(E,F) Increased AVOs in 3-D HeyA8 (E) and SKOV3 (F) spheroids induced by PRLR knockdown was not enhanced by G129R. n=3 *p < 0.01 (CTL vs. G129R+shRNA-prlr; E), *p < 0.05 (CTL vs. G129R+shRNA-prlr; F).
(G) FACS analysis for RFP-LC3+ cells showed the shift of the RFP+ population for HeyA8 3-D cells transfected with shRNA-PRLR (orange) or esiRNA-GHR (blue) from that of controls (red).
(H) PRLR was introduced into IG10 spheroids by transfection of a GFP-tagged pCMV-prlr-ORF. (Upper panel) Representative images indicate the expression of GFP-prlr-ORF in IG10-3D spheroids; scale bar = 5 μm. (Lower panel) Immunoblotting with anti-PRLR (clone BP774) validated PRLR expression.
(I) In prlr-transfected IG10 spheroids, G129R increased the percentages of AVOs. Differences between groups were compared by unpaired two-tailed t-test: n=6 *p < 0.0001 (CTL vs. G129R+prlr-ORF); **p <0.05 (CTL vs. PRL/G129R+prlr-ORF).
We then validated this finding by measuring the LC3-I/II conversion in HeyA8 3-D spheroids. Levels of LC3-II increased within 48 hours of treatment with G129R or Ad.PEA-15 in comparison with controls (Fig. 4E). Fold-densities of LC3-II immunoblotting bands were normalized with β-actin and showed substantial increases in cells treated with G129R, PRL+G129R, Ad.PEA-15 alone, or Ad.PEA-15+G129R (Fig. 4E). Neither A2780 nor IG10 cells showed such an increase in LC3-II accumulation (data not shown), indicating that expression of both PEA-15 and PRLR is necessary for G129R-induced autophagy.
Next, we investigated the regulatory machinery between PEA-15 and PRLR antagonism. Protein kinase C zeta (PKC-ζ), a downstream factor of PRLR-LF activation (Crowe et al., 1991), phosphorylates PEA-15 at serine 104 (S104), enhancing its stability (Kubes et al., 1998). Levels of PEA-15 and PKC-ζ in HeyA8 3-D spheroids increased along with duration of G129R treatment and decreased along with PRL treatment. Both total and activated forms of PEA-15 and PKC-ζ reached the highest levels at 72 hours of G129R treatment (Fig. 4F). The accumulation of LC3-II also reached the highest level at 72 hours following G129R treatment. Expression of SQSTM1/p62, an adaptor protein involved in the autophagy process (Dikic et al., 2010), varied slightly with duration of treatment, possibly because of the ubiquitin-protein degradation processes linked with autophagy progression. These results indicate that G129R-induced autophagy is mediated by activation of the PEA-15/PKC-ζ interactome.
PRLR depletion induces autophagy
To determine whether blockade of PRLR through genetic ablation induces autophagy, we used shRNA-PRLR and a heterogeneous mixture of siRNAs (esiRNA) against growth hormone receptor (GHR) to achieve substantial knockdown of PRLR and GHR in HeyA8 (Fig. 5A) and SKOV3 cells (Fig. 5B). The inclusion of GHR was based on the knowledge that G129R blocks partial activities of GHR by interfering with the PRLR/GHR heterodimer (Xu et al., 2011). The conversion of LC3-I to LC3-II was enhanced in 3-D HeyA8 and SKOV3 spheroids under PRLR or GHR knockdown. The percentages of AVOs were significantly increased when PRLR or GHR was completely depleted (HeyA8, Fig. 5C; SKOV3, Fig. 5D). Similar results were observed in the shRNA-PRLR cells treated with G129R (HeyA8, Fig. 5E; SKOV3, Fig. 5F), which indicates that G129R did not alter the PRLR depletion–induced autophagy. We then analyzed the autophagic flux by FACS through quantitation of the subpopulation of RFP+LC3 cells among pGFP-RFP-LC3-HeyA8 spheroids. Both shRNA-PRLR and esi-GHR induced a shift of RFP+ expression in comparison with controls (Fig. 5G).
Meantime, we transfected a prlr-open reading frame (ORF) construct driven by a GFP-tagged pCMV promoter to increase the expression of PRLR in IG10 cells to levels equivalent to those in HeyA8 and SKOV3 cells (Fig. 5H). The overexpression of GFP-PRLR in IG10 3-D spheroids was shown by confocal microscopy and immunoblots (Fig. 5H). Treatment of PRLR-positive IG10 spheroids with G129R resulted in significant increases in percentage of AVOs (G129R+prlr-ORF vs controls, p<0.05; PRL+G129R+prlr-ORF vs controls, p<0.05, Fig. 5I). No significant changes in percentage of AVOs were observed in parental IG10 cells treated with G129R in either 2-D or 3-D conditions (Fig. S2E), implying that sufficient PRLR is necessary for G129R to induce the autophagic vacuoles.
Next, we used PRLR-deficient, GHR-positive LNcap cells (Xu et al., 2011) to analyze the specificity of G129R in antagonizing PRLR activity. G129R alone did not induce an increase in the percentage of AVOs in these cells (Fig. S5A), while GHR antagonist B2036, alone or in combination with PRL, increased the AVOs. This is further supported by the response to G129R treatment of LNcap cells sensitized by exogenous expression of prlr-ORF; knockdown of endogenous PEA-15 reduced AVOs induced by G129R and prlr-ORF (unpaired two-tailed p<0.001, Fig S5B). We further determined the in vivo effect of G129R in LNcap cells using a mouse xenograft model. After 28 days of treatment, the G129R group had no significant tumor reduction or loss of body weight compared to the control group (p>0.05; Fig. S5C & S5D). The expression of PRLR-LF was validated in HeyA8, SKOV3, LNcap, and HEK293T cells (Fig. S5E). These data indicate that blockade of PRLR activity by G129R or by genetic depletion of prlr leads to inducible autophagy.
To determine if this mechanism of autophagy by G129R is cancer type specific, we measured the percentage of AVOs in two human pancreatic cancer cell lines cultured under 3-D settings: Panc-1 (expressing PEA-15 and PRLR-LF) and HPAF2 (expressing PEA-15 but not PRLR-LF) (Fig. S6). The percentage of AVOs increased significantly upon treatment with G129R in Panc-1 cells but not in HPAF2 cells, which indicated that expression of PRLR-LF is also necessary for G129R-induced autophagy in pancreatic cancer.
High tumoral PRLR and low autophagic factor correlates with worse clinical outcome
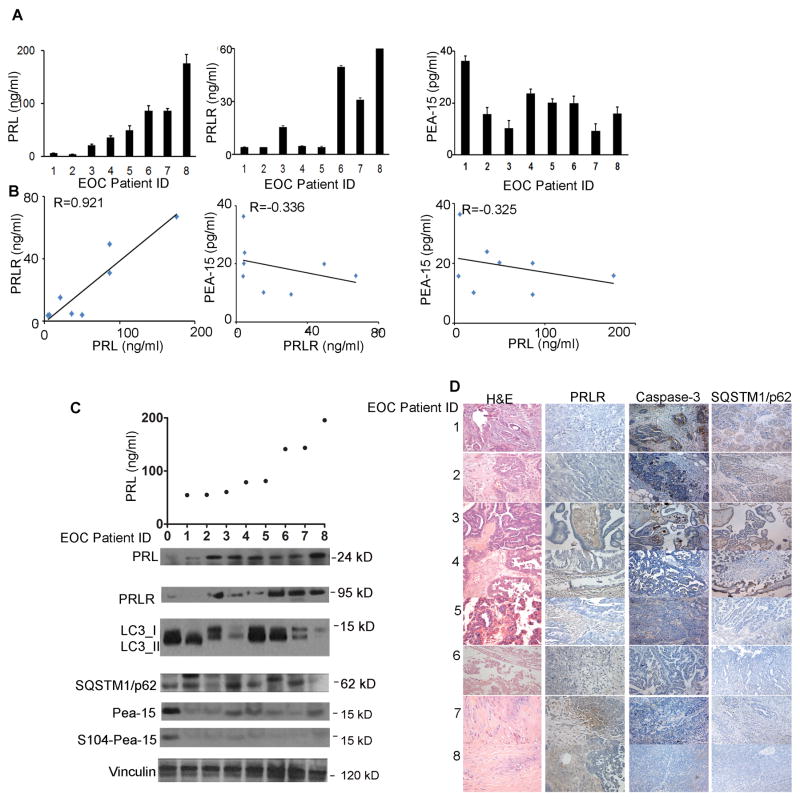
We next analyzed the clinical relevance of the PRL/PRLR axis and autophagic factors. First, we measured levels of PRL, PRLR, and PEA-15 in serum samples from eight patients with epithelial ovarian cancer by ELISA; results indicated an inverse correlation between PRL/PRLR and PEA-15 (PRL/PRLR, R=0.92 with high precision; PRLR/PEA-15, R=−0.33 with moderate precision; PRL/PEA-15, R=−0.32 with moderate precision, Fig. 6A&B). We then collected serum and tumor samples from another eight women with epithelial ovarian cancer. We measured levels of circulating PRL in serum samples by ELISA and levels of PRL/PRLR protein in tumor homogenates with immunoblotting. Results showed that PRL/PRLR was inversely correlated with LC3-II, SQSTM1/p62, and S104-phosphorylated and total PEA-15 (Fig. 6C). Immunohistochemical staining confirmed this finding and also showed an inverse correlation between PRLR and cleaved caspase-3 (Fig. 6D).
Figure 6.
Expression of PRL/PRLR axis and PEA-15 was inversely correlated in tumor samples from women with epithelial ovarian cancer.
(A) Levels of PRL (left), PRLR (middle), and PEA-15 (right) were measured by ELISA in tumor samples from eight patients with epithelial ovarian cancer. The Y-axis represents the concentration of the proteins (ng/mL) in serum (A) or in tumor homogenates (B,C). Six individual replicates were performed for every ELISA in each patient.
(B) Pearson correlation of PRL, PRLR, and PEA-15 concentrations in epithelial ovarian cancer patients. Correlation coefficient for PRL vs PRLR, R=0.921, p<0.001 (left); correlation coefficient for PRLR vs. PEA-15, R=−0.036, p<0.001 (middle); correlation coefficient for PRL vs. PEA-15, R=−0.325, p<0.05 (right).
(C) (Upper panel) Levels of PRL (ng/mL) were validated from serum samples of another eight patients with epithelial ovarian cancer. (Lower panel). Expression of PRL, PRLR, LC3, SQSTM1/p62, and PEA-15 (S104-phosphorylated and total) were further measured by immunoblotting from the tumors homogenates of these patients.
(D) Representative images of immunohistochemical staining of tumor tissues of the eight patients shown in panel C show the inverse PRL-caspase-3, PRL-SQSTM1/p62 correlations. Hematoxylin & eosin–stained images are included to show the presence of tumor. Representative images of each staining were selected from 15 high-power fields, magnification 200×.
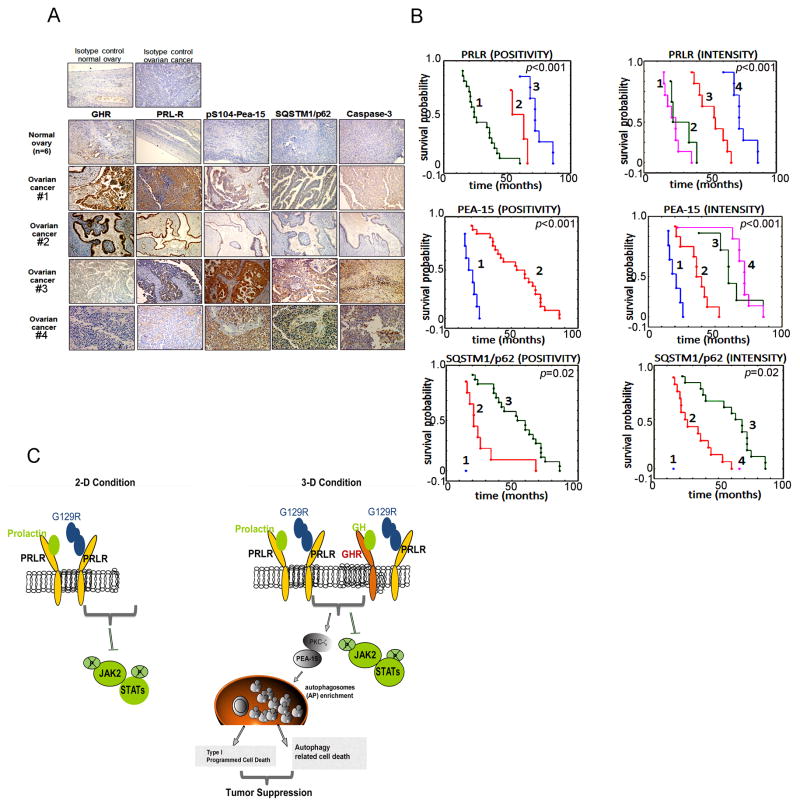
We further analyzed tumor samples from a larger cohort of patients (n=32) with epithelial ovarian cancer (Table S1). Immunohistochemical analysis showed that, in comparison with normal ovary, which expressed low levels of GHR and PRLR, epithelial ovarian tumors expressed higher levels of GHR and PRLR and lower levels of S104-phosphorylated PEA-15, SQSTM1/p62, and activated caspase-3 (Fig. 7A). Patients whose tumors expressed relatively lower PRLR and higher S104-phosphorylated PEA-15 and SQSTM1/p62 had a better overall survival rate (Fig. 7B). These data point to the potential clinical importance of targeting PRL/PRLR during development of epithelial ovarian cancer. To extend our finding, we compared the levels of S104-phosphorylated PEA-15, LC3-II, and cleaved caspase-3 in ovarian tissues from 4-week-old BL6 wild-type and prl−/− mice (Cruz-Soto et al., 2002) to determine their contribution during ovarian development. Levels of all three factors were higher in ovarian tissues from prl−/− mice than in those from the wild-type mice (Fig. S7).
Figure 7.
High levels of PRLR and low levels of autophagic factors correlated with worse prognosis.
(A) Representative images of immunohistochemical analysis for GHR, PRLR, S104-phosphorylated PEA-15, SQSTM1/p62, and cleaved caspase-3 on tissue sections from a larger cohort of patients with epithelial ovarian cancer (n=32) are shown. Magnification 100×. (Upper panels) isotype control in the normal ovary (left); isotype control in the tumor (right). (Lower panels) Expression of GHR, PRLR, S104-phosphorylated PEA-15, SQSTM1/p62, and cleaved caspase-3 in normal ovaries and ovarian cancers are shown in representative images. Patients #1 and #2 represent advanced stages of disease (FIGO IIIb and FIGO IIIc, respectively) and #3 and #4 represent early stages of disease (FIGO Ia and FIGO Ib, respectively).
(B) Patients with a low level of PRLR and high levels of PEA-15 and SQSTM1/p62 had a better survival rate. Analysis of PRLR staining patterns and overall survival: (POSITIVITY) 1=positive, 2=heterogenous, 3=negative; (INTENSITY) 1=strong, 2=moderate, 3=weak, 4=none. Analysis of PEA-15 staining pattern and overall survival: (POSITIVITY) 1=negative, 2=positive; (INTENSITY) 1=none, 2=weak, 3=moderate, 4=strong. Analysis of SQSTM1/p62 staining pattern and overall survival: (POSITIVITY) 1=negative, 2=heterogenous, 3=positive; (INTENSITY) 1=none, 2=weak, 3=moderate, 4=strong.
(C) Schematic illustration of divergent mechanisms induced by blockade of PRL/PRLR in tumor cells.
In summary, our studies reveal a novel mechanism for programmed cell death induced by blockade of tumoral PRL/PRLR in cancer cells. This cell death occurs through sustained autophagy and leads to inhibition of tumor growth (depicted schematically in Fig. 7C).
Discussion
Here, we report a previously unrecognized mechanism by which blockade of the tumoral PRL/PRLR axis induced sustained autophagy that led to programmed cell death. We demonstrated an inverse correlation between tumoral PRL/PRLR levels and autophagic factors in patients with epithelial ovarian cancer and showed evidence of clinical potential that provides a rationale for targeting tumoral PRL/PRLR.
Recent studies have shown that levels of PRL were substantially elevated in ovarian cancer patients and concomitantly enhanced proliferation, migration, and invasion and promoted angiogenesis of ovarian carcinoma cells (Asai-Sato et al., 2005; Levina et al., 2009). Our results indicate that antagonism of PRL/PRLR axis enriched late-stage autophagic vacuoles in cancer cells cultured in 3-D conditions, which led to programmed cell death. This might explain the robust inhibitory effects of G129R on tumor growth that we observed in vivo but not in 2-D cultured cancer cells.
Persistent metabolic stress, such as that caused by chemotherapeutic agents, can induce excessive and sustained autophagy by which tumor cells are eliminated (Bursch et al., 1996; Castino et al., 2010; Mathew et al., 2007; Thongon et al., 2008). Our studies showed that blockade of PRL/PRLR caused accumulation of substantial RFP+onlyLC3-containing autolysosomes in 3-D cancer spheroids. The abundant late-stage autophagic vacuoles were death-switch signals that led to programmed death of cancer cells.
We further showed that autophagy induced by blockade of PRL/PRLR is mediated by the PEA-15/ PKC-ζ interactome. This finding is consistent with recent reports regarding the role of PEA-15 in activating cell death in breast cancer cells (Bartholomeusz et al., 2010). (Bartholomeusz et al., 2010). We discovered that the function of PEA-15 in mediating G129R-induced autophagy requires stabilization of the protein. Kinase PKC-ζ reportedly enhances PEA-15 stability (Trencia et al., 2003). We showed that levels of PKC-ζ and S104-phosphorylated PEA-15 increased following G129R treatment, suggesting that S104-phosphorylated PEA-15 is tumor suppressive by contributing to G129R-induced growth inhibition. This finding diverges from a recent report that S104 phosphorylation is a switch to turn PEA-15 from tumor suppressor to tumor promoter (Sulzmaier et al., 2012). Further studies are needed to define the role of S104-PEA-15 in the tumor-inhibitory effect of blockade of PRL/PRLR.
Until now, there has been no report on the clinical correlation of autophagic factors with ovarian cancer progression. Our studies revealed an inverse correlation between expression of tumoral PRL/PRLR and of autophagy markers (e.g., LC3-II) and regulatory factors (e.g., SQSTM1/p62 and S104-PEA-15) in tumor samples from women with epithelial ovarian cancer. We demonstrated the prognostic significance of these markers: higher levels of tumoral PRL/PRLR axis and lower levels of PEA-15, SQSTM1/p62, and cleaved caspase-3 were associated with lower survival rate. This finding provides the first clinical rationale for targeting the tumoral PRL/PRLR axis and promoting cell death through autophagy as an alternative strategy for treating malignant tumors, particularly those with apoptotic resistance.
Experimental Procedures
Reagents and plasmids
G129R was supplied by Oncolix, Inc. (Houston, TX). The PEA-15 siRNA (ggaagacauccccagcgaatt) was described previously (Bartholomeusz et al., 2010). Adenoviral PEA-15 (Ad.PEA-15) was constructed by inserting PEA-15 cDNA into the adenoviral vector pAdTrack-CMV (He et al., 1998). The esiRNA against GHR was purchased from Sigma Aldrich (St. Louis, MO). The shRNA against BECN-1 or ATG7 and siRNA against ATG5 were purchased from Origene, Inc., (Rockville, MD). The siRNAs were applied to cells at a final concentration of 100nmol/L for 48 hours. Transfections of the pEGFP-tagged pCMV-prlr-ORF clone of PRLR, shRNA-PRLR, shATG7, or shRNA-BECN1 4 29-mer mix (human) were performed according to the manufacturer’s instructions. The pEGFP-RFP-LC3 plasmid was derived from Addgene plasmid 21074. Anti-PRLR (clone BP774) recognizes the epitope of PRLR 1-14 from humans and cross-reacts with PRLR from murine or rat origins.
Cultured cells
For the 3-D models, ovarian, pancreatic, and prostate cancer cells were grown on non-adhesive optical plates (MatTek Corporation, MA) then brought to the glass surface that was pre-coated with 200 μL of growth factor–reduced Matrigel (BD Biosciences, San Diego, CA). Approximately 18,000~20,000 spheroids-like cells were plated in each well of a 24-well plate and incubated at 37°C for 1 hour to allow cells to seed gradually. Spheroids were cultured with full medium containing 2% Matrigel as previously described (Li and Lu, 2011).
Autophagic flux analysis
For FACS analysis of AVOs, cells were first stained with AO. Green (510–530 nm) and red (>650 nm) fluorescence emissions from 1×104 cells illuminated with blue (488 nm) excitation light were measured with a FACSCalibur from Becton Dickinson (Franklin Lakes, NJ). pGFP-RFP-LC3 plasmids were stably transfected into epithelial ovarian cancer cells, and cell were visualized with a multiphoton confocal microscope (TCS SP5 MP; Leica, Buffalo Grove, IL) or analyzed by LSR Fortessa Flow Cytometry (BD Biosciences).
Clinical data
Tissue samples from 32 patients with histopathology-confirmed serous-type primary epithelial ovarian cancer were obtained under a protocol (214/11) approved by the Institutional Review Board at Poznan University of Medical Sciences. The clinical and pathological characteristics recorded for the patients enrolled in the study include menopausal status, tumor stage and grade, details of surgical debulking, and any tumor recurrence. Median follow-up duration for overall survival was 4.7 years. The plasma and tumor tissues were analyzed for the proteins indicated.
Statistical analysis
We performed Student’s t test (between two groups) or ANOVA (for all groups) if normally distributed to compare continuous variables, and the Mann-Whitney rank-sum test or Kruskal-Wallis test (for all groups) if nonparametric. Pairwise differences in normally distributed variables in our treatment groups were compared by the Tukey-Kramer statistic for multiple comparisons. A Bonferoni adjustment to a (default value 0.05) was made based on the number of pairwise comparisons within a treatment experiment using the formula: a(a) = 0.05/k, where k = number of comparisons against control. A p-value of <0.05 was considered significant. Error bars represent standard error of the mean.
Supplementary Material
Highlights.
PRLR antagonism by G129R inhibits growth of ovarian cancer or cancer spheroids
Autophagy related cell death is induced by G129R in cancer spheroids
Pea-15 functions as a tumor suppressor to mediate G129R-induced autophagy
Tumoral PRL/PRLR correlates inversely with autophagic factors in clinical samples
Acknowledgments
Portions of this work were supported by grants from the U.S. National Institutes of Health (P50CA083639, P50CA098258, CA109298, RC2GM092599, U54 CA151668, CA140933, CA16672), the Department of Defense (OC073399, W81XWH-10-1-0158, OC100237, BC085265), a Program Project Development Grant from the Ovarian Cancer Research Fund, Inc., the Bettyann Asche Murray Distinguished Professorship, the Marcus Foundation, and the Blanton-Davis Ovarian Cancer Research Program. Y.W. is supported, in part, by the NIH 5 P50 SPORE CDP Award CA116199, the Marsha Rivkin Center Pilot Grant, and a research grant from the Foundation for Women’s Cancer.
We thank Robert Langley and Donna M. Reynolds for imaging and Kathryn L. Hale, J. Donald Payne, and Michael Redman for editorial review.
Abbreviations
- PRL
prolactin
- PRLR
prolactin receptor
- PRLR(LF)
prolactin receptor long form
- EOC
epithelial ovarian cancer
- AVOs
acidic vesicle organelle
- AO
acridine orange
- PEA-15
astrocytic phosphoprotein (15kD)
- GHR
growth hormone receptor
- CTL
controls
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- AnV
annexin V
- FACS
fluorescence-activated cell sorting
- ORF
opening reading frame
Footnotes
Disclosure
We report no potential conflicts of interest.
Extended Methods, 7 Figures, 6 movies, and 1 table, are available in the online version of the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akar U, Chaves-Reyez A, Barria M, Tari A, Sanguino A, Kondo Y, Kondo S, Arun B, Lopez-Berestein G, Ozpolat B. Silencing of Bcl-2 expression by small interfering RNA induces autophagic cell death in MCF-7 breast cancer cells. Autophagy. 2008;4:669–679. doi: 10.4161/auto.6083. [DOI] [PubMed] [Google Scholar]
- Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, DiPaola RS, Lotze MT, White E. Principles and current strategies for targeting autophagy for cancer treatment. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:654–666. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai-Sato M, Nagashima Y, Miyagi E, Sato K, Ohta I, Vonderhaar BK, Hirahara F. Prolactin inhibits apoptosis of ovarian carcinoma cells induced by serum starvation or cisplatin treatment. International journal of cancer Journal international du cancer. 2005;115:539–544. doi: 10.1002/ijc.20810. [DOI] [PubMed] [Google Scholar]
- Bartholomeusz C, Gonzalez-Angulo AM, Kazansky A, Krishnamurthy S, Liu P, Yuan LX, Yamasaki F, Liu S, Hayashi N, Zhang D, et al. PEA-15 inhibits tumorigenesis in an MDA-MB-468 triple-negative breast cancer xenograft model through increased cytoplasmic localization of activated extracellular signal-regulated kinase. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16:1802–1811. doi: 10.1158/1078-0432.CCR-09-1456. [DOI] [PubMed] [Google Scholar]
- Bartholomeusz C, Rosen D, Wei C, Kazansky A, Yamasaki F, Takahashi T, Itamochi H, Kondo S, Liu J, Ueno NT. PEA-15 induces autophagy in human ovarian cancer cells and is associated with prolonged overall survival. Cancer research. 2008;68:9302–9310. doi: 10.1158/0008-5472.CAN-08-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursch W, Ellinger A, Kienzl H, Torok L, Pandey S, Sikorska M, Walker R, Hermann RS. Active cell death induced by the anti-estrogens tamoxifen and ICI 164 384 in human mammary carcinoma cells (MCF-7) in culture: the role of autophagy. Carcinogenesis. 1996;17:1595–1607. doi: 10.1093/carcin/17.8.1595. [DOI] [PubMed] [Google Scholar]
- Castino R, Bellio N, Follo C, Murphy D, Isidoro C. Inhibition of PI3k class III-dependent autophagy prevents apoptosis and necrosis by oxidative stress in dopaminergic neuroblastoma cells. Toxicol Sci. 2010;117:152–162. doi: 10.1093/toxsci/kfq170. [DOI] [PubMed] [Google Scholar]
- Crowe PD, Buckley AR, Zorn NE, Rui H. Prolactin activates protein kinase C and stimulates growth-related gene expression in rat liver. Mol Cell Endocrinol. 1991;79:29–35. doi: 10.1016/0303-7207(91)90092-7. [DOI] [PubMed] [Google Scholar]
- Cruz-Soto ME, Scheiber MD, Gregerson KA, Boivin GP, Horseman ND. Pituitary tumorigenesis in prolactin gene-disrupted mice. Endocrinology. 2002;143:4429–4436. doi: 10.1210/en.2002-220173. [DOI] [PubMed] [Google Scholar]
- Dalby KN, Tekedereli I, Lopez-Berestein G, Ozpolat B. Targeting the prodeath and prosurvival functions of autophagy as novel therapeutic strategies in cancer. Autophagy. 2010;6:322–329. doi: 10.4161/auto.6.3.11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nature reviews Cancer. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- Demarchi F, Bertoli C, Copetti T, Eskelinen EL, Schneider C. Calpain as a novel regulator of autophagosome formation. Autophagy. 2007;3:235–237. doi: 10.4161/auto.3661. [DOI] [PubMed] [Google Scholar]
- Dikic I, Johansen T, Kirkin V. Selective autophagy in cancer development and therapy. Cancer research. 2010;70:3431–3434. doi: 10.1158/0008-5472.CAN-09-4027. [DOI] [PubMed] [Google Scholar]
- He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci U S A. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamat AA, Coffey D, Merritt WM, Nugent E, Urbauer D, Lin YG, Edwards C, Broaddus R, Coleman RL, Sood AK. EphA2 overexpression is associated with lack of hormone receptor expression and poor outcome in endometrial cancer. Cancer. 2009;115:2684–2692. doi: 10.1002/cncr.24335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubes M, Cordier J, Glowinski J, Girault JA, Chneiweiss H. Endothelin induces a calcium-dependent phosphorylation of PEA-15 in intact astrocytes: identification of Ser104 and Ser116 phosphorylated, respectively, by protein kinase C and calcium/calmodulin kinase II in vitro. J Neurochem. 1998;71:1307–1314. doi: 10.1046/j.1471-4159.1998.71031307.x. [DOI] [PubMed] [Google Scholar]
- Levina VV, Nolen B, Su Y, Godwin AK, Fishman D, Liu J, Mor G, Maxwell LG, Herberman RB, Szczepanski MJ, et al. Biological significance of prolactin in gynecologic cancers. Cancer Res. 2009;69:5226–5233. doi: 10.1158/0008-5472.CAN-08-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4:600–606. doi: 10.4161/auto.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Lu Y. Optimizing a 3D Culture System to Study the Interaction between Epithelial Breast Cancer and Its Surrounding Fibroblasts. Journal of Cancer. 2011;2:458–466. doi: 10.7150/jca.2.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Lin S, Long HX, Xiang T, Zhu B, Xie RK. Isolation and identification of cancer stem cells from primary human ovarian cancer tissues. Zhonghua zhong liu za zhi [Chinese journal of oncology] 2011;33:896–899. [PubMed] [Google Scholar]
- Llovera M, Pichard C, Bernichtein S, Jeay S, Touraine P, Kelly PA, Goffin V. Human prolactin (hPRL) antagonists inhibit hPRL-activated signaling pathways involved in breast cancer cell proliferation. Oncogene. 2000;19:4695–4705. doi: 10.1038/sj.onc.1203846. [DOI] [PubMed] [Google Scholar]
- Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nature reviews Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng N, Wu L, Gao J, Zhao J, Su L, Su H, Zhang S, Miao J. Lipopolysaccharide induces autophagy through BIRC2 in human umbilical vein endothelial cells. Journal of cellular physiology. 2010;225:174–179. doi: 10.1002/jcp.22210. [DOI] [PubMed] [Google Scholar]
- Mor G, Visintin I, Lai Y, Zhao H, Schwartz P, Rutherford T, Yue L, Bray-Ward P, Ward DC. Serum protein markers for early detection of ovarian cancer. Proc Natl Acad Sci U S A. 2005;102:7677–7682. doi: 10.1073/pnas.0502178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinsay MN, Thomas RL, Lee Y, Gustafsson AB. Bnip3-mediated mitochondrial autophagy is independent of the mitochondrial permeability transition pore. Autophagy. 2010;6:855–862. doi: 10.4161/auto.6.7.13005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007;6:304–312. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- Rui H, Kirken RA, Farrar WL. Activation of receptor-associated tyrosine kinase JAK2 by prolactin. J Biol Chem. 1994;269:5364–5368. [PubMed] [Google Scholar]
- Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, Tsujimoto Y. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nature cell biology. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- Sulzmaier F, Opoku-Ansah J, Ramos JW. Phosphorylation is the switch that turns PEA-15 from tumor suppressor to tumor promoter. Small GTPases. 2012;3:173–177. doi: 10.4161/sgtp.20021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D, Chen KE, Khoo T, Walker AM. Prolactin increases survival and migration of ovarian cancer cells: importance of prolactin receptor type and therapeutic potential of S179D and G129R receptor antagonists. Cancer letters. 2011;310:101–108. doi: 10.1016/j.canlet.2011.06.014. [DOI] [PubMed] [Google Scholar]
- Thongon N, Nakkrasae LI, Thongbunchoo J, Krishnamra N, Charoenphandhu N. Prolactin stimulates transepithelial calcium transport and modulates paracellular permselectivity in Caco-2 monolayer: mediation by PKC and ROCK pathways. Am J Physiol Cell Physiol. 2008;294:C1158–1168. doi: 10.1152/ajpcell.00020.2008. [DOI] [PubMed] [Google Scholar]
- Trencia A, Perfetti A, Cassese A, Vigliotta G, Miele C, Oriente F, Santopietro S, Giacco F, Condorelli G, Formisano P, Beguinot F. Protein kinase B/Akt binds and phosphorylates PED/PEA-15, stabilizing its antiapoptotic action. Mol Cell Biol. 2003;23:4511–4521. doi: 10.1128/MCB.23.13.4511-4521.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. Lyophilization and development of solid protein pharmaceuticals. International journal of pharmaceutics. 2000;203:1–60. doi: 10.1016/s0378-5173(00)00423-3. [DOI] [PubMed] [Google Scholar]
- White E, Karp C, Strohecker AM, Guo Y, Mathew R. Role of autophagy in suppression of inflammation and cancer. Current opinion in cell biology. 2010;22:212–217. doi: 10.1016/j.ceb.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, LeBaron MJ, Nevalainen MT, Rui H. Role of tyrosine kinase Jak2 in prolactin-induced differentiation and growth of mammary epithelial cells. J Biol Chem. 2002;277:14020–14030. doi: 10.1074/jbc.M112399200. [DOI] [PubMed] [Google Scholar]
- Xu J, Zhang Y, Berry PA, Jiang J, Lobie PE, Langenheim JF, Chen WY, Frank SJ. Growth hormone signaling in human T47D breast cancer cells: potential role for a growth hormone receptor-prolactin receptor complex. Mol Endocrinol. 2011;25:597–610. doi: 10.1210/me.2010-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell structure and function. 1998;23:33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, Baehrecke EH, Lenardo MJ. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.